Abstract
Metabolic reprogramming contributes to tumor development and introduces metabolic liabilities that can be exploited to treat cancer. Chemotherapies targeting metabolism have been effective cancer treatments for decades, and the success of these therapies demonstrates that a therapeutic window exists to target malignant metabolism. New insights into the differential metabolic dependencies of tumors have provided novel therapeutic strategies to exploit altered metabolism, some of which are being evaluated in pre-clinical models or clinical trials. Here, we review our current understanding of cancer metabolism and discuss how this might guide treatments targeting the metabolic requirements of tumor cells.
Introduction
In 1947, Sidney Farber, one of the pioneers of modern chemotherapy, discovered that aminopterin could cause disease remission in children with acute lymphoblastic leukemia (Dayton et al., 2016; Farber and Diamond, 1948). Aminopterin is the precursor of the currently used drugs methotrexate and pemetrexed, both of which are folate analogues that inhibit one-carbon transfer reactions required for de novo nucleotide synthesis (Figure 1, Figure 2a)(Walling, 2006). The early clinical success of antifolates led to the development of an entire class of drugs known as ‘antimetabolites.’ Antimetabolites are small molecules that resemble nucleotide metabolites and inhibit the activity of enzymes involved in nucleotide base synthesis (Table 1). Notable examples include the purine analogues 6-mercaptopurine (6-MP) and 6-thioguanine (6-TG), which inhibit 5-phosphoribosyl-1-pyrophosphatase (PRPP) amidotransferase, the first enzyme in de novo purine biosynthesis (Figure 1a, Figure 2b). 6-MP and 6-TG have been successful in treating many cancers including childhood leukemia (Elion, 1989). The pyrimidine analogue 5-fluorouracil (5-FU) is a synthetic analogue of uracil that inhibits thymidylate synthase, limiting the availability of thymidine nucleotides for DNA synthesis (Figure 1b, Figure 2c). 5-FU and the related 5-FU-prodrug capecitabine remain widely used chemotherapies today and are an important treatment for gastrointestinal cancers (Heidelberger et al., 1957; Wagner et al., 2006). Other antimetabolite nucleoside analogues, such as gemcitabine and cytarabine, are incorporated into DNA, resulting in inhibition of DNA polymerases, and are commonly used to treat select cancers (Parker, 2009).
Figure 1. Nucleotide Biosynthesis.
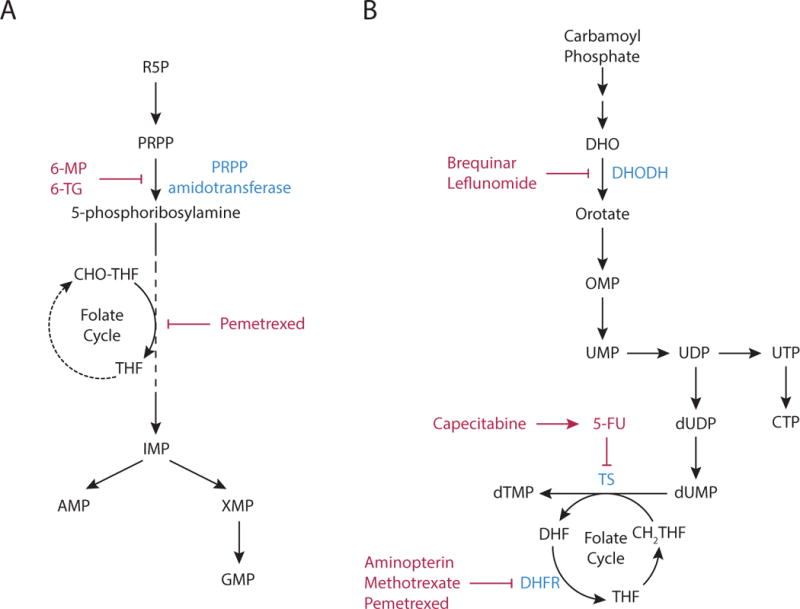
(A) Purine nucleotide synthesis. The first reaction in purine production generates 5-phosphoribosyl-1-pyrophosphatae (PRPP) from ribose 5-phosphate (R5P). The second step is catalyzed by PRPP amidotransferase, and commits PRPP to purine synthesis. This step can be inhibited by the antimetabolites, 6-mercaptopurine (6-MP) and 6-thioguanine (6-TG). Subsequent steps in the pathway assemble the purine ring and result in the formation of inosine monophosphate (IMP), which in turn can be converted to either adenosine monophosphate (AMP) or guanosine monophosphate (GMP) by distinct reactions. The synthesis of the purine ring requires N10-formyl-tetrahydrofolate (CHO-THF) via a reaction that can be inhibited by pemetrexed.
(B) Pyrimidine nucleotide synthesis. Pyrimidine nucleotide synthesis begins with the conversion of carbamoyl phosphate to the pyrimidine base orotate. One of the steps in pathway is catalyzed by dihydroorotate dehydrogenase (DHODH), which can be inhibited by brequinar sodium and leflunomide. Next, orotate is combined with PRPP to generate orotate monophosphate (OMP), which is subsequently converted to uridine monophosphate (UMP). UMP can be phosphorylated to form UDP and UTP, and the latter can be further converted to citidine triphosphate (CTP). Uridine nucleotides can also be used for de novo thymine nucleotide synthesis. UDP is converted to deoxy-UMP (dUMP), and the enzyme thymidylate synthase (TS) generates dTMP by catalyzing the methylation of dUMP using N5,N10-methylene-tetrahydrofolate (CH2-THF) as the methyl donor. TS activity is inhibited by the antipyrimidine 5-fluorouracil (5-FU) and the 5-FU pro-drug capecitabine. Thymidine synthesis can also be inhibited by the antifolates aminopterin, methotrexate, and pemetrexed, as these drugs inhibit the enzyme dihydrofolate reductase (DHFR), limiting the availability of CH2-THF.
Figure 2. Antimetabolites.
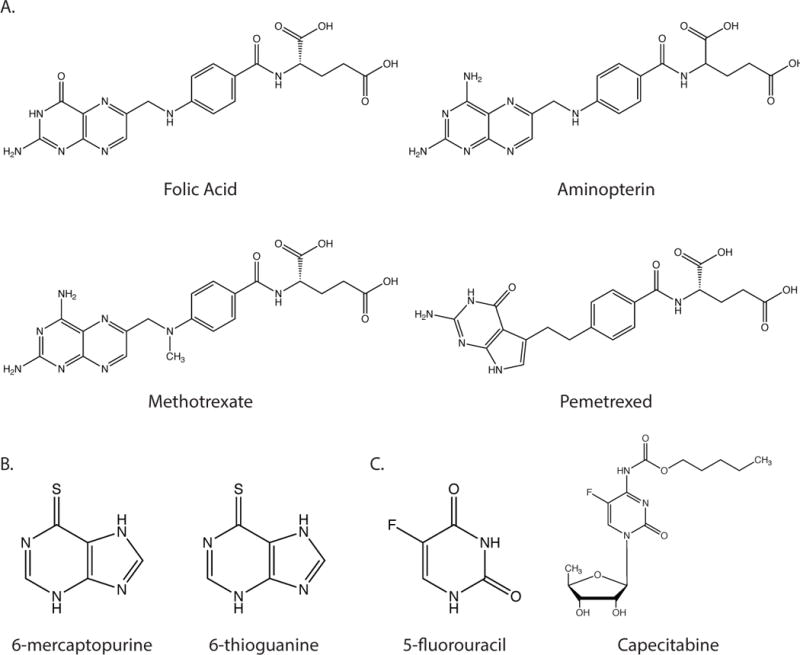
(A) Structures of folic acid and the antifolate compounds aminopterin, methotrexate and pemetrexed. (B) Structures f the purine analogues 6-mercaptopurine and 6-thioguanine (C) Structures of pyrimidine analogue 5-fluorouracil and the 5-fluorouracil prodrug capecitabine.
Table 1.
Select agents targeting metabolism that are approved, or are in trials, for the treatment of cancer, focusing on targets discussed in this review.
| Drug | Target Enzyme |
|---|---|
| Methotrexate | Dihydrofolate reductase (DHFR) |
| Pemetrexed | DHFR Thymidylate synthase (TS) Glycinamide ribonucleotide formyltransferase (GARFT) |
| 6-Mercaptopurine 6-Thioguanine |
PRPP amidotransferase |
| Capecitabine 5-Fluorouracil |
Thymidylate synthase (TS) |
| Gemcitabine Cytarabine |
DNA polymerase/ribonucleotide reductase (RnR) |
| Leflunomide | Dihydroorotate dehydrogenase (DHODH) |
| CB-839 | Glutaminase (GLS) |
| PEG-BCT-100 (ADI-PEG20) AEB-1102 |
Depletion of circulating arginine |
| L-Asparaginase | Depletion of circulating asparagine |
| TVB-2640 | Fatty-acid synthase (FASN) |
| AG-120 (Ivosidenib) IDH305 BAY1436032 FT-2102 AG-221 (Enasidenib) AG-881 |
mutant IDH1 mutant IDH2 mutant IDH1/2 |
| AZD3965 | Monocarboxylate transporter 1 (MCT1) |
| CPI-613 | Pyruvate dehydrogenase (PDH)/α-ketoglutarate dehydrogenase |
| Metformin | Mitochondrial complex I |
The clinical success of antimetabolites for treating cancer is attributed to the increased metabolic demand of neoplastic cells for nucleotide biosynthesis and DNA replication. However, nucleotide metabolism is only one of many metabolic dependencies altered to support cancer cell proliferation. Proliferating cells have different metabolic requirements from non-proliferating cells (Hsu and Sabatini, 2008; Lunt and Vander Heiden, 2011; Pavlova and Thompson, 2016). While non-proliferating cells have primarily catabolic demands, proliferating cells must balance the divergent catabolic and anabolic requirements of sustaining cellular homeostasis while duplicating cell mass, and thus engage in a metabolic program distinct from that of the normal tissue from which they arose. From a therapeutic perspective, the aberrant metabolism of proliferating cancer cells presents potential opportunities, and there has been a growing interest in studying how best to target cancer metabolism (Bobrovnikova-Marjon and Hurov, 2014; Galluzzi et al., 2013; Martinez-Outschoorn et al., 2017; Vander Heiden, 2011).
Targeting general proliferative metabolism may not offer an adequate therapeutic window since many non-malignant cells, including those in bone marrow, intestinal crypts, and hair follicles, are rapidly proliferating. Furthermore, the proliferation rates of normal cells are often greater than those of cancer cells (Vander Heiden and DeBerardinis, 2017), and prominent side effects of antimetabolite chemotherapy are caused by the destruction of normal rapidly proliferating cells. Myeloid suppression or gastrointestinal toxicity are often dose-limiting toxicities for these drugs.
In spite of toxicity, antimetabolites are standard in many modern chemotherapy regimens that increase patient survival and, in some cases, help cure disease. Factors other than proliferation rate may account for the efficacy of these drugs. Inducing DNA damage with the use of genotoxic chemotherapies can sensitize cells to inhibitors of nucleotide biosynthesis (Brown et al., 2017; Peters et al., 2000), suggesting that oncogenic mutations that reduce the DNA damage response is one explanation for why a therapeutic window exists for antimetabolite compounds. However, antimetabolite drugs are only effective against a subset of cancer types. Many resistant cancers have the same mutational spectrum as sensitive cancers, and defining genetic predictors of chemotherapy response for most malignancies has been difficult. Nevertheless, the fact that antimetabolite chemotherapies are clinically effective suggests that a metabolic therapeutic window exists beyond proliferation rate and response to DNA damage. While the precise mechanisms underlying the differential efficacies of existing antimetabolite therapies are unknown, a better understanding of these and other metabolic therapeutic windows may lead to the development of more effective and selective cancer treatments. Here, we discuss recent advances in cancer metabolism research that have identified metabolic targets and highlight features that might be exploited for improved cancer therapy.
Altered glucose metabolism
More than thirty years preceding Farber’s work on antifolates, Otto Warburg reported that cancer cells consume tremendous amounts of glucose and metabolize the majority of the glucose into lactate, even in the presence of oxygen (Warburg, 1924). This phenomenon is now referred to as aerobic glycolysis, or the Warburg effect, and represents a striking metabolic difference between cancer and most normal tissues. Substantial work has sought to target increased glycolysis including efforts to inhibit lactate production and excretion (Doherty and Cleveland, 2013; Hamanaka and Chandel, 2012; Hay, 2016; Pelicano et al., 2006; Zhao et al., 2013). One compound known to block glucose metabolism is 2-deoxyglucose (2-DG) (Wick et al., 1957). 2-DG is phosphorylated by hexokinase to produce 2-deoxyglucose-6-phosphate, which cannot be further metabolized by cells. It therefore accumulates intracellularly and competitively inhibits hexokinase to slow glucose uptake (Figure 3). Numerous preclinical studies have demonstrated anti-proliferative effects of 2-DG (Zhang et al., 2014a). Although early clinical testing yielded responses in some patients, the use of this drug was limited by toxicity associated with hypoglycemia symptoms (Landau et al., 1958). Recent clinical trials have revisited use of 2-DG at lower doses, but these doses are insufficient to inhibit disease progression (Raez et al., 2013; Stein et al., 2010). The relative lack of 2-DG clinical efficacy at tolerable doses has been echoed by most other attempts to directly target aerobic glycolysis. Though efforts to target glucose uptake or lactate production have found some success in preclinical models (Hay, 2016; Shim et al., 1997), clinical success has been limited (Vander Heiden and DeBerardinis, 2017).
Figure 3. Glycolysis.
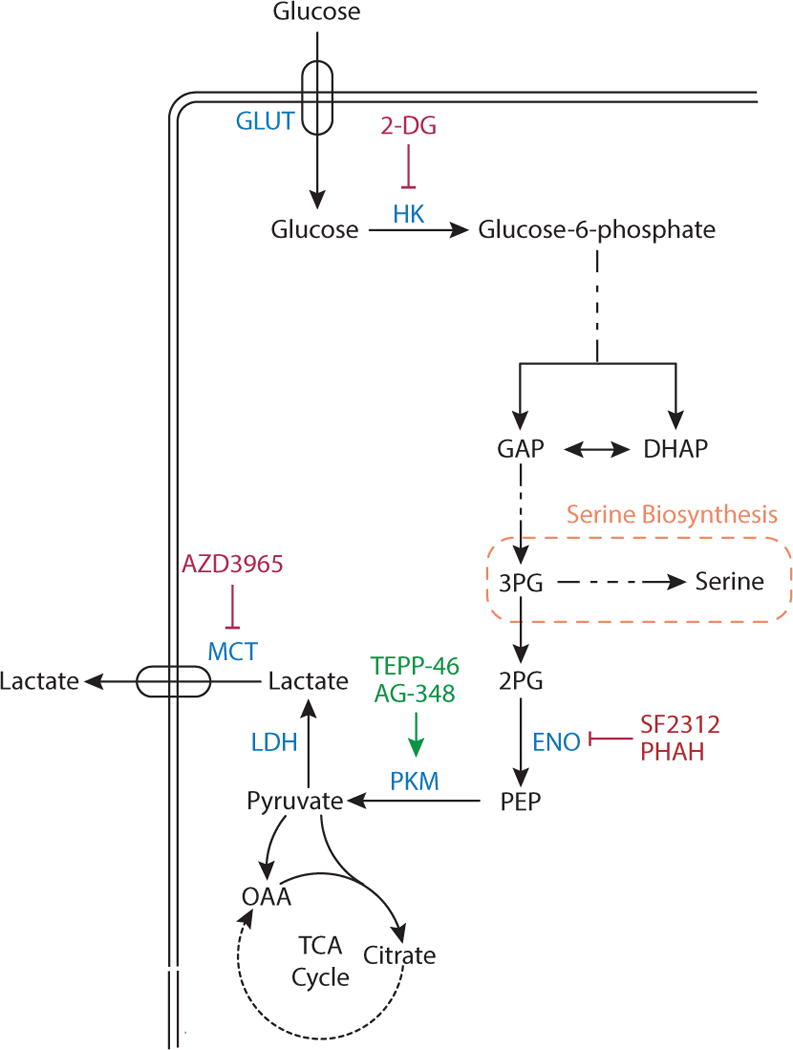
Glucose is imported in cells by one of several glucose transporters (GLUT). Glucose is phosphorylated by hexokinase (HK), a step that can be competitively inhibited by the compound 2-deoxyglucose (2-DG). In a later step of glycolysis, glucose carbon is cleaved into two interconvertible three-carbon units, dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (GAP). Downstream of GAP, 3-phosphoglycerate (3-PG) is converted to 2-phosphoglycerate (2-PG), and the enzyme enolase (ENO) generates phosphoenolpyruvate (PEP) from 2-PG. ENO activity can be inhibited by the compounds phosphonoacetohydroxamate (PHAH) and SF-2312. PEP is converted to pyruvate by pyruvate kinase (PKM), which can be activated by the drugs TEPP-46 and AG-348. Pyruvate can be oxidized in the TCA cycle, or it can produce lactate via lactate dehydrogenase (LDH), with lactate excreted by monocarboxylate transporters (MCT). MCT isoform 1 can be inhibited by the compound AZD3965.
Regulation of pyruvate kinase activity can influence aerobic glycolysis (Dayton et al., 2016). Paradoxically, decreased pyruvate kinase activity is associated with increased aerobic glycolysis, suggesting the activation of pyruvate kinase activity might be a way to target cancer (Christofk et al., 2008). Indeed, activation of pyruvate kinase can inhibit cancer cell proliferation and tumor growth in some settings (Anastasiou et al., 2012; Kung et al., 2012; Walsh et al., 2010), but because pyruvate kinase expression is dispensable for the growth of some tumors (Cortes-Cros et al., 2013; Israelsen et al., 2013), whether pyruvate kinase activation will lead to durable responses remains an area of active study (Israelsen et al., 2013; Israelsen and Vander Heiden, 2015)
Despite challenges associated with targeting glucose metabolism directly, glucose uptake by cancer cells has been successfully exploited in patients through the use of the fluoro-deoxyglucose positron-emission-tomography (FDG-PET) imaging to stage cancers and assess response to therapy (d’Amico, 2015; Farwell et al., 2014; Zhu et al., 2011). Notably, many noncancerous tissues, including the brain, are FDG-PET avid (Berti et al., 2014; Cohade, 2010), illustrating that high glucose uptake is not a unique feature of tumors and offering a potential explanation for the relative lack of success in directly targeting glucose metabolism for cancer treatment. Doses of 2-DG that inhibit glycolysis enough to limit cancer growth may not be tolerated due to similar effects in normal tissues that also rely on glucose metabolism.
Altered Metabolic Enzyme Expression
The expression of metabolic genes is frequently altered in cancer. Some changes in metabolic enzyme expression result from gene amplification or deletion, while others are downstream of growth signaling pathways or are the consequences of epigenetic changes. The activity of metabolic enzymes can also be affected by mutations in the genes encoding these enzymes. Regardless of the underlying mechanism, alterations in basal enzymatic activity of a given reaction present potential vulnerabilities that might be targeted for cancer therapy.
Oncogenic isocitrate dehydrogenase mutations
Recurrent somatic point mutations in the genes coding for isocitrate dehydrogenase 1 and 2 (IDH1, IDH2) are found in a wide variety of cancers, including glioblastoma multiforme (GBM) (Yan et al., 2009) and acute myeloid leukemia (AML) (Dang et al., 2016; Mardis et al., 2009). Cancers expressing mutant IDH represent a unique case in which a metabolic enzyme can act as an oncogene and contribute to tumor development. Wild-type IDH1 and IDH2 catalyze the reversible oxidative decarboxylation of isocitrate to alpha-ketoglutarate (αKG) and CO2 (Figure 4a). Cancer associated mutations in IDH1 and IDH2 eliminate this function and confer a neomorphic activity to the enzyme, generating D-2-hydroxyglutarate (D-2HG) via the reduction of αKG (Figure 4b)(Dang et al., 2009; Ward et al., 2010). Though it is found at low levels in normal cells, D-2HG can accumulate to millimolar levels in cancer cells expressing mutant IDH. At these high concentrations, D-2HG can inhibit αKG-dependent dioxygenases, including enzymes involved in histone and DNA demethylation (Chowdhury et al., 2011; Janke et al., 2017; Koivunen et al., 2012; Xu et al., 2011). As a result, D-2HG accumulation in cancer cells expressing mutant IDH results in hypermethylation of histones and CpG islands in DNA (Figueroa et al., 2010; Lu et al., 2012; Turcan et al., 2012). These epigenetic changes caused by D-2HG contribute to cancer phenotypes (Losman et al., 2013; Rohle et al., 2013; Saha et al., 2014; Wang et al., 2013), and have been proposed to promote oncogenesis by preventing normal cellular differentiation (Losman and Kaelin, 2013).
Figure 4. Mutant IDH.
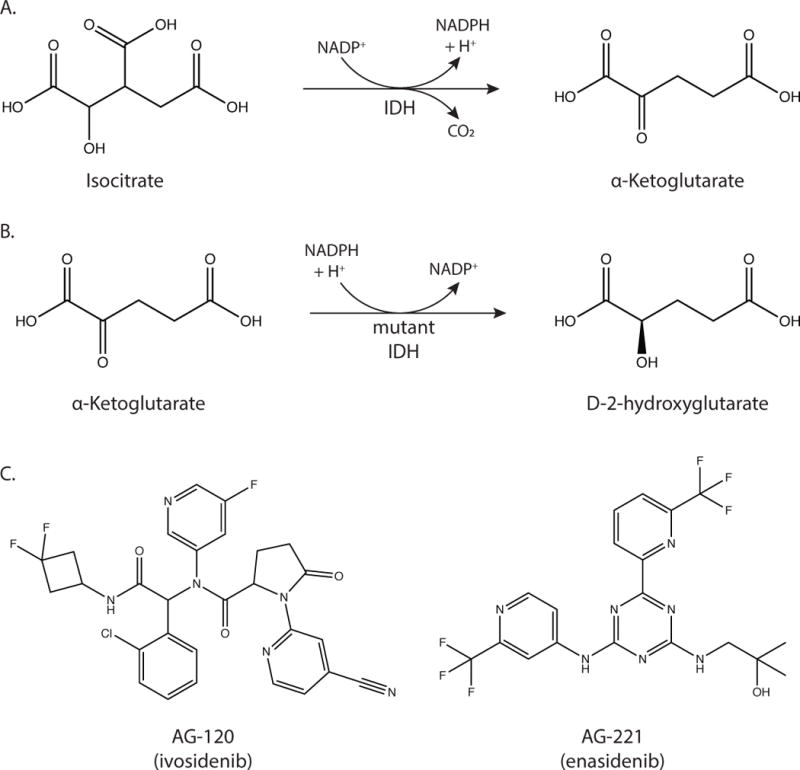
(A) The reaction catalyzed by wild-type IDH is the decarboxylation of isocitrate to α-ketoglutarate. (B) The reaction catalyzed by mutant IDH1 and IDH2 is the reduction of α-ketoglutarate to D-2-hydroxyglutarate. (C) Structures of AG-120 (ivosidenib) and AG-221 (enasidenib), which inhibit mutant IDH1 and IDH2 respectively.
Pharmacological agents that inhibit mutant IDH1 and IDH2 enzyme activity are being developed and assessed for antitumor efficacy (Figure 4c)(Dang et al., 2016). One of the first compounds reported was AGI-5198, which targets mutant IDH1. AGI-5198 reduces intratumoral D-2HG levels, induces expression of genes involved in glial cell differentiation, and suppresses growth of IDH1-mutant human glioma cells in a xenograft model (Rohle et al., 2013). A specific inhibitor of mutant IDH2, AG-221 (enasidenib) confers survival benefit in a mouse model of IDH2-mutant AML (Quivoron et al., 2014; Yen et al., 2017) and became the first compound targeting mutant IDH to enter clinical trials in 2014. Early results in IDH2-mutant AML patients have suggested enasidenib can provide clinical benefit (DiNardo et al., 2015; Stein et al., 2014), and this drug is also being evaluated in solid tumors. Mutant IDH1 inhibitors including AG-120 (ivosidenib) and IDH305, as well as the pan-mutant IDH inhibitor AG-881 (Table 1), are also in clinical trials to treat both hematologic malignancies and solid tumors, and will further inform whether targeting mutant IDH can control disease where IDH mutations are prevalent.
Despite success in some preclinical models and patients with AML, mutant IDH inhibitors may not be effective in all IDH-mutant cancers. For example, these drugs are unable to reverse epigenetic changes or inhibit tumor proliferation in many models of IDH mutant glioma, despite a robust ability to lower 2-HG in cells and tumors (Tateishi et al., 2015; Turcan et al., 2013). IDH1 mutations are early events in the development of glial cancer (Watanabe et al., 2009), raising the possibility that IDH mutations are important for tumor initiation, but accumulation of additional oncogenic mutations renders GBM tumors less dependent on constitutive expression of mutant IDH for tumor proliferation and progression (Johnson et al., 2014; Wakimoto et al., 2014). Nevertheless, the presence of an IDH mutation and high levels of 2-HG might still drive dependencies on some pathways and introduce therapeutic vulnerability. For example, tumors harboring an IDH mutation have increased sensitivity to hypomethylating agents (Turcan et al., 2013), electron transport chain inhibitors (Grassian et al., 2014), depletion of the coenzyme NAD+ (Tateishi et al., 2015) and chemoradiotherapy (Cairncross et al., 2014). However, some evidence suggests that inhibiting mutant IDH could confer resistance to some therapies, as inhibitors targeting mutant IDH1 can antagonize the effects of radiation therapy in glioma (Molenaar et al., 2015). Thus, testing whether combination therapies are synergistic or antagonistic with inhibition of mutant enzyme function is needed to guide treatment.
Upregulated glutaminolysis
Glutamine is a non-essential amino acid, and yet cancer cells proliferating in vitro consume glutamine far in excess of any other amino acid and are often dependent on extracellular glutamine for survival (DeBerardinis and Cheng, 2010; Eagle, 1955; Jain et al., 2012). Glutamine is an important nitrogen donor for amino acids and nucleotides (Hosios et al., 2016), but glutamine uptake can exceed the nitrogen requirement of some cancer cells (DeBerardinis et al., 2007). Glutamine carbon has been found to contribute to aspartate, glutamate and tricarboxylic acid (TCA) cycle metabolites via glutaminolysis (Figure 5) (Altman et al., 2016). High rates of glutaminolysis has been proposed to support rapid proliferation by supplying precursors to low-flux biosynthetic pathways (Newsholme et al., 1985). Providing cells with αKG, oxaloacetate, or pyruvate is sufficient to rescue cancer cell proliferation in conditions of glutamine starvation, confirming that glutamine supports proliferation by replenishing depleted TCA cycle intermediates, a process termed anaplerosis (Altman et al., 2016; Weinberg et al., 2010; Yuneva et al., 2007).
Figure 5. TCA Cycle.
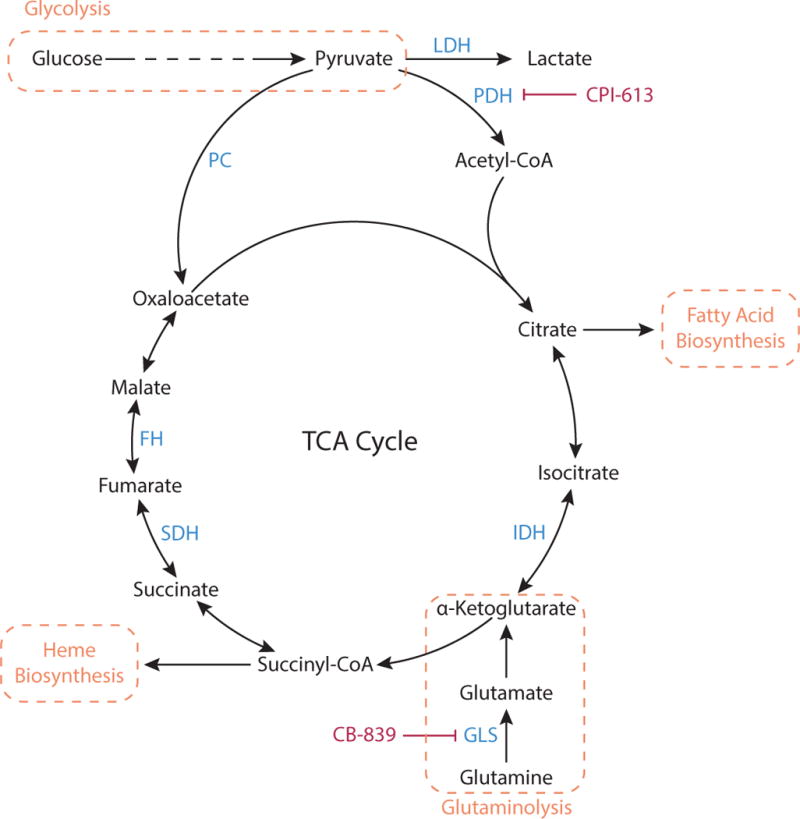
Pyruvate produced by glycolysis can be metabolized by the TCA cycle. Pyruvate is oxidized via pyruvate dehydrogenase (PDH) to the two-carbon unit acetyl-CoA, and subsequently combined with the four-carbon oxaloacetate to generate citrate. The cycle regenerates oxaloacetate while generating precursors that are important for biosynthetic processes including heme and fatty acid synthesis. When molecules are removed from the TCA cycle to feed these pathways, TCA intermediates must be replenished in a process termed anaplerosis. Pyruvate can serve as an anaplerotic substrate when converted to oxaloacetate by the enzyme pyruvate carboxylase (PC). Glutaminolysis, the conversion of glutamine to glutamate via glutaminase (GLS), can also support anaplerosis via production of α-ketoglutarate. PDH activity is inhibited by the lipoic acid derivative CPI-613 and GLS activity is inhibited by CB-839.
Glutamine metabolism is upregulated by various oncogenic signaling pathways (Altman et al., 2016). In certain contexts, MYC-transformed cancers become glutamine dependent and undergo apoptosis in the absence of glutamine (Yuneva et al., 2007). MYC has been found to increase mRNA and protein levels of glutamine transporters as well as expression of the enzyme glutaminase, which catalyzes the first step in glutaminolysis (Gao et al., 2009; Wise et al., 2008; Yuneva et al., 2012). Importantly, inhibiting glutamine entry into the TCA cycle can blunt tumor progression in a MYC-driven cancer model of liver cancer (Xiang et al., 2015; Yuneva et al., 2012) and a MYC-inducible Burkitt lymphoma model (Le et al., 2012; Xiang et al., 2015).
The dependence of cancer cells on glutamine has made glutaminolysis an attractive cancer therapy target (Altman et al., 2016; Daye and Wellen, 2012; DeBerardinis and Cheng, 2010; Vander Heiden, 2011). Clinical trials using glutamine analogues to treat cancers were initiated decades ago, but these trials were abandoned due to lack of efficacy and/or severe patient toxicity (Livingston et al., 1970; O’Dwyer et al., 1984). The absence of a therapeutic window for these studies can likely be attributed to fact that these drugs were relatively non-specific and a panoply of glutamine utilizing enzymes were likely affected.
Current attempts to target glutaminolysis clinically have largely focused on inhibiting glutaminase. Mammals have two glutaminase genes, GLS and GLS2, and targeting the enzymes encoded by these genes with chemical inhibitors has been found to decrease cancer cell proliferation in both in vitro and in vivo models (Gross et al., 2014; Jacque et al., 2015; Le et al., 2012; Xiang et al., 2015; Yuneva et al., 2012). One potent glutaminase inhibitor, CB-839, is currently being evaluated in cancer trials in patients (Table 1), although the exact disease context where glutaminase inhibition will be most effective remains an area of active investigation. There is also data that GLS2 activity can be tumor suppressive (Hu et al., 2010), underscoring the importance of defining the patient population likely to benefit from glutaminase inhibition.
Increased dependence on serine
Increased activity of de novo serine synthesis enzymes in cancer has been observed for more than 30 years (Snell, 1984; Snell et al., 1988). More recently, it was found that increased expression of the serine synthesis enzyme phosphoglycerate dehydrogenase (PHGDH) in some cancers can result from copy number gain of a region on chromosome 1p (Beroukhim et al., 2010; Locasale et al., 2011; Possemato et al., 2011) or a consequence of oncogenic signaling, including NRF2 and ATF4 signaling (DeNicola et al., 2015), or hypoxia responses (Samanta et al., 2016). The PHGDH gene encodes the enzyme that catalyzes conversion of the glycolytic intermediate 3-phosphoglycerate into 3-phosphohydroxypyruvate, and 3-phosphohydroxypyruvate is converted to serine via two subsequent reactions (Figure 6). Increased PHGDH gene expression leads to greater production of serine from glucose and is associated with specific subsets of breast cancer, lung adenocarcinoma, and melanoma (Locasale et al., 2011; Possemato et al., 2011; Zhang et al., 2017). Serine is present in plasma and can be taken up by cells via amino acid transporters, yet PHGDH expression and increased serine biosynthesis have been shown to be important for supporting cancer cell proliferation and survival in both in vitro and in vivo settings (Locasale et al., 2011; Possemato et al., 2011). However, PHGDH activity may not be a requirement for proliferation for all PHGDH-amplified cancers, as expression has been shown to be dispensable in a breast cancer xenograft model (Chen et al., 2013).
Figure 6. Serine Biosynthesis and Folate Cycle.
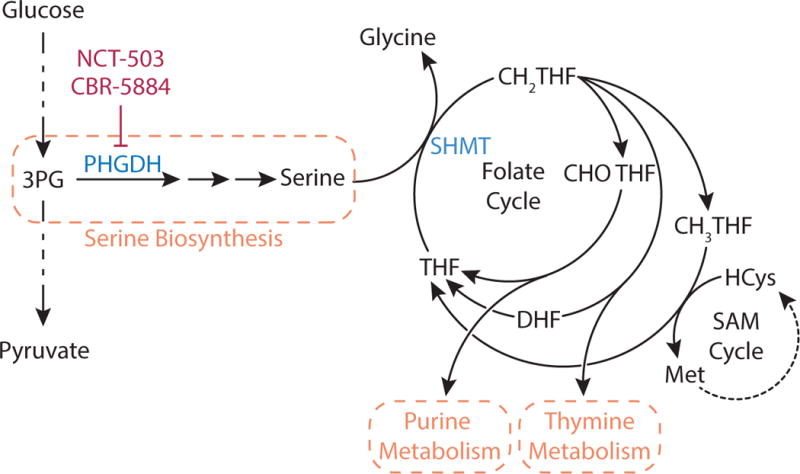
The glycolytic intermediate 3-phophoglycerate (3-PG) is metabolized to serine in a three-step pathway where the first step is catalyzed by the enzyme phosphoglycerate dehydrogenase (PHGDH), which can be inhibited by NCT-503 and CBR-5884. Serine metabolism to glycine by serine hydroxymethyltransferase (SHMT) transfers a one carbon unit to tetrahydrofolate (THF) to form N5,N10-methylene-tetrahydrofolate (CH2-THF). CH2-THF is the methyl donor for thymidine nucleotides, or it can be converted to N10-formyl-tetrahydrofolate (CHO-THF) for purine biosynthesis or to N5-methyl-THF (CH3-THF) to support methionine production and numerous methylation reactions via the S-adenosyl methionine (SAM) cycle.
Why some cancers are dependent on increased serine production is unknown, but increased flux through this pathway may serve to maintain adequate intracellular serine levels. Serine is an amino acid, and thus is required for protein synthesis, but serine can also support many other important critical metabolic processes including synthesis of glycine, glutathione, and phospholipids. With respect to proliferating cells such as cancer, serine is the primary carbon donor to the tetrahydrofolate (THF) cycle, which is required for both purine and pyrimidine nucleotide biosynthesis (Figure 6)(Snell et al., 1987). Serine can also contribute to NADPH production via the folate cycle, which serves to maintain redox homeostasis and support anabolic reactions (Fan et al., 2014; Lewis et al., 2014; Ye et al., 2014). Emerging work suggests that increased serine synthesis may be particularly important for maintaining redox homeostasis during metastasis (Piskounova et al., 2015).
Given the cancer cell requirement for serine, de novo serine synthesis is a potential target for cancer therapy. Functional PHGDH loss is toxic to tumor cells with PHGDH amplification or high serine biosynthetic flux (Locasale et al., 2011; Mattaini et al., 2015; Possemato et al., 2011; Zhang et al., 2017), and small-molecule inhibitors targeting PHGDH have been shown to inhibit serine synthesis and tumor proliferation in vitro and in xenograft cancer models (Mullarky et al., 2016; Pacold et al., 2016; Wang et al., 2017). However, inhibitors of PHGDH might have a limited therapeutic index, as de novo serine synthesis has an important physiological role in the central nervous system (Furuya, 2008) and PHGDH-deficient mice exhibit severe brain morphogenesis defects (Yoshida et al., 2004). Compounds with decreased distribution in the brain may be more effective for cancer therapy.
Certain tumors are dependent on uptake of environmental serine (Jain et al., 2012; Maddocks et al., 2013; Maddocks et al., 2017), and limiting plasma serine availability can be beneficial for patients with these cancers. Removing serine from culture media limits incorporation of one-carbon units into nucleotides and impairs proliferation (Labuschagne et al., 2014). Additionally, serine deprivation by dietary restriction is sufficient to slow growth of both xenograft (Gravel et al., 2014; Maddocks et al., 2013) and autochthonous cancer models, although efficacy of serine deprivation appears to be influenced by both the oncogenic driver mutation and tissue context (Maddocks et al., 2017). Combining serine restriction with other drugs may also potentiate antitumor responses (Gravel et al., 2014; Maddocks et al., 2013; Maddocks et al., 2017). Further understanding the roles of de novo serine synthesis and serine uptake in different tumor contexts can yield important insights into how to target serine metabolism.
FH and SDH loss in heritable cancer syndromes
In addition to genetic events that increase metabolic enzyme expression, some cancers select for deletion of metabolic enzymes. Familial cancer syndromes caused by fumarate hydratase (FH) or succinate dehydrogenase (SDH) deletion suggest that these TCA enzymes can behave as classical tumor suppressors. Affected families inherit one defective copy of either FH or SDH, and develop an aggressive form of cancer upon loss of heterozygosity (Baysal et al., 2000; Tomlinson et al., 2002). Loss of FH activity or SDH activity results in disruption of the TCA cycle and accumulation of fumarate or succinate, respectively. Like D-2HG, fumarate and succinate accumulation can inhibit some αKG-dependent dioxygenases, resulting in hypermethylation of DNA and histones in tumors exhibiting loss of FH or SDH (Hoekstra et al., 2015; Letouze et al., 2013; Xiao et al., 2012). Additionally, succinate and fumarate inhibit the prolyl hydroxylase domain-containing (PHD) enzymes that regulate stability of hypoxia-inducible factors (HIFs), such that FH and SDH deficient cancers activate the hypoxic gene expression program even under normoxic conditions (Hewitson et al., 2007; Pollard et al., 2005; Selak et al., 2005). Both epigenetic changes and aberrant HIF activation can contribute to tumor initiation and progression in these cancers.
Loss of FH or SDH introduces vulnerabilities that may be amenable to therapeutic targeting. In silico modeling of metabolic networks suggests that FH-null cells can upregulate heme metabolism to enable their survival (Frezza et al., 2011). The heme biosynthesis pathway uses succinyl-CoA to generate heme (Figure 5), which can be degraded to bilirubin and excreted from cells, allowing cells to dispose of excess TCA cycle carbon. Thus, FH-null cells may increase flux through this pathway as a means to allow partial TCA cycle activity. Consequently, FH deletion renders mouse and human cells more sensitive to genetic and pharmacological inhibition of heme oxygenase 1 (Hmox1), an enzyme involved in heme degradation.
Another potentially targetable metabolic liability of renal cell cancers that have lost FH expression is a dependence on exogenous arginine. The high levels of fumarate resulting from FH deficiency drives argininosuccinate lyase (ASL) and argininosuccinate synthase (ASS1) in a direction that consumes arginine (Figure 7). Depletion of intracellular arginine causes these cells to become arginine auxotrophs (Adam et al., 2013; Zheng et al., 2013), and therapies that deplete exogenous arginine (Table 1) may be effective in treating malignancies where FH is lost (Phillips et al., 2013).
Figure 7. Urea Cycle.
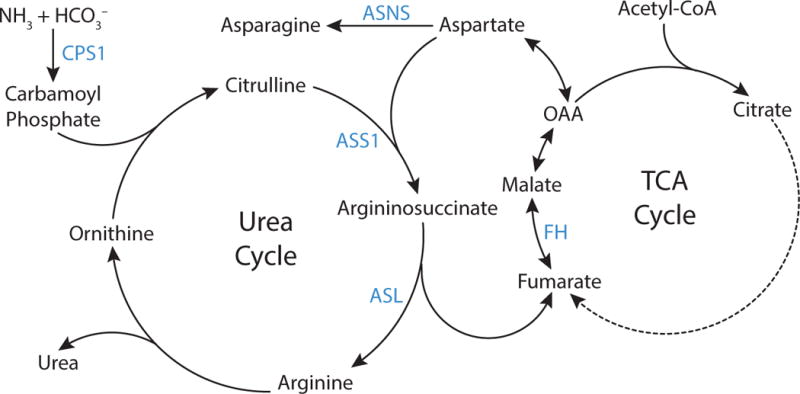
The urea cycle permits excretion of excess nitrogen as urea. Carbamoyl phosphate is synthesized from ammonia and bicarbonate by the enzyme carbamoyl phosphate synthetase 1 (CPS1), and subsequently carbamoyl phosphate and ornithine are used to produce citrulline. The enzyme argininosuccinate synthase (ASS1) condenses citrulline with aspartate to form argininosuccinate, which is then cleaved to arginine and fumarate by argininosuccinate lyase (ASL). Arginine is then hydrolyzed to produce urea and regenerate ornithine, completing the cycle. Fumarate produced by the urea cycle can regenerate aspartate via enzymes involved in the tricarboxylic acid (TCA) cycle as shown. Asparagine synthetase (ASNS) catalyzes production of asparagine from aspartate.
Loss of SDH also results in TCA cycle dysfunction, leading to the accumulation of succinate and depletion of aspartate, fumarate, citrate, and malate. In this context, aspartate cannot be synthesized from glutamine or other sources of αKG, and aspartate production is dependent on pyruvate carboxylase (PC) activity. PC catalyzes carboxylation of pyruvate to oxaloacetate (Figure 5), which can then undergo transamination to form aspartate (Figure 7). To cope with TCA cycle truncation and still produce aspartate, SDH defective cells upregulate PC protein expression in culture and in tumors to synthesize aspartate from glucose (Cardaci et al., 2015; Lussey-Lepoutre et al., 2015). PC ablation impairs SDH-deficient cell proliferation and tumor formation, and thus targeting PC might be exploited to treat these types of cancers.
Loss of argininosuccinate synthase 1 expression
Somatic loss of metabolic enzyme expression may be selected for in other tumor types. For example, some melanoma, lymphoma, glioma, and prostate cancers reduce or lose the expression of the urea cycle enzyme argininosuccinate synthase 1 (ASS1) (Delage et al., 2010). As noted above, ASS1 is involved in arginine synthesis, catalyzing the conversion of citrulline and aspartate to the arginine precursor argininosuccinate (Figure 7). Genetic or epigenetic silencing of ASS1 expression provides an advantage to tumor cells by allowing them to preserve cellular aspartate (Rabinovich et al., 2015), a poorly transported nutrient that can be a critical output of the TCA cycle to support de novo nucleotide synthesis and cell proliferation (Birsoy et al., 2015; Sullivan et al., 2015).
A potential liability of ASS1 deficient cancers is that, like FH null cancers, they are unable to synthesize arginine de novo. As functional arginine auxotrophs, these cells are reliant on exogenous arginine for proliferation and survival and may be sensitive to therapies that lower arginine availability (Table 1). Arginine deiminase (ADI) is a microbial enzyme that catabolizes arginine and can be used to deplete extracellular arginine levels. Recombinant pegylated arginine deiminase (ADI-PEG20) has been tested in clinical trials to treat melanoma and hepatocellular carcinoma with some therapeutic benefit (Ascierto et al., 2005; Izzo et al., 2004; Ott et al., 2013; Szlosarek et al., 2013; Yang et al., 2010). However, the use of arginine-catabolizing enzymes may not be an effective therapeutic strategy for all ASS1-deficient cancers, as some ADI-treated tumors have been found to re-express ASS1 (Feun et al., 2012; Long et al., 2013; Shen et al., 2003). Whether some tumors are more dependent on loss of ASS1 expression to proliferate is an area of active investigation, as these may be more responsive to arginine depleting drugs. The combination of arginine depletion with other therapies might also limit resistance, and the identification of synthetic lethal targets with ASS1-loss is another approach being evaluated to increase the clinical efficacy of therapies that deplete circulating arginine (Bean et al., 2016; Kremer et al., 2017; Locke et al., 2016).
Metabolic collateral lethality
Loss of metabolic enzyme expression can also occur as a passenger event. For example, genomic deletions leading to loss of tumor suppressor genes can also lead to loss of adjacent non-essential metabolic genes. Because cells often exhibit redundancy in essential pathways, this phenomenon can introduce a therapeutic opportunity to target cancer cells that has been termed collateral lethality (Muller et al., 2015). Pancreatic ductal adenocarcinoma (PDAC) cells exhibiting homozygous deletion of the tumor suppressor SMAD4 often lose malic enzyme 2 (ME2) expression due to the chromosomal proximity of the two genes. Targeting the malic enzyme 3 (ME3) isoform was found to impair tumor proliferation of PDAC xenografts lacking ME2 expression, but had no effect on tumors with intact ME2 (Dey et al., 2017). Similarly, the gene encoding enolase 1 (ENO1) is on the tumor-suppressor locus 1p36, and undergoes homozygous deletion in 1–5% of GBM cancers. Knockdown of enolase 2 (ENO2) in ENO1-null GBM cells resulted in significant inhibition of proliferation and intracranial tumorigenesis, whereas ENO1 expressing cancer cells were insensitive to ENO2 ablation. Furthermore, ENO1 loss results in extreme sensitivity to the pan-enolase inhibitor phosphonoacetohydroxamate (PHAH) and SF2312 (Leonard et al., 2016; Muller et al., 2012). In both examples, decreased metabolic enzyme redundancy rendered the cells dependent on a specific isoform of an enzyme that could be selectively targeted. It also decreases total cellular levels of enzymatic activity for a given reaction, thereby lowering the threshold for toxicity for targeting the corresponding enzyme.
Passenger deletion of metabolic genes can introduce vulnerabilities involving other pathways as well. Deletion of the tumor suppressor CDKN2A results in concomitant deletion of the methionine salvage pathway enzyme methylthioadenosine phosphorylase (MTAP) in many cancers, including 53% of glioblastomas and 26% of pancreatic cancers (Mavrakis et al., 2016). MTAP cleaves methylthioadenosine (MTA), a product of polyamine biosynthesis, into 5-methylthioribose-1-phosphate (MTR) and adenine, which are further metabolized to methionine and adenosine monophosphate (AMP), respectively (Figure 8). MTAP deficient cells are more reliant on de novo purine synthesis to generate AMP, since they are unable to cleave MTA to salvage adenine, and thus MTAP loss makes cells more susceptible to inhibitors of purine biosynthesis as well as to methionine depletion (Hori et al., 1996). Co-administration of MTA with toxic adenosine analogues has been shown to be selectively lethal to MTAP deficient cancer cells, since MTAP-expressing normal tissues are able to convert MTA to adenine to competitively inhibit the effects of the analogues (Lubin and Lubin, 2009). L-alanosine inhibits conversion of inosine monophosphate (IMP) to AMP, and shows selective toxicity towards MTAP-null cancer cells (Batova et al., 1999; Efferth et al., 2003; Harasawa et al., 2002), but was found to be clinically ineffective in patients with advanced MTAP-deficient tumors in a Phase II trial (Kindler et al., 2009). Pharmacokinetic analyses confirming successful purine biosynthesis inhibition by doses of L-alanosine used in the study were not reported, so further exploration of why this approach failed may yield insight into how best to exploit MTAP deficiency.
Figure 8. Methionine Cycle.
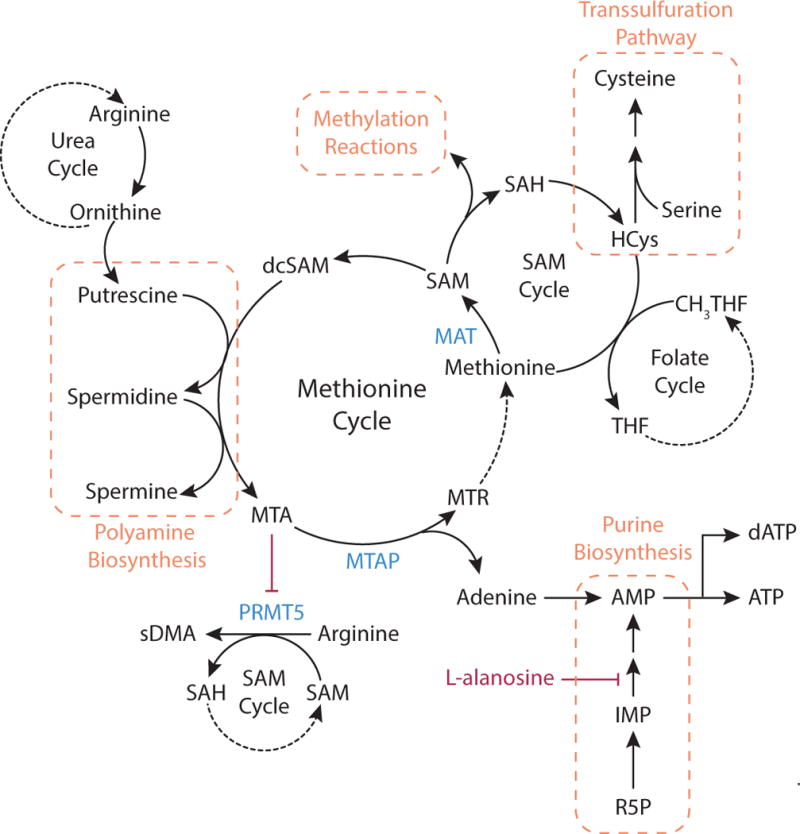
Methionine is an essential amino acid that can be used for methylation reactions, cysteine synthesis, and polyamine generation. Methionine is converted to S-adenosylmethionine (SAM) by methionine adenosyltransferase (MAT). Methyl transferases utilize SAM as the methyl donor for many methylation reactions in cells. Demethylation of SAM generates S-adenosylhomocysteine (SAH), which is hydrolyzed to homocysteine (HCys) and adenosine. Methionine is regenerated from homocysteine by transfer of the methyl group from N5-methyl-THF (CH3-THF). Homocysteine is also an intermediate of cysteine synthesis, where serine and homocysteine condense and form cysteine via the transsulfuration pathway. SAM can also support polyamine synthesis when SAM is decarboxylated to form S-adenosylmethioninamine, also known as decarboxylated S-adenosylmethionine (dcSAM). Together with the urea cycle metabolite ornithine, this compound generates putrescine and subsequently spermine and spermidine. In these reactions, dcSAM is converted to 5′-methylthioadenosine (MTA). Accumulation of MTA inhibits the enzyme protein arginine N-methyltrasferase 5 (PRMT5), which uses SAM as a methyl donor to synthesize symmetrical dimethylarginine (sDMA) from arginine. In the methionine cycle, MTA is cleaved to 5-methylthioribose-1-phosphate (MTR) and adenine by the enzyme methylthioadenosine phosphorylase (MTAP). Adenine can be converted to AMP, and the adenosine analogue L-alanosine can limit AMP production via the purine synthesis pathway. MTR can be converted to methionine, completing the methionine cycle.
Loss of MTAP expression can result in another vulnerability for cancer cells because the MTA that accumulates following MTAP loss can act as a potent inhibitor of the enzyme arginine methyltransferase 5 (PRMT5). MTAP loss results in reduced PRMT5 activity and renders MTAP-null cancer cell more sensitive to PRMT5 depletion than isogenic counterparts that express MTAP (Kryukov et al., 2016; Marjon et al., 2016; Mavrakis et al., 2016). Additionally, ablation of methionine adenosyltransferase II alpha (MAT2A), the enzyme that produces the canonical and high-affinity PRMT5 substrate, S-adenosylmethionine (SAM), also reduces PRMT5-dependent methylation and proliferation in MTAP-deleted cancer cells (Marjon et al., 2016). However, PRMT5 inhibitors in current clinical trials may not be effective in MTAP null cancers, in part because the high levels of MTA in these cells compete for binding of the inhibitors to the enzyme. Future studies will better define whether reducing PRMT5 activity using drugs that act via a different mechanism could be used as effective treatments in tumors where MTAP is co-deleted with CDKN2A.
Emerging Metabolic Targets
Numerous metabolic differences between cancer cells and normal cells have been described. Some recent examples to illustrate how these differences might be exploited for therapy are highlighted, but many other targets have been proposed and are discussed in detail elsewhere (Bobrovnikova-Marjon and Hurov, 2014; Galluzzi et al., 2013; Martinez-Outschoorn et al., 2017).
Targeting de novo lipid synthesis
Several lines of evidence suggest that targeting de novo fatty acid synthesis might be effective in the treatment of some cancers. Fatty acids are a key component of cell membranes and can also act as signaling molecules or store energy. It was first discovered in the 1950s that tumors are able to synthesize lipids, and a subsequent study determined that the large majority of lipids in tumors cells are synthesized de novo, rather being consumed from exogenous sources (Medes et al., 1953; Ookhtens et al., 1984). Since then, numerous studies have identified de novo fatty acid biosynthesis as a key metabolic requirement for some cancers, and it has been dubbed by some as a distinct metabolic “hallmark of the transformed phenotype” (Kuhajda et al., 1994; Menendez and Lupu, 2006; Rohrig and Schulze, 2016). With the exception of liver, adipose tissue, and lactating breast, adult tissues do not synthesize fatty acids de novo under normal physiological conditions (Menendez and Lupu, 2006), so inhibition of de novo fatty acid synthesis might have an adequate therapeutic window.
Fatty acid synthesis is a multi-step process that primarily occurs in the cytosol of cells. First, acetyl-CoA groups are converted to malonyl-CoA via the enzyme acetyl-CoA carboxylase (ACC). Next, the multidomain enzyme fatty acid synthase (FASN) assembles fatty acid chain palmitate from malonyl-CoA. Although fatty acid synthesis occurs in the cytosol, cytosolic acetyl-CoA is produced from mitochondrial citrate, which is exported from mitochondria and cleaved by cytosolic ATP-citrate lyase (ACLY) (Figure 9). Cancer cells are also able to generate acetyl-CoA from cytosolic acetate (Comerford et al., 2014; Gao et al., 2016; Kamphorst et al., 2014; Mashimo et al., 2014). However, acetate is not always abundant in blood and is not the predominate source of acetyl-CoA used for fatty acid synthesis in some cells (Hosios and Vander Heiden, 2014). Some cancers are nevertheless dependent on expression of acetyl-CoA synthetase 2 (ACSS2), the cytosolic enzyme that allows cells to synthesize acetyl-CoA from acetate, making this represents a potential therapeutic target (Comerford et al., 2014; Schug et al., 2015).
Figure 9. Fatty Acid Synthesis.
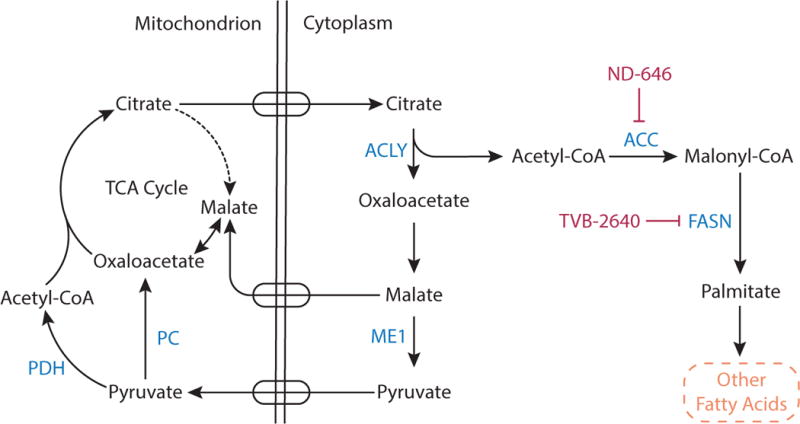
Citrate is a carrier of acetyl groups from the mitochondria to the cytoplasm to support fatty acid synthesis. Mitochondrial citrate is transported into the cytosol where ATP citrate lyase (ACLY) cleaves citrate to acetyl-CoA and oxaloacetate. Oxaloacetate is converted to malate, and malate can be transported back into the mitochondrial matrix or can be converted into pyruvate in the cytoplasm by malic enzyme 1 (ME1). Cytosolic acetyl-CoA contributes to fatty acid synthesis. First, the enzyme acetyl-CoA carboxylase (ACC) catalyzes carboxylation of acetyl CoA to malonyl CoA. Fatty acid synthase (FASN) then uses malonyl-CoA to sequentially add two carbon units to a growing acyl chain and synthesize the saturated 16-carbon fatty acid palmitate, which serves as a precursor to other fatty acids. The compound ND-464 and TVB-2640 are inhibitors of ACC and FASN, respectively.
Numerous inhibitors have been developed to target fatty acid synthesis, with attempts to limit cytosolic acetyl-CoA availability via ACLY inhibition as well as direct targeting of the enzymes ACC and FASN (Figure 9). ACLY activity is elevated in cancers (Migita et al., 2008), and targeting ACLY genetically or chemically prevents xenograft tumor formation and proliferation (Adam et al., 2013; Bauer et al., 2005; Hatzivassiliou et al., 2005; Migita et al., 2008). Genetic knockdown of ACC induces apoptosis in cancer cell lines (Brusselmans et al., 2005; Chajes et al., 2006), and an allosteric inhibitor of ACC, ND-646, has shown antitumor efficacy in autochthonous mouse lung tumor models (Svensson et al., 2016). Targeting FASN has been found to reduce palmitoylation of tubulin and disrupt microtubule organization, inhibiting tumor cell growth (Heuer et al., 2017). The compound TVB-2640 is the first compound targeting FASN to enter clinical trials (Table 1), and when combined with paclitaxel, can cause partial responses or prolonged stable disease (Brenner et al., 2017).
Differential requirements for NAD+/NADH homeostasis
Warburg’s observation that cancer cells have dramatically increased glucose consumption and lactate production, even in the presence of oxygen, led to the hypothesis that cancer cells have diminished mitochondrial function (Warburg, 1956). However, subsequent work determined that despite engaging in aerobic glycolysis, cancer cells consume oxygen at levels comparable to normal tissue (Weinhouse, 1956; Zu and Guppy, 2004). Moreover, inhibitors of cellular respiration block proliferation, suggesting that most cancer cells require respiration in order to proliferate (Harris, 1980; Howell and Sager, 1979; Kroll et al., 1983; Loffer and Schneider, 1982; Zhang et al., 2014b). Respiration is also needed for tumor initiation, as tumor cells with impaired oxidative phosphorylation due to depletion of mitochondrial DNA (mtDNA) exhibit increased tumor latency upon subcutaneous transplantation. In fact, cells derived from these tumors acquire host mtDNA to regain the ability to do respiration, providing compelling evidence that respiration is required and selected for in tumorigenesis (Tan et al., 2015a).
Most cells in the body generate ATP via respiration, so targeting respiration might be expected to be toxic with a limited therapeutic window. However, metformin, one of the commonly prescribed drugs for treating type II diabetes, is safe despite acting as a mitochondrial complex I inhibitor that impairs respiration (Bridges et al., 2014; El-Mir et al., 2000; Owen et al., 2000; Wheaton et al., 2014). Furthermore, retrospective clinical studies have found that metformin use is associated with improved cancer outcomes, reductions in cancer incidence, and decreased cancer mortality (Evans et al., 2005; Franciosi et al., 2013; Gandini et al., 2014; Lee et al., 2012; Noto et al., 2012). Metformin has also been found to cooperate with neo-adjuvant chemotherapy to result in complete tumor regression in some breast cancer patients (Jiralerspong et al., 2009).
The anti-tumorigenic properties of metformin and other biguanides have been modeled in various mouse cancer models (Buzzai et al., 2007; Huang et al., 2008; Shackelford et al., 2013; Wheaton et al., 2014). Although the precise mechanism of metformin action remains controversial (Luengo et al., 2014), recent work has shown that the anti-tumorigenic effect of metformin can at least be partially accounted for by direct mitochondrial complex I inhibition in tumors (Gui et al., 2016; Wheaton et al., 2014). Consistent with this notion, other complex I inhibitors have shown efficacy as anti-tumor agents (Appleyard et al., 2012; Schockel et al., 2015) and may show selective toxicity against oncogene-ablation resistant cells (Viale et al., 2014) and cancer stem cells (Sancho et al., 2015). Additional complex I inhibitors are under development (Bastian et al., 2017), and other inhibitors of respiration or mitochondrial metabolism, including the lipoic acid derivative CPI-613 (Table 1), are currently being assessed in clinical trials (Lycan et al., 2016; Pardee et al., 2014).
Recent work has shed some light on the potential therapeutic window for treating tumors with respiration inhibitors. Mitochondria are viewed as the powerhouse of the cell, with respiration considered primarily as an ATP-producing catabolic process. However, mitochondrial ATP production appears dispensable for many proliferating cells (Birsoy et al., 2015; Sullivan et al., 2015; Titov et al., 2016). Instead, respiration serves an alternative anabolic role for proliferating cells by regenerating the oxidized form of nicotinamide adenine dinucleotide (NAD+) from the reduced form (NADH) (Birsoy et al., 2015; Sullivan et al., 2015; Titov et al., 2016). Maintenance of intracellular NAD+ is required for many cellular processes, including protein deacetylation, ADP-ribosylation and calcium signaling (Chiarugi et al., 2012). Additionally, NAD+ serves as a critical redox cofactor required to generate oxidized molecules, such as amino acids and nucleotides necessary for biomass accumulation (Birsoy et al., 2015; Sullivan et al., 2015; Titov et al., 2016). Proliferating cells often require a high NAD+/NADH ratio to support anabolic reactions, and in some contexts the NAD+/NADH ratio directly correlates with proliferation rate (Gui et al., 2016). Higher NAD+/NADH ratios appear important for proliferation while lower ratios favor mitochondrial ATP production. Thus, one possible explanation for the potential therapeutic window of respiration inhibitors is that the NAD+/NADH ratio regime required for proliferation of tumor cells is distinct from that which is required to maintain ATP production in normal cells.
Targeting NAD+ synthesis could be another mechanism to limit NAD+ pools and target proliferative metabolism. NAD+ is produced via multiple synthesis and salvage pathways, but a major source of NAD+ in proliferating cells is salvage from nicotinamide. The rate-limiting enzyme in this pathway is nicotinamide phosphoribosyltransferase (NAMPT), which is highly expressed in tumors arising from diverse tissues. Small-molecules targeting NAMPT can be effective anti-tumor agents in vitro and in xenograft cancer models (Nahimana et al., 2009; Tan et al., 2015b; Watson et al., 2009). In clinical trials, use of these inhibitors as single agents has not caused tumor remission (von Heideman et al., 2010), but additional studies may uncover how best to safely use these drugs for patient benefit. For example, NAMPT inhibitors may be more effective in contexts of MYC-amplified glioblastoma (Tateishi et al., 2016) or when used in combination with other drugs known to deplete NAD+ pools (Bajrami et al., 2012; Chan et al., 2014).
Targeting nucleotide acquisition and synthesis
Acquiring nucleotides is critical for cell proliferation. Nucleic acids can be synthesized de novo or scavenged from the environment. Though nucleotide salvage pathways can support tumor cell proliferation in certain contexts (Tabata et al., 2017), concentrations of circulating nucleotides are likely too low for many tumors to satisfy their nucleotides demand via salvage alone (Traut, 1994). Nevertheless, the ability of some tumor cells to scavenge nucleotides is exploited for cancer therapy. While most cells are able to scavenge the epigenetically modified cytosine bases 5-hydroxymethyl-2′deoxycytidine (5hmdC) and 5-formy-2′deoxycytidine (5fdC) and incorporate these bases into the DNA without compromising genome integrity, cancer cell lines overexpressing cytidine deaminase (CDA) convert these bases to the corresponding modified uracil analogues, which induce cytotoxicity when incorporated into DNA (Zauri et al., 2015). Thus, administering 5hmdC and 5fdC may selectively target tumors overexpressing CDA.
The clinical success of some antimetabolites argues select tumors are reliant on de novo nucleotide synthesis. Nucleotide biosynthesis interfaces with other metabolic pathways; ribose is synthesized from glucose via the pentose phosphate pathway and nucleotide bases require carbon atoms from amino acids and the one-carbon pool as well as nitrogen atoms from aspartate and glutamine. Consequently, targeting amino acid and folate metabolism can affect nucleotide production. For example, eliminating environmental serine, limiting aspartate synthesis via mitochondrial inhibition, and ablating the mitochondrial folate pathway impairs purine biosynthesis (Ducker et al., 2016; Labuschagne et al., 2014; Sullivan et al., 2015). This raises the possibility that these interventions can widen the therapeutic window of established antimetabolites, and future studies will determine whether targeting these pathways is synergistic with existing antimetabolite therapies in certain contexts.
Nucleotide production is downstream of some oncogenic signaling pathways. Activation of mTOR can increase ribose synthesis via the oxidative pentose phosphate pathway (Duvel et al., 2010) and enhance flux through the pyrimidine and purine synthesis pathways (Ben-Sahra et al., 2013; Ben-Sahra et al., 2016; Robitaille et al., 2013), suggesting that targeting nucleotide metabolism in tumor contexts with mTOR hyperactivation may be more effective. PTEN-deficient cells, which have active mTOR signaling, are more sensitive to inhibition of DHODH by brequinar and leflunomide (Mathur et al., 2017). Conversely, inhibiting oncogenic signaling using PI3K inhibitors, has been found to deplete DNA nucleotides, decrease DNA synthesis, and cause DNA damage (Juvekar et al., 2016). KRAS/LKB1-null lung cancer cells are more reliant on the urea cycle enzyme carbamoyl phosphate synthetase-1 (CPS1) for pyrimidine biosynthesis, although this dependency is independent of mTOR signaling (Kim et al., 2017).
Increased requirement for detoxification of reactive metabolites
Cancer cells generally have increased levels of reactive metabolites (DeBerardinis and Chandel, 2016; Sullivan et al., 2016). These metabolites can be formed as error products of metabolic enzymes or can be formed non-enzymatically due to the intrinsic reactivity of certain metabolic intermediates. Some of these byproducts, including reactive oxygen species (ROS) and methylglyoxal, are themselves chemically unstable and, thus, can covalently modify both amino acids and nucleic acids. While elevation of reactive metabolites can support oncogenesis in some contexts (Arnold et al., 2001; Irani et al., 1997; Suh et al., 1999; Weinberg et al., 2010), at high levels reactive metabolites become toxic and kill cells due to the resulting damage to the proteome and genome (Karisch et al., 2011; Rabbani and Thornalley, 2014). Cancer cells rely on several pathways to prevent reactive metabolite accumulation. For example, the transcription factor Nrf2, which mediates an antioxidant response program to detoxify ROS, is downstream of several oncogenic drivers, and loss of Nrf2 in this context can impair tumor proliferation (Chio et al., 2016; DeNicola et al., 2011). Disruption of this and other detoxification pathways is predicted to have a more deleterious effect on cancer cells than normal cells, and thus serves as a potential targetable metabolic liability.
An alternative strategy for exploiting the increased production of reactive metabolites observed in cancer cells is to target the synthesis or utilization of antioxidants such as glutathione. Glutathione is a cysteine-containing tripeptide that plays a crucial role in many detoxification pathways. It is one of the most abundant metabolites in the cell and serves to reduce the levels of reactive metabolites by serving as a substrate for electrophilic attack. Several screens for small-molecules that selectively kill transformed cells have identified compounds that reduce glutathione levels (Dolma et al., 2003; Raj et al., 2011; Trachootham et al., 2006), and genetic or pharmacological inhibition of glutathione synthesis results in ROS accumulation and impedes cancer initiation and cell proliferation (Harris et al., 2015; Lien et al., 2016). Therapeutic interventions that decrease intracellular glutathione levels, such as administration of the oxidized form of vitamin C, dehydroascorbate (DHA), can result in cancer cell death and impaired tumor progression (Yun et al., 2015)
Another way to deplete intracellular glutathione is by limiting availability of the amino acid cysteine, the key reactive residue of glutathione that is required for its synthesis. Many cancer cells are auxotrophic for this amino acid, and lowering whole body cysteine levels with bacterial cysteineases is a potential cancer treatment being evaluated in clinical trials (Graczyk-Jarzynka et al., 2017). Targeting the cystine and glutamate transporter xCT (SLC7A11) is another approach to limit intracellular cysteine availability and prevent cysteine-dependent glutathione synthesis. Inhibitors of this transporter have been found to deplete intracellular glutathione in cancer cells, resulting in iron-dependent accumulation of ROS and a form of cell death termed ferroptosis (Dixon et al., 2012; Dixon et al., 2014; Yang et al., 2014).
Lineage and Environment Specific Vulnerabilities
Analysis of gene expression across different cancer types has determined that metabolic enzyme expression is heterogeneous across tumors, and there is no universal metabolic transformation common to all cancers. In fact, tumors retain many features that correspond to their parental normal tissue (Gaude and Frezza, 2016; Hu et al., 2013). It is not known whether this effect results from rigid lineage-specific metabolic expression programs or from the similar local microenvironment experienced by the tumor and normal tissue. However, these analyses suggest that metabolic vulnerabilities may not be universal across all cancers and highlight the possibility that some metabolic dependencies of cancer may be modulated by environment or defined by tumor lineage.
Differential utilization of amino acids
Acquisition of amino acids is an important biosynthetic requirement for cancer cells to support proliferation (Hosios et al., 2016). How cells acquire amino acids can vary according to cell type and environment, and therefore targeting amino acid metabolism could offer a therapeutic window for cancer treatment (Yuneva et al., 2012). Recently, it has been determined that mouse lung and pancreatic tumors exhibit differences in amino acid metabolism, even when harboring the same genetic lesion. Lung tumors support their nitrogen requirement by catabolizing free branched chain amino acids (BCAA), whereas pancreatic tumors expressing the same driver mutation do not. Targeting BCAA transaminases inhibits lung tumor formation better than pancreatic tumor formation, suggesting that dependence on branched chain amino acid catabolism is determined by cell lineage rather than tumor site (Mayers et al., 2016). Tumors of other lineages are also dependent on BCAA transaminases, including some glioblastomas and blast crisis chronic myeloid leukemia (Hattori et al., 2017; Tonjes et al., 2013). Together, these observations suggest that tissue-of-origin can influence metabolic requirements and vulnerabilities.
Asparagine auxotrophy in acute lymphoblastic leukemia
The non-essential amino acid asparagine is abundant in plasma and can also be synthesized by many cell types, so either source of asparagine is dispensable for most cells. Acute lymphoblastic leukemia (ALL) and related lymphomas, however, are auxotrophic for asparagine (Neuman and McCoy, 1956). Bacterial L-asparaginase deamidates asparagine to aspartic acid, thereby limiting the availability of asparagine for cancer cells. L-asparaginase has been found to be effective in the treatment of ALL and is included in standard chemotherapy regimens for this disease (Table 1)(Egler et al., 2016). Though the mechanism of asparagine auxotrophy in ALL cells was assumed to be genetically defined, studies have shown that clinical response to L-asparaginase treatment is independent of asparagine synthase (ASNS) expression, the enzyme that converts aspartate to asparagine (Figure 7) (Appel et al., 2006; Stams et al., 2003). In fact, genome-wide analysis has shown that there is no consistent gene expression pattern that dictates sensitivity to L-asparaginase (Fine et al., 2005). The absence of a genetic driver for asparagine auxotrophy, combined with the fact that L-asparaginase has little known clinical utility outside of the context of ALL, suggests that this metabolic vulnerability may be lineage dependent.
Inducing differentiation in myeloid cells
A common feature of acute myeloid leukemia (AML) is that leukemic myeloblasts arrest at an immature stage of differentiation. This differentiation blockade is a hallmark of AML even though there are multiple driver mutations for this disease. Inhibitors of mutant IDH1 or IDH2 have been shown to induce differentiation in AML cells harboring mutant IDH (Okoye-Okafor et al., 2015; Wang et al., 2013). The transcription factor HoxA9 is overexpressed in 70% of AML cases, and overexpression of this transcription factor alone is sufficient to immortalize murine bone marrow derived myeloid cells (Ayton and Cleary, 2003; Golub et al., 1999; Kroon et al., 1998). Recently, a myeloid cell screen involving HoxA9-enforced differentiation arrest found that inhibition of dihydroorotate dehydrogenase (DHODH), an enzyme in the pyrimidine biosynthesis pathway (Figure 1b), can induce differentiation (Sykes et al., 2016). Targeting DHODH depletes uridine and downstream metabolites, and providing uridine is sufficient to maintain myeloid arrest, even in the presence of compounds targeting DHODH. Treatment with brequinar sodium (BRQ), an inhibitor of DHODH that is approved to treat autoimmune disease, is able to overcome myeloid differentiation blockade and reduces leukemic cell burden in patient-derived xenografts and syngeneic mouse AML models of diverse genetic subtypes. BRQ was previously evaluated to treat advanced solid tumors but was not shown to be effective in clinical trials (Arteaga et al., 1989; Noe et al., 1990; Peters et al., 1990). However, the effect of BRQ on patients with hematologic malignancies has not yet been evaluated. Future trials will determine whether targeting DHODH could exploit a cell lineage-dependent liability in hematological malignancies.
Non-tumor cells can influence tumor cell metabolism
Metabolic symbiosis likely exists between tumor cells and the surrounding stroma, and this interaction could also contribute to tumor growth and proliferation. Typically, lactate is excreted at high rates by tumor cells, but lactate has also been described as an oxidative substrate for some cancers. It has been reported that lactate produced by cancer-associated fibroblasts can enter the TCA cycle of cancer cells (Bonuccelli et al., 2014; Witkiewicz et al., 2012), and lactate produced in hypoxic regions of a tumor can fuel respiration in well-oxygenated tumor cells (Kennedy et al., 2013; Sonveaux et al., 2008). Intraoperative isotopically-labeled glucose infusions performed on non-small cell lung cancer patients revealed higher contribution of glucose carbon to TCA intermediates than glycolytic intermediates in tumors, leading the authors to conclude that lactate could serve as an anaplerotic substrate in this context as well (Hensley et al., 2016). Lactate utilization by tumors is dependent on monocarboxylate transporters (MCTs) (Figure 3), which are highly expressed in a variety of tumors (Pinheiro et al., 2012). Compounds targeting MCT1 are currently being evaluated for antitumor efficacy (Table 1)(Marchiq and Pouyssegur, 2016).
Alanine is rarely limiting for proliferation, as cancer cells typically excrete, rather than consume, alanine (DeBerardinis et al., 2007; Hosios et al., 2016; Jain et al., 2012). However, in some contexts cells may not synthesize alanine in excess of their biosynthetic requirements. Pancreatic ductal adenocarcinoma cells can consume alanine produced by pancreatic stellate cells in vitro, and the secreted alanine appears to help promote tumor cell proliferation and survival under nutrient-limited conditions (Sousa et al., 2016). An improved understanding of the interaction between PDAC tumors and stroma can help determine the metabolic limitations of pancreatic tumor growth, and guide target selection to exploit this symbiotic relationship for therapy.
Metabolic dependencies can be modulated by environment
Differences in nutrient availability may contribute to sensitivity or resistance of cancer cells to some drugs. For example, most cells in culture rely on glutamine metabolism, but targeting glutaminase is not an effective cancer therapy in all contexts. Glutamine tracing studies have demonstrated that in some tumor tissues, glutamine contributes minimally to TCA cycle intermediates, and those cancers can be resistant to glutaminase inhibition (Davidson et al., 2016; Hensley et al., 2016; Marin-Valencia et al., 2012; Sellers et al., 2015; Tardito et al., 2015). Importantly, cell lines derived from resistant lung tumors become sensitive to glutaminase inhibition in vitro, highlighting that environment can be a contributor to glutaminase sensitivity (Davidson et al., 2016). What factors determine the differential use of glutamine in different environments remains an active question. Targeting redox homeostasis with the drug metformin is another example where environment can influence drug efficacy (Gui et al., 2016). Altering culture media conditions is sufficient to change cancer cell sensitivity to metformin, highlighting the importance of environment in modulating metabolic dependencies.
What Dictates Metabolic Dependencies?
One of the biggest hurdles for cancer drug development is identifying the patients most likely to respond to a given therapy. With the recent success of some targeted cancer therapies, patient selection for drugs is often predicated on expression of an oncogene or the increased activity of a downstream signaling pathway. However, this same approach may not identify responders for all metabolic cancer therapies, and indeed, response to chemotherapies that target metabolism has not been well predicted by this approach. The protein targets of methotrexate and 5-FU are present in almost all cancers, but the efficacy of these drugs varies dramatically across malignancies. Many of the emerging metabolic targets discussed in this review are also expressed widely in cancer, but dependency on these metabolic pathways is not always universal. For example, while targeting ACC to limit fatty synthesis can suppress lung tumor progression in some models (Svensson et al., 2016), loss of ACC activity can accelerate tumor growth in other cancer models (Jeon et al., 2012). Similarly, many PHGDH-amplified tumors and cell lines are sensitive to genetic or pharmacological inhibition of the enzyme (Locasale et al., 2011; Mullarky et al., 2016; Pacold et al., 2016; Possemato et al., 2011; Wang et al., 2017; Zhang et al., 2017), but for some tumor models PHGDH expression can be dispensable for growth (Chen et al., 2013). Even among cancers where mutant IDH is an oncogenic driver, sensitivity to mutant IDH inhibitors appears to differ between leukemia and solid tumor models (Tateishi et al., 2015; Turcan et al., 2013).
There is growing evidence that metabolic dependencies in cancer are influenced by tissue environment, cancer lineage, as well as genetic events. Cancer tissue-of-origin has served as a successful way to select patients for chemotherapies targeting metabolism for decades, and environment can also influence the efficacy of drugs targeting DHODH, mitochondrial complex I, and glutaminase (Davidson et al., 2016; Gui et al., 2016; Marin-Valencia et al., 2012; Sellers et al., 2015; Sykes et al., 2016; Tardito et al., 2015). Unraveling the complexity for how lineage, environment and genetics interact to create metabolic dependencies will be challenging, but may provide a path to exploit metabolism in a way that could be transformative for patients. Additionally, immunotherapies are playing an increasing role in cancer therapy, and immune cell fate can also be influenced by metabolism. Determining how immune cell metabolism interacts with tumor metabolism, and how this is modulated by drugs targeting metabolic enzymes, might aid in the design of more effective immunotherapies (Buck et al., 2017; Mockler et al., 2014).
Many drugs targeting metabolism are among the most effective clinical drugs for particular diseases, and many newer metabolic therapies are limited more by toxicities than by their ability to kill cancer cells. A better understanding of the metabolic dependencies in specific tumor tissues holds the key for defining the aspects of metabolism most limiting for tumor growth and finding a therapeutic window to exploit those vulnerabilities for better cancer treatment.
The success of chemotherapies targeting metabolism illustrate that metabolic liabilities can be exploited to treat cancer. We review new insights into the differential metabolic dependencies of tumors and discuss how understanding cancer metabolism might guide the development of new drugs that target the metabolic requirements of tumor cells.
Acknowledgments
The authors thank members of the Vander Heiden laboratory for thoughtful discussions and comments on the manuscript. M.G.V.H. acknowledges support from the Lustgarten Foundation, SU2C, the Ludwig Center at MIT, the NCI, and an HHMI Faculty Scholar’s award. A.L. was supported by the Ludwig Center for Molecular Oncology Fund and NSF (GRFP DGE-1122374). D.Y.G received support from NIH (T32 GM007753).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
M.G.V.H. is a consultant and Scientific Advisory Board member for Agios Pharmaceuticals and Aeglea Biotherapeutics.
References
- Adam J, Yang M, Bauerschmidt C, Kitagawa M, O’Flaherty L, Maheswaran P, Ozkan G, Sahgal N, Baban D, Kato K, et al. A role for cytosolic fumarate hydratase in urea cycle metabolism and renal neoplasia. Cell Rep. 2013;3:1440–1448. doi: 10.1016/j.celrep.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel IM, den Boer ML, Meijerink JP, Veerman AJ, Reniers NC, Pieters R. Up-regulation of asparagine synthetase expression is not linked to the clinical response L-asparaginase in pediatric acute lymphoblastic leukemia. Blood. 2006;107:4244–4249. doi: 10.1182/blood-2005-06-2597. [DOI] [PubMed] [Google Scholar]
- Appleyard MV, Murray KE, Coates PJ, Wullschleger S, Bray SE, Kernohan NM, Fleming S, Alessi DR, Thompson AM. Phenformin as prophylaxis and therapy in breast cancer xenografts. Br J Cancer. 2012;106:1117–1122. doi: 10.1038/bjc.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold RS, Shi J, Murad E, Whalen AM, Sun CQ, Polavarapu R, Parthasarathy S, Petros JA, Lambeth JD. Hydrogen peroxide mediates the cell growth and transformation caused by the mitogenic oxidase Nox1. Proc Natl Acad Sci U S A. 2001;98:5550–5555. doi: 10.1073/pnas.101505898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga CL, Brown TD, Kuhn JG, Shen HS, O’Rourke TJ, Beougher K, Brentzel HJ, Von Hoff DD, Weiss GR. Phase I clinical and pharmacokinetic trial of Brequinar sodium (DuP 785; NSC 368390) Cancer Res. 1989;49:4648–4653. [PubMed] [Google Scholar]
- Ascierto PA, Scala S, Castello G, Daponte A, Simeone E, Ottaiano A, Beneduce G, De Rosa V, Izzo F, Melucci MT, et al. Pegylated arginine deiminase treatment of patients with metastatic melanoma: results from phase I and II studies. J Clin Oncol. 2005;23:7660–7668. doi: 10.1200/JCO.2005.02.0933. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajrami I, Kigozi A, Van Weverwijk A, Brough R, Frankum J, Lord CJ, Ashworth A. Synthetic lethality of PARP and NAMPT inhibition in triple-negative breast cancer cells. EMBO Mol Med. 2012;4:1087–1096. doi: 10.1002/emmm.201201250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian A, Matsuzaki S, Humphries KM, Pharaoh GA, Doshi A, Zaware N, Gangjee A, Ihnat MA. AG311, a small molecule inhibitor of complex I and hypoxia-induced HIF-1alpha stabilization. Cancer Lett. 2017;388:149–157. doi: 10.1016/j.canlet.2016.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batova A, Diccianni MB, Omura-Minamisawa M, Yu J, Carrera CJ, Bridgeman LJ, Kung FH, Pullen J, Amylon MD, Yu AL. Use of alanosine as a methylthioadenosine phosphorylase-selective therapy for T-cell acute lymphoblastic leukemia in vitro. Cancer Res. 1999;59:1492–1497. [PubMed] [Google Scholar]
- Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- Bean GR, Kremer JC, Prudner BC, Schenone AD, Yao JC, Schultze MB, Chen DY, Tanas MR, Adkins DR, Bomalaski J, et al. A metabolic synthetic lethal strategy with arginine deprivation and chloroquine leads to cell death in ASS1-deficient sarcomas. Cell Death Dis. 2016;7:e2406. doi: 10.1038/cddis.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti V, Mosconi L, Pupi A. Brain: normal variations and benign findings in fluorodeoxyglucose-PET/computed tomography imaging. PET Clin. 2014;9:129–140. doi: 10.1016/j.cpet.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrovnikova-Marjon E, Hurov JB. Targeting metabolic changes in cancer: novel therapeutic approaches. Annu Rev Med. 2014;65:157–170. doi: 10.1146/annurev-med-092012-112344. [DOI] [PubMed] [Google Scholar]
- Bonuccelli G, Avnet S, Grisendi G, Salerno M, Granchi D, Dominici M, Kusuzaki K, Baldini N. Role of mesenchymal stem cells in osteosarcoma and metabolic reprogramming of tumor cells. Oncotarget. 2014;5:7575–7588. doi: 10.18632/oncotarget.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner A, Falchook G, Patel M, Infante J, Arkenau HT, Dean E, Borazanci E, Lopez J, Moore K, Schmid P, et al. Abstract P6-11-09: Heavily pre-treated breast cancer patients show promising responses in the first in human study of the first-In-class fatty acid synthase (FASN) inhibitor, TVB-2640 in combination with paclitaxel. Cancer Research. 2017;77:6-11-09–16-11-09. [Google Scholar]
- Bridges HR, Jones AJ, Pollak MN, Hirst J. Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. Biochem J. 2014;462:475–487. doi: 10.1042/BJ20140620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KK, Spinelli JB, Asara JM, Toker A. Adaptive Reprogramming of De Novo Pyrimidine Synthesis Is a Metabolic Vulnerability in Triple-Negative Breast Cancer. Cancer Discov. 2017;7:391–399. doi: 10.1158/2159-8290.CD-16-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusselmans K, De Schrijver E, Verhoeven G, Swinnen JV. RNA interference-mediated silencing of the acetyl-CoA-carboxylase-alpha gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer Res. 2005;65:6719–6725. doi: 10.1158/0008-5472.CAN-05-0571. [DOI] [PubMed] [Google Scholar]
- Buck MD, Sowell RT, Kaech SM, Pearce EL. Metabolic Instruction of Immunity. Cell. 2017;169:570–586. doi: 10.1016/j.cell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, Viollet B, Thompson CB. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67:6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- Cairncross JG, Wang M, Jenkins RB, Shaw EG, Giannini C, Brachman DG, Buckner JC, Fink KL, Souhami L, Laperriere NJ, et al. Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol. 2014;32:783–790. doi: 10.1200/JCO.2013.49.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardaci S, Zheng L, MacKay G, van den Broek NJ, MacKenzie ED, Nixon C, Stevenson D, Tumanov S, Bulusu V, Kamphorst JJ, et al. Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis. Nat Cell Biol. 2015;17:1317–1326. doi: 10.1038/ncb3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chajes V, Cambot M, Moreau K, Lenoir GM, Joulin V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res. 2006;66:5287–5294. doi: 10.1158/0008-5472.CAN-05-1489. [DOI] [PubMed] [Google Scholar]
- Chan M, Gravel M, Bramoulle A, Bridon G, Avizonis D, Shore GC, Roulston A. Synergy between the NAMPT inhibitor GMX1777(8) and pemetrexed in non-small cell lung cancer cells is mediated by PARP activation and enhanced NAD consumption. Cancer Res. 2014;74:5948–5954. doi: 10.1158/0008-5472.CAN-14-0809. [DOI] [PubMed] [Google Scholar]
- Chen J, Chung F, Yang G, Pu M, Gao H, Jiang W, Yin H, Capka V, Kasibhatla S, Laffitte B, et al. Phosphoglycerate dehydrogenase is dispensable for breast tumor maintenance and growth. Oncotarget. 2013;4:2502–2511. doi: 10.18632/oncotarget.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarugi A, Dolle C, Felici R, Ziegler M. The NAD metabolome–a key determinant of cancer cell biology. Nat Rev Cancer. 2012;12:741–752. doi: 10.1038/nrc3340. [DOI] [PubMed] [Google Scholar]
- Chio II, Jafarnejad SM, Ponz-Sarvise M, Park Y, Rivera K, Palm W, Wilson J, Sangar V, Hao Y, Ohlund D, et al. NRF2 Promotes Tumor Maintenance by Modulating mRNA Translation in Pancreatic Cancer. Cell. 2016;166:963–976. doi: 10.1016/j.cell.2016.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Cohade C. Altered biodistribution on FDG-PET with emphasis on brown fat and insulin effect. Semin Nucl Med. 2010;40:283–293. doi: 10.1053/j.semnuclmed.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, Walters H, Tantawy MN, Fu A, Manning HC, et al. Acetate dependence of tumors. Cell. 2014;159:1591–1602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Cros M, Hemmerlin C, Ferretti S, Zhang J, Gounarides JS, Yin H, Muller A, Haberkorn A, Chene P, Sellers WR, et al. M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proc Natl Acad Sci U S A. 2013;110:489–494. doi: 10.1073/pnas.1212780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Amico A. Review of clinical practice utility of positron emission tomography with 18F-fluorodeoxyglucose in assessing tumour response to therapy. Radiol Med. 2015;120:345–351. doi: 10.1007/s11547-014-0446-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol. 2016;27:599–608. doi: 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daye D, Wellen KE. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Semin Cell Dev Biol. 2012;23:362–369. doi: 10.1016/j.semcdb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Dayton TL, Jacks T, Vander Heiden MG. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17:1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delage B, Fennell DA, Nicholson L, McNeish I, Lemoine NR, Crook T, Szlosarek PW. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int J Cancer. 2010;126:2762–2772. doi: 10.1002/ijc.25202. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P, Baddour J, Muller F, Wu CC, Wang H, Liao WT, Lan Z, Chen A, Gutschner T, Kang Y, et al. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature. 2017;542:119–123. doi: 10.1038/nature21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo CD, Ravandi F, Agresta S, Konopleva M, Takahashi K, Kadia T, Routbort M, Patel KP, Mark B, Pierce S, et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am J Hematol. 2015;90:732–736. doi: 10.1002/ajh.24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Patel DN, Welsch M, Skouta R, Lee ED, Hayano M, Thomas AG, Gleason CE, Tatonetti NP, Slusher BS, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife. 2014;3:e02523. doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagle H. The specific amino acid requirements of a human carcinoma cell (Stain HeLa) in tissue culture. J Exp Med. 1955;102:37–48. doi: 10.1084/jem.102.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efferth T, Gebhart E, Ross DD, Sauerbrey A. Identification of gene expression profiles predicting tumor cell response to L-alanosine. Biochem Pharmacol. 2003;66:613–621. doi: 10.1016/s0006-2952(03)00341-1. [DOI] [PubMed] [Google Scholar]
- Egler RA, Ahuja SP, Matloub Y. L-asparaginase in the treatment of patients with acute lymphoblastic leukemia. J Pharmacol Pharmacother. 2016;7:62–71. doi: 10.4103/0976-500X.184769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem. 2000;275:223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- Elion GB. The purine path to chemotherapy. Science. 1989;244:41–47. doi: 10.1126/science.2649979. [DOI] [PubMed] [Google Scholar]
- Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- Farwell MD, Pryma DA, Mankoff DA. PET/CT imaging in cancer: current applications and future directions. Cancer. 2014;120:3433–3445. doi: 10.1002/cncr.28860. [DOI] [PubMed] [Google Scholar]
- Feun LG, Marini A, Walker G, Elgart G, Moffat F, Rodgers SE, Wu CJ, You M, Wangpaichitr M, Kuo MT, et al. Negative argininosuccinate synthetase expression in melanoma tumours may predict clinical benefit from arginine-depleting therapy with pegylated arginine deiminase. Br J Cancer. 2012;106:1481–1485. doi: 10.1038/bjc.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine BM, Kaspers GJ, Ho M, Loonen AH, Boxer LM. A genome-wide view of the in vitro response to l-asparaginase in acute lymphoblastic leukemia. Cancer Res. 2005;65:291–299. [PubMed] [Google Scholar]
- Franciosi M, Lucisano G, Lapice E, Strippoli GF, Pellegrini F, Nicolucci A. Metformin therapy and risk of cancer in patients with type 2 diabetes: systematic review. PLoS One. 2013;8:e71583. doi: 10.1371/journal.pone.0071583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Zheng L, Folger O, Rajagopalan KN, MacKenzie ED, Jerby L, Micaroni M, Chaneton B, Adam J, Hedley A, et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477:225–228. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- Furuya S. An essential role for de novo biosynthesis of L-serine in CNS development. Asia Pac J Clin Nutr. 2008;17(Suppl 1):312–315. [PubMed] [Google Scholar]
- Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nat Rev Drug Discov. 2013;12:829–846. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- Gandini S, Puntoni M, Heckman-Stoddard BM, Dunn BK, Ford L, DeCensi A, Szabo E. Metformin and cancer risk and mortality: a systematic review and meta-analysis taking into account biases and confounders. Cancer Prev Res (Phila) 2014;7:867–885. doi: 10.1158/1940-6207.CAPR-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Tashiro Y, Wang Q, Sakai K, Sonomoto K. High acetone-butanol-ethanol production in pH-stat co-feeding of acetate and glucose. J Biosci Bioeng. 2016;122:176–182. doi: 10.1016/j.jbiosc.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude E, Frezza C. Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival. Nat Commun. 2016;7:13041. doi: 10.1038/ncomms13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Graczyk-Jarzynka A, Zagozdzon R, Muchowicz A, Siernicka M, Firczuk M, Juszczynski P. New insights into redox homeostasis as a therapeutic target in B-cell malignancies. Curr Opin Hematol. 2017 doi: 10.1097/MOH.0000000000000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassian AR, Parker SJ, Davidson SM, Divakaruni AS, Green CR, Zhang X, Slocum KL, Pu M, Lin F, Vickers C, et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014;74:3317–3331. doi: 10.1158/0008-5472.CAN-14-0772-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel SP, Hulea L, Toban N, Birman E, Blouin MJ, Zakikhani M, Zhao Y, Topisirovic I, St-Pierre J, Pollak M. Serine deprivation enhances antineoplastic activity of biguanides. Cancer Res. 2014;74:7521–7533. doi: 10.1158/0008-5472.CAN-14-2643-T. [DOI] [PubMed] [Google Scholar]
- Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, Janes JR, Laidig GJ, Lewis ER, Li J, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- Gui DY, Sullivan LB, Luengo A, Hosios AM, Bush LN, Gitego N, Davidson SM, Freinkman E, Thomas CJ, Vander Heiden MG. Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab. 2016;24:716–727. doi: 10.1016/j.cmet.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Chandel NS. Targeting glucose metabolism for cancer therapy. J Exp Med. 2012;209:211–215. doi: 10.1084/jem.20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harasawa H, Yamada Y, Kudoh M, Sugahara K, Soda H, Hirakata Y, Sasaki H, Ikeda S, Matsuo T, Tomonaga M, et al. Chemotherapy targeting methylthioadenosine phosphorylase (MTAP) deficiency in adult T cell leukemia (ATL) Leukemia. 2002;16:1799–1807. doi: 10.1038/sj.leu.2402570. [DOI] [PubMed] [Google Scholar]
- Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, Yung KY, Brenner D, Knobbe-Thomsen CB, Cox MA, et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27:211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- Harris M. Pyruvate blocks expression of sensitivity to antimycin A and chloramphenicol. Somatic Cell Genet. 1980;6:699–708. doi: 10.1007/BF01538969. [DOI] [PubMed] [Google Scholar]
- Hattori A, Tsunoda M, Konuma T, Kobayashi M, Nagy T, Glushka J, Tayyari F, McSkimming D, Kannan N, Tojo A, et al. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature. 2017;545:500–504. doi: 10.1038/nature22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer. 2016;16:635–649. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger C, Chaudhuri NK, Danneberg P, Mooren D, Griesbach L, Duschinsky R, Schnitzer RJ, Pleven E, Scheiner J. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature. 1957;179:663–666. doi: 10.1038/179663a0. [DOI] [PubMed] [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer TS, Ventura R, Mordec K, Lai J, Fridlib M, Buckley D, Kemble G. FASN Inhibition and Taxane Treatment Combine to Enhance Anti-tumor Efficacy in Diverse Xenograft Tumor Models through Disruption of Tubulin Palmitoylation and Microtubule Organization and FASN Inhibition-Mediated Effects on Oncogenic Signaling and Gene Expression. EBioMedicine. 2017;16:51–62. doi: 10.1016/j.ebiom.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson KS, Lienard BM, McDonough MA, Clifton IJ, Butler D, Soares AS, Oldham NJ, McNeill LA, Schofield CJ. Structural and mechanistic studies on the inhibition of the hypoxia-inducible transcription factor hydroxylases by tricarboxylic acid cycle intermediates. J Biol Chem. 2007;282:3293–3301. doi: 10.1074/jbc.M608337200. [DOI] [PubMed] [Google Scholar]
- Hoekstra AS, de Graaff MA, Briaire-de Bruijn IH, Ras C, Seifar RM, van Minderhout I, Cornelisse CJ, Hogendoorn PC, Breuning MH, Suijker J, et al. Inactivation of SDH and FH cause loss of 5hmC and increased H3K9me3 in paraganglioma/pheochromocytoma and smooth muscle tumors. Oncotarget. 2015;6:38777–38788. doi: 10.18632/oncotarget.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H, Tran P, Carrera CJ, Hori Y, Rosenbach MD, Carson DA, Nobori T. Methylthioadenosine phosphorylase cDNA transfection alters sensitivity to depletion of purine and methionine in A549 lung cancer cells. Cancer Res. 1996;56:5653–5658. [PubMed] [Google Scholar]
- Hosios AM, Hecht VC, Danai LV, Johnson MO, Rathmell JC, Steinhauser ML, Manalis SR, Vander Heiden MG. Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev Cell. 2016;36:540–549. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosios AM, Vander Heiden MG. Acetate metabolism in cancer cells. Cancer Metab. 2014;2:27. doi: 10.1186/s40170-014-0027-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N, Sager R. Cytoplasmic genetics of mammalian cells: conditional sensitivity to mitochondrial inhibitors and isolation of new mutant phenotypes. Somatic Cell Genet. 1979;5:833–845. doi: 10.1007/BF01542645. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Hu J, Locasale JW, Bielas JH, O’Sullivan J, Sheahan K, Cantley LC, Vander Heiden MG, Vitkup D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat Biotechnol. 2013;31:522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wullschleger S, Shpiro N, McGuire VA, Sakamoto K, Woods YL, McBurnie W, Fleming S, Alessi DR. Important role of the LKB1-AMPK pathway in suppressing tumorigenesis in PTEN-deficient mice. Biochem J. 2008;412:211–221. doi: 10.1042/BJ20080557. [DOI] [PubMed] [Google Scholar]
- Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen WJ, Vander Heiden MG. Pyruvate kinase: Function, regulation and role in cancer. Semin Cell Dev Biol. 2015;43:43–51. doi: 10.1016/j.semcdb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo F, Marra P, Beneduce G, Castello G, Vallone P, De Rosa V, Cremona F, Ensor CM, Holtsberg FW, Bomalaski JS, et al. Pegylated arginine deiminase treatment of patients with unresectable hepatocellular carcinoma: results from phase I/II studies. J Clin Oncol. 2004;22:1815–1822. doi: 10.1200/JCO.2004.11.120. [DOI] [PubMed] [Google Scholar]
- Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L, Saland E, Decroocq J, Maciel TT, Lambert M, et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015;126:1346–1356. doi: 10.1182/blood-2015-01-621870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke R, Iavarone AT, Rine J. Oncometabolite D-2-Hydroxyglutarate enhances gene silencing through inhibition of specific H3K36 histone demethylases. Elife. 2017;6 doi: 10.7554/eLife.22451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiralerspong S, Palla SL, Giordano SH, Meric-Bernstam F, Liedtke C, Barnett CM, Hsu L, Hung MC, Hortobagyi GN, Gonzalez-Angulo AM. Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer. J Clin Oncol. 2009;27:3297–3302. doi: 10.1200/JCO.2009.19.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvekar A, Hu H, Yadegarynia S, Lyssiotis CA, Ullas S, Lien EC, Bellinger G, Son J, Hok RC, Seth P, et al. Phosphoinositide 3-kinase inhibitors induce DNA damage through nucleoside depletion. Proc Natl Acad Sci U S A. 2016;113:E4338–4347. doi: 10.1073/pnas.1522223113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Chung MK, Fan J, Rabinowitz JD. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab. 2014;2:23. doi: 10.1186/2049-3002-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karisch R, Fernandez M, Taylor P, Virtanen C, St-Germain JR, Jin LL, Harris IS, Mori J, Mak TW, Senis YA, et al. Global proteomic assessment of the classical protein-tyrosine phosphatome and “Redoxome”. Cell. 2011;146:826–840. doi: 10.1016/j.cell.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy KM, Scarbrough PM, Ribeiro A, Richardson R, Yuan H, Sonveaux P, Landon CD, Chi JT, Pizzo S, Schroeder T, et al. Catabolism of exogenous lactate reveals it as a legitimate metabolic substrate in breast cancer. PLoS One. 2013;8:e75154. doi: 10.1371/journal.pone.0075154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hu Z, Cai L, Li K, Choi E, Faubert B, Bezwada D, Rodriguez-Canales J, Villalobos P, Lin YF, et al. CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature. 2017;546:168–172. doi: 10.1038/nature22359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler HL, Burris HA, 3rd, Sandler AB, Oliff IA. A phase II multicenter study of L-alanosine, a potent inhibitor of adenine biosynthesis, in patients with MTAP-deficient cancer. Invest New Drugs. 2009;27:75–81. doi: 10.1007/s10637-008-9160-1. [DOI] [PubMed] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JC, Prudner BC, Lange SE, Bean GR, Schultze MB, Brashears CB, Radyk MD, Redlich N, Tzeng SC, Kami K, et al. Arginine Deprivation Inhibits the Warburg Effect and Upregulates Glutamine Anaplerosis and Serine Biosynthesis in ASS1-Deficient Cancers. Cell Rep. 2017;18:991–1004. doi: 10.1016/j.celrep.2016.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll W, Loffler M, Schneider F. Energy parameters, macromolecular synthesis and cell cycle progression of in vitro grown Ehrlich ascites tumor cells after inhibition of oxidative ATP synthesis by oligomycin. Z Naturforsch C. 1983;38:604–612. [PubMed] [Google Scholar]
- Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, Vazquez F, Weir BA, Fitzgerald ME, Tanaka M, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351:1214–1218. doi: 10.1126/science.aad5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhajda FP, Jenner K, Wood FD, Hennigar RA, Jacobs LB, Dick JD, Pasternack GR. Fatty acid synthesis: a potential selective target for antineoplastic therapy. Proc Natl Acad Sci U S A. 1994;91:6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C, Hixon J, Choe S, Marks K, Gross S, Murphy E, DeLaBarre B, Cianchetta G, Sethumadhavan S, Wang X, et al. Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chem Biol. 2012;19:1187–1198. doi: 10.1016/j.chembiol.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- Landau BR, Laszlo J, Stengle J, Burk D. Certain metabolic and pharmacologic effects in cancer patients given infusions of 2-deoxy-D-glucose. J Natl Cancer Inst. 1958;21:485–494. [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim TI, Jeon SM, Hong SP, Cheon JH, Kim WH. The effects of metformin on the survival of colorectal cancer patients with diabetes mellitus. Int J Cancer. 2012;131:752–759. doi: 10.1002/ijc.26421. [DOI] [PubMed] [Google Scholar]
- Leonard PG, Satani N, Maxwell D, Lin YH, Hammoudi N, Peng Z, Pisaneschi F, Link TM, Lee GRt, Sun D, et al. SF2312 is a natural phosphonate inhibitor of enolase. Nat Chem Biol. 2016;12:1053–1058. doi: 10.1038/nchembio.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien EC, Lyssiotis CA, Juvekar A, Hu H, Asara JM, Cantley LC, Toker A. Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat Cell Biol. 2016;18:572–578. doi: 10.1038/ncb3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston RB, Venditti JM, Cooney DA, Carter SK. Glutamine antagonists in chemotherapy. Adv Pharmacol Chemother. 1970;8:57–120. doi: 10.1016/s1054-3589(08)60594-3. [DOI] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke M, Ghazaly E, Freitas MO, Mitsinga M, Lattanzio L, Lo Nigro C, Nagano A, Wang J, Chelala C, Szlosarek P, et al. Inhibition of the Polyamine Synthesis Pathway Is Synthetically Lethal with Loss of Argininosuccinate Synthase 1. Cell Rep. 2016;16:1604–1613. doi: 10.1016/j.celrep.2016.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffer M, Schneider F. Further characterization of the growth inhibitory effect of rotenone on in vitro cultured Ehrlich ascites tumour cells. Mol Cell Biochem. 1982;48:77–90. doi: 10.1007/BF00227608. [DOI] [PubMed] [Google Scholar]
- Long Y, Tsai WB, Wangpaichitr M, Tsukamoto T, Savaraj N, Feun LG, Kuo MT. Arginine deiminase resistance in melanoma cells is associated with metabolic reprogramming, glucose dependence, and glutamine addiction. Mol Cancer Ther. 2013;12:2581–2590. doi: 10.1158/1535-7163.MCT-13-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG., Jr (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin M, Lubin A. Selective killing of tumors deficient in methylthioadenosine phosphorylase: a novel strategy. PLoS One. 2009;4:e5735. doi: 10.1371/journal.pone.0005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luengo A, Sullivan LB, Heiden MG. Understanding the complex-I-ty of metformin action: limiting mitochondrial respiration to improve cancer therapy. BMC Biol. 2014;12:82. doi: 10.1186/s12915-014-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Lussey-Lepoutre C, Hollinshead KE, Ludwig C, Menara M, Morin A, Castro-Vega LJ, Parker SJ, Janin M, Martinelli C, Ottolenghi C, et al. Loss of succinate dehydrogenase activity results in dependency on pyruvate carboxylation for cellular anabolism. Nat Commun. 2015;6:8784. doi: 10.1038/ncomms9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycan TW, Pardee TS, Petty WJ, Bonomi M, Alistar A, Lamar ZS, Isom S, Chan MD, Miller AA, Ruiz J. A Phase II Clinical Trial of CPI-613 in Patients with Relapsed or Refractory Small Cell Lung Carcinoma. PLoS One. 2016;11:e0164244. doi: 10.1371/journal.pone.0164244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks ODK, Athineos D, Cheung EC, Lee P, Zhang T, van den Broek NJF, Mackay GM, Labuschagne CF, Gay D, Kruiswijk F, et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature. 2017;544:372–376. doi: 10.1038/nature22056. [DOI] [PubMed] [Google Scholar]
- Marchiq I, Pouyssegur J. Hypoxia, cancer metabolism and the therapeutic benefit of targeting lactate/H(+) symporters. J Mol Med (Berl) 2016;94:155–171. doi: 10.1007/s00109-015-1307-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjon K, Cameron MJ, Quang P, Clasquin MF, Mandley E, Kunii K, McVay M, Choe S, Kernytsky A, Gross S, et al. MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell Rep. 2016;15:574–587. doi: 10.1016/j.celrep.2016.03.043. [DOI] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, Nannepaga S, Piccirillo SG, Kovacs Z, Foong C, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D, Stratikopoulos E, Ozturk S, Steinbach N, Pegno S, Schoenfeld S, Yong R, Murty VV, Asara JM, Cantley LC, et al. PTEN Regulates Glutamine Flux to Pyrimidine Synthesis and Sensitivity to Dihydroorotate Dehydrogenase Inhibition. Cancer Discov. 2017;7:380–390. doi: 10.1158/2159-8290.CD-16-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaini KR, Brignole EJ, Kini M, Davidson SM, Fiske BP, Drennan CL, Vander Heiden MG. An epitope tag alters phosphoglycerate dehydrogenase structure and impairs ability to support cell proliferation. Cancer Metab. 2015;3:5. doi: 10.1186/s40170-015-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis KJ, McDonald ER, 3rd, Schlabach MR, Billy E, Hoffman GR, deWeck A, Ruddy DA, Venkatesan K, Yu J, McAllister G, et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351:1208–1213. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, Lau AN, Ji BW, Dixit PD, Hosios AM, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353:1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medes G, Thomas A, Weinhouse S. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 1953;13:27–29. [PubMed] [Google Scholar]
- Menendez JA, Lupu R. Oncogenic properties of the endogenous fatty acid metabolism: molecular pathology of fatty acid synthase in cancer cells. Curr Opin Clin Nutr Metab Care. 2006;9:346–357. doi: 10.1097/01.mco.0000232893.21050.15. [DOI] [PubMed] [Google Scholar]
- Migita T, Narita T, Nomura K, Miyagi E, Inazuka F, Matsuura M, Ushijima M, Mashima T, Seimiya H, Satoh Y, et al. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008;68:8547–8554. doi: 10.1158/0008-5472.CAN-08-1235. [DOI] [PubMed] [Google Scholar]
- Mockler MB, Conroy MJ, Lysaght J. Targeting T cell immunometabolism for cancer immunotherapy; understanding the impact of the tumor microenvironment. Front Oncol. 2014;4:107. doi: 10.3389/fonc.2014.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar RJ, Botman D, Smits MA, Hira VV, van Lith SA, Stap J, Henneman P, Khurshed M, Lenting K, Mul AN, et al. Radioprotection of IDH1-Mutated Cancer Cells by the IDH1-Mutant Inhibitor AGI-5198. Cancer Res. 2015;75:4790–4802. doi: 10.1158/0008-5472.CAN-14-3603. [DOI] [PubMed] [Google Scholar]
- Mullarky E, Lucki NC, Beheshti Zavareh R, Anglin JL, Gomes AP, Nicolay BN, Wong JC, Christen S, Takahashi H, Singh PK, et al. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc Natl Acad Sci U S A. 2016;113:1778–1783. doi: 10.1073/pnas.1521548113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller FL, Aquilanti EA, DePinho RA. Collateral Lethality: A new therapeutic strategy in oncology. Trends Cancer. 2015;1:161–173. doi: 10.1016/j.trecan.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller FL, Colla S, Aquilanti E, Manzo VE, Genovese G, Lee J, Eisenson D, Narurkar R, Deng P, Nezi L, et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488:337–342. doi: 10.1038/nature11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahimana A, Attinger A, Aubry D, Greaney P, Ireson C, Thougaard AV, Tjornelund J, Dawson KM, Dupuis M, Duchosal MA. The NAD biosynthesis inhibitor APO866 has potent antitumor activity against hematologic malignancies. Blood. 2009;113:3276–3286. doi: 10.1182/blood-2008-08-173369. [DOI] [PubMed] [Google Scholar]
- Neuman RE, McCoy TA. Dual requirement of Walker carcinosarcoma 256 in vitro for asparagine and glutamine. Science. 1956;124:124–125. doi: 10.1126/science.124.3212.124. [DOI] [PubMed] [Google Scholar]
- Newsholme EA, Crabtree B, Ardawi MS. The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Biosci Rep. 1985;5:393–400. doi: 10.1007/BF01116556. [DOI] [PubMed] [Google Scholar]
- Noe DA, Rowinsky EK, Shen HS, Clarke BV, Grochow LB, McGuire WB, Hantel A, Adams DB, Abeloff MD, Ettinger DS, et al. Phase I and pharmacokinetic study of brequinar sodium (NSC 368390) Cancer Res. 1990;50:4595–4599. [PubMed] [Google Scholar]
- Noto H, Goto A, Tsujimoto T, Noda M. Cancer risk in diabetic patients treated with metformin: a systematic review and meta-analysis. PLoS One. 2012;7:e33411. doi: 10.1371/journal.pone.0033411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dwyer PJ, Alonso MT, Leyland-Jones B. Acivicin: a new glutamine antagonist in clinical trials. J Clin Oncol. 1984;2:1064–1071. doi: 10.1200/JCO.1984.2.9.1064. [DOI] [PubMed] [Google Scholar]
- Okoye-Okafor UC, Bartholdy B, Cartier J, Gao EN, Pietrak B, Rendina AR, Rominger C, Quinn C, Smallwood A, Wiggall KJ, et al. New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat Chem Biol. 2015;11:878–886. doi: 10.1038/nchembio.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookhtens M, Kannan R, Lyon I, Baker N. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am J Physiol. 1984;247:R146–153. doi: 10.1152/ajpregu.1984.247.1.R146. [DOI] [PubMed] [Google Scholar]
- Ott PA, Carvajal RD, Pandit-Taskar N, Jungbluth AA, Hoffman EW, Wu BW, Bomalaski JS, Venhaus R, Pan L, Old LJ, et al. Phase I/II study of pegylated arginine deiminase (ADI-PEG 20) in patients with advanced melanoma. Invest New Drugs. 2013;31:425–434. doi: 10.1007/s10637-012-9862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348(Pt 3):607–614. [PMC free article] [PubMed] [Google Scholar]
- Pacold ME, Brimacombe KR, Chan SH, Rohde JM, Lewis CA, Swier LJ, Possemato R, Chen WW, Sullivan LB, Fiske BP, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12:452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee TS, Lee K, Luddy J, Maturo C, Rodriguez R, Isom S, Miller LD, Stadelman KM, Levitan D, Hurd D, et al. A phase I study of the first-in-class antimitochondrial metabolism agent, CPI-613, in patients with advanced hematologic malignancies. Clin Cancer Res. 2014;20:5255–5264. doi: 10.1158/1078-0432.CCR-14-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker WB. Enzymology of purine and pyrimidine antimetabolites used in the treatment of cancer. Chem Rev. 2009;109:2880–2893. doi: 10.1021/cr900028p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- Peters GJ, Nadal JC, Laurensse EJ, de Kant E, Pinedo HM. Retention of in vivo antipyrimidine effects of Brequinar sodium (DUP-785; NSC 368390) in murine liver, bone marrow and colon cancer. Biochem Pharmacol. 1990;39:135–144. doi: 10.1016/0006-2952(90)90657-7. [DOI] [PubMed] [Google Scholar]
- Peters GJ, van der Wilt CL, van Moorsel CJ, Kroep JR, Bergman AM, Ackland SP. Basis for effective combination cancer chemotherapy with antimetabolites. Pharmacol Ther. 2000;87:227–253. doi: 10.1016/s0163-7258(00)00086-3. [DOI] [PubMed] [Google Scholar]
- Phillips MM, Sheaff MT, Szlosarek PW. Targeting arginine-dependent cancers with arginine-degrading enzymes: opportunities and challenges. Cancer Res Treat. 2013;45:251–262. doi: 10.4143/crt.2013.45.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro C, Longatto-Filho A, Azevedo-Silva J, Casal M, Schmitt FC, Baltazar F. Role of monocarboxylate transporters in human cancers: state of the art. J Bioenerg Biomembr. 2012;44:127–139. doi: 10.1007/s10863-012-9428-1. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, Wortham NC, Hunt T, Mitchell M, Olpin S, Moat SJ, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet. 2005;14:2231–2239. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, David M, Straley K, Travins J, Kim H, Chen Y, Zhu D, Saada V, Bawa O, Opolon P, et al. AG-221, an Oral, Selective, First-in-Class, Potent IDH2-R140Q Mutant Inhibitor, Induces Differentiation in a Xenotransplant Model. Blood. 2014;124:3735–3735. [Google Scholar]
- Rabbani N, Thornalley PJ. Dicarbonyl proteome and genome damage in metabolic and vascular disease. Biochem Soc Trans. 2014;42:425–432. doi: 10.1042/BST20140018. [DOI] [PubMed] [Google Scholar]
- Rabinovich S, Adler L, Yizhak K, Sarver A, Silberman A, Agron S, Stettner N, Sun Q, Brandis A, Helbling D, et al. Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature. 2015;527:379–383. doi: 10.1038/nature15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raez LE, Papadopoulos K, Ricart AD, Chiorean EG, Dipaola RS, Stein MN, Rocha Lima CM, Schlesselman JJ, Tolba K, Langmuir VK, et al. A phase I dose-escalation trial of 2-deoxy-D-glucose alone or combined with docetaxel in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2013;71:523–530. doi: 10.1007/s00280-012-2045-1. [DOI] [PubMed] [Google Scholar]
- Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X, Tolliday NJ, Golub TR, Carr SA, Shamji AF, et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. 2011;475:231–234. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- Saha SK, Parachoniak CA, Ghanta KS, Fitamant J, Ross KN, Najem MS, Gurumurthy S, Akbay EA, Sia D, Cornella H, et al. Mutant IDH inhibits HNF-4alpha to block hepatocyte differentiation and promote biliary cancer. Nature. 2014;513:110–114. doi: 10.1038/nature13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D, Park Y, Andrabi SA, Shelton LM, Gilkes DM, Semenza GL. PHGDH Expression Is Required for Mitochondrial Redox Homeostasis, Breast Cancer Stem Cell Maintenance, and Lung Metastasis. Cancer Res. 2016;76:4430–4442. doi: 10.1158/0008-5472.CAN-16-0530. [DOI] [PubMed] [Google Scholar]
- Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, Barneda D, Sellers K, Campos-Olivas R, Grana O, et al. MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab. 2015;22:590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- Schockel L, Glasauer A, Basit F, Bitschar K, Truong H, Erdmann G, Algire C, Hagebarth A, Willems PH, Kopitz C, et al. Targeting mitochondrial complex I using BAY 87-2243 reduces melanoma tumor growth. Cancer Metab. 2015;3:11. doi: 10.1186/s40170-015-0138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, Goodwin LM, Smethurst E, Mason S, Blyth K, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Sellers K, Fox MP, Bousamra M, 2nd, Slone SP, Higashi RM, Miller DM, Wang Y, Yan J, Yuneva MO, Deshpande R, et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Invest. 2015;125:687–698. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23:143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen LJ, Lin WC, Beloussow K, Shen WC. Resistance to the anti-proliferative activity of recombinant arginine deiminase in cell culture correlates with the endogenous enzyme, argininosuccinate synthetase. Cancer Lett. 2003;191:165–170. doi: 10.1016/s030-43835(02)00693-6. [DOI] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell K. Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Adv Enzyme Regul. 1984;22:325–400. doi: 10.1016/0065-2571(84)90021-9. [DOI] [PubMed] [Google Scholar]
- Snell K, Natsumeda Y, Eble JN, Glover JL, Weber G. Enzymic imbalance in serine metabolism in human colon carcinoma and rat sarcoma. Br J Cancer. 1988;57:87–90. doi: 10.1038/bjc.1988.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell K, Natsumeda Y, Weber G. The modulation of serine metabolism in hepatoma 3924A during different phases of cellular proliferation in culture. Biochem J. 1987;245:609–612. doi: 10.1042/bj2450609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stams WA, den Boer ML, Beverloo HB, Meijerink JP, Stigter RL, van Wering ER, Janka-Schaub GE, Slater R, Pieters R. Sensitivity to L-asparaginase is not associated with expression levels of asparagine synthetase in t(12;21)+ pediatric ALL. Blood. 2003;101:2743–2747. doi: 10.1182/blood-2002-08-2446. [DOI] [PubMed] [Google Scholar]
- Stein EM, Altman JK, Collins R, DeAngelo DJ, Fathi AT, Flinn I, Frankel A, Levine RL, Medeiros BC, Patel M, et al. AG-221, an Oral, Selective, First-in-Class, Potent Inhibitor of the IDH2 Mutant Metabolic Enzyme, Induces Durable Remissions in a Phase I Study in Patients with IDH2 Mutation Positive Advanced Hematologic Malignancies. Blood. 2014;124:115–115. [Google Scholar]
- Stein M, Lin H, Jeyamohan C, Dvorzhinski D, Gounder M, Bray K, Eddy S, Goodin S, White E, Dipaola RS. Targeting tumor metabolism with 2-deoxyglucose in patients with castrate-resistant prostate cancer and advanced malignancies. Prostate. 2010;70:1388–1394. doi: 10.1002/pros.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh YA, Arnold RS, Lassegue B, Shi J, Xu X, Sorescu D, Chung AB, Griendling KK, Lambeth JD. Cell transformation by the superoxide-generating oxidase Mox1. Nature. 1999;401:79–82. doi: 10.1038/43459. [DOI] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Vander Heiden MG. Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nat Rev Cancer. 2016;16:680–693. doi: 10.1038/nrc.2016.85. [DOI] [PubMed] [Google Scholar]
- Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, Lombardo PS, Van Nostrand JL, Hutchins A, Vera L, et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med. 2016;22:1108–1119. doi: 10.1038/nm.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes DB, Kfoury YS, Mercier FE, Wawer MJ, Law JM, Haynes MK, Lewis TA, Schajnovitz A, Jain E, Lee D, et al. Inhibition of Dihydroorotate Dehydrogenase Overcomes Differentiation Blockade in Acute Myeloid Leukemia. Cell. 2016;167:171–186 e115. doi: 10.1016/j.cell.2016.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szlosarek PW, Luong P, Phillips MM, Baccarini M, Stephen E, Szyszko T, Sheaff MT, Avril N. Metabolic response to pegylated arginine deiminase in mesothelioma with promoter methylation of argininosuccinate synthetase. J Clin Oncol. 2013;31:e111–113. doi: 10.1200/JCO.2012.42.1784. [DOI] [PubMed] [Google Scholar]
- Tabata S, Yamamoto M, Goto H, Hirayama A, Ohishi M, Kuramoto T, Mitsuhashi A, Ikeda R, Haraguchi M, Kawahara K, et al. Thymidine Catabolism as a Metabolic Strategy for Cancer Survival. Cell Rep. 2017;19:1313–1321. doi: 10.1016/j.celrep.2017.04.061. [DOI] [PubMed] [Google Scholar]
- Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015a;21:81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Tan B, Dong S, Shepard RL, Kays L, Roth KD, Geeganage S, Kuo MS, Zhao G. Inhibition of Nicotinamide Phosphoribosyltransferase (NAMPT), an Enzyme Essential for NAD+ Biosynthesis, Leads to Altered Carbohydrate Metabolism in Cancer Cells. J Biol Chem. 2015b;290:15812–15824. doi: 10.1074/jbc.M114.632141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, Miletic H, Sakariassen PO, Weinstock A, Wagner A, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol. 2015;17:1556–1568. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Iafrate AJ, Ho Q, Curry WT, Batchelor TT, Flaherty KT, Onozato ML, Lelic N, Sundaram S, Cahill DP, et al. Myc-Driven Glycolysis Is a Therapeutic Target in Glioblastoma. Clin Cancer Res. 2016;22:4452–4465. doi: 10.1158/1078-0432.CCR-15-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Wakimoto H, Iafrate AJ, Tanaka S, Loebel F, Lelic N, Wiederschain D, Bedel O, Deng G, Zhang B, et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell. 2015;28:773–784. doi: 10.1016/j.ccell.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov DV, Cracan V, Goodman RP, Peng J, Grabarek Z, Mootha VK. Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science. 2016;352:231–235. doi: 10.1126/science.aad4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Tonjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, Pleier SV, Bai AHC, Karra D, Piro RM, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19:901–908. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, Chiao PJ, Achanta G, Arlinghaus RB, Liu J, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- Turcan S, Fabius AW, Borodovsky A, Pedraza A, Brennan C, Huse J, Viale A, Riggins GJ, Chan TA. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT Inhibitor Decitabine. Oncotarget. 2013;4:1729–1736. doi: 10.18632/oncotarget.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heideman A, Berglund A, Larsson R, Nygren P. Safety and efficacy of NAD depleting cancer drugs: results of a phase I clinical trial of CHS 828 and overview of published data. Cancer Chemother Pharmacol. 2010;65:1165–1172. doi: 10.1007/s00280-009-1125-3. [DOI] [PubMed] [Google Scholar]
- Wagner AD, Grothe W, Haerting J, Kleber G, Grothey A, Fleig WE. Chemotherapy in advanced gastric cancer: a systematic review and meta-analysis based on aggregate data. J Clin Oncol. 2006;24:2903–2909. doi: 10.1200/JCO.2005.05.0245. [DOI] [PubMed] [Google Scholar]
- Wakimoto H, Tanaka S, Curry WT, Loebel F, Zhao D, Tateishi K, Chen J, Klofas LK, Lelic N, Kim JC, et al. Targetable signaling pathway mutations are associated with malignant phenotype in IDH-mutant gliomas. Clin Cancer Res. 2014;20:2898–2909. doi: 10.1158/1078-0432.CCR-13-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling J. From methotrexate to pemetrexed and beyond. A review of the pharmacodynamic and clinical properties of antifolates. Invest New Drugs. 2006;24:37–77. doi: 10.1007/s10637-005-4541-1. [DOI] [PubMed] [Google Scholar]
- Walsh MJ, Brimacombe KR, Anastasiou D, Yu Y, Israelsen WJ, Hong BS, Tempel W, Dimov S, Veith H, Yang H, et al. ML265: A potent PKM2 activator induces tetramerization and reduces tumor formation and size in a mouse xenograft model. Probe Reports from the NIH Molecular Libraries Program (Bethesda (MD) 2010 [PubMed] [Google Scholar]
- Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, Straley K, Kernytsky A, Liu W, Gliser C, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liberti MV, Liu P, Deng X, Liu Y, Locasale JW, Lai L. Rational Design of Selective Allosteric Inhibitors of PHGDH and Serine Synthesis with Anti-tumor Activity. Cell Chem Biol. 2017;24:55–65. doi: 10.1016/j.chembiol.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. Über den Stoffwechsel der Carcinomzelle. Naturwissenschaften. 1924;12:1131–1137. [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M, Roulston A, Belec L, Billot X, Marcellus R, Bedard D, Bernier C, Branchaud S, Chan H, Dairi K, et al. The small molecule GMX1778 is a potent inhibitor of NAD+ biosynthesis: strategy for enhanced therapy in nicotinic acid phosphoribosyltransferase 1-deficient tumors. Mol Cell Biol. 2009;29:5872–5888. doi: 10.1128/MCB.00112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhouse S. On respiratory impairment in cancer cells. Science. 1956;124:267–269. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick AN, Drury DR, Nakada HI, Wolfe JB. Localization of the primary metabolic block produced by 2-deoxyglucose. J Biol Chem. 1957;224:963–969. [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkiewicz AK, Whitaker-Menezes D, Dasgupta A, Philp NJ, Lin Z, Gandara R, Sneddon S, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Using the “reverse Warburg effect” to identify high-risk breast cancer patients: stromal MCT4 predicts poor clinical outcome in triple-negative breast cancers. Cell Cycle. 2012;11:1108–1117. doi: 10.4161/cc.11.6.19530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Stine ZE, Xia J, Lu Y, O’Connor RS, Altman BJ, Hsieh AL, Gouw AM, Thomas AG, Gao P, et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J Clin Invest. 2015;125:2293–2306. doi: 10.1172/JCI75836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TS, Lu SN, Chao Y, Sheen IS, Lin CC, Wang TE, Chen SC, Wang JH, Liao LY, Thomson JA, et al. A randomised phase II study of pegylated arginine deiminase (ADI-PEG 20) in Asian advanced hepatocellular carcinoma patients. Br J Cancer. 2010;103:954–960. doi: 10.1038/sj.bjc.6605856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4:1406–1417. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Travins J, Wang F, David MD, Artin E, Straley K, Padyana A, Gross S, DeLaBarre B, Tobin E, et al. AG-221, a First-in-Class Therapy Targeting Acute Myeloid Leukemia Harboring Oncogenic IDH2 Mutations. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-16-1034. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Furuya S, Osuka S, Mitoma J, Shinoda Y, Watanabe M, Azuma N, Tanaka H, Hashikawa T, Itohara S, et al. Targeted disruption of the mouse 3-phosphoglycerate dehydrogenase gene causes severe neurodevelopmental defects and results in embryonic lethality. J Biol Chem. 2004;279:3573–3577. doi: 10.1074/jbc.C300507200. [DOI] [PubMed] [Google Scholar]
- Yun J, Mullarky E, Lu C, Bosch KN, Kavalier A, Rivera K, Roper J, Chio II, Giannopoulou EG, Rago C, et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350:1391–1396. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauri M, Berridge G, Thezenas ML, Pugh KM, Goldin R, Kessler BM, Kriaucionis S. CDA directs metabolism of epigenetic nucleosides revealing a therapeutic window in cancer. Nature. 2015;524:114–118. doi: 10.1038/nature14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zheng A, Hydbring P, Ambroise G, Ouchida AT, Goiny M, Vakifahmetoglu-Norberg H, Norberg E. PHGDH Defines a Metabolic Subtype in Lung Adenocarcinomas with Poor Prognosis. Cell Rep. 2017;19:2289–2303. doi: 10.1016/j.celrep.2017.05.067. [DOI] [PubMed] [Google Scholar]
- Zhang D, Li J, Wang F, Hu J, Wang S, Sun Y. 2-Deoxy-D-glucose targeting of glucose metabolism in cancer cells as a potential therapy. Cancer Lett. 2014a;355:176–183. doi: 10.1016/j.canlet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Zhang X, Fryknas M, Hernlund E, Fayad W, De Milito A, Olofsson MH, Gogvadze V, Dang L, Pahlman S, Schughart LA, et al. Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments. Nat Commun. 2014b;5:3295. doi: 10.1038/ncomms4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Mackenzie ED, Karim SA, Hedley A, Blyth K, Kalna G, Watson DG, Szlosarek P, Frezza C, Gottlieb E. Reversed argininosuccinate lyase activity in fumarate hydratase-deficient cancer cells. Cancer Metab. 2013;1:12. doi: 10.1186/2049-3002-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Lee D, Shim H. Metabolic positron emission tomography imaging in cancer detection and therapy response. Semin Oncol. 2011;38:55–69. doi: 10.1053/j.seminoncol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun. 2004;313:459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]


