Abstract
Limited expression and distribution of nectin-1, the major herpes simplex virus (HSV) type-1 entry-receptor, within tumors has been proposed as an impediment to oncolytic HSV (oHSV) therapy. To determine whether resistance to oHSVs in malignant peripheral nerve sheath tumors (MPNSTs) was explained by this hypothesis, nectin-1 expression and oHSV viral yields were assessed in a panel of MPNST cell lines using γ134.5-attenuated (Δγ134.5) oHSVs and a γ134.5 wild-type (wt) virus for comparison. Although there was a correlation between nectin-1 levels and viral yields with the wt virus (R = 0.75, P = 0.03), there was no correlation for Δγ134.5 viruses (G207, R7020 or C101) and a modest trend for the second-generation oHSV C134 (R = 0.62, P = 0.10). Nectin-1 overexpression in resistant MPNST cell lines did not improve Δγ134.5 oHSV output. While multistep replication assays showed that nectin-1 overexpression improved Δγ134.5 oHSV cell-to-cell spread, it did not confer a sensitive phenotype to resistant cells. Finally, oHSV yields were not improved with increased nectin-1 in vivo. We conclude that nectin-1 expression is not the primary obstacle of productive infection for Δγ134.5 oHSVs in MPNST cell lines. In contrast, viruses that are competent in their ability to counter the antiviral response may derive benefit with higher nectin-1 expression.
INTRODUCTION
Malignant peripheral nerve sheath tumors (MPNSTs) are a highly aggressive cancer of the peripheral nervous tissue believed to originate within the Schwann cell lineage1 and are most commonly associated with the genetic condition neurofibromatosis type-1. Treatment options for MPNSTs beyond surgery are inadequate, resulting in a median survival of only 26 months.2 Oncolytic virotherapy by attenuated herpes simplex type-1 viruses (oHSVs) has been proposed as an alternative to chemotherapy and radiotherapy for the treatment of MPNSTs.3–7 HSVs with γ134.5 neurovirulence gene deletions are safe in humans and have been shown to selectively replicate in tumor cells.8 These attenuated Δγ134.5 oHSVs have a clinically verified safety profile in patients with malignant glioma and have been associated with measurable antitumor responses.9–13 However, these patient responses have varied widely, likely due to tumor susceptibility. Therefore, we have sought to further elucidate the mechanisms of oHSV resistance.
In our initial investigation into potential oHSV resistance mechanisms within MPNSTs, we have tested the hypothesis that oHSV resistance is attributable to the insufficient expression of HSV-1 entry receptors by tumor cells.14–18 Four viral glycoproteins (gD, gB and gH/gL) and a cellular glycoprotein D (gD)-interacting receptor have been demonstrated as necessary and sufficient to trigger cellular entry.19–23 Of the three cellular HSV-1 gD-interacting receptors, nectin-1, a cellular adhesion protein expressed in epithelial cells,24 fibroblasts and neurons,25 has been proposed as the major HSV-1 entry receptor.26 Herpes virus entry mediator (HVEM) 27 and 3-O-sulfated heparan sulfate (3-OS-HS)28 have also been demonstrated to facilitate HSV-1 entry. Additional cell-surface molecules that interact with other viral glycoproteins have been identified, though the broad necessity of these in permitting HSV-1 infection and spread remains to be determined and the lack of these molecules has not yet been implicated in limiting the oncolytic capacity of oHSV.
Here, we have investigated the hypothesis that HSV entry-receptor expression is a determinant of oHSV efficacy in MPNST cells and have identified whether an increase in entry-receptor expression improves the viral yield and spread of oHSVs. The influence of entry-receptor expression was examined in the context of an array of viral genotypes, including a representative wild-type (wt) γ134.5 HSV-1, a fully attenuated Δγ134.5 oHSV, and an attenuated second-generation oHSV capable of host antiviral evasion. We report the following conclusions: (1) correlation of nectin-1 expression with viral production capacity appears more important in viruses which are genetically competent to counter the intrinsic antiviral response, (2) increased expression of entry-receptor molecules modestly improves cell-cell spread of Δγ134.5 oHSVs, but yields little benefit to viral production and (3) increases in entry-receptor expression do not render resistant MPNST cell lines permissive to Δγ134.5 oHSV infection.
RESULTS AND DISCUSSION
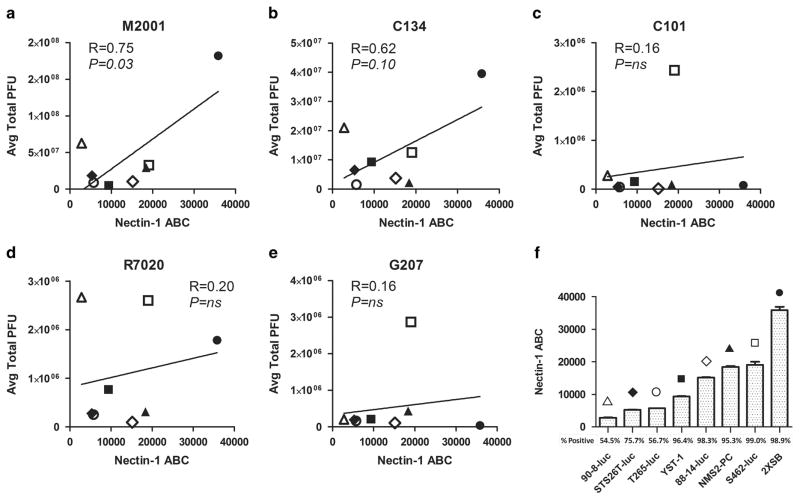
MPNST cell lines have been previously identified as susceptible to oHSV infection and cytotoxicity.3 To examine the correlation between viral production capacity and entry-receptor expression, human MPNST cell lines STS-26T, T265-2c, NMS2-PC, S462, YST-1, 90-8, ST88-14 and 2XSB, or their luciferase-expressing derivatives (‘ − luc’), were first infected at a multiplicity of infection (MOI) of 10 (single-step replication assay) with a panel of genetically modified HSV-1 and cellular lysates collected 24 h post infection (h.p.i.) for viral recovery analysis (Supplementary Figure 1). The viral yields from this assay are presented in correlation with nectin-1 expression in Figures 1a–e.
Figure 1.
Correlation of nectin-1 expression with viral titers. Pearson’s correlation coefficients (a–e) were calculated between the viral titering data from M2001, C134, C101, R7020, G207 (Supplementary Figure 1) and the nectin-1 expression levels (f) from cell lines 90-8-luc (open triangle), STS26T-luc (closed diamond), T265-luc (open circle), YST-1 (closed square), 88-14-luc (open diamond), NMS2-PC (closed triangle), S462-luc (open square), and 2xSB (closed circle). A strong and significant correlation was noted for M2001. Cells were infected in triplicate by a single-step replication assay (MOI =10) and the lysates collected and titered at 24 h.p.i. Nectin-1 expression was quantified by flow cytometry after incubation with phycoerythrin (PE)-conjugated mouse monoclonal antibody with subsequent quantification using antibody quantification beads. The percentage of the cell population staining above the isotype control is also reported. Receptor quantification was performed in triplicate with the standard deviation reported.
HSV-1 entry receptors have not been previously identified in MPNSTs or cells of the Schwann cell lineage. Upon examination of nectin-1 and HVEM expression in our panel of MPNST cell lines, we found detectable levels of nectin-1 in all of the lines, with population-wide (>95%) expression of nectin-1 in five of eight cell lines (Figure 1f). Population-wide expression of HVEM in MPNST cell lines was observed in only one of eight lines (Supplementary Figure 2D); therefore, HVEM was excluded as a candidate for the major entry receptor in MPNSTs. The other established entry receptor 3-OS-HS was not examined in this study due to the lack of a commercially available antibody. However, HSV-1 infection of the resistant cell lines selected for further study (STS26T-luc and T265-luc) was found to be dependent on nectin-1 expression alone by nectin-1 neutralization assays (Supplementary Figure 2).
Pearson’s correlation coefficients were calculated between viral recovery data and nectin-1 expression levels. Possible positive associations were found for the wt M2001 (R = 0.75; P = 0.03) and attenuated second-generation oHSV C134 (R = 0.62, P = 0.10) viruses (Figures 1a and b). While prior studies involving thyroid cancer14 and head and neck squamous cell carcinoma15 cell lines demonstrated correlation between viral yields of oHSV NV1023 (a derivative of R7020) and nectin-1 expression, no such associations were observed for G207, C101 or R7020 oHSVs suggesting that the specific viral genotype influences the outcome of infection.
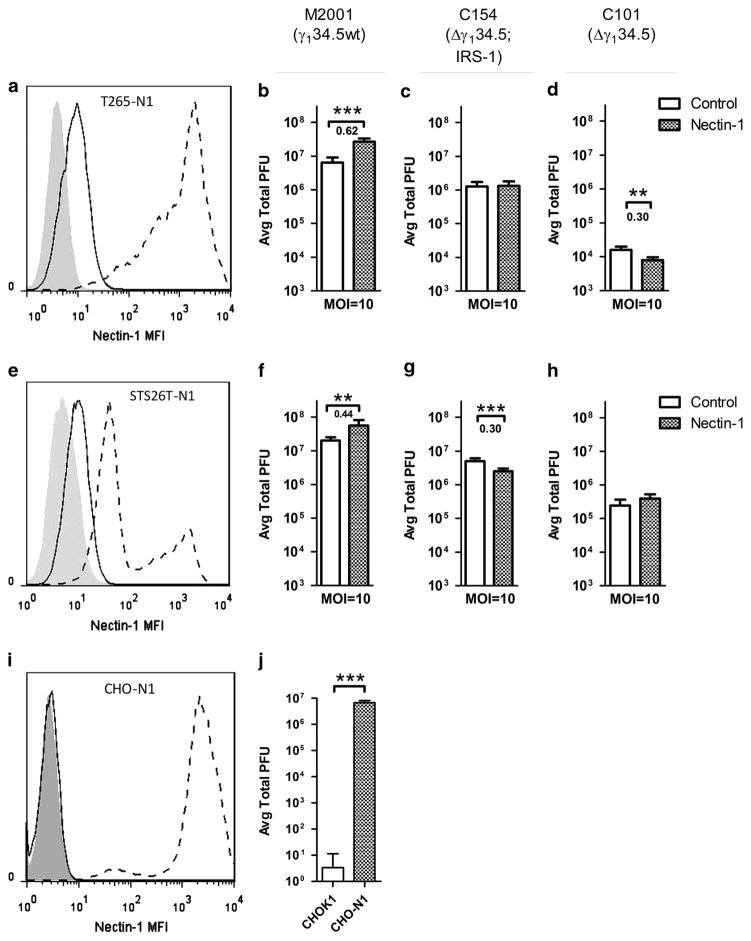
The lack of a clear association of nectin-1 expression with attenuated Δγ134.5 oHSV viral yields led us to further evaluate the functional impact of increased entry-receptor expression on oHSV sensitivity. To assess this, the oHSV-resistant cell lines, T265-luc and STS26T-luc were transduced with full-length human nectin-1 (nectin-1α) using lentivirus LV2114CK. An mCherry expressing lentivirus was used as a control and confirmed that transduction alone did not alter viral production (data not shown).
If low entry-receptor expression diminishes the ability of oHSV to establish an initial infection, we would predict that increased nectin-1 expression would increase the initial opportunity for entry, resulting in replication within a greater number of cells and an increase in the total production of virus. To determine the impact of increased nectin-1 expression on viral yields in MPNSTs, single-step (MOI= 10, 24 h.p.i.) and multistep (MOI= 0.1, 24, 48, 72 h.p.i.; Supplementary Figure 3) viral recovery assays were performed using the parent and nectin-1 transduced cell lines. Because the reliable titering repeatability of HSV is within approximately 0.5 log, only changes in titer greater than 0.5 log are considered to be biologically relevant. The nectin-1 transduction of T265-luc and STS26T-luc resulted in abundant nectin-1 expression in T265-N1 and STS26T-N1 cell lines, respectively (Figures 2a and e). While increased entry-receptor expression improved the yields of a representative wt virus (Figures 2b and f), the increased expression did not improve the titers of a next-generation oHSV C154, an EGFP expressing variant of C134 (Figures 2c and g) or first-generation Δγ134.5 oHSV C101 (Figures 2d and h). To demonstrate that increased nectin-1 expression would be expected to improve viral production, the HSV receptor-deficient cell line CHO-K1 was transduced with nectin-1. A significant and greater than 5 log increase in wt HSV-1 titers was observed (Figures 2i and j).
Figure 2.
Overexpression of nectin-1 in resistant cells and impact on single-step replication assays. Nectin-1 was transduced via lentivirus into oHSV-resistant cell lines T265-luc (a) and STS26T-luc (e) as well as control cell line CHO-K1. Isotype control (shaded), parent (solid line) and transduced (dashed line) cell lines are shown. Transduction of the nectin-1-deficient cell line (i) demonstrated function as an entry receptor as apparent by M2001 replication (j). The impact of nectin-1 overexpression in resistant cell lines was tested by single-step (MOI =10) replication by viruses M2001 (b, f), C154 (c, g) and C101 (d, h) and compared with control cell lines. Significance was determined by two-tailed Student’s t-test with unequal variance. Significance was set at P<0.05. For cells with significant changes in titer, the logarithm of the absolute value of the increase was reported below the significance marking. Changes in titer greater than 0.5 log are considered to be biologically relevant. **P>0.01 and ***P>0.001.
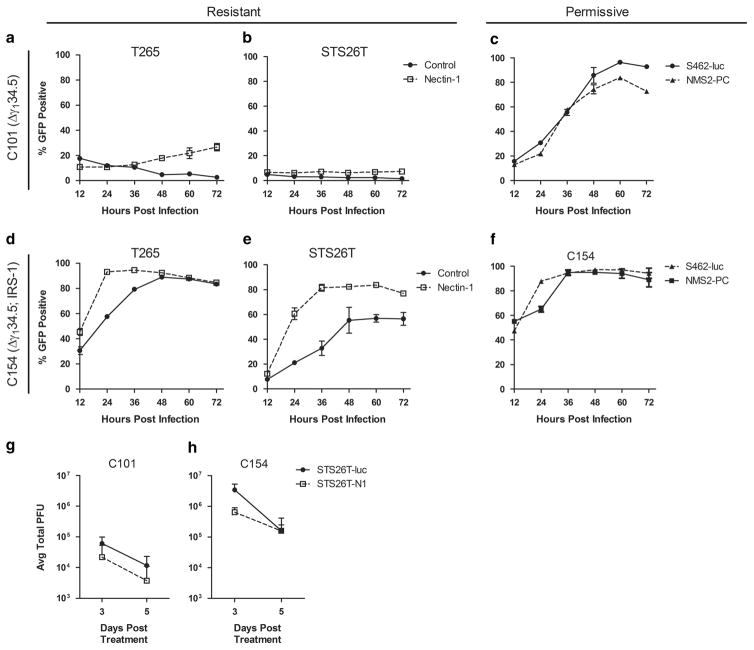
Interaction with HSV entry receptors is essential for initial HSV entry as well as the subsequent cell-to-cell spread of HSV.29 To assess the effect of increased nectin-1 expression on viral spread, we measured viral GFP expression in MPNST cells over the time in multistep assays following infection with GFP expressing C101 and C154 (Figure 3) or M2001 (Supplementary Figure 4). The results show that nectin-1 overexpression improved the ability of C101 to undergo cell-to-cell spread and increased the proportion of cells infected from 3 to 27% and from 1 to 7% of the cell population in T265-N1 and STS26T-N1 in multistep replication assays (MOI = 0.1, 24, 48 and 72 h.p.i.) respectively (Figures 3a and b). Despite this improved spread in resistant lines, the maximum spread was much less than that observed in the naturally permissive S462-luc and NMS-2PC MPNST cell lines, where C101 was capable of infecting >80% of the cells (Figure 3c). This suggests that endogenous levels of entry receptors are sufficient to permit infection and sustain Δγ134.5 oHSV spread in these lines and that increased entry-receptor expression is not sufficient to render resistant cell lines with a permissive phenotype. Of note, the overexpressed nectin-1 levels far exceeded the highest endogenous levels in the permissive lines (Supplementary Figure 5), suggesting that restricted entry is not an explanation for MPNST resistance to oHSVs. This conclusion is further supported by the fact that infection of the same cell lines with a second-generation oHSV (that is, C134 or C154) capable of evading the antiviral response30 resulted in approximately 10- 100 fold increase in viral titers and notably greater cell-to-cell spread as compared with C101 (Figures 3d and e).
Figure 3.
Impact of increased nectin-1 expression on oHSV spread in vitro and viral recovery in vivo. Resistant cell lines STS26T-luc and T265-luc and their nectin-1 transduced variants, as well as permissive cell lines S462-luc and NMS2-PC, were infected in a multistep assay (MOI =0.1) with fully attenuated oHSV C101 (a–c) or second-generation C154 expressing HCMV IRS1 (d–f) and monitored by flow cytometry over time for viral infection as evident by expression of viral GFP. STS26T-luc and STS26T-N1 cells were engrafted in the flanks of nude mice and following tumor formation were injected with 1×107 plaque forming units (PFU) of C101 or C154. Tumors were harvested and viral titers determined at days 3 and 5 following infection (g and h). Data are representative of four tumors with standard deviation reported.
To determine the extent to which in vivo studies recapitulated these results, athymic nude mice were engrafted with either parent or nectin-1 expressing cell lines. Of the resistant cell lines, only STS26T-luc and the nectin-1 overexpressing variant established flank tumors. Tumors were injected with 1×107 plaque forming units of C101 or C154, and viral recovery was measured on days 3 and 5 post injection. Similar to the in vitro results (Figure 3g and h), the next-generation virus had a >10-fold viral production advantage over the Δγ134.5 oHSV C101, however neither virus demonstrated an increased viral titer between days 3 and 5 even with increased nectin-1 expression. Tumors were also collected for immunohistochemistry and staining for HSV-1 confirmed that increased nectin-1 expression did not benefit oHSV spread between days 3 and 5 (data not shown). The in vivo results therefore confirmed that neither the first-nor the second-generation oHSVs derived a benefit to viral output from increased entry-receptor expression.
In summary, the work presented here provides insight into one of the proposed determinants of oHSV therapeutic efficacy. We conclude that the primary mode of MPNST resistance to Δγ134.5 oHSVs is not due to limited expression of nectin-1. Despite the primary conclusions of previously published work that entry-receptor expression is predictive of a productive infection by oHSV, we suggest that the use of viruses in these previous studies which contained at least one functional copy of the γ134.5 gene (NV1023)14,15,18 or γ134.5 under a nestin promoter (rQnestin34.5)16 is in line with our conclusions that viruses which are genetically competent to counter the intrinsic antiviral response benefit the most from increased entry-receptor expression. Similarly in our work, the wt HSV-1 and C134 viruses derived greater benefit from higher entry-receptor expression than did the first-generation Δγ134.5 oHSVs. Furthermore, the work of Wang et al16 showed that only the γ134.5 containing virus was able to substantially benefit from increased nectin-1 expression while the Δγ134.5 control virus did not. Future work should therefore include the characterization of the capacity for an intrinsic antiviral response as the major mechanism for oHSV resistance in MPNSTs.
MATERIALS AND METHODS
Cell lines
MPNST cell lines STS26T-luc, T265-luc, ST88-14-luc, S462-luc, 90-8- luc, NMS2-PC, YST-1 and 2XSB were provided by Dr Steve Carroll (University of Alabama at Birmingham). Cell lines STS26T-luc, T265- luc, and ST88-14-luc express firefly luciferase and have been previously described.31 S462-luc and 90-8-luc were transduced via lentivirus to express Renilla luciferase. HSV-1 entry receptor-deficient cell line CHO-K1 was generously provided by Dr Yancey Gillespie (University of Alabama, Birmingham). All MPNST cell lines were maintained in DMEM, 10% FBS, and 1% P/S. CHO-K1 cells were maintained in Ham’s F12, 10% FBS and 1% P/S. Vero cells were obtained from American Type Culture Collection (Manassas, VA, USA) and maintained in MEM and 5% BGS. All cell lines were confirmed to be free of Mycoplasma by DAPI staining and PCR detection.
Viruses
All viruses have been previously described. Briefly, M2001 was constructed by insertion of the gene encoding EGFP under the control of the CMV immediate early promoter into the UL3-UL4 intergenic region of the prototypical wt HSV-1 (F) strain.32 C101 and C134 were derived from the Δγ134.5 mutant HSV-1 R3616 by insertion respectively of the EGFP or HCMV IRS1 genes under the control of the CMV immediate early promoter in the UL3-UL4 intergenic region.33 C154 is derived from C134 by insertion of EGFP into the deletion loci of γ134.5. G207 (Medigene, Inc., San Diego, CA, USA) is a clinical grade oHSV derived from R3616 with the additional insertion of lacZ in the UL39 region.34 R7020 (kindly provided by Bernard Roizman; University of Chicago, Chicago, IL, USA), is a clinical grade oHSV derived from HSV-1 (F) strain by insertion of a region of the HSV-2 genome encoding glycoproteins G, D, I and a portion of E into one of the internal repeat regions of HSV-1 (F) disrupting one copy of the neurovirulence gene γ134.5.35
Viral titering assays
Viral titers were determined by limiting dilution plaque formation assays as previously described.33 MPNST cells were incubated for 2 h with virus diluted in 100 μl infection media (DMEM + 1% FBS) and replaced with growth media after infection. An equivalent volume of sterile milk was added and the plate subjected to three cycles of freeze-thaw at − 80 °C. Lysate was collected, sonicated, serially diluted in Vero infection media (MEM + 1% BGS), and incubated on Vero monolayers. Infection media was replaced with growth media containing 0.01% human AB serum (Corning Cellgro, Corning, NY, USA). After 48 h, plaques were counted following May-Grunwald/methanol staining as previously described. All experiments were performed in triplicate and the average total plaque forming units reported with standard deviation.
Viral entry-receptor quantification
Expression of entry receptors was quantified by flow cytometry using either phycoerythrin-conjugated mouse monoclonal antibodies to nectin-1 (R1.302) (Biolegend, San Diego, CA, USA), HVEM (Biolegend), or isotype control (BD Biosciences, San Jose, CA, USA). Antibody concentrations used were confirmed to be saturating. Cells analyzed using a FACSCaliber flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). Concurrently, Quantum Simply Cellular beads (Bangs Laboratories, Fisher, IN, USA) were used to determine the antibody binding capacity of each cell line. Mean fluorescence analysis was performed using FlowJo (v 7.6.1; Tree Star, Ashland, OR, USA) and the receptor-mean fluorescence intensity data converted to antibody binding capacity using a script provided with the Quantum Simply Cellular kit. All measurements were averaged from three independently seeded wells, and the final antibody binding capacity reported as the difference above the isotype control.
Correlation of nectin-1 and viral recovery
Pearson’s correlation coefficients between nectin-1 expression and viral recovery were determined by analysis of the data in Prism 5 (GraphPad Software, La Jolla, CA, USA). Cutoff for statistical significance was set at P<0.05.
Nectin-1 overexpression
A self-inactivating lentiviral vector was used to overexpress nectin-1 or control mCherry in oHSV-resistant cell lines. Human nectin-1 clone (Clone ID: 8322523) was obtained from Open Biosytems (Thermo Scientific, Waltham, MA, USA). Nectin-1 cDNA was PCR amplified using primers 5′-CGGATCCCGGGTCGACCCGATGGCTC GGATGGGGCTT-3′ and 5′-CCGGGTCGAGCGGCCGCGCTACACGTAC CACTCCTTCTTGGAA-3′ (IDT, Coralville, IA, USA) in a T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA). The recipient vector has been previously described.36 An intermediate lenti-vector was first constructed by insertion of an IRES-puromycin N-acetyl-transferase cassette into the NotI- and EcoRI-digested pLVmnd lentivirus. The intermediate construct was then digested with SalI and NotI for subsequent insertion of the PCR-amplified Nectin-1 cDNA. The sequence of the coding region of the resulting lentiviral vector pCK2114 was verified. The control lentivirus (pLVmnd.CIP) was similarly constructed by insertion of mCherry upstream of the IRES-puromycin N-acetyl-transferase cassette in the intermediate virus. Lentiviruses were produced by co-transfection of pCK2114 or pLVmnd.CIP with pMD.G (VSVG pseudotype), and pCMV. deltaR8.91 (HIV packaging) in 293T cells using Lipofectamine 2000 and OptiMEM media (Gibco, Carlsbad, CA, USA). After 12 h, transfection media was replaced with DMEM/F12 with 10% FBS. Lentiviral-enriched supernatant was collected 48 h post transfection, filtered through a 0.22-micron filter, and mixed with polybrene (8 μgml − 1). STS26T-luc and T265-luc cells were subsequently transduced with the resulting lentiviruses enriched by puromycin selection (5 μgml − 1), followed by fluorescence activated cell sorting to obtain pure populations (UAB Comprehensive Flow Cytometry Core, Birmingham, AL, USA).
HSV titers in nectin-1 overexpressing cell lines
The impact of increased nectin-1 expression on viral titers in resistant cell lines was determined as described above with the exception of replacing infection media with MPNST growth media and 0.01% human AB serum to minimize extracellular spread of the virus. Statistical significance was determined by two-tailed Student’s T-test assuming equal variance. Star notation indicating significant differences is as follows: (*) for P<0.05, (**) for P<0.01 and (***) for P<0.001.
HSV spread as measured by GFP expression
Cells were incubated with virus or mock infected for 2 h at MOI = 0.1 in infection media. Infection media was replaced with growth media and 0.01% human AB serum following incubation. Cells were harvested at 12 h intervals and analyzed by flow cytometry. The percent GFP-positive measurement was assessed by defining the GFP (FL1) gate at 1% positive of the mocktreated cells. The percentage of the infected cell population expressing GFP was then recorded. All data points were performed in triplicate, averaged, and the standard deviation reported.
In vivo viral recovery
Six-week-old athymic nude (nu/nu) mice (NCI-Frederick, Frederick, VA, USA) were obtained and allowed to adjust for a period of 2 weeks. Bilateral, subcutaneous tumors were engrafted in the flank by injection of 5 × 106 cells suspended in 50:50 BD Matrigel (Becton Dickinson) and serum-free DMEM. Four tumors were used for each virus and timepoint. When tumors reached an average size of 300mm3, 1×107 plaque forming units of oHSV suspended in saline-buffered solution was injected intratumorally. At days 3 and 5 following infection, mice were euthanized and tumors recovered in DMEM and kept on ice. After tumors were mechanically dissociated, an equivalent volume of sterile milk was added to the homogenate and viral titering was performed as described above. Titers are plotted with standard deviation.
Supplementary Material
Acknowledgments
Funding for this research was provided through DOD-W81XWH-10-NFRP-IIRA-NF1001157, P20 CA151129-03, P01 CA71933-15 and P50 CA97247-05. Special thanks to Enid Keyser in the UAB Comprehensive Flow Cytometry Core for assistance in cell sorting and Dr Bernard Roizman for providing R7020.
Footnotes
CONFLICT OF INTEREST
JMM, GYG, and RJW are co-founders, stockholders and consultants for Catherex, Inc., which holds intellectual property related to oncolytic HSV.
Supplementary Information accompanies this paper on Gene Therapy website (http://www.nature.com/gt)
References
- 1.Carroll SL, Ratner N. How does the Schwann cell lineage form tumors in NF1? Glia. 2008;56:1590–1605. doi: 10.1002/glia.20776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolberg M, Høland M, Ågesen TH, Brekke HR, Liestøl K, Hall KS, et al. Survival metaanalyses for> 1800 malignant peripheral nerve sheath tumor patients with and without neurofibromatosis type 1. Neuro-oncology. 2013;15:135–147. doi: 10.1093/neuonc/nos287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahller YY, Rangwala F, Ratner N, Cripe TP. Malignant peripheral nerve sheath tumors with high and low Ras-GTP are permissive for oncolytic herpes simplex virus mutants. Pediatric Blood Cancer. 2006;46:745–754. doi: 10.1002/pbc.20565. [DOI] [PubMed] [Google Scholar]
- 4.Mahller YY, Vaikunth SS, Currier MA, Miller SJ, Ripberger MC, Hsu Y-H, et al. Oncolytic HSV and erlotinib inhibit tumor growth and angiogenesis in a novel malignant peripheral nerve sheath tumor xenograft model. Mol Ther. 2007;15:279–286. doi: 10.1038/sj.mt.6300038. [DOI] [PubMed] [Google Scholar]
- 5.Farassati F, Pan W, Yamoutpour F, Henke S, Piedra M, Frahm S, et al. Ras signaling influences permissiveness of malignant peripheral nerve sheath tumor cells to oncolytic herpes. Am J Pathol. 2008;173:1861–1872. doi: 10.2353/ajpath.2008.080376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maldonado AR, Klanke C, Jegga AG, Aronow BJ, Mahller YY, Cripe TP, et al. Molecular engineering and validation of an oncolytic herpes simplex virus type 1 transcriptionally targeted to midkine-positive tumors. J Gene Med. 2010;12:613–623. doi: 10.1002/jgm.1479. [DOI] [PubMed] [Google Scholar]
- 7.Mahller Y, Sakthivel B, Baird W, Aronow B, Hsu Y, Cripe T, et al. Molecular analysis of human cancer cells infected by an oncolytic HSV-1 reveals multiple upregulated cellular genes and a role for SOCS1 in virus replication. Cancer Gene Ther. 2008;15:733–741. doi: 10.1038/cgt.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou J, Kern ER, Whitley RJ, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to gamma 134. 5, a gene nonessential for growth in culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 9.Harrow S, Papanastassiou V, Harland J, Mabbs R, Petty R, Fraser M, et al. HSV1716 injection into the brain adjacent to tumour following surgical resection of high-grade glioma: safety data and long-term survival. Gene Therapy. 2004;11:1648–1658. doi: 10.1038/sj.gt.3302289. [DOI] [PubMed] [Google Scholar]
- 10.Markert J, Medlock M, Rabkin S, Gillespie G, Todo T, Hunter W, et al. Conditionally replicating herpes simplex virus mutant, G207 for the treatment of malignant glioma: results of a phase I trial. Gene Therapy. 2000;7:867–874. doi: 10.1038/sj.gt.3301205. [DOI] [PubMed] [Google Scholar]
- 11.Rampling R, Cruickshank G, Papanastassiou V, Nicoll J, Hadley D, Brennan D, et al. Toxicity evaluation of replication-competent herpes simplex virus (ICP 34. 5 null mutant 1716) in patients with recurrent malignant glioma. Gene Therapy. 2000;7:859. doi: 10.1038/sj.gt.3301184. [DOI] [PubMed] [Google Scholar]
- 12.Markert JM, Liechty PG, Wang W, Gaston S, Braz E, Karrasch M, et al. Phase Ib trial of mutant herpes simplex virus G207 inoculated pre-and post-tumor resection for recurrent GBM. Mol Ther. 2008;17:199–207. doi: 10.1038/mt.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markert JM, Razdan SN, Kuo H-C, Cantor A, Knoll A, Karrasch M, et al. A phase I trial of oncolytic HSV-1, G207, given in combination with radiation for recurrent GBM demonstrates safety and radiographic responses. Mol Ther. 2014;22:1048–1055. doi: 10.1038/mt.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y-Y, Yu Z, Lin S-F, Li S, Fong Y, Wong RJ. Nectin-1 is a marker of thyroid cancer sensitivity to herpes oncolytic therapy. J Clin Endocrinol Metab. 2007;92:1965–1970. doi: 10.1210/jc.2007-0040. [DOI] [PubMed] [Google Scholar]
- 15.Yu Z, Adusumilli PS, Eisenberg DP, Darr E, Ghossein RA, Li S, et al. Nectin-1 expression by squamous cell carcinoma is a predictor of herpes oncolytic sensitivity. Mol Ther. 2007;15:103–113. doi: 10.1038/sj.mt.6300009. [DOI] [PubMed] [Google Scholar]
- 16.Wang P, Currier M, Hansford L, Kaplan D, Chiocca E, Uchida H, et al. Expression of HSV-1 receptors in EBV-associated lymphoproliferative disease determines susceptibility to oncolytic HSV. Gene Therapy. 2012;20:761–769. doi: 10.1038/gt.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman GK, Langford CP, Coleman JM, Cassady KA, Parker JN, Markert JM, et al. Engineered herpes simplex viruses efficiently infect and kill CD133+ human glioma xenograft cells that express CD111. J Neuro-oncology. 2009;95:199–209. doi: 10.1007/s11060-009-9926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C-h, Chen W-Y, Lin S-F, Wong RJ. Epithelial mesenchymal transition enhances response to oncolytic herpes viral therapy through nectin-1. Human Gene Ther. 2014;25:539–551. doi: 10.1089/hum.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ligas MW, Johnson DC. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pertel PE, Fridberg A, Parish ML, Spear PG. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology. 2001;279:313–324. doi: 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- 23.Roop C, Hutchinson L, Johnson DC. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rikitake Y, Mandai K, Takai Y. The role of nectins in different types of cell-cell adhesion. J Cell Sci. 2012;125:3713–3722. doi: 10.1242/jcs.099572. [DOI] [PubMed] [Google Scholar]
- 25.Mizoguchi A, Nakanishi H, Kimura K, Matsubara K, Ozaki-Kuroda K, Katata T, et al. Nectin an adhesion molecule involved in formation of synapses. J Cell Biol. 2002;156:555–565. doi: 10.1083/jcb.200103113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richart SM, Simpson SA, Krummenacher C, Whitbeck JC, Pizer LI, Cohen GH, et al. Entry of herpes simplex virus type 1 into primary sensory neurons in vitro is mediated by nectin-1/HveC. J Virol. 2003;77:3307–3311. doi: 10.1128/JVI.77.5.3307-3311.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 28.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 29.Even DL, Henley AM, Geraghty RJ. The requirements for herpes simplex virus type 1 cell–cell spread via nectin-1 parallel those for virus entry. Virus Res. 2006;119:195–207. doi: 10.1016/j.virusres.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Shah A, Parker J, Gillespie G, Lakeman F, Meleth S, Markert J, et al. Enhanced antiglioma activity of chimeric HCMV/HSV-1 oncolytic viruses. Gene Therapy. 2007;14:1045–1054. doi: 10.1038/sj.gt.3302942. [DOI] [PubMed] [Google Scholar]
- 31.Byer SJ, Eckert JM, Brossier NM, Clodfelder-Miller BJ, Turk AN, Carroll AJ, et al. Tamoxifen inhibits malignant peripheral nerve sheath tumor growth in an estrogen receptor–independent manner. Neuro-oncology. 2011;13:28–41. doi: 10.1093/neuonc/noq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman GK, Haas MC, Kelly VM, Markert JM, Gillespie GY, Cassady KA. Hypoxia moderates γ134. 5-deleted herpes simplex virus oncolytic activity in human glioma xenoline primary cultures. Transl Oncol. 2012;5:200. doi: 10.1593/tlo.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cassady KA. Human cytomegalovirus TRS1 and IRS1 gene products block the double-stranded-RNA-activated host protein shutoff response induced by herpes simplex virus type 1 infection. J Virol. 2005;79:8707–8715. doi: 10.1128/JVI.79.14.8707-8715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mineta T, Rabkin SD, Yazaki T, Hunter WD, Martuza RL. Attenuated multi-mutated herpes simplex virus-1 for the treatment of malignant gliomas. Nat Med. 1995;1:938–943. doi: 10.1038/nm0995-938. [DOI] [PubMed] [Google Scholar]
- 35.Meignier B, Longnecker R, Roizman B. In vivo behavior of genetically engineered herpes simplex viruses R7017 and R7020: construction and evaluation in rodents. J Infect Dis. 1988;158:602–614. doi: 10.1093/infdis/158.3.602. [DOI] [PubMed] [Google Scholar]
- 36.Gerson SL. Cotransduction with MGMT and ubiquitous or erythroid-specific GFP lentiviruses allows enrichment of dual-positive hematopoietic progenitor cells in vivo. ISRN Hematol. 2012:2012. doi: 10.5402/2012/212586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.