Abstract
Nuclear organization of genomic DNA affects DNA damage and repair processes, and yet its impact on mutational landscapes in cancer genomes remains unclear. Here we analyzed genome-wide somatic mutations from 366 samples of 6 cancer types. We found that lamina-associated regions, which are typically localized at the nuclear periphery, displayed higher somatic mutation frequencies compared to the inter-lamina regions at the nuclear core. This effect remained even after adjusting for features such as GC%, chromatin, and replication timing. Furthermore, mutational signatures differed between the nuclear core and periphery, indicating differences in the patterns of DNA damage and/or DNA repair processes. For instance, smoking and UV-related signatures were more enriched in the nuclear periphery. Substitutions at certain motifs were also more common in the nuclear periphery. Taken together, we found that the nuclear architecture influences mutational landscapes in cancer genomes beyond the effects already captured by chromatin and replication timing.
Emerging evidence indicates that somatic mutations in cancer genomes are non-randomly distributed, influenced by factors such as genomic context and DNA secondary structures, chromatin organization, transcriptional activity, and replication timing1–11. Local variation in the mutation burden stems from variability in DNA damage and/or repair processes3,5,12,13, and has implications for identification of potential cancer driver genes14 and clinical management of cancer patients, e.g. radio-sensitivity and immunotherapy15. However, the factors identified so far do not explain the entire extent of regional variation of the mutational burden in cancer genomes, suggesting that other factors are yet to be identified.
Genomic DNA is folded into higher-order domains, which occupy different territories in the three-dimensional architecture of the nucleus16–18, and nuclear lamina-binding regions are usually at the nuclear periphery16,19,20. Nuclear organization of genetic material plays an important role in DNA replication21 as well as DNA damage and repair processes22–24. For instance, the nuclear lamina-associated regions are refractory to homologous recombination-mediated repair and utilize an error-prone alternative end-joining mechanism to repair DNA double strand breaks25. Oct-1 and p53 dependent pathways link lamin functions to oxidative stress response26. Indeed, a previous multivariate analysis suggests that nuclear lamina association significantly contributes to germ line mutation rate variation27. Furthermore, it was recently reported that regulatory domain boundaries are frequently disrupted in cancer28, and in some cases such boundaries and the chromatin loops that underlie them are associated with unusual mutational spectra29. Here, we hypothesized that the nuclear organization of genomic DNA modulates the somatic mutational landscapes in cancer genomes, and that its effects might go beyond the variations due to known covariates such as chromatin domains and DNA replication timing4,6.
To test these hypotheses, we obtained somatic point mutation data from 366 completely sequenced genomes of 6 different cancer types: melanoma (SKCA, 25 samples)30, lung squamous cell carcinoma (LUSC, 31 samples)31, gastric cancer (STAD, 100 samples)32, diffuse large B cell lymphoma (DLBCL, 40 samples)33, chronic lymphocytic leukemia (CLL, 150 samples)34, and prostate cancer (PRAD, 20 samples)35,36. The somatic mutation frequencies for these cancer cohorts were comparable to published estimates of the mutation burden for the respective cancer type14 (Supplementary Fig. 1). We chose these cancer types because they have distinct etiologies, different patterns of DNA damage and repair, and a difference of several orders of magnitude in somatic mutation frequencies14,37, enabling us to identify effects of nuclear localization on somatic mutational patterns across diverse cancer types. We focused on the noncoding, non-repetitive, non-conserved regions of the genome and analyzed somatic mutations therein to minimize biases due to selection during clonal evolution as well as sequencing and mapping artifacts (see Online Methods for details). We denoted the mutation detection frequency per base pair in these regions, when normalized by the mutation detection frequency per base pair in the genome, as adjusted mutation rate (AMR).
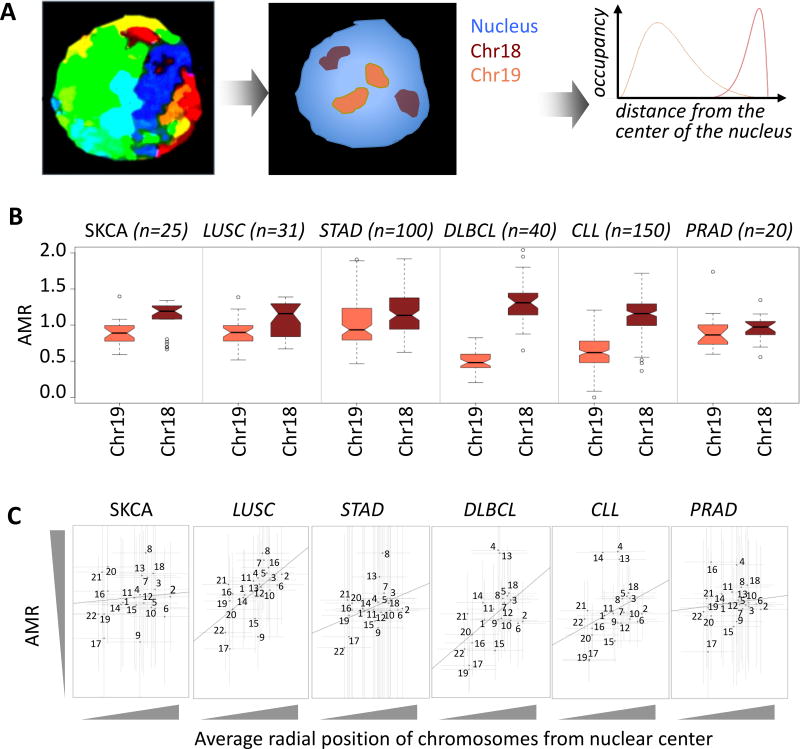
First, we investigated whether nuclear localization of chromosomes correlates with their AMR. We used chr18 and chr19 as classic examples since it has long been known that human chr18 is preferentially localized close to the nuclear periphery, while chr19 is primarily at the nuclear core38 (Figure 1A). Indeed, the AMR for chr18 was significantly higher compared to that for chr19 across all 6 cancer types analyzed (Figure 1B; Mann Whitney U test p-value < 1e-02 for all cohorts). Integrating paired copy number data when available (e.g. LUSC; Supplementary Fig. 2), we established that the difference was not due to proportionally more copy number deletion events on chr19. Extending this investigation to all other autosomes, whose nuclear positioning was determined using 3D FISH (fluorescence in situ hybridization), we observed a similar association between the overall nuclear positioning of chromosomes and their AMR – those that are predominantly in the nuclear periphery have a higher AMR compared to those in the core (Figure 1C). The coefficient of determination was weak (< 0.1) in all cohorts, which was, at least partly, due to the fact that chromosomes are large nuclear entities that typically span multiple nuclear domains; i.e. some parts of the same chromosome could be localized at the periphery while other parts at the relative interior of the nucleus38. Therefore, we chose to investigate whether more precise measures of the nuclear localization of genomic regions within and across chromosomes would be able to explain to the observed differences in chromosome-level variations in AMR.
Figure 1. Somatic mutation frequencies differ between chromosomes that are at the nuclear core versus periphery.
A) Eukaryotic chromosomes occupy different radial positions from the center of the nucleus. Classic examples are human chr18 and chr19, which are located at the nuclear periphery and core, respectively. B) The adjusted mutation rate (AMR) tends to be significantly higher for chr18 relative to that for chr19. Cancer types compared are as follows: melanoma (SKCA, n = 25), lung squamous cell carcinoma (LUSC, n = 31), gastric cancer (STAD, n = 100), diffuse large B cell lymphoma (DLBCL, n = 40), chronic lymphocytic leukemia (CLL, n = 150), and prostate cancer (PRAD, n = 20). Mann Whitney U test p-value < 1e-02 for all cohorts. In the boxplots, upper whisker is defined to be 1.5×IQR more than the third quartile or the maximal value of the adjusted mutation rate (depending on which value is greater) and the lower whisker is defined to be 1.5×IQR lower than the first quartile or the minimum value of the adjusted mutation rate (depending on which value is smaller) respectively, where IQR is the difference between the third quartile and the first quartile, i.e. the box length. C) AMR for chr1 to chr22 is plotted against their average normalized radial distances from the center of the nucleus. Average and standard deviation of normalized radial distances of chromosomes from the center of the nucleus were estimated from 54 measurements, as described in38. The number of samples used for AMR estimation is identical to panel b. Standard deviations of AMR and radial positions are shown with vertical and horizontal error bars, respectively. Coefficient of determination was < 0.1 in all cohorts.
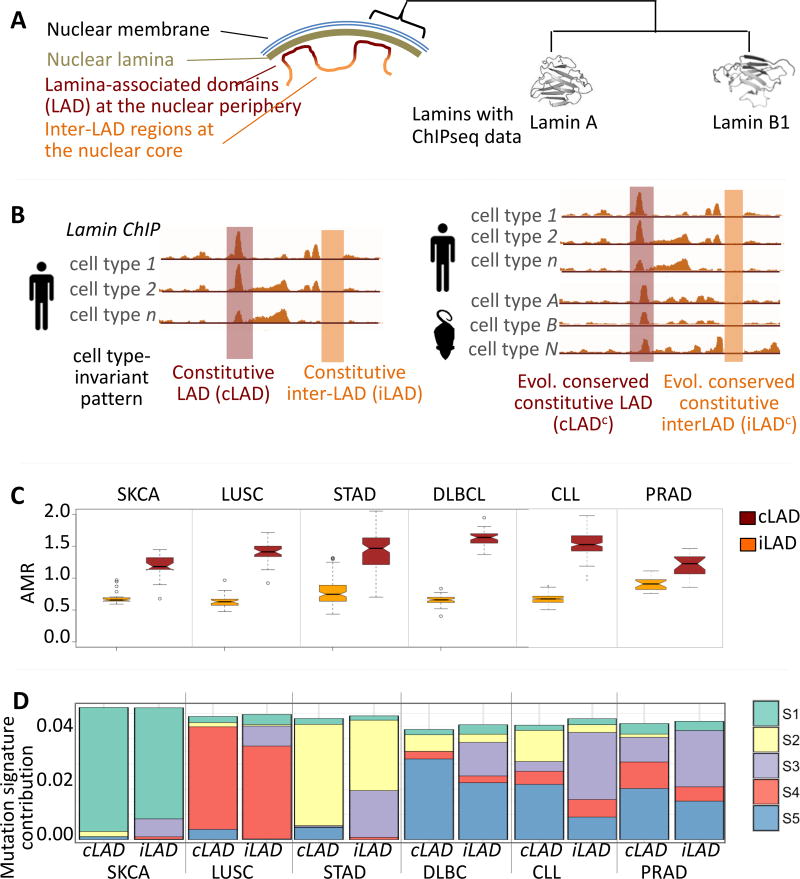
We obtained chromatin immuno-precipitation data for the lamin-family proteins Lamin A and B1 (Figure 2A), and classified a region as constitutively in the nuclear periphery if the region was associated with lamins in all cell types examined; conversely, a region was categorized as constitutively in the nuclear core if it did not overlap with lamin-associated domains in any of the cell types analyzed (Figure 2B). As before, we prioritized non-coding, non-repetitive, non-conserved segments of genomic regions, which are constitutively at the periphery (constitutive lamina-associated domains; cLAD) and core (inter-lamina-associated domains; iLAD), respectively. We then integrated somatic mutation data from each cancer cohort and calculated the AMR for these two types of regions for each sample. We found that the AMR for cLADs was significantly higher compared to that for iLADs, and once again, this observation was consistent across all 6 cancer cohorts (Figure 2C; Mann Whitney U test p-value < 1e-05 for all cohorts). Within respective chromosomes, cLAD and iLAD regions displayed a systematic difference in their AMR, regardless of the average nuclear localization of the chromosomes. A minor subset of lamins accumulates away from the nuclear periphery, usually in nucleoli-associated domains (NADs)39, and we found consistent results after excluding NADs (Supplementary Fig. 3). We also repeated the experiments more conservatively by analyzing only the cLADs and iLADs that have evolutionarily conserved patterns of nuclear localization, after integrating data on lamina-associated regions from multiple cell types in mouse (see Online Methods for details), and found similar results (Supplementary Fig. 3). Therefore, our findings are not sensitive to our choice of definition for cLADs and iLADs, and indicate that lamina-associated regions localized at the periphery have higher somatic mutation frequencies compared to the inter-lamina regions at the nuclear core.
Figure 2. Somatic mutation patterns differ between genomic regions that are at the nuclear core versus periphery.
A) Genomic regions interacting with lamina proteins such as Lamin A, and B1 are predominantly localized at the nuclear periphery (with some exceptions). B) Identification of genomic regions that are predominantly at the nuclear core (iLAD) and periphery (cLAD), respectively, in a cell type-invariant manner. Lamin chromatin immuno-precipitation was used to identify genomic regions interacting with lamins in individual cell types. We classified a region as constitutively in the nuclear periphery if the region was associated with lamins in all cell types examined; conversely, a region was categorized as constitutively in the nuclear core if it did not overlap with lamin-associated domains in any of the cell types analyzed. A subset of these regions also shows preferential positioning at the nuclear core (iLADc) and periphery (cLADc) in an evolutionarily conserved manner (Supplementary Figure 3). C) cLADs tend to have a significantly higher AMR compared to iLADs. The number of samples in each cohort is as described in Figure 1B. Mann Whitney U test p-value < 1e-05 for all cohorts. Similar results are observed when mutations in iLADc and cLADc regions are considered (Supplementary Fig. 3). In the boxplots, upper whisker is defined to be 1.5×IQR more than the third quartile or the maximal value of the adjusted mutation rate (depending on which value is greater) and the lower whisker is defined to be 1.5×IQR lower than the first quartile or the minimum value of the adjusted mutation rate (depending on which value is smaller) respectively, where IQR is the difference between the third quartile and the first quartile, i.e. the box length. (D) Mutational signatures differ between the nuclear core and periphery across different cancer types. Somatic mutational signatures were identified based on the non-negative matrix factorization and principal component analysis, using the somaticSignature40 R package. Details of the mutation signatures S1–S5 are provided in Supplementary Fig. 4.
We next focused on mutational signature differences between the nuclear core and periphery. In the SKCA cohort UV-induced C>T substitutions, including those in the pi-pyrimidine context, were proportionally more common in the cLADs compared to the iLADs (Mann Whitney U test p-value < 1e-08) (Figure 2D; Supplementary Fig. 4). Similarly, C>A substitutions displayed a higher enrichment in the cLADs in the LUSC cohort, indicating that smoking-associated oxidative DNA damage was greater in the nuclear periphery compared to the nuclear core (Mann Whitney U test p-value < 1e-02, Figure 2D; Supplementary Fig. 4). For LUSC patients, data on their smoking history and the number of pack years was available. We calculated the AMR for cLAD and iLAD considering only C:G>A:T mutations, and plotted AMR(cLAD)/AMR(iLAD) against the number of pack years smoked. Indeed, we found that the number of pack years smoked was weakly correlated with the AMR(cLAD)/AMR(iLAD) ratio (Spearman correlation coefficient: 0.29; Supplementary Fig. 4), suggesting that the relative strength of the signature of oxidative damage induced by smoking in the nuclear periphery was higher for heavy smokers compared to light smokers. Therefore, in cancer types driven by external carcinogens, the nuclear periphery had a proportionally higher burden of corresponding mutation signatures.
Even though the patterns of DNA damage and response in the cancer cohorts were dominated by disease etiology, there were some other differences in mutational signatures between cLAD and iLADs, which were tissue type-invariant (Figure 2D; Supplementary Fig. 4). For instance, when we summarized the tri-nucleotide substitution patterns into mutational signatures using non-negative matrix factorization40, mutation signatures 3 and 5 had a proportionally larger contribution in the iLAD and cLADs, respectively, in most cancer types as compared to the other signatures. Translating the mutational signatures into substitution patterns, it became clear that a majority of the cancer types had a proportional increase in the contribution of mutations in the WNW context (W: A or T; N: A, G, C, or T) in the cLADs at the periphery compared to the iLADs in the core. Different cancer types, however, showed subtle variation in the preference for specific sub-motifs; for instance, in the DLBCL and CLL cohorts, W[T>G]W and also W[T>C]W mutations were relatively more common in the cLADs than in the iLADs (Mann Whitney U test p-value < 1e-10). There were other differences in mutational signatures that were dominated by the biology of the cancer type. For instance, in the SKCA cohort, T[C>T]W substitutions were more common in the cLADs relative to the iLADs (Mann Whitney U test p-value < 1e-08; Supplementary Fig. 4).
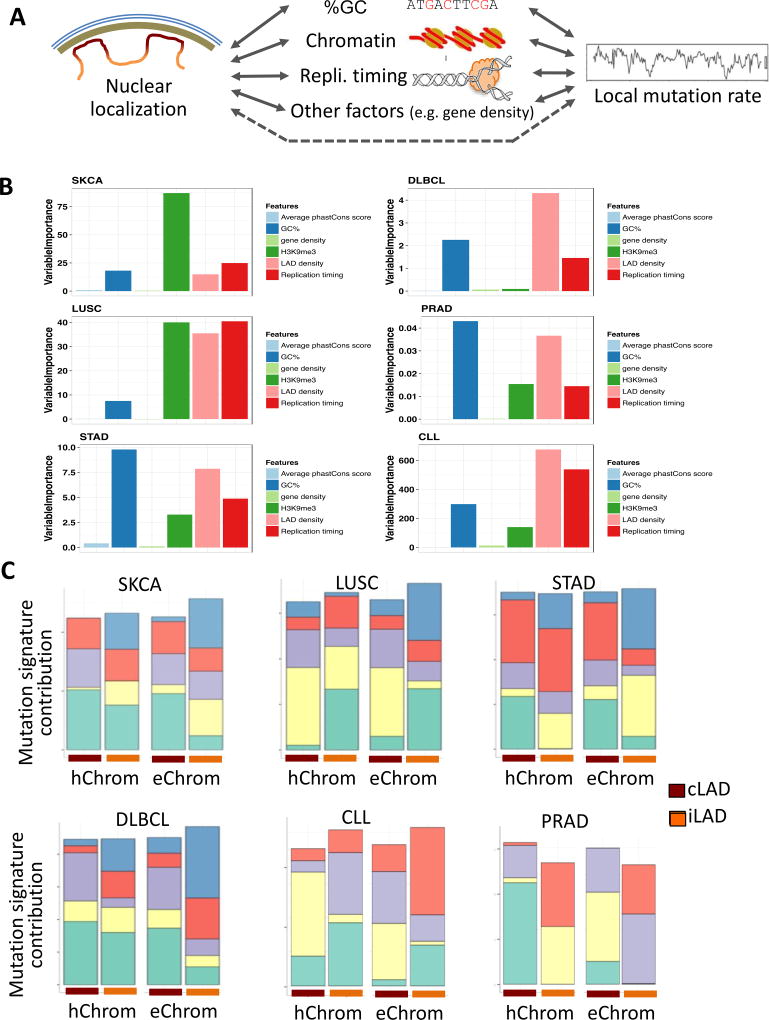
Nuclear localization of genomic DNA is coupled with many genomic and epigenomic features: regions in the nuclear periphery tend to be, on average, AT-rich, gene-poor, more heterochromatic, and have late replication timing compared to genomic regions in the core16,18–20. Features such as replication timing and chromatin influence DNA damage and repair processes, affecting mutational frequencies and signatures4,6,41–43(Figure 3A). However, not all point mutations arise during replication, and nuclear lamins play a key role in DNA double strand break repair, such that preference for repair mechanisms in the nuclear periphery is different from that in the nuclear interior25. We thus assessed whether nuclear localization influences the mutational landscapes in cancer genomes beyond what is already captured by chromatin and replication timing. Using a multiple linear regression including chromatin, replication timing, gene density, and GC content as covariates, we observed that the cLAD density was significantly associated with somatic mutation frequency even after adjusting for other features in all cancer type tested (Supplementary Fig. 5, Supplementary Tab. 1–2). After normalizing all features to zero mean and unitary variance, we also computed the variable importance metrics using random forest regression (Fig. 3B) and the effect sizes using multiple linear regression (Supplementary Fig. 5) for all features including the cLAD density in each 1MB bin. In general, the variable importance metrics of the cLAD density computed from the random forest regression are of similar magnitudes as that for the H3K9me3 signal and replication timing. We also computed the approximate conditional variable importance metrics to address the multicollinearities among the features (Online methods). We found that the cLAD density had a similar metric magnitude as H3K9me3 and replication timing in most cases (Online methods). We also ascertained that the influence of the sample size in the collected cohort on the results was not significant based on a sub-sampling analysis of the lymphoma cohort.
Figure 3. Certain differences in somatic mutation patterns between nuclear core versus periphery are not due to chromatin and other factors.
A) Nuclear localization of genomic DNA is, at least partly, associated with chromatin, and also other features such as GC content, replication timing and gene density, which modulate the local mutation rate. Effects of nuclear localization beyond that explained via these known covariates are investigated. B) Marginal variable importance metrics for different genomic features computed from random forest regression were compared for six cancer types. C) Mutational signatures differ between genomic material localized at the nuclear core and periphery even when assessed within similar euchromatic or heterochromatic contexts. Note that color codes of mutation signatures and their relative contributions are comparable only within respective cancer cohorts. See Supplementary Fig. 5 for additional comparative assessments.
Key differences between the nuclear core and periphery in detected mutational signatures also persisted even when we adjusted for both chromatin (Figure 3C) and replication timing (Supplementary Note; Methods). In the SKCA cohort, we found a proportionally higher burden of UV-mediated DNA damage and trans-lesion synthesis errors in the pyrimidine dimer context in the nuclear periphery relative to that in the core, even when controlling for replication timing and chromatin. We also found that cLADs had a larger contribution of the mutational signature SSKCA1, dominated by T[C>T]W substitutions, while iLADs had a relative enrichment of mutational signature SSKCA2, representing C[C>T]Y; these preferences were observed even after adjusting for both chromatin and replication timing. Indeed, there is evidence that nuclear lamin B1 is critical for the nucleotide excision repair (NER) pathway for effective repair of DNA damage response to UV irradiation44. The preference for C[C>T]N (where N: A, T, G, or C) in iLADs over cLADs was detectable in other cancer types including LUSC (signature SLUSC1). Moreover, in the LUSC cohort, the signature of oxidative DNA damage marked by C>A substitutions, especially W[C>A]W, was more common in the cLADs even after adjusting for chromatin and replication timing (Mann Whitney U test p-value < 1e-10). Therefore, a higher burden of mutation signatures arising due to external mutagens in the nuclear periphery was, at least partly, attributable to nuclear localization even when adjusting for replication timing and chromatin context. The increased incidence of somatic mutations in the WNW context was also detected across most cancer types regardless of replication timing and chromatin context. In the DLBCL and CLL cohorts, we observed an increase in C>T transitions in iLADs and an increase in T>G transversions in the WTN tri-nucleotide context in cLADs (Mann Whitney U test p-value < 1e-05) (Supplementary Note). The former signature is similar to COSMIC signature 2 and therefore might be due to deamination of cytosine mediated by off-target effects of AICDA/APOBEC family enzymes37,45,46. This hypothesis is also consistent with the observation that AICDA is predominantly localized in nucleoli and cajal bodies in the nuclear core47. The latter signature is similar to COSMIC signature 9, and a variant of this signature, N[T>G]T, was also observed in cLADs in the STAD cohort (Mann Whitney U test p-value < 1e-07; Supplementary Note). Based on the interpretation of COSMIC signature 9, we suspect that the signature arises primarily due to mutations attributed to polymerase η37, but other factors could also play a role.
Nuclear pores are large multi-protein channels that are conduits for nuclear transport of many small molecules and proteins, including DNA damage response and repair factors, and nuclear pores play key role in DNA repair24,48. Extending our analysis further, we investigated whether nuclear pore proximal regions (Figure 4A) display mutational patterns different from those observed for nuclear core and periphery regions. Nup98 is a component of the nuclear pore complex (Figure 4B); it is predominantly localized in the nuclear periphery, but can also be detected in the nuclear interior, and its dynamics of interaction with genomic regions depend on the developmental trajectory of the cell49. Using Nup98 ChIP-Seq data from multiple cell types49, we identified genomic regions that bind to Nup98 in one or more cell types. Accordingly we identified cLAD and iLADs that are localized in the neighborhood of Nup98-bound regions (NBR) or distal from it in a cell type-invariant manner (see Methods for details). cLADs at the nuclear periphery that are also close to NBRs in a cell-type invariant manner are likely to be nuclear pore-proximal. Unfortunately the number of mutations in these sub-regions was small; nonetheless, cLADs that were nuclear pore-proximal had a relatively lower AMR compared to those that were distal (Figure 4C) in the STAD, lymphoma, and CLL cohorts (FDR adjusted Mann Whitney U test p-value < 5e-02). The tri-nucleotide contexts of the substitution patterns in NBRs did not show any prominent, cancer type-invariant mutational signatures (Supplementary Note). Interaction of genomic DNA with the nuclear pore is dynamic, and DNA breaks are shunted to nuclear pores for a repair pathway controlled by a conserved SUMO-dependent E3 ligase50. Therefore, the effects of nuclear pore-assisted repair may not be restricted to just NBRs. Nonetheless, DNA lesions in the NBRs could be relocated to the nuclear pore complex more quickly for repair, which might play a role in lowering AMR in NBRs; further evidence is required to establish this conjecture conclusively.
Figure 4. Nuclear pore-proximal genomic regions have characteristic somatic mutation patterns.
A) Schematic representation of nuclear pores that are large, multi-protein complexes on the nuclear envelope, regulating nuclear transport of biomolecules including some mutagens and DNA repair factors. B) Nup98 is a key component of the nuclear pore complex, and based on Nup98 ChIP data, Nup98-bound regions were identified, using an approach similar to that in Fig 1b. We classified genomic regions as nuclear pore-proximal if they were within 50kb of Nup58 ChIP peaks in all cell types examined. Conversely, genomic regions that were at least 50kb from Nup58 ChIP peaks in all cell types were considered distal to nuclear pores. C) AMR of cLAD and iLADs that are proximal to and away from nuclear pore regions were compared. FDR-adjusted Mann Whitney U test p-value < 5e-02 in the STAD, lymphoma, and CLL cohorts. In the boxplots, the upper whisker is defined as 1.5×IQR more than the third quartile or the maximal value of the adjusted mutation rate (depending on which value is greater) and the lower whisker is defined as 1.5×IQR lower than the first quartile or the minimum value of the adjusted mutation rate (depending on which value is smaller) respectively, where IQR is the difference between the third quartile and the first quartile, i.e. the box length.
Taken together, our mutational signatures and multivariate analyses indicate that the nuclear localization of genomic DNA could potentially modulate somatic mutational patterns of cancer genomes, and that the effect attributed to nuclear localization on mutational landscapes in cancer is of similar magnitude to the already captured features such as chromatin and replication timing. This fact probably arises because a subset of mutations do not emerge during replication, and nuclear lamina plays a role in DNA damage recognition and repair21–24. Our observations are consistent with the reported effects of nuclear lamina on the germline mutation rate variation27. Even benign somatic tissue samples, albeit having considerably fewer somatic mutations, show similar patterns as well (Supplementary Fig. 6; p-value > 5e-02). However, our results should be interpreted with caution: (1) the LAD information used in our paper does not match with the (potentially unknown) cell type of origin of the six cancer types studied in this paper. To identify the effect of cell type-specific LADs on mutation frequencies requires matched data which is not yet available; (2) the multicollinearities among features such as replication timing, chromatin, and nuclear localization pose a statistical challenge in order to dissect their individual effects. Here, we performed our analyses from multiple angles – only looking at ‘neutrally’ evolving genomic regions and investigating the data using different multivariate models (Supplementary Fig. 7–8, Supplementary Tab. 3). Even though the results from different analyses are in general consistent with each other, further experiments/analyses are still needed to confirm the effect of nuclear localization on somatic mutations in somatic tissues.
There are multiple biological processes that might contribute to the observed differences in the mutation burden between the nuclear core and periphery. In 1975, Hsu proposed the “bodyguard hypothesis”, suggesting that constitutive heterochromatin is used by the cell as a bodyguard to protect the vital euchromatin by forming a layer of dispensable shield on the outer surface of the nucleus51. In agreement with this hypothesis, in the melanoma and lung squamous cell carcinoma cohorts we found that the nuclear periphery had a larger mutation burden and also displayed mutation signatures consistent with greater exposure to external mutagens. In addition, some of the DNA damage recognition and repair processes also depend on lamina association or nuclear localization. For instance, lamin B1 controls oxidative stress responses through sequestration of Oct-1 at the nuclear periphery52, which also leads to slow repair of DNA lesions. Furthermore, competing DNA repair mechanisms may recruit different DNA polymerases or their co-factors with variable fidelity and signature error profiles53, depending on nuclear localization. For instance, XPC and XPA are two damage recognition proteins associated with the nucleotide excision repair pathway, and after UV radiation, both XPC and XPA quickly accumulate in the border region of condensed chromatin called perichromatin of the nuclear core, but in condensed heterochromatin domains only accumulation of XPC was observed54. Another possibility could be that competing DNA repair mechanisms recruit different DNA polymerases or their co-factors with variable fidelity and signature error profiles53, depending on nuclear localization and cancer type. Furthermore, there is substantial evidence that DNA double strand break repair is nuclear localization-dependent -- repair in the nuclear interior or at the nuclear pores occur through the classical homologous recombination and non-homologous end-joining-mediated repair pathways, but the nuclear lamina-proximal regions tend to be refractory to HR and to allow repair primarily by the error-prone alternative end-joining mechanism25, which could be a source of point mutations in the nuclear periphery. In any case, our findings advocate for analyzing somatic mutations in tumor and benign tissues the context of their 3D nuclear architecture.
ONLINE METHODS
Somatic mutation data
We obtained somatic point mutation data from 366 completely sequenced genomes from melanoma (SKCA, 25 samples)30, lung squamous cell carcinoma (LUSC, 31 samples)31, gastric cancer (STAD, 100 samples)32, diffuse large B cell lymphoma (DLBCL, 40 samples)33, chronic lymphocytic leukemia (CLL, 150 samples)34, and prostate cancer (PRAD, 20 samples)35,36. Somatic mutation and other data types were mapped to the human reference genome (hg19). Mutation frequencies for the samples in these cohorts were comparable to published literature14, and there were no outlier subsets of samples with excessive mutations and skewed mutational signatures that dominated the overall patterns observed in our analyses.
Annotation of non-coding, non-repetitive, non-conserved regions
Since the mutational landscape of cancer genomes is shaped by both the incidence of mutations as well as natural selection during clonal evolution acting on the variability thus generated55,56, and since variant calling is technically challenging in some genomic regions (e.g. centromere, telomere, and repetitive region), we focused only on the non-coding, non-repetitive, non-conserved regions (tier III annotation obtained from Mardis et al.57). In brief, such regions were identified after excluding repeat-masked regions, coding regions of annotated exons, canonical splice sites, and RNA genes, conserved genomic elements (cutoff: conservation score greater than or equal to 500 based on either the phastConsElements28way table or the phastConsElements17way table from UCSC genome browser), and regions with regulatory potential (Regulatory annotations included are targetScanS, ORegAnno, tfbsConsSites, vistaEnhancers, eponine, firstEF, L1 TAF1 Valid, Poly(A), switchDbTss, encodeUViennaRnaz, cpgIslandExt)57. Such regions are generally expected to evolve in the absence of strong (positive or negative) selective pressures58, and should have no major issues with next generation sequencing or mappability.
Annotation of nuclear core and periphery regions
Data on nuclear localization of human chromosomes was obtained from Bolzer et al.38. We obtained genome-wide data on lamina-associated domains for multiple human and mouse cell types19,20. In these datasets, lamina-associated domains were identified using DamID treatment by a chimeric protein consisting of DNA adenine methyltransferase fused to lamin A or B1. DamID maps of (i) lamin B1 in mouse embryonic stem cells (ESCs), astrocytes (ACs), neuronal precursor cells (NPCs), and embryonic fibroblasts in mouse (MEFs) were obtained from Peric-Hupkes et al.20, (i) of lamin B1 in human Tig3 fibroblasts from Guelen et al19, and (iii) of lamin B1 in human ESCs and HT1080 cells and in mouse POU2F1−/− and matching wild-type MEFs; and of lamin A in human HT1080 cells and in mouse NPCs and ACs from Mueleman et al.59 Genomic regions associated with lamins are predominantly at the nuclear periphery, although some nucleoplasmic lamina-associated domains accumulate around nucleoli in the interior19,20,39, while those at the core were distinguished by the absence of interactions with nuclear lamina. Genome-wide distributions of lamina-associated regions are largely similar (73%–87%) between different cell types in higher eukaryotes20.
Overlaying Lamin A and B1 data, we identified the regions that overlap lamin-associated regions in (i) all the human cell line tested, and (ii) none of the human cell line tested, and denoted them as being constitutively at the nuclear periphery (dubbed constitutive lamina-associated domains, cLAD) and core (dubbed constitutive inter-lamina-associated domains, iLAD), respectively, in a cell-type invariant manner (Figure 2B). Genomic regions in the nuclear core and periphery have difference in gene density, repetitive elements, and evolutionarily conserved elements, and those features can influence selection on the somatic mutations (e.g. gene region), mutation calling (e.g. repetitive regions). Therefore, to minimize biases in our analysis, for all analyses presented in Figure 1, 2, and 4, we only considered tier-III segments57 (i.e. noncoding, non-repetitive, non-conserved genomic segments) of the cLAD and iLAD regions. In the multivariate analysis presented in Figure 3B, we used gene density, repetitive elements, evolutionary conservation, and other features as covariates.
As an even more conservative approach, by integrating human and mouse lamina-associated domains data in the similar manner, we also identified tier-III segments of cLAD and iLADs that have evolutionarily conserved patterns of localization in the nuclear periphery (denoted as conserved and constitutive cLAD regions, cLADc) and nuclear core (denoted as conserved and constitutive iLAD regions, iLADc; Figure 2B), respectively, and compared AMR between them (Supplementary Fig. 3).
Annotation of nuclear pore proximal regions
Nucleoporins are key components of nuclear pore complexes that control nucleo-cytoplasmic trafficking. Liang et al. examined genomic regions bound to NUP98, a nucleoporin family nuclear pore protein, by chromatin immuno-precipitation (ChIP) using multiple Nup98 antibodies in four cell types, three of which are related by direct lineage49. In tissue stem and progenitor cell populations, NUP98 bound regions (NBR) are predominantly at the nuclear periphery, but some NUP98 bound regions also exist at the nuclear core, and NUP98 binding dynamically changes between cell types and during development49. We classified the cLAD and iLAD genomic regions as nuclear pore proximal if those were within 50kb of NUP58 ChIP peaks in all cell types examined. We observed similar results using 20kb and 100kb windows.
Annotation of replication timing, chromatin, and other covariates
Repli-Seq signals were downloaded for multiple tissue types60 from the ENCODE data portal (Supplementary Tab. 1) and, following the approach used in a previous study61, we only kept one GM12878 cell line dataset to decrease the bias towards blood. Similarly, H3K9me3 histone modification marks across different tissue types were obtained from the Epigenomic Roadmap project62, including tissues such as liver, lung, and etc (Supplementary Tab. 3). The transcripts, GC% information and phastCons conservation scores for the human genome (hg19) calculated from multiple alignments with other 99 vertebrates were extracted from UCSC genome browser database63. For each 1MB bin, the GC%, number of genes overlapping with the bin, the proportion of nucleotides located in gene region, the average phastCons conservation scores were computed. For replication timing and H3K9me3 signals, we first calculated the average signal for each 1MB within each cell type, and then averaged across different cell types. Since in general the cell of origin of different cancer types are unknown, the average signal across different cell types can be used as a more robust measure of such signals, with the trade-off of loss of cell type-specific information.
Statistical analysis
We conducted both random forest regression and multiple linear regression to analyze the effect of lamina-associated domains on the average mutation frequency over different tumors within a certain cancer type adjusting for conservation score, GC%, gene density, average replication timing signals (higher indicating more enriched with early replication timing on average), and the heterochromatin mark H3K9me3 average signal across multiple cell lines (Supplementary Fig. 7–8, Supplementary Tab. 1–3). The adjusted R2 for the linear model and the variance explained by the features of the random forest regression are shown in Supplementary Tab. 3. The use of linear regression was justified using the residual plots and central limit theorem when averaging the mutation frequencies of each 1Mb bin over different tumors (Supplementary Fig. 7). To account for potential correlation among 1MB bins, we calculated the robust sandwich standard error64 in all regression analyses. When analyzing the mutation frequency averaging across different tumors within the same cancer type, the appropriateness of a linear model with additive effects of different genomic features can be justified using residual plots (Supplementary Fig. 7). To make the scale of coefficients of different features comparable, we normalized all the features to zero mean and unitary variance.
For the random forest regression, the function cforest() in the R package ‘party’ was used. The variable importance metrics for the genomic features were computed based on permutation methods using the varimp() function in the same package (Fig. 3B). The same set of features was included when performing random forest regression, again with average mutation frequencies in 1MB windows across samples as the dependent variable. The goodness of fit of random forest regression was again justified using the residual plots (Supplementary Fig. 7). Since the genomic features analyzed are in general correlated, we also computed the conditional variable importance metric65, which aims to remove some of the bias due to multicollinearities among the features (Supplementary Fig. 8). Because of the computational complexity, we were not able to compute the genome-wide metrics. As an alternative approach, we randomly divided the genome into 10 groups 50 times, computed the metrics within each group, calculated the median metrics across groups, and eventually plotted the distribution of these median scores across 50 randomizations. However, as outlined in Strobl et al. 200865, such an attempt cannot guarantee the complete removal of the multicollinearity bias. Therefore, even though Supplementary Fig. 8 shows that LAD has a similar, and sometimes even stronger, conditional variable importance metric compared to H3K9me3 and replication timing, this does not necessarily mean that we can interpret such results as “LAD is a more important factor than H3K9me3 in DLBCL”.
Finally, since different cancer cohorts have different sample sizes, it is worth exploring how the sample size influences our key results. To test the robustness of our findings over different sample sizes, we computed the variable importance metrics for the genomic features based on sample size equal to 10, 20, 30, and 40, respectively, in the lymphoma cohort, and found that the patterns are very similar across different sample sizes (Supplementary Fig. 8).
Mutational signatures are patterns in the occurrence of somatic single-nucleotide variants that can reflect underlying mutational and/or repair processes. We applied non-negative matrix factorization (NMF) and principal component analysis (PCA) to define mutation signatures, and then evaluated their contribution to each sample’s mutational spectrum using somaticSignature R package40. To examine significance of nuclear localization for mutagenic and repair processes, we partitioned the genome according to their chromatin or replication timing context, and then analyzed difference in mutation signatures between cLAD and iLAD regions within respective context. P-values for respective cohorts were calculated by way of Mann Whitney U tests. COSMIC mutational signatures were obtained from the COSMIC: Catalogue of Somatic Mutations in Cancer (http://cancer.sanger.ac.uk/cosmic), and were based on published report37.
DATA AVAILABILITY
Publicly available datasets were used for this analysis, as mentioned in above sections. Nonetheless, all data will be made available upon request.
Supplementary Material
Acknowledgments
The authors acknowledge financial support from T15LM009451 (KS), U54CA193461 (FM), P30CA072720, American Cancer Society, and Boettcher Foundation (SD). The authors thank other members of Michor and De laboratories for helpful discussions. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
AUTHOR CONTRIBUTIONS
SD conceived the project with FM. KS, LLL, FM, SD designed the experiments. KS, LLL, SD performed the experiments. KS, LLL, SG, FM, SD interpreted the results. FM, SD wrote the manuscript with input from other authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat. Struct. Mol. Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De S, Michor F. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat. Biotechnol. 2011;29:1103–1108. doi: 10.1038/nbt.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, De S, Michor F. DNA replication timing and higher-order nuclear organization determine single-nucleotide substitution patterns in cancer genomes. Nat. Commun. 2013;4:1502. doi: 10.1038/ncomms2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 7.Polak P, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perera D, et al. Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature. 2016;532:259–263. doi: 10.1038/nature17437. [DOI] [PubMed] [Google Scholar]
- 9.Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, Lopez-Bigas N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature. 2016;532:264–267. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]
- 10.Smith KS, et al. Signatures of accelerated somatic evolution in gene promoters in multiple cancer types. Nucleic Acids Res. 2015;43:5307–17. doi: 10.1093/nar/gkv419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen BS, De S. Loss of heterozygosity preferentially occurs in early replicating regions in cancer genomes. Nucleic Acids Res. 2013;41:7615–24. doi: 10.1093/nar/gkt552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nat Rev Genet. 2013;14:703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De S, Ganesan S. Looking beyond drivers and passengers in cancer genome sequencing data. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. mdw677. 2016 doi: 10.1093/annonc/mdw677. [DOI] [PubMed] [Google Scholar]
- 16.Bickmore WA. The spatial organization of the human genome. Annu Rev Genomics Hum Genet. 2013;14:67–84. doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- 17.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 20.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meister P, Taddei A, Gasser SM. In and out of the replication factory. Cell. 2006;125:1233–1235. doi: 10.1016/j.cell.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Ball AR, Jr, Yokomori K. Damage site chromatin: open or closed? Curr Opin Cell Biol. 2011;23:277–283. doi: 10.1016/j.ceb.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell O, Tiwari VK, Thoma NH, Schubeler D. Determinants and dynamics of genome accessibility. Nat Rev Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 24.Lemaître C, Bickmore WA. Chromatin at the nuclear periphery and the regulation of genome functions. Histochem. Cell Biol. 2015;144:111–122. doi: 10.1007/s00418-015-1346-y. [DOI] [PubMed] [Google Scholar]
- 25.Lemaître C, et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 2014;28:2450–63. doi: 10.1101/gad.248369.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimi T, Goldman RD. Nuclear lamins and oxidative stress in cell proliferation and longevity. Adv. Exp. Med. Biol. 2014;773:415–30. doi: 10.1007/978-1-4899-8032-8_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ananda G, Chiaromonte F, Makova KD. A genome-wide view of mutation rate co-variation using multivariate analyses. Genome Biol. 2011;12:R27. doi: 10.1186/gb-2011-12-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weischenfeldt J, et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 2016;49:65–74. doi: 10.1038/ng.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiser VB, Taylor MS, Semple CA. Mutational Biases Drive Elevated Rates of Substitution at Regulatory Sites across Cancer Types. PLoS Genet. 2016;12:e1006207. doi: 10.1371/journal.pgen.1006207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 33.Morin RD, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122:1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puente XS, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–24. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 35.Abeshouse A, et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolzer A, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nemeth A, et al. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gehring JS, Fischer B, Lawrence M, Huber W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics. 2015;31:3673–3675. doi: 10.1093/bioinformatics/btv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kazanov MD, et al. APOBEC-Induced Cancer Mutations Are Uniquely Enriched in Early-Replicating, Gene-Dense, and Active Chromatin Regions. Cell Rep. 2015;13:1103–1109. doi: 10.1016/j.celrep.2015.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morganella S, et al. The topography of mutational processes in breast cancer genomes. Nat Commun. 2016;7:11383. doi: 10.1038/ncomms11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo YH, Li WH. DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes. Nat Commun. 2012;3:1004. doi: 10.1038/ncomms1982. [DOI] [PubMed] [Google Scholar]
- 44.Butin-Israeli V, et al. Regulation of Nucleotide Excision Repair by Nuclear Lamin B1. PLoS One. 2013;8:e69169. doi: 10.1371/journal.pone.0069169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 46.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Y, et al. Activation-induced cytidine deaminase (AID) is localized to subnuclear domains enriched in splicing factors. Exp. Cell Res. 2014;322:178–192. doi: 10.1016/j.yexcr.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genet. 2013;9:e1003308. doi: 10.1371/journal.pgen.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagai S, et al. Functional Targeting of DNA Damage to a Nuclear Pore-Associated SUMO-Dependent Ubiquitin Ligase. Science (80-.) 2008;322 doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu TC. A possible function of constitutive heterochromatin: the bodyguard hypothesis. Genetics. 1975;79(Suppl):137–150. [PubMed] [Google Scholar]
- 52.Malhas AN, Lee CF, Vaux DJ. Lamin B1 controls oxidative stress responses via Oct-1. J Cell Biol. 2009;184:45–55. doi: 10.1083/jcb.200804155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solimando L, et al. Spatial organization of nucleotide excision repair proteins after UV-induced DNA damage in the human cell nucleus. J Cell Sci. 2009;122:83–91. doi: 10.1242/jcs.031062. [DOI] [PubMed] [Google Scholar]
- 55.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 56.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohta T. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 1992;23:263–286. [Google Scholar]
- 59.Meuleman W, et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–80. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci U S A. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Speir ML, et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44:D717–25. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freedman DA. On the so-called ‘Huber sandwich estimator’ and ‘robust standarderrors’. Am. Stat. 2006;60:299–302. [Google Scholar]
- 65.Strobl C, Boulesteix A-L, Kneib T, Augustin T, Zeileis A. Conditional Variable Importance for Random Forests. BMC Bioinformatics. 2008;9:307. doi: 10.1186/1471-2105-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were used for this analysis, as mentioned in above sections. Nonetheless, all data will be made available upon request.