Abstract
Intragenic recombination is one of the most important sources of genetic variability. In our previous study, RI92 a tall line (160 cm of plant height) was observed in the cross progeny between two semi-dwarf indica cultivars Zhenshan 97 and Minghui 63. Genome-wide genotyping and sequencing indicated that the genome constitution of RI92 was completely from both parents. Bulk segregant analysis in a BC3F2 population revealed that “green revolution gene” semi-dwarf 1 (sd1) was most likely the gene controlling the tall plant height in RI92. Sequencing analysis of SD1 revealed that an intragenic recombination occurred between two parental non-functional sd1 alleles and generated a functional SD1 in RI92. Four-fold high recombination rate in SD1 located bins to the genome-wide average was observed in two RIL populations, indicating recombination hotspot in the SD1 region. Intragenic recombination creates new alleles in the progeny distinct from parental alleles and diversifies natural variation.
Introduction
Recombination and mutation are the motivation of evolution. Recombination is critical for repairing DNA lesions and for chromosomal pairing and exchange during meiosis [1]. The well-studied yeast meiotic recombination revealed that non-uniformity of recombination is observed when the frame of reference is an entire chromosome, a multi-gene region, a pair of genes, or a small region upstream of a gene [2]. The ratio of genetic distance, measured by meiotic recombination frequencies, to physical distance, measured by molecular markers, is used for evaluating recombination rate and varies within different species and genome regions. The genome regions with higher recombination rate than the genome average is called recombination hotspots, which enhance the resolution of gene mapping and help introduce the excellent alleles into breeding lines by broken linkage. Intragenic recombination, the recombination among different varying sites within the same gene locus, can create novel alleles with more genetic variability, and benefit to evolution which selection could act [3]. To identify intragenic recombination, restriction fragment length polymorphism (RFLP) markers within target genes were used to distinct two parental alleles in traditional genetics [4]. The intragenic recombination causes a distinct fragment pattern from both parents. Intragenic recombination hotspots have been observed at a number of gene loci in the recombination hotspots in plants, such as the wx locus in rice (Oryza sativa) and maize (Zea mays); the maize A1 locus and the bronze locus [4–7]. The recombination rate at A1 locus was from 12.5 kb/cM to 25 kb/cM, which is two orders of magnitude higher than the average rate of whole maize genome [5]. However, not all the intragenic recombination was occurred in the recombination hotspots, only by chance. For example, the intragenic recombination rate at csr1 locus in Arabidopsis is almost the same as the genome average [8]. To easily discover the intragenic recombination, two individuals carried non-functional alleles of targeted genes; whose intragenic recombination causes a functional allele and obtained a distinct phenotype change, are priority used to develop a population for estimation of intragenic recombination rate [9, 10]. The functional alleles/plants in the progeny are resulted from intragenic recombination, and the ratio of functional allele plants to mutation allele plants was deemed as intragenic recombination rate. For example, the wx gene encodes a granule-bound starch synthase required for the synthesis of amylose in both the endosperm and pollen of maize and rice Wx pollen grains stain deep blue in I2-KI solution, whereas wx pollen grains stain light brown. The large sample of pollen and the easily phenotype distinguishing with I2-KI staining make wx frequently used for estimating intragenic recombination rate [4]. The progeny of the hybrid between two wx mutants each from different mutation events produces Wx pollen, which is the result of intragenic recombination of wx alleles. The recombination frequency within the wx gene was 10 times higher than the genome average [6]. With the advent of next generation sequencing technique, high-density bin maps were constructed with RILs populations in maize and rice [11–13]. Intergenic and intragenic recombination could be detected on the basis of allele sequence, and significant associations between intragenic recombination events and agronomic traits were observed [13].
Several studies support the hypothesis that recombination occurs primarily in genes: (1) comparing to gene-poor chromosomal regions, the gene-rich regions are recombination active in wheat and barley [14, 15], (2) in Arabidopsis, maize and rice, recombination is suppressed in the regions nearby centromere [16–18]. The higher recombination frequencies in maize were predictable on the broad scale, following the distribution of gene density and CpG methylation; meanwhile, the inversions also suppress recombination in distinct regions [18]. Rice genome exhibits recombination hot spot and cold spot, where recombination rates per kb are significantly higher and lower than the genome average [19, 20].
Plant height, one of the most important traits affecting rice production, was widely researched in the recent years. There are various factors regulating plant height, but gibberellin (GA) is the most important factor. SD1 as the green revolution gene encoded gibberellin 20-oxidase and involved in GA synthesis[21]. It is a key enzyme in gibberellin biosynthesis that catalyzes the three steps; GA53→GA44→GA19→GA20. Impaired GA 20-oxidase activity will accumulate the content of GA53, and the reduced amount of GA20 resulted in a decreased plant height. At least, there are seven sd1 alleles have been used in the breeding of semi-dwarf rice varieties in China, USA and Japan [22]. Two functional nucleotide polymorphisms for the short culm of Nipponbare (O. sativa ssp. japonica) were identified in the coding region of SD1, and the dramatic nucleotide diversity reduction around SD1 was only occurring in the japonica landraces which indicated SD1 had been subjected to the japonica domestication [23]. These results indicated that SD1 was not only involved in the green revolution of modern breeding but also in the early steps of rice domestication.
In our previous research, we identified an extraordinary tall line, RI92 (160 cm), in the recombinant inbred line (RIL) population during the cross between Zhenshan 97 (ZS97) and Minhui 63 (MH63). To uncover why RI92 was much taller than parents, we constructed a near-isogenic line with tall plant height using RI92 as a donor to backcross with ZS97. Map-based cloning revealed that SD1 is the player responsible for such tall plant height in RI92. Comparative sequencing showed that intragenic recombination between two non-functional sd1 alleles in parents resulted in a functional SD1 in RI92.
Materials and methods
Plant materials and phenotypic data collection
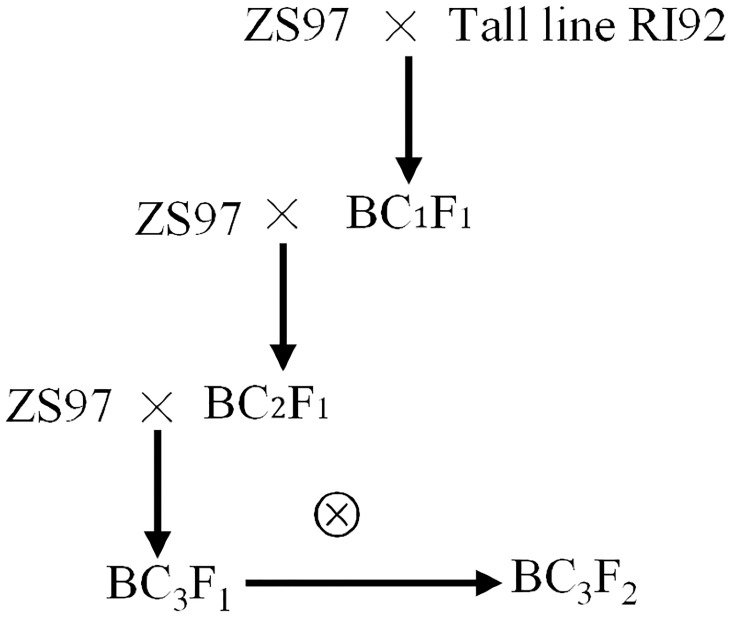
RI92 was an extraordinary tall line of the RIL population from the cross between ZS97 and MH63 [24]. The F1 between RI92 and ZS97 exhibited a similar plant height to RI92. The tall plant at each backcross generation was continuously backcrossed to ZS97 two times to generate a BC3F1 plant, and then self-crossing to obtain a BC3F2 population was done (Fig 1).
Fig 1. The construction of near isogenic line population used for mapping plant height.
The seeds of 96 BC3F2 plants were sown on 26 May 2015, then the seedlings were transplanted individually in the field at distance of 20*25 cm on 15 June 2015 in the bird net-equipped field in the experimental farm of Huazhong Agricultural University, Wuhan, China (114° 21΄ E, 30° 28΄ N). Plant height was measured in both seedling stage and maturing stage as the distance from the ground surface to the tip of the tallest leaf and the tip of the tallest panicle, respectively. At maturation stage, the whole plants individually harvested, panicle length was measured as the length from first branch node to the tip of panicle, spikelets per panicle (SPP), grains per panicle (GPP) and yield per plant (YP) were measured using automatic phenotyping machine [25]. Then harvest index was calculated as the ratio of grain weight per plant to biomass per plant.
DNA extraction and sequencing
DNA was extracted from fresh leaves at the seedling stage using the CTAB method with minor modified [26]. SD1 Genomic DNA including exon and intron were amplified from genomic DNA using LA Taq (Takara). Polymerase chain reactions (PCR) performed with the following conditions for thermal cycling: 94°C for 4 min; then 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min per 1000-base; followed by 72°C for seven min. All primers used for PCR and sequencing listed in S1 Table. For sequencing, 5 μL PCR product digested with five U EXOI (Biolabs) and 0.13 U Shrimp Alkaline Phosphatase (Takara) together with 1×PCR buffer and incubated at 37°C for 1 hour; the reaction was stopped by maintaining the PCR product in 80°C for 20 min. Sequencing was independently performed three times in both forward and reverse primers on ABI 3730 with BigDye terminator sequencing kits (Applied Biosystems). Sequence contigs assembled by SEQUENCHER 4.1.2 (Gene Codes Corporation).
Gibberellin acid and PBZ treatment in the seedling stage
The GA3 was first dissolved in a small volume of ethanol and diluted to 1 mM using double-distilled water (DDW). The GA3 solution was added to the rooting culture medium with a final concentration 0.2 uM, 0.4 uM, 0.8 uM and 1.6 uM. The rooting culture medium containing an equivalent amount of ethanol was used as a control. PBZ, acting by inhibiting gibberellin biosynthesis, dissolved in DDW, and the solution added to the rooting culture medium with a final concentration 0.2, 0.6 and 1.8 uM. The rooting culture with equivalent amount of DDW was used as a control.
The seeds of NIL-SD1 and ZS97 germinated on the same rooting culture medium with a different concentration of GA3 and PBZ for each treatment benches (25 ± 2°C, 55 ± 3% RH, 4800 lux and ten hours photoperiod). Each experiment performed twice per each treatment. The controls were treated with the same conditions free of GA3 and PBZ. Fifteen days later, the length of plantlets height measured from the bottom of the stem to the leaf tip.
Results
Characterization of RI92
At the seedling stage of 35 days old, plant height recorded 56 and 40 cm on RI92 and ZS97, respectively (Table 1). RI92 showed a much higher plant height (161 cm) than its parents; ZS97 (86 cm) and MH63 (110 cm) at the maturation stage.
Table 1. Plant height (cm) of RI92 and its NIL and parents at seedling and maturation stages.
| Stages | RI92 | NIL | Zhenshan 97 | Minghui 63 |
|---|---|---|---|---|
| Seedling | 56.2±5.1 | 52.7±4.8 | 40.4±3.0 | 46±3.5 |
| Maturation | 161.0±5.2 | 140.1±6.6 | 86.1±4.3 | 110.2±4.7 |
10 plants were investigated each genotype.
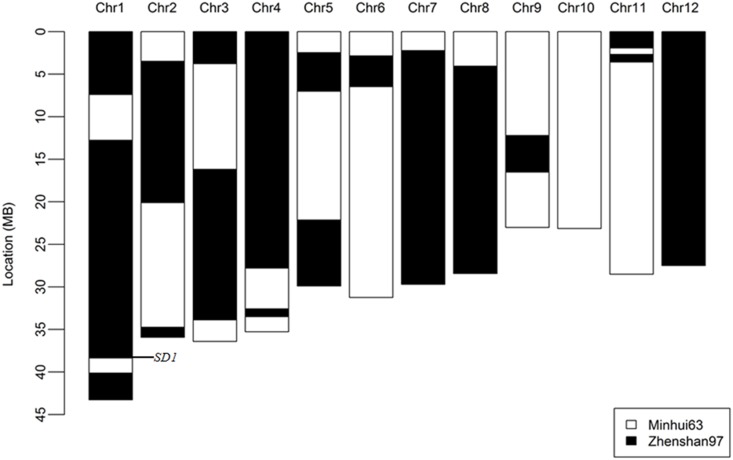
Because of its extraordinary plant height in the RIL population, it is possible that RI92 contaminated by alien pollen from uncertain genotypes. We proved that the genetic constitution is the mixture of both parents ZS97 and MH63, and no alien genotypes using 221 RFLP and SSR markers [27], meanwhile, we compared its genome constitution with both parents by low-genome coverage re-sequencing [28]. All genome sequences came from both parents without exception (Fig 2). The RI92 genetic background constituted with 34 bins; 55% and 45% of genomic regions were coming from ZS97 and MH63, respectively. At least, one recombination event of each chromosome occurred in RI92 except chromosomes 10 and 12 (Fig 2) which entire originate from ZS97 and MH63 respectively. These data confirmed that RI92 is the offspring of ZS97 and MH63.
Fig 2. The genetic constitute of RI92.
Development of the tall near isogenic lines (NIL)
The plant height of F1 derived from the crossing between RI92 and ZS97 was similar to RI92, indicating a dominant gene controlling the tall plant height trait. A BC3F2 population consisted of 96 plants; the tall NIL in this population was about 10 and 60 cm higher than the recurrent parent (ZS97) at the seedling and mature stage, respectively (Table 1). The NIL had six internodes; two more than ZS97 (each internode of NIL was longer than ZS97). Also the panicle length of NIL was longer than ZS97 panicle length (Table 2).
Table 2. Internode length of NIL, ZS97 and RI92.
| Terms | RI92 | NIL | Zhenshan 97 |
|---|---|---|---|
| Panicle length | 26.2±3.1 | 24.0±2.8 | 22.4±2.3 |
| 1st internode | 45.8±5.6 | 40.2±5.2 | 33.2±2.2 |
| 2nd internode | 30.5±5.2 | 27.3±4.7 | 21.6±1.0 |
| 3rd internode | 24.9±3.4 | 21.1±4.5 | 7.0±1.8 |
| 4th internode | 18.4±4.3 | 14.0±6.1 | 2.6±0.5 |
| 5th internode | 10.1±4.8 | 8.4±5.0 | - |
| 6th internode | 4.5±2.2 | 4.2±1.8 | - |
10 plants were investigated each genotype.
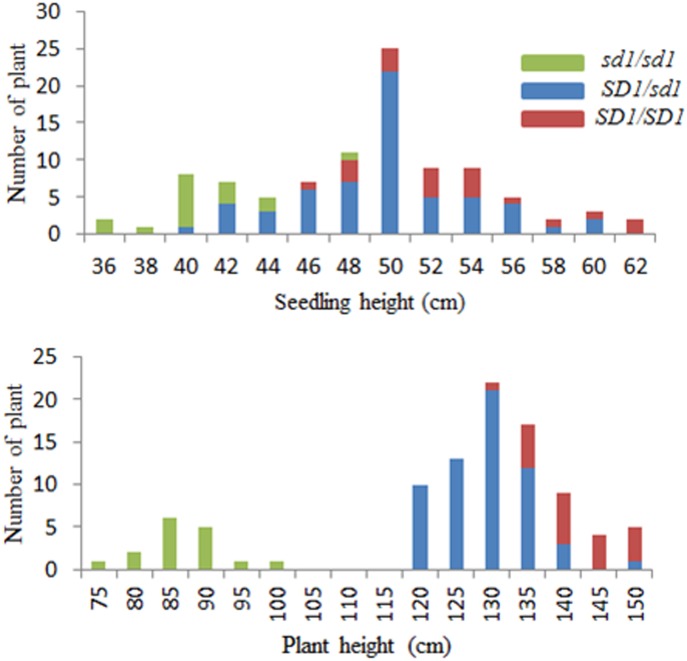
At the seedling stage, the segregation of seedling height in the BC3F2 population exhibited a continuous distribution (Fig 3A), while the plant height in the mature stage showed a discontinuous distribution with a gap from 100–120 cm (Fig 3B). Obviously, these plants easily classified into two groups: short and tall. The segregation of 16 short plants to 80 tall plants was fit to the segregation ratio of the single Mendelian gene (χ2 = 3.56< 3.84, P = 0.05), which indicated that a major gene controlling the variation of plant height. Only 2–3 days’ variation of heading date was observed in the BC3F2 population, suggesting the plant height gene did not impact on heading date.
Fig 3. Frequency distribution of seedling height (A) and plant height (B) in the random BC3F2 population.
The red, blue and green bars mean the homozygotes of SD1/SD1 and sd1/sd1 types and heterozygote identified by the functional SNP (FNP) in the third exon of SD1.
Quick mapping for the plant height QTL
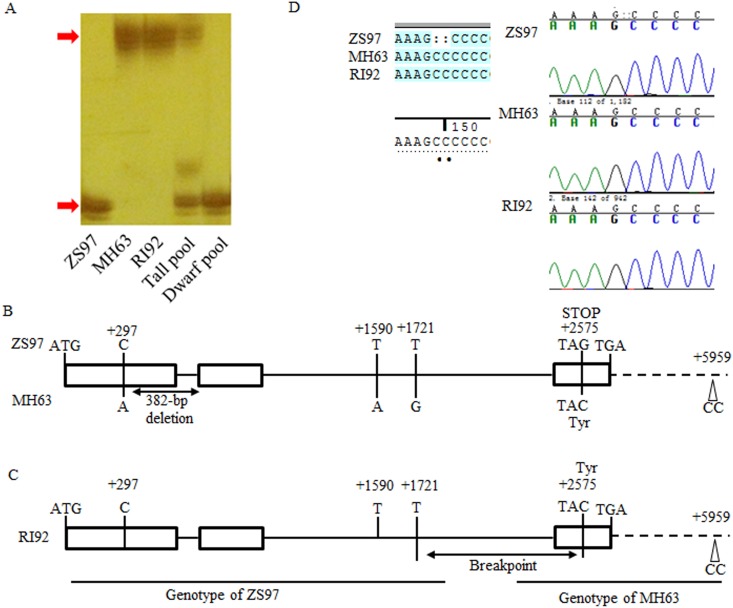
The difference in plant height is of 60 cm between the tall NIL and ZS97 indicated that the unknown gene had a large effect. In the previous study, many QTLs for plant height mapped in the RILs and Chromosome Segment Substitution Lines (CSSLs) between ZS97 and MH63 [24, 29]. But only one QTL qPH7/qPH7.2 expressed a large effect on plant height in the RILs and CSSLs. Thus, we exclusively focused on the major QTL qPH7/qPH7.2 in the short arm of chromosome 7. But the chromosome region contained qPH7/qPH7.2 was fixed with homozygous ZS97 in RI92. So, the variation of plant height in the BC3F2 population is not the result of qPH7/qPH7.2. Then bulk segregant analysis used to quickly identify the major gene. We constructed two DNA bulks for tall and short plants, each bulk consisting of ten plants. SD1, a gibberellin synthetase gene in rice, has tremendous effects on plant height. SD1 was regarded as the possible target gene considering its high effect on plant height in rice. Then the polymorphic SSR markers RM3523 linked to SD1 were used to genotype the two bulks. PCR results showed two bands and one band for tall and short bulk, respectively (Fig 4A), which suggested that the major gene linked to RM3523, and SD1 as the most likely candidate.
Fig 4. Comparison of the allele sequences of SD1 among ZS97, MH63 and RI92.
(A) Bulk segregant analysis of 10 tall and dwarf plants in BC3F2 population. The red arrows indicate the target band for ZS97 and MH63 genotypes, respectively (B) Comparison of SD1 sequences between ZS97 and MH63 alleles. (C) The intragenic recombination of sd1, (D) and the same 2-bp insertion in MH63 and RI92.
Comparative sequencing of SD1 between parents
Therefore, we sequenced SD1 for both two parents and RI92. Sequence analysis revealed that there were four SNPs and a 382-bp deletion in the SD1 gene body between ZS97 and MH63 allele. Two of these four SNPs located in the exon and the other placed in the intron (Fig 4B). The first SNP (+297) which found in the first exon did not lead to any amino acid change. While the second SNP (+2575) of ZS97 in the exon 3 produced a premature termination code, indicating the parent ZS97 possesses non-function SD1, that corresponding to the semi-dwarf plant height of ZS97.
Comparing to ZS97, MH63 allele had a 382-bp deletion and the large deletion including the entire first intron and part of the first and second exon. This data indicated that the MH63 allele possessed a non-function SD1 allele. Therefore, SD1 was non-functional in both parents. However, unlike both parents, RI92 contained a strong functional SD1 allele (SD1-GR) same as indica Kasalath, which had 2 amino acids, differ from Nipponbare or Taichung65 (SD1-EQ) (S1 Fig) [21, 23, 30]. The only difference between RI92 and ZS97 allele was the SNP (+2575) guanine (G) substituted to Cytosine (C) (Fig 4C), but the RI92 allele can produce complete gibberellin 20-oxidase protein. The unique SNP at site +2575 between RI92 and ZS97 puzzled us whether the point mutation occurred in the ZS97 allele of SD1 or a recombination within SD1 happened between two parents in the process of developing RILs, and resulted in the functional SD1 allele in RI92. To answer the assumption, 4-kb fragment downstream of SD1 were sequenced and distinguished between MH63 and ZS97 by a 2-bp insertion (Fig 4C and 4D), while RI92 allele carried the same 2-bp insertion as MH63 contained. Also, the sequence of 2-kb promoter region showed no differences between ZS97 and MH63 alleles.
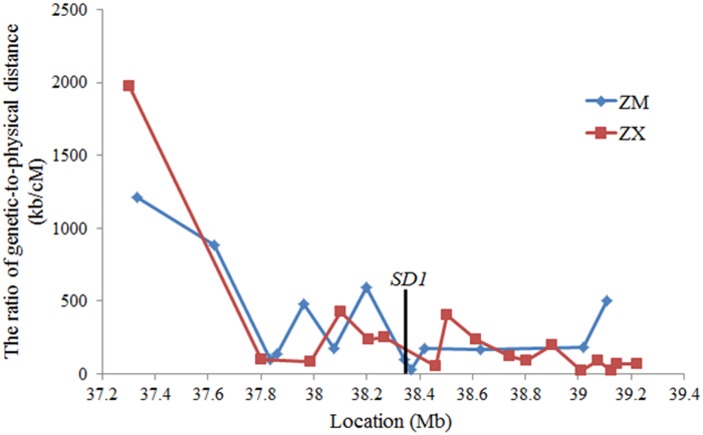
Recombination rate in the 2-Mb chromosome region surrounding SD1 calculated based on the bins in the RIL population of 210 lines between two indica cultivars ZS97 and MH63 [11]. The average recombination ratio of physical-to-genetical distance in this region was 220 kb/cM (Fig 5), which was similar to the genome-wide averaged recombination ratio of 250~300 kb/cM [11, 31, 32]. But the ratio for sd1 located bin was 64 kb/cM. We accordingly estimated the recombination rate in the same 2-Mbregion using a RIL population of 197 lines between indica cultivar ZS97 and japonica cultivar Xizang 2 [12]. The averaged recombination ratio in the whole 2-Mb region was 132kb/cM, while the ratio of SD1 located bin was 55 kb/cM, this is very similar to that in the indica RIL populations.
Fig 5. The ratio of genetic-to-physical distance in the 2-Mb region surrounding SD1 in the RILs of ZM (derived from the cross between Zhenshan 97 and Minghui 63) and ZX (derived from the cross between Zhenshan 97 and Xizang 2).
Genetic effects of SD1 on yield related traits
Now, the possible candidate gene for plant height is SD1, and we sequenced it for the 96 BC3F2 plants. The only SNP G-C at +2575 was the functional nucleotide polymorphism (FNP) of SD1 [22]. According to the genotype of the FNP, the BC3F2 population in the maturation stage divided into three classes of CC (SD1/SD1), CG (SD1/sd1) and GG (sd1/sd1). The homozygote sd1/sd1 plants showed the similar plant height of ZS97 (on average plants 85 cm), whereas the average of homozygote SD1/SD1 plants and heterozygote SD1/sd1 plants scored the similar plant height, approximately 140 cm, about 55 cm higher than sd1/sd1 genotypes (Table 3). Significant differences in plant height between SD1/SD1 and sd1/sd1 plants were observed in seedling and maturing stages with P values of 1.7E-25 and 2.2E-10 by ANOVA, respectively. We also measured the yield per plant (YP), spikelets per panicle (SPP), grains per panicle (GPP), 1000-grain weight (KGW), panicle length (PL) and biomass per plant. The genotype SD1/SD1 always had higher significant phenotypic values than the genotypes sd1/sd1 in all traits except the harvest index (Table 3). Subsequently, the tall plant produces more biomass and grain yield.
Table 3. The genetic effects of sd1 on grain yield related traits in the BC3F2 population.
| Traits | sd1/sd1 | SD1/sd1 | SD1/SD1 | P value | Additive | Dominance | D/A |
|---|---|---|---|---|---|---|---|
| SH (cm) | 40.4±3.0 | 49.3±4.3 | 52.7±4.8 | 2.2E-10 | 6.1 | 2.7 | 0.45 |
| PH (cm) | 85±5.8 | 127.5±6.0 | 139±5.4 | 1.7E-25 | 27.0 | 15.5 | 0.57 |
| SPP | 102±17.7 | 113±14.2 | 126±20.6 | 0.0008 | 12.0 | 1.1 | 0.09 |
| GPP | 75±13.9 | 94±12.7 | 102±18.6 | 3.1E-05 | 13.4 | 5.3 | 0.39 |
| PL (cm) | 20.2±0.8 | 22.2±1.0 | 23.9±1.2 | 7.5E-12 | 1.8 | 0.1 | 0.08 |
| BM (g) | 35.6±11.5 | 50.5±19.0 | 57.4±13.6 | 1.2E-05 | 10.9 | 4.0 | 0.36 |
| YP (g) | 17.4±5.5 | 23.9±9.2 | 25.8±5.9 | 0.0001 | 4.2 | 2.2 | 0.54 |
| HI | 0.49±0.06 | 0.47±0.04 | 0.45±0.04 | 0.02 | 0.02 | 0.002 | 0.1 |
SH seedling height, PH plant height, SPP spikelets per panicle, GPP grains per panicle, PL panicle length, BM biomass per plant, HI harvest index, YP yield per plant. The P value indicated the significance of difference between homozygote sd1/sd1 and SD1/SD1.
Functional SD1 plants show sensitive to PBZ
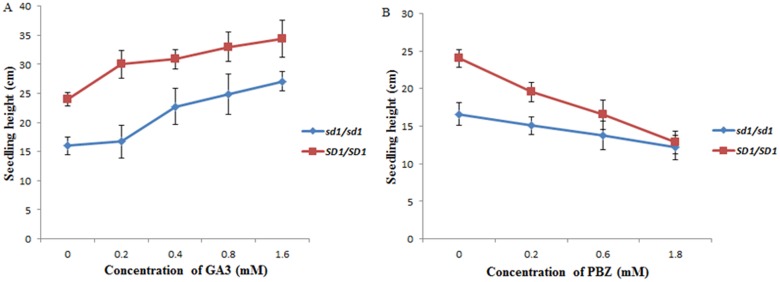
GAs biosynthesis inhibitor, Paclobutrazol (PBZ), was widely used for decreased vegetative growth of rice and increased the chlorophyll content [33]. All treated plants with PBZ showed decreasing seedling heights as compared to the control. With rising of PBZ concentration, the seedling height was further reduced (Fig 6). However, the height reduction rate of SD1/SD1 plants was higher than those of sd1/sd1 plants at all concentrations of PBZ while achieving the same reduction at 1.8 mM. These data indicated that the function of SD1 in SD1/SD1 plants was completed inhibited by PBZ. On the other hand, the seedling height of SD1/SD1 plants was rapidly increased after treated with 0~0.2 mM GA3, whereas the seedling height of sd1/sd1 plants was enhanced sharply with 0.2~0.4 mM GA3.
Fig 6. GA3 and PBZ treatment of homozygote sd1/sd1 and SD1/SD1 plants.
Discussion
Gain-of-function SD1 allele produced by intragenic recombination led to extraordinary tall plant height in RI92
Plant height as a major factor influencing rice production was well studied in the last two decades. Dozens of QTL for plant height in rice were identified and cloned. The green revolution gene (sd1) was widely adopted in semi-dwarf rice breeding for several decades, although the function and mechanization of sd1 was not clear in that time. Nitrogen fertilization is essential for high yield production, but the unfavourable stem elongation and tall plant height increased the risk of lodging under high levels of nitrogen fertilization. Thus, breeding of semi-dwarf varieties overcome the disadvantage of the nitrogen fertilization, and greatly increased rice production in the world during 1960s-1970s. At least seven types of non-function sd1 alleles were identified. In this study, the rice cultivar ZS97 possesses the same allele as cultivar 9311 with a premature termination in the third exon [22], and MH63 possesses the same allele as IR8 with a large deletion. Both parents carry non-functional sd1 alleles. The co-segregation of plant height with the FNP genotype of SD1 in the BC3F2 population indicated that SD1 was the gene underlying the extraordinary tall plant in RI92. GA3 increased the seedling heights of both SD1/SD1 and sd1/sd1; however, the SD1/SD1 was more sensitive to a lower GA3 concentration. In contrast, PBZ treatment decreased the seedling heights of both SD1/SD1 and sd1/sd1, however, the more decreased heights of SD1/SD1 with PBZ treatment indicated the higher level of endogenous GAs in SD1/SD1. Therefore, endogenous GAs played a critical role in regulating seedling height in SD1/SD1 plants. According to these results, it is SD1 that contributes to the tall plant in RI92, although both parents carried non-functional alleles of sd1. Hence, gain-of-function of SD1 occurred in RI92. Most japonica cultivars possessed the weak function SD1-EQ allele, which resulted either two mutations occurred simultaneously in SD1-GR or single mutation happened independently resulting SD1-ER and SD1-GQ and subsequently occurred intragenic recombination between SD1-ER and SD1-GQ [23]. In this study, the intragenic recombination between two non-functional sd1 alleles generated SD1-GR allele in RI92, which encoded GA20-oxidase with a higher enzyme activity than the one encoded by SD1-EQ allele [23]. That is why RI92 is much taller than both parents.
Sequencing of SD1 in RI92 showed a functional SD1 allele, which presented the unique SNP at +2575 in the ZS97 allele. The SNP in ZS97 resulted in a non-functional sd1 allele by a premature stop codon. Thus, it is a mutant type of non-functional ZS97 allele. On the other hand, ZS97 carried the 5’ end of functional SD1, and MH63 carried the 3’ end of functional SD1. In theory, the intragenic recombination of sd1 between parents could produce a functional SD1 allele. Consequently, either point mutation or intragenic recombination is a possible reason for producing the functional SD1 allele in RI92. Sequencing of 4-kb fragment downstream of SD1 identified the same 2-bp insertion in RI92 and MH63 as compared to ZS97. Therefore, the genotypes of RI92 at site +2575 and the 2-bp insertion are the same as MH63. It is almost impossible that both mutations in two sites have simultaneously occurred in one plant. Therefore, we supposed that an intragenic recombination of ZS97 and MH63 sd1 alleles resulted in a functional SD1 allele in RI92. Moreover, the recombination breakpoint was located in the region between the site +1721 and the site +2575 because RI92 carried the ZS97 allele at the site +1721 and the MH63 allele at the site +2575 (Fig 4). The 4-fold recombination rate was observed in the SD1 located bins in both intra-subspecific and inter-subspecific populations. The high recombination rate implemented the occurrence of SD1 intragenic recombination.
SD1 deceased harvest index and increased yield per plant
Since 1960s, the sd1 was widely used in breeding semi-dwarf varieties, because decreasing plant height can increase lodging-resistant ability and planting number in a given area [34, 35]. In the previous report, null sd1 with reduced plant height and biomass can increase harvest index. In our study, the SD1/SD1 increased panicle length by 3.7 cm in the BC3F2 population, spikelets per panicle and grains per panicle are also increased nearly 25 grains comparing to the sd1/sd1. Hence, the yield per plant was increased by 8.4 g in the SD1/SD1 plants. The SD1/SD1 plants have larger yield per plant and biomass. However, the harvest index is lower than sd1/sd1 plants. Therefore, functional SD1 derived from RI92 not only increases plant height but also yield per plant. Of course, such a tall plant faces to a high risk of suffering lodging.
High mapping resolution would be realized by inter-crossing F2 plants
There were 34 bins in RI92, nearly three bins each chromosome. Moreover, chromosomes 10 and 12 were entirely from MH63 and ZS97, respectively. This low number of recombination bins in each plant greatly limited the resolution of QTL mapping, and of course, enlargement of the population size is a way to enhance the mapping resolution. But, this method costs more labors and money. The alternative is increasing the number of bins in each plant, which enhances the mapping resolution in a given population size. The randomly crossing between F2 plants and sequentially intercrossing of each line could increase the recombination rate and result in a RIL construction with an improved resolution [36]. Also, the multi-parent advanced generation inter-crossed population may resolve this low recombination rate in a bi-parental RIL population [37].
Conclusions
We isolated a functional SD1 allele in a tall recombinant line, RI92, from the offsprings of two semi-dwarf indica cultivars. Sequencing of SD1 and its flanking region showed an intragenic recombination occurred and the recombination breakpoint located in the region between the site +1721 and the site +2575 of SD1. Genome re-sequencing identified only 34 bins in RI92, which limited the QTL mapping resolution in recombinant inbred lines. These results indicated that intragenic recombination could create new alleles in the progeny distinct from parental alleles and diversify natural variation, which provided more chance to select target traits; and increasing population size or changing the crossing style (intercross) would be alternatives to enhancing mapping resolution.
Supporting information
red triangles indicate the two amino acid difference between SD1-EQ and SD1-GR.
(TIF)
(DOCX)
Acknowledgments
We thank farm technician Mr. Jianbo Wang for his excellent work in the field.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31371248 and 91535301) http://www.nsfc.gov.cn/ and the Natural science foundation of Hubei province, China (2015CFA006) http://www.hbstd.gov.cn/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40(13):5795–818. doi: 10.1093/nar/gks270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lichten M, Goldman AS. Meiotic recombination hotspots. Annu Rev Genet. 1995;29(1):423–44. doi: 10.1146/annurev.ge.29.120195.002231 [DOI] [PubMed] [Google Scholar]
- 3.Schnable PS, Hsia AP, Nikolau BJ. Genetic recombination in plants. Curr Opin Plant Biol. 1998;1(2):123–9. [DOI] [PubMed] [Google Scholar]
- 4.Okagaki RJ, Weil CF. Analysis of recombination sites within the maize waxy locus. Genetics. 1997;147(2):815–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown J, Sundaresan V. A recombination hotspot in the maize A1 intragenic region. Theor Appl Genet. 1991;81(2):185–8. doi: 10.1007/BF00215721 [DOI] [PubMed] [Google Scholar]
- 6.Inukai T, Sako A, Hirano H-Y, Sano Y. Analysis of intragenic recombination at wx in rice: correlation between the molecular and genetic maps within the locus. Genome. 2000;43(4):589–96. [DOI] [PubMed] [Google Scholar]
- 7.Dooner HK, Martínez-Férez IM. Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. The Plant Cell. 1997;9(9):1633–46. doi: 10.1105/tpc.9.9.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mourad G, Haughn G, King J. Intragenic recombination in the CSR1 locus of Arabidopsis. Mol Gen Genet. 1994;243(2):178–84. [DOI] [PubMed] [Google Scholar]
- 9.Shure M, Wessler S, Fedoroff N. Molecular identification and isolation of the Waxy locus in maize. Cell. 1983;35(1):225–33. [DOI] [PubMed] [Google Scholar]
- 10.Wang ZY, Wu ZL, Xing YY, Zheng FG, Guo XL, Zhang WG, et al. Nucleotide sequence of rice waxy gene. Nucleic Acids Res. 1990;18(19):5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie W, Feng Q, Yu H, Huang X, Zhao Q, Xing Y, et al. Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc Natl Acad Sci U S A. 2010;107(23):10578–83. doi: 10.1073/pnas.1005931107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan C, Han Z, Yu H, Zhan W, Xie W, Chen X, et al. QTL scanning for rice yield using a whole genome SNP array. J Genet Genomics. 2013;40(12):629–38. doi: 10.1016/j.jgg.2013.06.009 [DOI] [PubMed] [Google Scholar]
- 13.Pan Q, Li L, Yang X, Tong H, Xu S, Li Z, et al. Genome-wide recombination dynamics are associated with phenotypic variation in maize. New Phytol. 2016;210(3):1083–94. doi: 10.1111/nph.13810 [DOI] [PubMed] [Google Scholar]
- 14.Gill KS, Gill BS, Endo TR, Taylor T. Identification and high-density mapping of gene-rich regions in chromosome group 1 of wheat. Genetics. 1996;144(4):1883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunzel G, Korzun L, Meister A. Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics. 2000;154(1):397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choo KA. Why is the centromere so cold? Genome Res. 1998;8(2):81–2. [DOI] [PubMed] [Google Scholar]
- 17.Chen M, Presting G, Barbazuk WB, Goicoechea JL, Blackmon B, Fang G, et al. An integrated physical and genetic map of the rice genome. The Plant Cell. 2002;14(3):537–45. doi: 10.1105/tpc.010485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodgers-Melnick E, Bradbury PJ, Elshire RJ, Glaubitz JC, Acharya CB, Mitchell SE, et al. Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc Natl Acad Sci U S A. 2015;112(12):3823–8. doi: 10.1073/pnas.1413864112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan X, Weng J, Zhai H, Wang J, Lei C, Liu X, et al. Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allele gw-5 in a recombination hotspot region on chromosome 5. Genetics. 2008;179(4):2239–52. doi: 10.1534/genetics.108.089862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–7. doi: 10.1038/ng.143 [DOI] [PubMed] [Google Scholar]
- 21.Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, et al. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416(6882):701–2. [DOI] [PubMed] [Google Scholar]
- 22.Asano K, Takashi T, Miura K, Qian Q, Kitano H, Matsuoka M, et al. Genetic and molecular analysis of utility of sd1 alleles in rice breeding. Breeding Sci. 2007;57(1):53–8. doi: 10.1270/jsbbs.57.53 [Google Scholar]
- 23.Asano K, Yamasaki M, Takuno S, Miura K, Katagiri S, Ito T, et al. Artificial selection for a green revolution gene during japonica rice domestication. Proc Natl Acad Sci U S A. 2011;108(27):11034–9. doi: 10.1073/pnas.1019490108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing Y, Xu C, Hua J, Tan Y, Sun X. Mapping and isolation of quantitative trait loci controlling plant height and heading date in rice. Acta Botanica Sinica. 2000;43(7):721–6. [Google Scholar]
- 25.Duan L, Yang W, Huang C, Liu Q. A novel machine-vision-based facility for the automatic evaluation of yield-related traits in rice. Plant Methods. 2011;7:44 doi: 10.1186/1746-4811-7-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8(19):4321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing Z, Tan F, Hua P, Sun L, Xu G, Zhang Q. Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor Appl Genet. 2002;105(2–3):248–57. doi: 10.1007/s00122-002-0952-y [DOI] [PubMed] [Google Scholar]
- 28.Xie W, Feng Q, Yu H, Huang X, Zhao Q, Xing Y, et al. Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proceedings of the National Academy of Sciences. 2010;107(23):10578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen G, Zhan W, Chen H, Xing Y. Dominance and epistasis are the main contributors to heterosis for plant height in rice. Plant Sci. 2014;215–216:11–8. doi: 10.1016/j.plantsci.2013.10.004 [DOI] [PubMed] [Google Scholar]
- 30.Ashikari M, Sasaki A, Ueguchi-Tanaka M, Itoh H, Nishimura A, Datta S, et al. Loss-of-function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox-2), led to the rice ‘green revolution’. Breeding Sci. 2002;52(2):143–50. doi: 10.1270/jsbbs.52.143 [Google Scholar]
- 31.Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009;19(6):1068–76. doi: 10.1101/gr.089516.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Wang A, Huang X, Zhao Q, Dong G, Qian Q, et al. Mapping 49 quantitative trait loci at high resolution through sequencing-based genotyping of rice recombinant inbred lines. Theor Appl Genet. 2011;122(2):327–40. doi: 10.1007/s00122-010-1449-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yim K-O, Kwon Y, Bayer D. Growth responses and allocation of assimilates of rice seedlings by paclobutrazol and gibberellin treatment. J Plant Growth Regul. 1997;16(1):35–41. [Google Scholar]
- 34.Sakamoto T, Matsuoka M. Generating high-yielding varieties by genetic manipulation of plant architecture. Curr Opin Biotechnol. 2004;15(2):144–7. doi: 10.1016/j.copbio.2004.02.003 [DOI] [PubMed] [Google Scholar]
- 35.Hedden P. The genes of the Green Revolution. Trends Genet. 2003;19(1):5–9. [DOI] [PubMed] [Google Scholar]
- 36.Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141(3):1199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavanagh C, Morell M, Mackay I, Powell W. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr Opin Plant Biol. 2008;11(2):215–21. doi: 10.1016/j.pbi.2008.01.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
red triangles indicate the two amino acid difference between SD1-EQ and SD1-GR.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.