Abstract
We screened the genomes of a broad panel of kinetoplastid protists for genes encoding proteins associated with the RNA interference (RNAi) system using probes from the Argonaute (AGO1), Dicer1 (DCL1), and Dicer2 (DCL2) genes of Leishmania brasiliensis and Crithidia fasciculata. We identified homologs for all the three of these genes in the genomes of a subset of these organisms. However, several of these organisms lacked evidence for any of these genes, while others lacked only DCL2. The open reading frames encoding these putative proteins were structurally analyzed in silico. The alignments indicated that the genes are homologous with a high degree of confidence, and three-dimensional structural models strongly supported a functional relationship to previously characterized AGO1, DCL1, and DCL2 proteins. Phylogenetic analysis of these putative proteins showed that these genes, when present, evolved in parallel with other nuclear genes, arguing that the RNAi system genes share a common progenitor, likely across all Kinetoplastea. In addition, the genome segments bearing these genes are highly conserved and syntenic, even among those taxa in which they are absent. However, taxa in which these genes are apparently absent represent several widely divergent branches of kinetoplastids, arguing that these genes were independently lost at least six times in the evolutionary history of these organisms. The mechanisms responsible for the apparent coordinate loss of these RNAi system genes independently in several lineages of kinetoplastids, while being maintained in other related lineages, are currently unknown.
Keywords: Kinetoplastea, Trypanosomatida, RNAi, Evolutionary loss of genes
Introduction
The phenomenon of double-stranded RNA-induced gene silencing known as RNA interference (RNAi) is widely spread among metazoans and plays critical roles in a myriad of life-associated processes in those organisms. The RNAi system has been hypothesized to have originally evolved to down-regulate dsRNA-borne genes in invading viruses, retrotransposons, or other less well-known mechanisms (Agrawal et al. 2003; Malone and Hannon 2009). However, in many eukaryotes, the system has been coopted for regulation of cellular processes (Ketting 2011; Hutzinger and Izaurralde 2011). Since the RNAi pathway is operational in most if not all modern eukaryotes, it is assumed to have been present in the common progenitors of these organisms. Therefore, the absence of genes central to the RNAi pathway from the genomes of several kinetoplastids, as well as some other protozoans (Cerutti and Casas-Mollano 2006), is intriguing. Since many kinetoplastids are parasitic and have important impacts on human, plant, and animal health, and the RNAi pathways have become important components of many therapeutic strategies (Vaishnaw et al. 2010; Davidson and McCray 2011), these processes have additional importance for this group of organisms. The major participants in the otherwise quite complex RNAi system are the RNAse III endonuclease Dicer, frequently represented by two distinct gene products (DCL1 and DCL2) that function in the nucleus and cytoplasm, respectively; and Argonaute (AGO1), which recruits RNAs to be digested by Dicer (Wilson and Doudna 2013). These proteins mediate cleavage of target dsRNA precursors to generate mature active populations of 20–26 nt small RNAs. These mature siRNAs are responsible for degradation of target mRNAs and for mediating the other regulatory activities of the RNAi pathway.
Some eukaryotes apparently lack the genes encoding Dicer and Argonaute, and are thought to lack the RNAi pathway (Cerutti and Casas-Mollano 2006). Where this has been empirically examined, organisms lacking the Dicer and Argonaute genes do not exhibit the expected panel of siRNAs, and are unable to degrade RNA targets of short complementary RNAs (Shi et al. 2004). Previous focused studies of some species of trypanosomatids demonstrated that those organisms apparently missing these genes lack significant RNAi activity (Ullu et al. 2004; Lye et al. 2010). In the study described herein, we have examined the genomes of a broad range of species from the class Kinetoplastea, including free-living species, plant pathogens, insect parasites, and vertebrate parasites, for the presence of Dicer and Argonaute. Our data suggest that the progenitor of the Kinetoplastea bore genomic copies of the Dicer and Argonaute genes, but that many modern descendants of these progenitors have lost these genes. Examination of current phylogenetic relationships of these organisms argues that loss of these genes has been largely coordinate and has occurred independently several times in the lineage of kinetoplastids, suggesting a possible sporadic exertion of a positive, selective pressure for loss of the RNAi system.
Materials and Methods
Genome and Gene Sequences
All organisms studied in this work together with the Gen-Bank accession numbers for their genomes and RNAi-related genes (where applicable) are listed in Table 1. Access to the unpublished draft genomes analyzed in this study can be obtained by contacting the corresponding author. The organisms included in this study are cryopreserved at the trypanosomatid culture collection (TCC) of the University of São Paulo, Brazil.
Table 1.
Genome and gene sequences of kinetoplastids tested for the presence of RNAi-associated genes
| Strain | Resource | Genome accession number | GenBank accession numbers for RNAi-related genes
|
||
|---|---|---|---|---|---|
| AGO1 | DCL1 | DCL2 | |||
| Leishmania major Friedlin. | Ref. strain | GCA_000002725 | |||
| L. braziliensis M2904 | Ref. strain | GCA_000002845 | ACI22628 | XP001565111 | XP_001565607 |
| Crithidia fasciculata Cf-Cl | Ref. strain | GCA_000331325 | ACD91648 | KY351826 | KY351827 |
| C. acanthocephali | TCC037E | AUXI00000000 | KT377047 | KT377048 | KT377049 |
| Trypanosoma cruzi G | pp | ||||
| T. cruzi CL | pp | ||||
| T. cruzi 1994 (Tcbat) | TCC1994 | pp | |||
| T. cruzi marinkellei | TCC 344 | pp | |||
| T. conorhini | TCC025E | pp | KT377056 | KT377057 | KT377058 |
| T. lewisi | TCC034 | pp | |||
| T. dionisii | TCC211 | pp | |||
| T. rangeli AM80 | TCC086 | pp | KT377059 | KT377060 | KT377061 |
| T. erneyi | TCC1946 | pp | |||
| T. brucei b. TREU927 | Ref. strain | GCA_000002445 | XP_823303 | XP_847070 | XP_843717 |
| T. b. gambiense DAL972 | Ref. strain | GCA_000210295 | XP_011778481 | XP_011775511 | XP_001565607 |
| Bodo sp | ATCC50149 | pp | KY305538 | KY308193 | |
| Endotrypanum schaudinni | TCC224 | pp | |||
| Leptomonas costaricensis BR | TCC169E | pp | KT377053 | KT377054 | KT377055 |
| Parabodo caudatus | ATCC30905 | pp | |||
| Phytomonas sp. Jma | TCCxxx | pp | |||
| Angomonas deanei | TCC036E | AUXM00000000 | KT377043 | KT377044 | |
| A. desouzai | TCC079E | AUXL00000000 | KT377045 | KT377046 | |
| Strigomonas galati | TCC219 | AUXN00000000 | |||
| S. culicis | TCC012E | AUXH00000000 | |||
| S. oncopelti | TCC 290E | AUXK00000000 | |||
| Herpetomonas muscarum | TCC001E | AUXJ00000000 | KT377050 | KT377051 | KT377052 |
Abbreviations used in the Table: pp—genome sequences were generously provided pre-publication by the Euglenozoan Genome Consortium (see Acknowledgements)
Genome sequences of strains with shown accession numbers were previously published (Alves et al. 2013a, b). Other sequences used herein were obtained from TriTrypDB (Aslett et al. 2010)
Strains of L. major (MHOM/IL/80/Friedlin), L. braziliensis (MHOM/MHOM/BR/75/M2904), C. fasciculata (CFACl_0070011200), T. brucei brucei TREU927 (clone derived from GPAL/KE/70/EATRO 1534) and T. brucei gambiense DAL972 (clone derived from MHOM/CI/86/DAL972) were used as a reference in this work
Identification of Gene and Protein Sequences
The AGO1, DCL1, and DCL2 gene sequences from each of the above genomes were identified by BLAST (tBLASTn) alignments using the experimentally confirmed gene sequences from L. braziliensis and C. fasciculata (Lye et al. 2010), kindly provided prior to their publication, and their ORFs were manually extracted. Probe sequences derived from these strains and used for identification of APRT (adenine phosphoribosyl transferase), GSH1 (γ-glutamylcysteine synthetase), and PTR1 (pteridine reductase 1), which were used for phylogenetic analysis of all organisms analyzed regardless of the RNAi system presence, were acquired from GeneDB (http://www.genedb.org). Whenever necessary, the integrity of the imperfect ORFs was confirmed by the PCR-amplifying problematic regions internal to the gene followed by sequencing. The completeness of ORFs thus obtained was confirmed by identifying the first in-frame termination codon upstream of the deduced gene sequence.
Phylogenetic Analysis
Protein sequences were aligned using ClustalW2 (Larkin et al. 2007), and alignments were edited using Gblocks (Castresana 2000) to remove regions of ambiguity. Maximum likelihood phylogenetic inferences were performed by means of RAxML v. 8.2.4 (Stamatakis 2006) using the PROTGAMMAWAGF substitution model (empirical model WAG) (Whelan and Goldman 2001), with gamma-distributed heterogeneity rate categories and estimated empirical residue frequencies, as determined by RAxML’s automatic model selection when used in preliminary runs. Two hundred best-tree searches were performed for each alignment, and the tree with best likelihood found was kept. RAxML’s rapid bootstrap was performed with 100 pseudoreplicates, and support is only shown for branches with support of at least 50. Trees were drawn using Dendroscope (Huson et al. 2007).
Structural Alignments and Conservation
The secondary structure and molecular surface of putative AGO1 and DCL1 proteins were generated and compared according to sequence conservation. In brief, proteins with significant structural homology were initially identified and aligned using the mGenTHREADER algorithm of the PSIPRED protein structure prediction server (McGuffin et al. 2000) and Clustal Omega (Sievers et al. 2011), respectively. The results of these comparisons were illustrated using the molecular visualization software Pymol (DeLano 2013). Sequence conservation (percent identity) for each residue within the reference template was calculated (gaps were not considered for alignment), and the secondary structure and corresponding molecular surface were colored as follows: residues that were highly conserved (81–100% identity) were colored red, less conserved (61–80%) colored blue, still less (41–60%) green, and poorly conserved residues (21–40%) are shown in gray. No residues were less than 20% conserved among the structural homologs.
Synteny Analyses
The genomic contexts of the RNAi genes analyzed in this work were determined by either analysis of precomputed orthology relationships and synteny maps present in the TriTrypDB database (Aslett et al. 2010), with manual adjustment of the final maps for presentation purposes, or by BLAST similarity analysis. The latter was performed to investigate synteny of the genomic regions for unpublished draft genome sequences analyzed herein. Annotated genes found flanking Argonaute or Dicer genes were selected in the published genomes of C. fasciculata Cf-C1 (http://tritrypdb.org/), or Trypanosoma brucei TREU927 (Berriman et al. 2005) (“guide genomes”) were used as probes against genomic contigs from draft genomes. The identified contigs were examined using TBLASTN to find all putative genes present. Any genes present in the draft genomes that were not present in the two guide genomes were then added to the set of probes and searched against all other genomes in the analysis. Finally, gene orders were manually compared and graphically displayed.
Results and Discussion
RNAi-related Gene Sequences in Kinetoplastid Protozoa
Initial similarity searches of genomic sequences of kinetoplastid protists using AGO1 and DCL1 and DCL2 gene sequences from a variety of eukaryotic species were unproductive (data not shown). However, probing of these genomes with the putative AGO and DCL genes (AGO1, DCL1, and DCL2) from L. braziliensis and C. fasciculata (Lye et al. 2010) permitted us to identify apparent AGO1 and DCL homologs in a broad spectrum of organisms (Table 2).
Table 2.
Distribution of RNAi-associated genes in the kinetoplastid genome
| Strain | Host | Argonaute | Dicer1 | Dicer2 | |||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| Aa | Expect | Aa | Expect | Aa | Expect | ||
| Bodo sp. | Free-living | Part | 6e-19 | Part | 2e-12 | N.F | |
| P. caudatus | Free-living | N.F | N.F | N.F | |||
| L. major | Human | N.F | N.F | N.F | |||
| L. braziliensis | Human | 898 | 0.0 | 2214 | 0.0 | 1330 | 0.0 |
| T. cruzi G | Opossum | N.F | N.F | N.F | |||
| T. brucei brucei | Diptera | 903 | 1e-110 | 1648 | 2e-76 | 948 | 1e-11 |
| T. b. gambiense | Human | 914 | 1e-103 | 1648 | 1e-76 | 946 | 8e-12 |
| T. cruzi 1994 | Bat | N.F | N.F | N.F | |||
| T. cruzi CL | Hemiptera | N.F | N.F | N.F | |||
| T.cruzi marinkellei | Bat | N.F | N.F | N.F | |||
| T.dionisii | Bat | N.F | N.F | N.F | |||
| T. erneyi | Bat | N.F | N.F | N.F | |||
| T. rangeli AM80 | Human | 873 | 1e-100 | 1642 | 5e-71 | 941 | 3e-11 |
| T. conorhini | Rat | 905 | 1e-102 | 1686 | 8e-74 | 937 | 1e-11 |
| T. lewisi | Rat | N.F | N.F | N.F | |||
| E. schaudinni | Sloth | N.F | N.F | N.F | |||
| C. fasciculata | Diptera | 965 | 0.0 | 2240 | 0.0 | 1251 | 0.0 |
| C. acanthocephali | Hemiptera | 931 | 0.0 | 2201 | 0.0 | 1499 | 0.0 |
| L. costaricensis BR | Hemiptera | 959 | 0.0 | 2379 | 0.0 | 1316 | 0.0 |
| H. muscarum | Diptera | 915 | 1e-144 | 2000 | 6e-84 | 1380 | 8e-14 |
| Phytomonas sp. Jma | Plant | N.F | N.F | N.F | |||
| A. deanei | Hemiptera | 882 | 1e-114 | 1412 | 9e-71 | N.F | |
| A. desouzai | Diptera | 893 | 1e-120 | 1336 | 3e-81 | N.F | |
| S. galati | Diptera | N.F | N.F | N.F | |||
| S. oncopelti | Hemiptera | N.F | N.F | N.F | |||
| S. culicis | Diptera | N.F | N.F | N.F | |||
Sequences of AGO1, DCL1 and DCL2 genes identified in Leishmania braziliensis and C. fasciculata genomes (Lye et al. 2010) were used as queries in searches of similarities; BLAST p values were based on amino acid sequences from L. braziliensis. In addition, T. brucei brucei TREU927, T. b. gambiense DAL972 and L. major Friedlin were used as reference strains (see Table 1)
Hosts are specified by the original source of organisms; however, some organisms, like T. cruzi strains G and CL, or, strains of Leishmania are pathogenic to humans. The organism of isolation for C. fasciculata, was likely a mosquito
This table includes incomplete data on Bodo sp. ATCC50149 proteins AGO1 and DCL1, whose genes have been identified in the genome. Due to the scarcity of the available material, it was not possible to obtain amino acid sequences that would form complete ORFs with presumably only small portions missing. It was found important though to include this organism, being the only free-living kinetoplastid with identified RNAi genes among those tested, in this work
Aa number of amino acids in the protein, Part partial open reading frame, NF not found
Thus, Crithidia acanthocephali, Leptomonas costaricensis BR, Trypanosoma conorhini, and T. rangeli AM80, like the African trypanosomes (Ngo et al. 1998), L. braziliensis, and C. fasciculata (Lye et al. 2010), harbor genes that encode proteins with significant similarity to all three targets; namely, AGO, DCL1, and DCL2. Herpetomonas muscarum also bears genes with high similarity to AGO and DCL1, but DCL2 aligns only poorly with a potential H. muscarum homolog. Similarly, we were only able to identify putative AGO1 and DCL1 genes in Angomonas deanei, A. desouzai, the African trypanosome Trypanosoma vivax, and a Bodo sp. isolate, suggesting the genomes of these organisms lack any genes with similarity to DCL2.
Other species of Kinetoplastea, notably T. cruzi, T. lewisi, T. dionisii, S. galati, S. culicis, S. oncopelti, Phytomonas sp., P. caudatus, and E. schaudinni (Table 2), exhibited no genes with significant similarity to any of the known AGO or DCL genes, including those identified in this study (Table 2), even with sensitive searches designed to detect low levels of similarity. Since we were readily able to identify these genes in taxa closely related to these organisms, these taxa apparently lack homologs for these genes and therefore apparently lack a functional classical RNAi pathway. A less-likely alternative is that the AGO and DCL genes have diverged at a higher rate in these organisms than they have in related taxa, and therefore are not detected in our similarity searches. Since we have no evidence for a differential rate of evolution of the genes in these organisms, and similar searches readily identify orthologs of many other genes in their genomes (data not shown), our data strongly favor the hypothesis that genomes in which we fail to identify AGO and DCL homologs actually lack genes encoding orthologs of these proteins.
To further confirm that the absence of RNAi genes in these organisms is a result of evolutionary loss, we examined the synteny of the immediate genomic environment of these genes in each of these organisms. The results obtained demonstrate an obvious synteny of the genes both upstream and downstream from the AGO1 (Fig. S1), DCL1 (Fig. S2), and DCL2 (not shown) in all kinetoplastid genomes examined, irrespective of the presence or absence of these genes. These results are supported by the data from TriTrypDB (Aslett et al. 2010) that have been performed with a number of genomes of organisms on that site and include AGO1 gene (Leishmania group, Fig. S3), AGO1 (Trypanosoma group, Fig. S4), DCL1 gene (Fig. S5), and Dicer 2 gene (Fig. S6).
Out of about a dozen of T. cruzi strains examined, in addition to those included in Table 2, none displayed the presence of genes encoding for Argonaute or any of the Dicer proteins (not shown). T. rangeli AM80 (TCC086) reported herein, possesses the major components of the RNAi system in intact open reading frames (Table 2). Interestingly, Stoco et al. (2014) reported that the genome of T. rangeli SC-58, which is a rodent isolate representative of a different lineage (TrD) of this species (Grisard et al. 2010), and which was isolated in a geographic area (Southern Brazil) distant from where (Brazilian Amazonia) T. rangeli AM80 was isolated from human (Maia da Silva et al. 2004), bears only pseudogenes of the RNAi-related genes. It is possible that this apparent difference is due to the incompleteness of the T. rangeli SC-58 assembly. However, Lye et al. (2010) reported that some species of Leishmania have functional RNAi systems, including intact AGO and DCL genes, whereas other species in the genus lack these genes and their activities. Thus, for reasons yet to be clarified, the evolutionary pressures, if any, responsible for abandonment of the RNAi functions can be expressed differentially within different species in a given genus. This conclusion is only minimally tempered by the fact that the definition of species among the kinetoplastid protozoa remains to be clarified.
In silico Structural Analysis of the Putative Argonaute and Dicer Homologs
The putative AGO and DCL protein sequences described above were aligned to homologous sequences from T. brucei and L. braziliensis and other available Argonaute and Dicer sequences from the protein database, using the conserved domain architecture retrieval tool (see “Materials and Methods” section). The alignment of the AGO1 orthologs revealed the presence of the most conserved PAZ and PIWI domains (Fig S7) typical of Argonaute proteins (Ullu et al. 2004; Hutvanger and Simard 2008; Ender and Meister 2010). Other domains, including the N-terminalor MID domain usually found in AGO proteins (Song and Joshua-Tor 2006; Boland et al. 2011), were not readily identified. In contrast, only the RNAse III terminal domain (RIBOc) was sufficiently conserved to be clearly identified on all trypanosomatid Dicer1 protein sequences (Fig. S8). Other components conferring functional activity of Dicer proteins (Cerutti and Casas-Mollano 2006) were not identifiable by this approach. These results argue that, as has been found in other eukaryotes, the RNAi components in trypanosomatids are quite divergent, and that Dicer1, the “simplistic” domain architecture of which has been demonstrated for T. brucei (Shi et al. 2006), is less conserved than AGO1.
To further confirm the identity of these proteins as Argonaute and Dicer homologs, we compared in silicogenerated 3D maps of the newly identified proteins to in vitro-generated 3D structures of known orthologs of these proteins. This analysis confirmed that both these proteins were Argonaute and Dicer homologs and also yielded results supporting a high level of divergence among these proteins. Thus, the best matches for the 3D structure of AGO1 were from bacterial or archaean Argonautes (Table 3), where this protein binds DNA preferentially to RNA (Swarts et al. 2014). However, this observation may be due to the relative paucity of the database of crystallized AGO1 and Dicer proteins.
Table 3.
Structural alignment hits for AGO1 within RNAipositive trypanosomatids (organism/group/net score/pvalue)
| Angomonas deaneí | Angomonas desouzai |
| Thermus thermophilus/Bacteria/144.745/1e-13 | Thermus thermophilus/Bacteria/132.324/2e-12 |
| Aquifex aeolicus/Bacteria/133.144/2e-12 | Aquifex aeolicus/Bacteria/121.194/3e-11 |
| Pyrococcus furiosus/Archaea/119.083/5e-11 | Pyrococcus furiosus/Archaea/110.411/4e-10 |
| Archaeoglobus fulgidus/Archaea/88.365/6e-08 | Neurospora crassa/Fungi/74.942/1e-06 |
| Neurospora crassa/Fungi/85.92/1e-07 | Archaeoglobus fulgidus/Archaea/73.297/2e-06 |
| Crithidia acanthocephali | Herpetomonas muscarum |
| Thermus thermophilus/Bacteria/133.862/2e-12 | Thermus thermophilus/Bacteria/139.978/4e-13 |
| Aquifex aeolicus/Bacteria/116.165/9e-11 | Aquifex aeolicus/Bacteria/118.653/5e-11 |
| Pyrococcus furiosus/Archaea/113.010/2e-10 | Pyrococcus furiosus/Archaea/116.433/9e-11 |
| Neurospora crassa/Fungi/81.393/3e-07 | Archaeoglobus fulgidus/Archaea/76.701/9e-07 |
| Archaeoglobus fulgidus/Archaea/77.951/7e-07 | Neurospora crassa/Fungi/72.209/3e-06 |
| Leishmanía brazilíensis | Leptomonas costarícensis BR |
| Thermus thermophilus/Bacteria/135.176/1e-12 | Thermus thermophilus/Bacteria/140.450/3e-13 |
| Pyrococcus furiosus/Archaea/104.827/1e-09 | Pyrococcus furiosus/Archaea/103.086/2e-09 |
| Aquifex aeolicus/Bacteria/98.940/5e-09 | Aquifex aeolicus/Bacteria/97.137/8e-09 |
| Neurospora crassa/Fungi/61.799/3e-05 | Neurospora crassa/Fungi/78.719/6e-07 |
| Homo sapiens/Mammalia/61.407/3e-05 | Archaeoglobus fulgidus/Archaea/72.818/2e-06 |
| Trypanosoma conorhini | Trypanosoma rangeli AM80 |
| Thermus thermophilus/Bacteria/165.365/1e-15 | Thermus thermophilus/Bacteria/171.037/3e-16 |
| Pyrococcus furiosus/Archaea/131.683/3e-12 | Pyrococcus furiosus/Archaea/142.483/2e-13 |
| Aquifex aeolicus/Bacteria/131.350/3e-12 | Aquifex aeolicus/Bacteria/138.705/5e-13 |
| Archaeoglobus fulgidus/Archaea/104.903/1e-09 | Archaeoglobus fulgidus/Archaea/101.867/3e-09 |
| Neurospora crassa/Fungi/88.926/5e-08 | Neurospora crassa/Fungi/94.733/1e-08 |
Profile-profile alignments, fold recognition and identification performed with mGenTHREADER, as provided by Protein Structure Prediction Server (psipred), http://bioinf.cs.ucl.ac.uk/psipred/
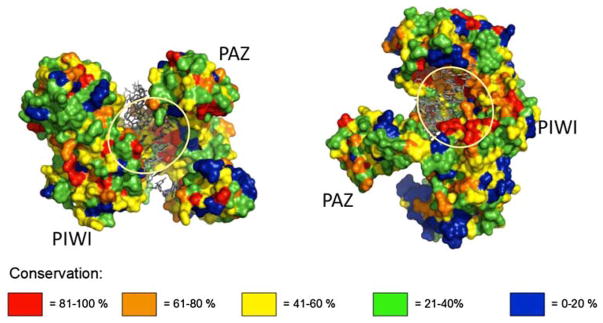
Figure 1 illustrates an overlay of the aligned protein sequences of the AGO1 genes on the 3D crystal structure of the AGO1 protein from the archaean Thermus thermophilus. The overlay is color coded according to the level of conservation of the amino acid sequence. Continuous regions of high (40–100%) structural conservation are observed on both lobes of the AGO1 protein, coincident with those sequences known to interact with the guide RNA. These sequences correspond to the nucleic-acidbinding pocket located between PAZ- and PIWI-containing lobes, as described for Argonaute proteins from both prokaryotic and eukaryotic origins (Wang et al. 2008; Boland et al. 2011; Nakanishi et al. 2012). Since both domains were identified for all trypanosomatids, this conservation pattern further strongly supports the conclusion that these genes are orthologs of the Argonaute genes in the genomes of these protozoans.
Fig. 1.
Structural conservation of Argonaute proteins within kinetoplastid strains. AGO1 protein sequences from nine trypanosomatid strains aligned with the corresponding sequence from T. thermophilus and superimposed on its 3D model derived from the crystal structure. Conservation levels shown in different colors. The proposed nucleic-acid-binding pocket is indicated
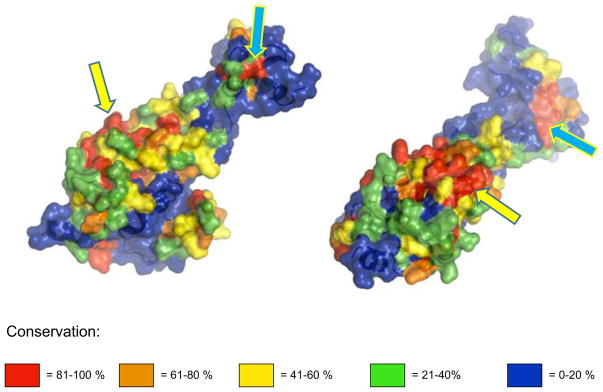
In contrast to the putative AGO1 homologs, no direct structural match was observed for the Dicer proteins, probably because of the poor representation of structures within the available data and the generally higher level of divergence of the corresponding genes. Therefore, we performed a superimposition of multiple-aligned sequences on the 3D structure of the human Dicer (Fig. 2). Regions of high conservation were observed in those parts of the molecule that, according to the reconstruction of the architecture (Lau et al. 2012), should harbor RNAseIII- and dsRNA-binding domains (Fig. 2, lower middle portion of the model, which is the proposed nuclease core of the enzyme), while the high-conservation area illustrated on the upper part of the model as shown may represent PAZ-like or a Platform-like domain, necessary for the capture and cleavage control of the dsRNA precursor (MacRae et al. 2007). The peripheral sequences of the putative trypanosomatid Dicer proteins exhibit low conservation, on the one hand, confirming the high divergence suggested for this enzyme, and on the other hand, highlighting the biased conservation pattern of the proposed PAZ (Platform) positioning. Overall, these structural analyses strongly support the identification of these genes as Argonaute and Dicer homologs.
Fig. 2.
Structural conservation of Dicer1 proteins within kinetoplastid strains. Dicer1 protein sequences from nine trypanosomatid strains aligned with the corresponding sequence (human) and superimposed on its 3D model derived from the crystal structure. Conservation levels shown in different colors. Arrows point at the proposed nuclease core (yellow) and PAZ-like or a platform-like domain (blue)
Phylogenetic Analysis of RNAi-Associated Genes From Kinetoplastid Protozoa
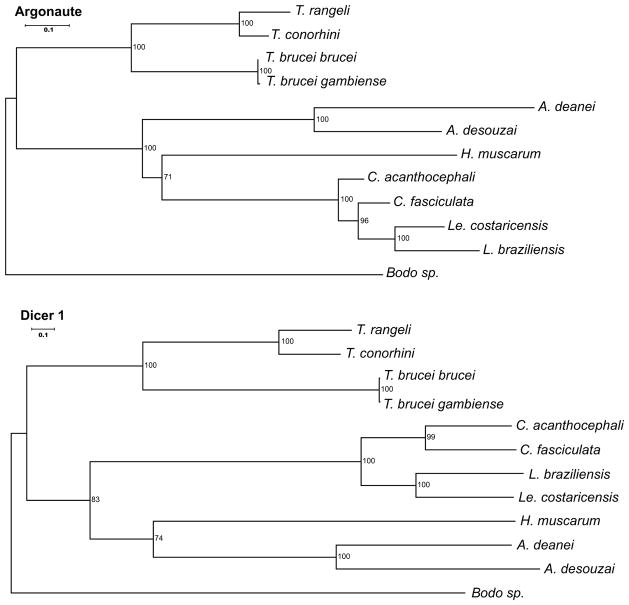
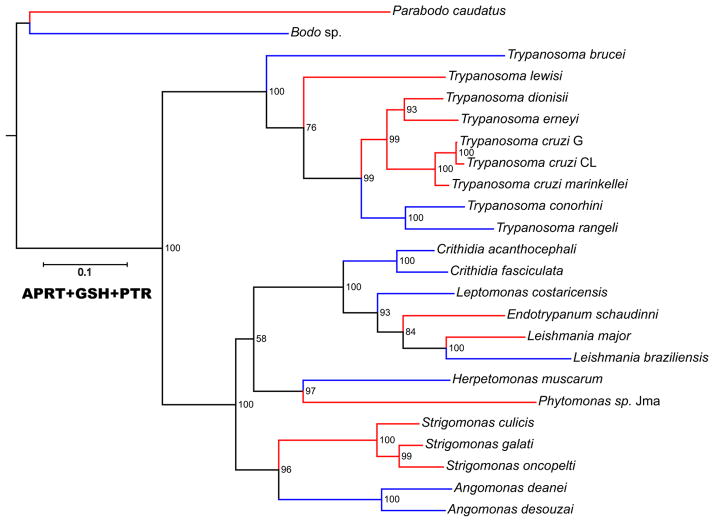
Putative AGO1 and DCL1 homologs from all species from which they have been identified were extracted. The protein sequences of these genes were generated in silico, aligned, and then subjected to phylogenetic analysis (Fig. 3). In general, the phylogenies of the RNAi-associated proteins that were identified as AGO and DCL1 orthologs were quite similar to the previously published phylogenies (e.g., Teixeira et al. 2011; Lima et al. 2012), as well as to those based on phylogenies of the housekeeping proteins APRT (adenine phosphoribosyl transferase), GSH1 (γ-glutamylcysteine synthetase), and PTR1 (pteridine reductase 1) (Fig. 4). Together, these results argue that the RNAi-associated genes AGO and DCL1 likely evolved in parallel with their ‘host’ organisms from orthologs of these genes present in their last common progenitor.
Fig. 3.
Evolutionary trees of trypanosomatid RNAi-related genes Argonaute (AGO1) and Dicer 1 (DCL1) genes. Molecular trees were generated as described in Materials and Methods and include gene sequences from seven sequenced trypanosomatids plus two gene sequences each from L. braziliensis and C. fasciculata, used as probes. Numbers on nodes are bootstrap support values (50 or greater shown)
Fig. 4.
Evolutionary trees based on trypanosomatid housekeeping genes APRT(adenine phosphoribosyl transferase), GSH1 (γ-glutamylcysteine synthetase), and PTR1 (pteridine reductase 1). The scale corresponds to the degree of evolutionary divergence among these organisms. Lineages lacking RNAi genes are colored red, while RNAi-proficient lineages are colored blue
Loss of RNAi Pathway Components in the Evolution of the Kinetoplastida
Despite previous work demonstrating that T. brucei (Shi et al. 2006; Patrick et al. 2009) and some species of Leishmania (Lye et al. 2010) bear homologs for DCL1, DCL2, and AGO1, our analyses indicated that the presence or absence of these genes varies across the species and genera of the Order Kinetoplastida. Our results showed that all T. cruzi isolates examined, T.cruzi-like from bats T. c. marinkellei, T. erneyi, and T. dionisii (Lima et al. 2012), T. lewsii, and species of Strigomonas, Phytomonas Endotrypanum, and Parabodo caudatus, lack AGO1, DCL1, and DCL2 (see Table 2). Our findings suggest that the RNAi system is missing entirely in the trypanosomes of the subgenus Schizotrypanum. Other species, including Bodo sp., T. rangeli AM80, T. conorhini, C. acanthocephali, L. costaricensis BR, H. muscarum, A. deanei, and A. desouzai, maintain complete or nearly complete RNAi systems. The role of Argonaute-like proteins for a spectrum of RNAi-associated pathways is well documented (Hutvanger and Simard 2008; Ketting 2011), and the rare event of a natural loss of an Argonaute gene by a yeast strain is apparently associated with a total absence of siRNAs even in the presence of a functional Dicer gene (Drinnenberg et al. 2009). Thus, we can assume that the loss of such an important gene-regulatory function as the RNAi system would have immediate effects on the gene expression program of that organism.
A careful analysis of the placement of these species on the phylogenetic trees generated from the housekeeping and the rRNA genes (see above) suggests that closely related organisms differ with regard to the presence of the RNAi system. Thus, Leishmania sp. (but not Leishmania major), C. fasciculata, and C. acanthocephali seem to have competent RNAi systems, whereas the closely related Endotrypanum schaudinni lacks it. Similarly, Angomonas and Strigomonas spp. (Teixeira et al. 2011), a clade of closely related insect trypanosomatids, also differ with respect to the presence of the AGO and DCL genes and presumably the RNAi system (Fig. 4).
Our phylogenetic survey also demonstrated that, in addition to the two evolutionary divergences mapped previously as leading to the Leishmania/Crithidia branch and the Trypanosoma (Lye et al. 2010), and extended herein for the Trypanosoma, the RNAi system is apparently also absent in the lineages leading to the Phytomonas/Herpetomonas (Borghesan et al. 2013) and the Angomonas/Strigomonas (Teixeira et al. 2011) phylogenetic lineages (Fig. 4). The Leishmania/Crithidia lineage likely includes Endotrypanum, which also apparently lacks RNAi homologs. Although this parasite, isolated from sloths and sand flies, clusters with Leishmania, its final classification may still require confirmation (Cupolillo et al. 2000; Hughes and Piontkivska 2003).
An additional point of possible RNAi loss was determined outside the trypanosomatid family, although still within the Kinetoplastea, on the lineage leading to Bodo sp. ATCC 50149 and P. caudatus (Fig. 4). Using the protein probes derived from L. braziliensis, AGO1 and DCL1, but not DCL2 were identified as very likely encoded by the corresponding genes from the free-living Bodo sp. genome. However, P. caudatus, another distantly related free-living kinetoplastid, seems to lack both Argonaute and Dicer homologs (Fig. 4) even when searches using the homologs from Bodo are performed. This organism is sufficiently distant from the parasitic kinetoplastids, so that it is possible that extensive sequence divergence has led to the inability to detect homologous genes using simple similarity searches. However, this would suggest that AGO and DCL genes in this microbe are more divergent than most other genes, including those used for generation of phylogenetic trees herein (Fig. 4) and elsewhere (Lye et al. 2010), which are readily detected in similarity searches. Thus, it seems most likely that this free-living kinetoplastid also lacks a functional RNAi system.
To further examine the presence or absence of the Argonaute and Dicer genes in these organisms, we examined the genomic contexts of these genes. The results (Fig. S1–S2) show that, first, the genes within the genomic segments upstream and downstream from the loci bearing these genes are highly syntenic. Moreover, this structure is generally conserved even when the Argonaute and Dicer genes have been deleted. These observations suggest that when these genes are eliminated, they are eliminated completely largely coordinately, possibly by a single deletion event. However, since Dicer 2 is occasionally deleted first, and there is evidence that some organisms, e.g., T. rangeli SC58 (Stoco et al. 2014), still carry pseudogenes of the RNAi genes, these genes may also be lost in a rapid incremental process. The mechanisms by which these changes are driven are currently a matter of speculation.
The results presented here strongly confirm the previous observation that several clades of kinetoplastids have independently and discretely lost the RNAi system. The close relationships of some of these organisms, like Leptomonas, the Leishmania, and Endotrypanum, or T. cruzi, T. rangeli, and T. conorhini, argue that the loss has been recent in some of these organisms. In contrast, the presence of Dicer and Argonaute genes in many branches of the evolutionary tree, including probably at least some of the free-living Bodo species, argues that the progenitors of these organisms were RNAi-proficient and had both Dicer and Argonaute genes.
There is no immediately obvious answer to the question of what, if any, selective advantage could be attributed to the loss of the RNAi system in the trypanosomatid strains studied in this work, as well as in other protozoans, including the malaria-causing Plasmodium (Baum et al. 2009). Several hypotheses about these selective advantages that have been previously promoted invoke viruses and/or transposable elements and enhanced pathogenesis (Lye et al. 2010). However, there is as of yet no hard evidence to support these hypotheses, and the cause of the loss of these activities remains elusive. An alternative explanation might be that the RNAi system in these organisms is, or became, nonessential, and that we are observing a random loss of the genes in the different taxa. It is striking that, in most instances where the system is lost, it is apparently lost completely, i.e., there are no lingering Argonaute or Dicer genes or pseudogenes, suggesting that the loss occurs largely coordinately. Interestingly, the selective advantage of acquiring a dsRNA virus at the expense of functional RNAi was well demonstrated for several yeast-like fungal species (Drinnenberg et al. 2011), i.e., the satellite dsRNA of the virus encodes a protein toxin lethal to neighboring cells but innocuous to cells harboring the virus. Further study of the sporadic loss of the RNAi system in the Kinetoplastida will be required to identify selective mechanisms, if any.
Loss of one of the two dicer genes may represent a transient intermediate state that will eventually lead to loss of both genes. The two dicers are active in different intracellular compartments (DCL1 in the cytoplasm; DCL2—in the nucleus), target different RNA substrates, and are seemingly serving distinct RNAi functions, resulting in production of either siRNAs or, presumably, miRNAs. Despite the differences in activity, which may be determined more by the cellular microenvironment than by the biochemical specificity of proteins themselves (Cenik et al. 2011), Dicer1 and Dicer2 have been shown to share similar functions or even to cooperate in substrate processing (Patrick et al. 2009). If so, the impact of the loss of one of the Dicers might be less dramatic, as the remaining enzyme could at least partially compensate its functions. As was recently reported (Catta-Preta et al. 2016), the RNAi system remains functional in A. deanei that, according to our data, also seems to have lost Dicer2. In favor of this suggestion may be the fact that RNAi in the organisms defective in Dicer can be complemented by a Dicer from a distantly related host (MacRae et al. 2006). In addition, some organisms, e.g., yeasts and mammals, possess just a single Dicer.
Our current observations suggest a plasticity of the function of the Dicer protein, also favoring the different complexity levels proposed for RNAi systems in general (Cerutti and Casas-Mollano 2006; Ketting 2011). Thus, it is of note that in the process of evolutionary fading, Dicer2 “goes first” (Table 1), at least among the relatively small number of genomes examined. It has been recently demonstrated that, in addition to its function in the classical RNAi pathway, the Dicer2 protein in T. brucei is also required for silencing of small nucleolar RNAs (snoRNAs: Gupta et al. 2010) via the process that was previously called snoRNAi (Liang et al. 2003). The snoRNAs are required for processing and modification of rRNAs (Hartshorne and Toyofuku 1999; Barth et al. 2005). Thus, the contribution of nuclear Dicer2 to the regulation of essential cellular functions may seem more complex than that of cytoplasmic Dicer1, making the former a more significant player that is still seemingly easier to lose. However, the recent observation demonstrates that dcl2 remains the only RNAi-related gene in T. rangeli SC-58 that is potentially functional (Stoco et al. 2014), which may point to a possible involvement in some alternative process, nevertheless, important for the host.
In addition to the defense function provided by the siRNAs, the RNAi pathway is also involved in production of microRNAs that perform numerous regulatory activities by gene silencing at the posttranscriptional level (Chen and Rajewsky 2007). Evidence for the presence of functional microRNAs in RNAi-positive kinetoplastid species is scarce. Thus, although putative microRNAs have been identified in the genome sequence of T. brucei (Mallick et al. 2008), empirical evidence of the function of these sequences is absent. Moreover, despite the recent demonstration of the contribution of an epigenetic factor to transcription activation (Ekanayake and Sabatini 2011), regulation of gene expression in trypanosomatids is believed to be primarily posttranscriptional (c.f.; Clayton 2002; Palenchar and Bellofatto 2006; Clayton and Shapira 2007). In other organisms, microRNAs play important roles in the regulation of differentiation and developmental processes (c.f.; He and Hannon 2004; Hutzinger and Izaurralde 2011). Thus, microRNAs could be effective mediators of the gene expression patterns associated with complex life cycles of the trypanosomatids, involving multiple stages of differentiation of dixenous species in vertebrate and invertebrate hosts (Tyler and Engman 2001; Maia da Silva et al. 2004; de Souza et al. 2010; Teixeira et al. 2011; Lima et al. 2012; Borghesan et al. 2013). The inevitable and intriguing question that still remains is, if in fact progenitors of the current protozoans used the RNAi system extensively and broadly for regulation of gene expression, how could those trypanosomatids that have lost the RNAi potential still manage to maintain their complex life cycle? The existence of compensatory mechanisms here seems to be an obvious answer to this question and may suggest a certain flexibility of the parasites’ life process in general. Alternatively, RNA-mediated silencing of an unknown nature may also be hypothesized, and such a mechanism could involve alternative RNAse-driven maturation processes, resulting in different inhibitory RNA factors. The unique RNA-processing activities of guide RNA biogenesis in the kinetoplastids (Madina et al. 2011), and the recently observed seemingly site-specific cleavage of tRNA in RNAi-deficient T. cruzi (Garcia-Silva et al. 2010), remain to be explained. These or other related unknown mechanisms may represent an alternative RNA-processing machinery aimed at generating small RNA-driven silencing function.
Supplementary Material
Acknowledgments
This work has been funded by the Grant from the National Science Foundation’s Assembling the Tree of Life program (G.A. Buck, Award #080056); the grant from the NIH (S.M. Beverley, award #RO1AI029646), and grants from the Brazilian agencies CNPq and CAPES to MMGT. JMPA is supported by FAPESP, Brazil (Grant #2013/14622-3, São Paulo Research Foundation). AGCM has PhD fellowship from CNPq (PROTAX). The authors are grateful to Marta Campaner for cultures of parasites, Flávia Maia da Silva and Luciana Lima for the preparation of DNAs used for genome sequencing, and the personnel of the Nucleic Acids Research Facilities (NARF) at VCU.
Footnotes
Electronic Supplementary Material The online version of this article (doi:10.1007/s00239-017-9780-1) contains supplementary material, which is available to authorized users.
References
- Agrawal N, Dasaradhi PVN, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNAi interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 2003;67:657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves JMP, Klein CC, da Silva FM, Costa-Martins AG, Serrano MG, Buck GA, Vasconcelos AT, Sagot MF, Teixeira MM, Motta MC, Camargo EP. Endosymbiosis in trypanosomatids: the genomic cooperation between bacterium and host in the synthesis of essential amino acids is heavily influenced by multiple horizontal gene transfers. BMC Evol Biol. 2013a;13:190. doi: 10.1186/1471-2148-13-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves JMP, Serrano MG, Maia da Silva F, Voegtly LJ, Matveyev AV, Teixeira MMG, Camargo EP, Buck GA. Genome evolution and phylogenomic analysis of Candidatus Kinetoplastibacterium, the betaproteobacterial endosymbionts of Strigomonas and Angomonas. Genome Biol Evol. 2013b;5:338–350. doi: 10.1093/gbe/evt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, Gardner MJ, Gingle A, Grant G, Harb OS, Heiges M, Hertz-Fowler C, Houston R, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Logan FJ, Miller JA, Mitra S, Myler PJ, Nayak V, Pennington C, Phan I, Pinney DF, Ramasamy G, Rogers MB, Roos DS, Ross C, Sivam D, Smith DF, Srinivasamoorthy G, Stoeckert CJ, Jr, Subramanian S, Thibodeau R, Tivey A, Treatman C, Velarde G, Wang H. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Hury A, Liang XH, Michaeli S. Elucidating the role of H/ACA-like RNAs in trans-splicing and rRNA processing via RNA interference silencing of the Trypanosoma brucei CBF5 pseudouridine synthase. J Biol Chem. 2005;280:34558–34568. doi: 10.1074/jbc.M503465200. [DOI] [PubMed] [Google Scholar]
- Baum J, Papenfuss AT, Mair GR, Janse CJ, Vlachou D, Waters AP, Cowman AF, Crabb BS, de Koning-Ward TF. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 2009;37:3788–3798. doi: 10.1093/nar/gkp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Böhme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, Macleod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc Natl Acad Sci. 2011;108:10466–10471. doi: 10.1073/pnas.1103946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesan TC, Ferreira RC, Takata CSA, Campaner M, Borda CC, Paiva F, Milder RV, Teixeira MMG, Camargo EP. Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist. 2013;164:129–152. doi: 10.1016/j.protis.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Catta-Preta CMC, Pascoalino BD, de Souza W, Mottram JC, Motta MCM, Schenkman S. Reduction of tubulin expression in Angomonas deanei by RNAi modifies the ultrastructure of the trypanosomatid protozoan and impairs division of its endosymbiotic bacterium. J Eukaryot Microbiol. 2016;0:1–10. doi: 10.1111/jeu.12326. [DOI] [PubMed] [Google Scholar]
- Cenik ES, Fukunaga R, Lu G, Dutcher R, Wang Y, Tanaka Hall TM, Zamore PD. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Cell. 2011;42:172–184. doi: 10.1016/j.molcel.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Casas-Mollano JA. On the origin and function of RNA-mediated silencing: from protists to man. Curr Genet. 2006;50:81–89. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Cupolillo E, Medina-Acosta E, Noyes H, Momen H, Grimaldi G. A revised classification for Leishmania and Endotrypanum. Parasitol Today. 2000;16:142–144. doi: 10.1016/s0169-4758(99)01609-9. [DOI] [PubMed] [Google Scholar]
- Davidson BL, McCray PB. Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza W, de Carvalho TMU, Barrias ES. Review on Trypanosoma cruzi: host cell interaction. Int. J Cell Biol. 2010;2010:1–19. doi: 10.1155/2010/295394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. DeLano Scientific LLC; San Carlos: 2013. http://www.pymol.org. [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekanayake D, Sabatini R. Epigenetic regulation of polymerase II transcription initiation in Trypanosoma cruzi: modulation of nucleosome abundance, histone modification, and polymerase occupancy by o-linked thymine DNA glucosylation. Eukaryot Cell. 2011;10:1465–1472. doi: 10.1128/EC.05185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Meister G. Argonaute proteins at a glance. J Cell Sci. 2010;123:1819–1823. doi: 10.1242/jcs.055210. [DOI] [PubMed] [Google Scholar]
- Garcia-Silva MR, Frugier M, Tosara JP, Correa-Dominguez A, Ronalte-Alves L, Parodi-Talice A, Rovirae C, Robello C, Goldenberg S, Cayota A. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol. 2010;171:64–73. doi: 10.1016/j.molbiopara.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Grisard EC, Stoco PH, Wagner G, Sincero TC, Rotava G, Rodrigues JB, Snoeijer CQ, Koerich LB, Sperandio MM, Bayer-Santos E, Fragoso SP, Goldenberg S, Triana O, Vallejo GA, Tyler KM, Dávila AMR, Steindel M. Transcriptomic analyses of the avirulent protozoan parasite Trypanosoma rangeli. Mol Biochem Parasitol. 2010;174:18–25. doi: 10.1016/j.molbiopara.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Hury A, Ziporen Y, Shi H, Ullu E, Michaeli S. Small nucleolar RNA interference in Trypanosoma brucei: mechanism and utilization for elucidating the function of snoRNAs. Nucleic Acids Res. 2010;38:7236–7247. doi: 10.1093/nar/gkq599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne T, Toyofuku W. Two 5′-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei. Nucleic Acids Res. 1999;27:3300–3309. doi: 10.1093/nar/27.16.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Piontkivska H. Phylogeny of Trypanosomatidae and Bodonidae (Kinetoplastida) based on 18 S rRNA: evidence for paraphyly of Trypanosoma and six other genera. Mol Biol Evol. 2003;20:644–652. doi: 10.1093/molbev/msg062. [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinform. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvanger H, Simard MJ. Argonaute proteins: key players in RNA silencing. Nature Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Hutzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Ketting RF. The many faces of RNAi. Dev Cell. 2011;20:148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lau P-W, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ. The molecular architecture of human Dicer. Nature Struct Mol Biol. 2012;19:435–441. doi: 10.1038/nsmb.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Michaeli S. Small nucleolar RNA interference induced by antisense or double-stranded RNA in trypanosomatids. Proc Natl Acad Sci. 2003;100:7521–7526. doi: 10.1073/pnas.1332001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima L, Silva FM, Neves L, Attias M, Takata CS, Campaner M, de Souza W, Hamilton PB, Teixeira MMG. Evolutionary insights from bat trypanosomes: morphological, developmental and phylogenetic evidence of a new species, Trypanosoma (Schizotrypanum) erneyi sp. nov., in African bats closely related to Trypanosoma (Schizotrypanum) cruzi and allied species. Protist. 2012;163:856–872. doi: 10.1016/j.protis.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Lye L-F, Owens K, Shi H, Murta SMF, Vieira AC, Turco SJ, Tschudi C, Ullu E, Beverly SM. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6:e1001161. doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- Madina BR, Kuppan G, Vashisht AA, Liang YH, Downey KM, Wohlschlegel JA, Ji X, Sze SH, Sacchettini JC, Read LK, Cruz-Reyes J. Guide RNA biogenesis involves a novel RNase III family endoribonuclease in Trypanosoma brucei. RNA. 2011;17:1821–1830. doi: 10.1261/rna.2815911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia da Silva F, Noyes H, Campaner M, Junqueira AC, Coura JR, Añez N, Shaw JJ, Stevens JR, Teixeira MMG. Phylogeny, taxonomy and grouping of Trypanosoma rangeli isolates from man, triatomines and sylvatic mammals from widespread geographical origin based on SSU and ITS ribosomal sequences. Parasitology. 2004;129:549–561. doi: 10.1017/s0031182004005931. [DOI] [PubMed] [Google Scholar]
- Mallick B, Ghosh Z, Chakrabarti J. MicroRNA switches in Trypanosoma brucei. Biochem Biophys Res Commun. 2008;372:459–463. doi: 10.1016/j.bbrc.2008.05.084. [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenchar JB, Bellofatto V. Gene transcription in trypanosomes. Mol Biochem Parasitol. 2006;146:135–141. doi: 10.1016/j.molbiopara.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Patrick KL, Shi H, Kolev NG, Ersfeld K, Tschudi C, Ullu E. Distinct and overlapping roles for two Dicer-like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc Natl Acad Sci. 2009;106:17933–17938. doi: 10.1073/pnas.0907766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Djikeng A, Tschudi C, Ullu E. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol Cell Biol. 2004;24:420–427. doi: 10.1128/MCB.24.1.420-427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Tschudi C, Ullu E. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA. 2006;12:2063–2072. doi: 10.1261/rna.246906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J-J, Joshua-Tor L. Argonaute and RNA – getting into the groove. Curr Opinion Struct Biol. 2006;16:5–11. doi: 10.1016/j.sbi.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stoco PH, Wagner G, Travera-Lopez C, Gerber A, Zaha A, Thompson CE, Bartholomeu DC, Lückemeyer DD, Bahia D, Loreto E, Prestes EB, Lima FM, Rodrigues-Luiz G, Vallejo GA, da Silveira Filho JF, Schenkman S, Monteiro KM, Tyler KM, de Almeida LGP, Ortiz MF, Chiurillo MA, de Moraes MH, de Lima Cunha O, Mendonça-Neto R, Silva R, Teixeira SMR, Murta SMF, Sincero TCM, de Oliveira Mendes TA, Urmenyi TP, Silva VG, DaRocha WD, Andersson B, Romanha AJ, Steindel M, de Vasconcelos ATR, Grisard EC. Genome of the avirulent human-infective Trypanosome—Trypanosoma rangeli. PLoS Negl Trop Dis. 2014;8:e3176. doi: 10.1371/journal.pntd.0003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, Wang Y, Patel DJ, Berenguer J, Brouns SJJ, van der Oost J. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MMG, Borghesan TC, Ferreira RC, Santos MA, Takata CS, Campaner M, Nunes VL, Milder RV, de Souza W, Camargo EP. Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of proteobacterial symbionts. Protist. 2011;162:503–524. doi: 10.1016/j.protis.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Tyler KM, Engman DM. The life cycle of Trypanosoma cruzi revisited. Int J Parasitol. 2001;31:472–481. doi: 10.1016/s0020-7519(01)00153-9. [DOI] [PubMed] [Google Scholar]
- Ullu E, Tschudi C, Chakraborty T. RNA interference in protozoan parasites. Cell Microbiol. 2004;6:509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- Vaishnaw AK, Gollob J, Gamba-Vitalo C, Hutabarat R, Sah D, Meyers R, de Fougerolles T, Maraganore J. A status report on RNAi therapeutics. Silence. 2010;1:1–14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.