Abstract
Tubulin is important for a wide variety of cellular processes including cell division, ciliogenesis, and intracellular trafficking. To perform these diverse functions, tubulin is regulated by post-translational modifications (PTM), primarily at the C-terminal tails of both the α- and β-tubulin heterodimer subunits. The tubulin C-terminal tails are disordered segments that are predicted to extend from the ordered tubulin body and may regulate both intrinsic properties of microtubules and the binding of microtubule associated proteins (MAP). It is not understood how either interactions with the ordered tubulin body or PTM affect tubulin’s C-terminal tails. To probe these questions, we developed a method to isotopically label tubulin for C-terminal tail structural studies by NMR. The conformational changes of the tubulin tails as a result of both proximity to the ordered tubulin body and modification by mono- and polyglycine PTM were determined. The C-terminal tails of the tubulin dimer are fully disordered and, in contrast with prior simulation predictions, exhibit a propensity for β-sheet conformations. The C-terminal tails display significant chemical shift differences as compared to isolated peptides of the same sequence, indicating that the tubulin C-terminal tails interact with the ordered tubulin body. Although mono- and polyglycylation affect the chemical shift of adjacent residues, the conformation of the C-terminal tail appears insensitive to the length of polyglycine chains. Our studies provide important insights into how the essential disordered domains of tubulin function.
Graphical abstract
Intrinsically disordered domain containing proteins (IDPs) possess protein regions without a fixed structural conformation. These proteins have the unique advantage that their structural architecture rapidly moves between many conformations. The collection of conformations in rapid equilibrium is called the conformational ensemble. The conformational ensembles sampled by IDPs are thought to be a key determinant of protein functions—impacting, for example, the protein accessibility for binding, size, and electrostatic distributions.1,2 Disordered proteins can operate via either a conformational-selection mechanism (in which the bound conformation is present in the disordered conformational ensemble) or an induced-fit-type mechanism (in which the final bound protein achieves a significantly different conformation than found in the disordered conformational ensemble).3,4 An example of the conformational selection mechanism is the disordered nuclear pore proteins containing phenylalanineglycine repeats (FG Nups), which adopt conformations primed for interactions with transport factors. This conformational-selection-type mechanism allows for diffusion-limited association of Nups and transport factors, enabling rapid transport into and out of the nucleus.5,6 Alternatively, in some systems, significant changes in conformation occur only after binding (induced fit), so that the final conformation does not significantly contribute to the original ensemble, greatly slowing the association time. The c-Myb interaction with the KIX domain of the CREB-binding protein involves a transient encounter complex that enables the large conformational change between the unbound disordered state and the bound state.7 The intrinsic flexibility of disordered proteins allows for significant advantages including speed of interaction and versatility to interact with a large number of binding partners. How IDPs perform their functions depends on their conformational ensemble in both the bound and unbound states.
Despite the importance of disordered protein conformational ensembles, the physical ingredients that determine these ensembles remain poorly understood. Several lines of evidence support the importance of protein sequence in determining the conformational ensemble. First, chemical shift values for IDPs, a measure of the conformational ensemble, show a relatively narrow distribution once corrected for effects of amino acid sequence.8 The chemical shift is the difference relative to a reference of the nuclear spin of an atom. Chemical shifts depend on the environment of an amino acid, including its chemical bonding, motion, and interactions with nearby atoms. For an IDP in rapid exchange between many different conformations, the NMR chemical shift depends on the average environment provided by an ensemble of interactions. The narrow distribution of chemical shifts in disordered proteins implies that the average environment is strongly constrained by the amino acid sequence, though interactions and modifications can have a significant effect. Second, the NMR spectrum of isolated IDP fragments is typically reproduced when that fragment is studied in the context of the full length protein.5,9 This means that the local sequence, rather than interactions between domains, is the primary determinant of the domain’s conformational ensemble. Third, IDPs typically lack intramolecular interactions of sufficient strength to bias the conformational ensemble.
However, if the ensemble is actively regulated in cells, then factors other than primary sequence should play a significant role in determining the accessible conformations. Some IDPs exhibit significant changes upon interaction with binding partners, even while remaining disordered and highly dynamic. Thus, proximity to another protein surface can be an important determinant of the IDP conformational ensemble. Moreover, these cases make clear that small changes in chemical shift values for disordered proteins can signify significant interaction. In addition to protein binding, post-translational modifications (PTM) influence the structure of disordered proteins. Phosphorylation of the 4E-BP2 IDP causes the disordered domain to fold into a well-defined structure.10 Thus, both protein association and post-translational modifications significantly influence the conformations of IDPs. However, our understanding of these binding events and modifications remains limited because of the difficulties in studying IDPs; for example spectral overlap leading to ambiguities in NMR and their inaccessibility by X-ray crystallography or electron microscopy.
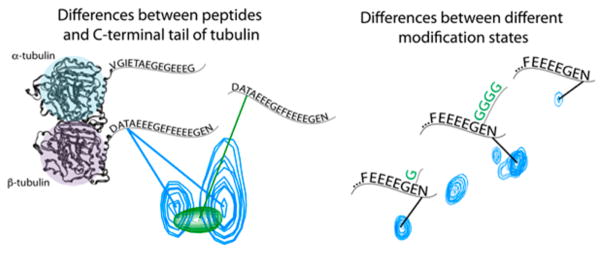
Microtubules are cytoskeletal polymers of α–β-tubulin heterodimers that form structural elements in all eukaryotic cells. The C-terminal tails of both the α- and β-tubulin monomers are important regulators of protein binding and are a primary site of PTM.11 The binding or activity of microtubule associated proteins (MAP) is altered when the C-terminal tails are cleaved by subtilisin or modified by PTM.12,13 Genetic deletion of the C-terminal tails from budding yeast sensitizes microtubules to destabilizing drugs and, in the case of the β-tubulin C-terminal tail, causes mitotic defects.14 In addition, glycylation of the tubulin C-terminal tails (see Figure 1) is generally found in the stable pool of tubulin residing in the cilia of Tetrahymena thermophila and is important for ciliary formation and function.15,16 Despite their importance and regulation, the intrinsic flexibility of the C-terminal tails of tubulin has hindered direct interrogation of their conformational ensemble, except in cases where the tails are bound to associated proteins. X-ray crystallography and cryo-electron microscopy reveal the conformation of the bound state of only a limited number of tubulin C-terminal tail residues in complex with a small number of binding partners.17–19 Several studies have used NMR to probe the conformations of peptides with a similar sequence to the C-terminal tails, though with limited chemical shift data or in an organic solvent.20–22 NMR has also been used to determine the structure of peptides bound to the CAP-Gly-2 domain of Clip-170.23
Figure 1.
NMR chemical shifts (peak position) of C-terminal tail residues differing from those same residues within peptides of the same sequences. Mono- and polyglycylation (green Gs) affect the chemical shift of adjacent residues but do not appear to have a significant influence on the conformational ensemble of the domain.
Until now, NMR has not been available for study of the C-terminal tails of the full tubulin dimer. This is because the quantities of heavy isotope labeled tubulin required for NMR studies have been limiting. Here, we use NMR to determine the effects of the adjacent ordered tubulin protein domain (which we call the “tubulin body”) on the disordered protein environment and of mono- and polyglycine PTM on the C-terminal tails of tubulin.11,24,25 We developed a technique for 15N, 13C labeling of tubulin, allowing for NMR studies of the C-terminal tails of tubulin in the context of the full dimer. The environment of the disordered tails, as determined by chemical shift values, differs from isolated C-terminal tail peptides, indicating that the interaction with the ordered tubulin body influences the conformation of the tubulin C-terminal tails. Moreover, the C-terminal tails show heterogenity in association with the tubulin body. Our work suggests that proximity to a protein surface significantly affects the disordered protein domain’s conformational ensemble and suggests that cells could regulate the ensemble by modulating accessibility to the adjacent protein surface. Finally, we show that mono- and polyglycine modifications appear to affect the chemical shift of residues adjacent to the modified residue, but not beyond. This suggests that glycine PTM do not have a significant effect on the tubulin C-terminal tails’ conformational ensemble. The biological regulation of tubulin function by mono- and polyglycylation could be due to blocking or creating binding surfaces, rather than directly modifying the C-terminal tail conformational ensemble.
RESULTS AND DISCUSSION
We purified polymerization-competent endogenous tubulin from T. thermophila using a TOG affinity column (Figure S1).26 A difficulty with endogenous tubulin purification is that the sample can be complex, containing a mixture of tubulin isotypes (unique sequences) and isoforms (unique PTM).25,27 To characterize the isotype and isoforms present in our samples, we performed liquid chromatography and tandem mass spectrometry (LC MS/MS) on both trypsin- (α-tubulin C-terminal tail) and chymotrypsin- (β-tubulin C-terminal tail) generated peptides. Although T. thermophila contain several tubulin isotypes, our growth and purification strategy produced a single isotype as detected by LC MS/MS (Atu1 and Btu1/2).27 The BTU1 and BTU2 genes encode for the same protein.27 Tubulin isolated using the TOG affinity column was entirely detyrosinated. Moreover, it contained primarily glycylated tubulin but no detectable glutamylation modifications. We presume that the TOG affinity purification strategy excludes glutamylated T. thermophila tubulin. A relatively homogeneous sample lacking C-terminal tail PTM has also been produced using a TOG purification strategy from human cells (tsA201).28,29 The lack of glutamylation in the purified T. thermophila tubulin was surprising, given that it was a prominent tubulin PTM in Tetrahymena cells30 and was not excluded by a TOG purification of Chlamydomonas tubulin.31 Our relatively homogeneous population of glycylated tubulin allowed us to quantify how the conformational ensemble was affected by the presence of glycylation.
To produce heavy labeled tubulin for NMR, we made use of the ability of T. thermophila to subsist by eating only bacteria. In their native environment, T. thermophila prey on bacteria and readily consume bacteria in culture. We grew bacteria on media containing the heavy isotopes 15N and 13C and then used these as the sole nitrogen and carbon source for a bacterized minimal media. T. thermophila cells were inoculated to this bacterized media at a cell density of 103 cells/mL and harvested when they grew to 105 cells/mL. Our labeling efficiency using 99% 15N or 13C starting material is expected to be 98% for every macromolecule within T. thermophila.
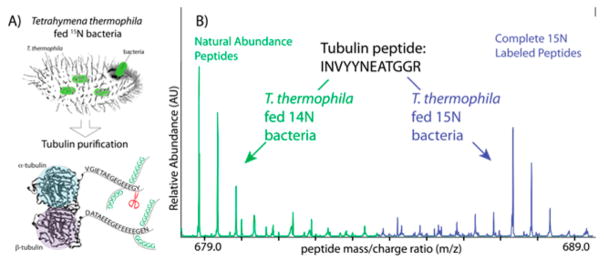
We confirmed the incorporation of 15N into our purified tubulin by mass spectrometry (Figure 2, blue peaks). Incorporation of 15C was expected to be similar and so was not independently confirmed. Figure 2 shows the spectra corresponding to a representative tubulin peptide (INVYYNEATGGR). The position of the peaks (mass to charge ratio, m/z) for each condition was determined by the amount of heavy isotope incorporation into the peptide. The green spectrum represent protein from T. thermophila containing natural abundance nitrogen and carbon (99.6% 14N and 98.9% 12C). The highest peaks correspond to the peptide containing entirely 14N and 12C atoms. We also observed peptides containing one or two atoms of either 13C or 15N, as expected from the small natural abundance of these isotopes. The blue spectra represent protein from T. thermophila fed 15N-labeled bacteria. We observed nearly complete labeling of the representative peptide with 15N. The highest blue peak shows that every nitrogen atom in the peptide was 15N labeled. As expected, we observed peptides containing one to two 14N atoms, since our starting material was only 99% 15N, as well as peaks from the natural abundance of 13C. In summary, 15N incorporation was essentially complete and comparable to that obtained in prokaryotic or yeast sources for protein production.32 As T. thermophila offers a robust protein overexpression system, this system should be useful for purification of a wide range of endogenous and exogenous proteins containing the isotopes necessary for NMR studies.33–35
Figure 2.
Labeling method and confirmation of heavy isotope incorporation. (A) We fed heavy bacteria grown on 15N to T. thermophila. We then purified endogenous T. thermophila tubulin; our protocol resulted in full incorporation of 15N into tubulin. (B) Mass spectrometry showing two degrees of incorporation of 15N into the tubulin peptide (INVYYNEATGGR). The highest green peak appears at the m/z position expected for the fully 14N, 12C peptide. The cluster of three peaks result from the incorporation of single heavy atoms, primarily 13C carbon, which has a natural abundance of 1.1%. In blue are the fully 15N labeled peptides, with a similar peptide distribution to the natural abundance peptides, but at a mass increased by the number of nitrogen atoms in the peptide.
To determine the average chemical environment and flexibility of the tubulin C-terminal tails attached to the tubulin body, we performed standard solution NMR experiments of the TOG-purified tubulin. These NMR experiments are specific to domains that rotate faster than a globular protein of approximately 40 kDa. The ~110 kDa tubulin dimer is too large for these types of experiments. The residues in the ordered tubulin body rotate slowly, and relaxation of the nuclear spins causes the corresponding peaks to be too broad to measure. However, regions of the tubulin dimer that are disordered or flexible rotate much faster than the macromolecular tumbling time, have reduced relaxation, and therefore appear as sharp, defined peaks in the spectrum. We observed the C-terminal tails in the 2D (H–N) plane of a 3D HNCO spectrum taken on 15N, 13C labeled tubulin dimers (Figure 3). In this projection, directly bonded H–N pairs (i.e., from backbone amines) appear as a single peak with the 1H chemical shift on the x-axis and the 15N chemical shift on the y-axis. The chemical shift is the difference in resonance frequency of each atom from the standard and gives information about the average environment. In this case, the peak positions were fully consistent with the supposition that the C-terminal tails are disordered.
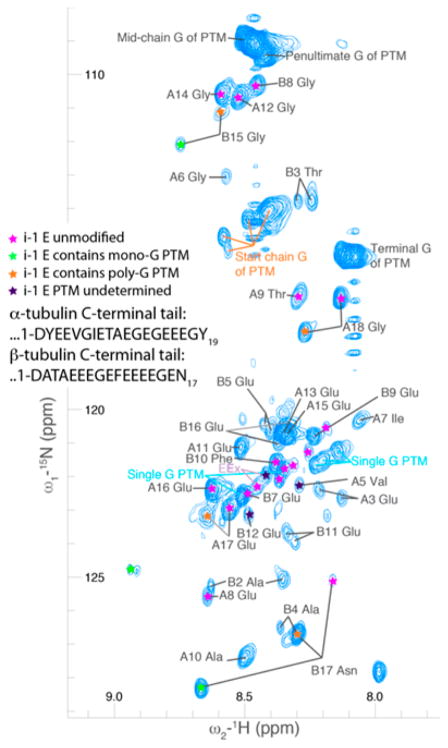
Figure 3.
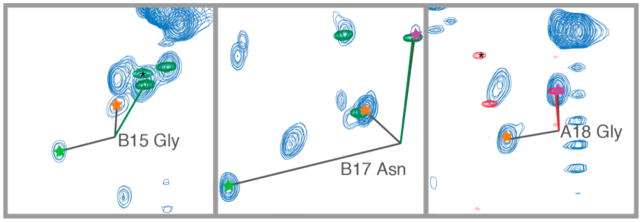
A 2D projection of a 3D HCNO spectrum of the C-terminal tails of tubulin in the context of the full TOG-purified tubulin dimer. Several modification states are represented (denoted by colored stars), resulting in more than one peak for several residues. Distinct clusters of peaks appeared for glycine residues in polyglycine modifications (the beginning, middle, penultimate, and final residues of the chain) as well as for monoglycine modifications. The residues at the beginning of the chain (those directly conjugated to a glutamate) were separated into two clusters, corresponding to monoglycine (light blue text) and polyglycine (orange text) modifications. Peaks are numbered relative to the start of the C-terminal tail, as indicated in the inset; the corresponding residues are β-tubulin D427 (B1) and α-tubulin D431 (A1) in the T. thermophila sequences for BTU1 and ATU1, respectively.
In order to assign which peak corresponds to which residue in the protein C-terminal tail, we utilized standard 3D NMR experiments to link sequential residues (HNCO, HNcaCO, HNCACB and CBCAcoNH, and HNN) on TOG-purified, 15N,13C labeled tubulin, to correlate each peak with residues on the C-terminal tails (Figure 5). We labeled the peaks beginning with β-tubulin D427 (B1) and α-tubulin D431 (A1) in the T. thermophila sequences for Btu1 and Atu1, respectively. Because the peaks corresponding to the repeated glutamate residues were similar, we were unable to resolve which of the peaks labeled EEx in Figure 3 (purple text) corresponded to amino acids B13 Glu or B14 Glu in the β-tubulin C-terminal tail. We were not able to resolve this ambiguity using carbon detection experiments due to the lack of sufficiently concentrated samples for these relatively insensitive experiments.36 Neighbor corrected chemical shift predictions8 indicated that the C-terminal tails have a slight propensity for β-sheet formation (Figure S2). This result is in contrast to computer simulations showing a bias of the C-terminal tails of human tubulin isoforms toward α-helical conformations when simulated in isolation.37 This could mean that the C-terminal tails would more readily bind proteins whose binding pocket constrains them to a β-sheet like conformation.
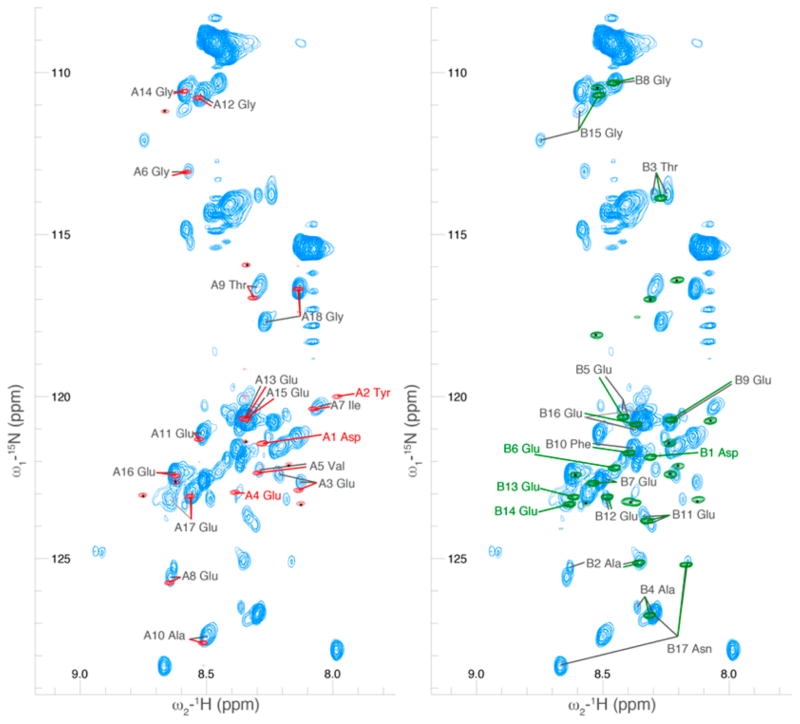
Figure 5.
A comparison of the 2D projection of a 3D HCNO spectrum of the C-terminal tails of tubulin in the context of the full TOG-purified dimer as compared to bacterially produced GST-fusion peptides of the C-terminal tail sequences. The spectrum of the GST-α fusion peptide is shown in red, overlaid with the tubulin spectrum; similarly, the spectrum of the GST-β fusion peptide is shown in green, overlaid with the tubulin spectrum. Significant differences between the tubulin and peptide spectrum could be due to the presence of post-translational modification on our endogenous tubulin samples or due to interactions of the C-terminal tails with the rest of the tubulin dimer. As the majority of our endogenous sample was unmodified, we expect interactions with the rest of the dimer played the dominant role. Peaks with a small black asterisk have been assigned to the linker region between the peptides and GST and thus do not overlay with the C-terminal tails.
In our NMR spectra, we identified peaks corresponding to a variety of modifications. We observed distinct peaks for monoglycine modifications. For polyglycine chains, we observed distinct peaks for the first, penultimate, and terminal glycine residues (Figure 3). All other glycines in the polyglycine chains (from the second glycine to the third to last in the chain) appeared as one cluster. The peaks from the monoglycine modifications were shifted downfield relative to the typical nitrogen chemical shift values for random coil glycine residues. The most dispersed peaks corresponded to glycine residues that were directly attached to the C-terminal tails, as expected, since these represent modifications of different C-terminal tail amino acids. However, we were unable to identify to which amino acids each particular glycine was conjugated.
For those amino acids with an i − 1 glutamate neighbor, we were able to directly identify the modification state of that neighbor (stars in Figure 3). This identification was possible because the modified side chain Cγ was shifted relative to the unmodified case, and this shift was different for mono- and polyglycine chains (Figure S3). For example, the terminal residues of both α- and β-tubulin C-terminal tails were present in our spectrum in multiple peaks, each corresponding to a different modification state of the i − 1 glutamate neighbor. The significant intensity of the modified form of these residues is consistent with our mass spectrometry results showing that the most C-terminal glutamate residues were the primary sites of modification. Glycylation had a significant effect on the chemical environment of neighboring residues, as expected due to nearest-neighbor interactions in the disordered domain of the tubulin protein. The relative simplicity of our spectra excluded large-scale conformational changes as a result of differing levels of polyglycylation. An extreme conformational change would be for the protein to fold into an ordered state upon PTM, as seen upon the phosphorylation of the 4E-BP2 IDP.10 In contrast, several other disordered proteins show only minimal changes upon post-translational modification.38,39 The C-terminal tails remained disordered upon glycylation and did not appear to show any shift in the conformation ensemble of the disordered domain upon modification. This spectrum would have many dispersed peaks corresponding to the range of modification states if the conformation depended strongly on the degree of modification. For example, the peak corresponding to the polyglycine modification on B17 or A18 would be a broad smear rather than a relatively sharp peak, because the sample include a wide range of polyglycine chain lengths (Table S2). However, we did not see this, indicating that polyglycylation does not appear to regulate the tubulin C-terminal tail’s conformational ensemble. However, mono- and polyglycine modifications do modulate tubulin function.29 T. thermophila containing mutations in the C-terminal tail residues that are modified, or deletions of the enzymes that add mono-and polyglycine modifications, exhibit ciliogenesis defects.15 We hypothesize that these glycine modifications affect MAP binding by blocking or creating binding surfaces, rather than directly modifying the C-terminal tail conformational ensemble.
We next probed the degree and location of PTM using LC MS/MS (Figure 4, Table S2). We detected detyrosinated α-tubulin, consistent with our NMR results and previous observations.29,40 As determined from label-free quantification, approximately 50% of the α-tubulin peptide in our sample was otherwise unmodified, with the remainder being both detyrosinated and glycylated. Glycylation was observed at levels ranging from 1 to 40 glycines for α-tubulin, and 2–26 glycines for β-tubulin. We used MS/MS to localize the glycine modifications for a range of modification states (Table S2). The primary glycine modifications of α-tubulin occurred on the final two glutamate residues (A16 Glu/A17 Glu). However, additional glycines were detected on the A15 Glu and A13 Glu for peptides with greater than 4 and 20 additional glycines, respectively. The glycylation on β-tubulin was dispersed over the final four glutamate residues. This direct mapping of glycylation on T. thermophila C-terminal tails is consistent with observations of polyglutamylation in Paramecium41 and mutagenesis studies of polyglycylation in Tetrahymena.16,29
Figure 4.
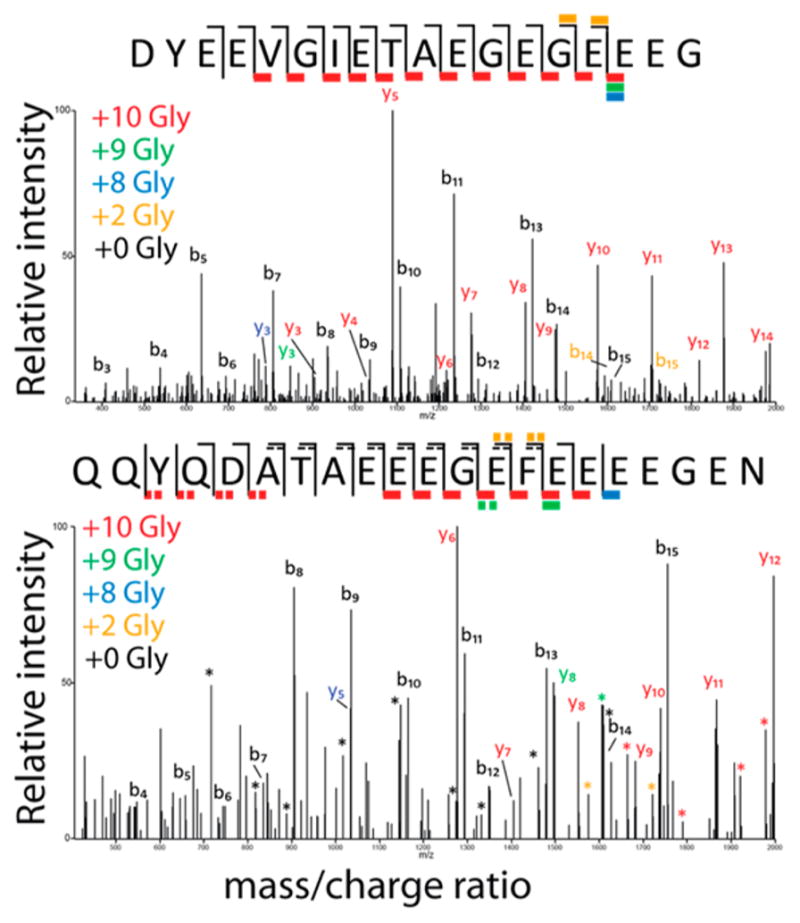
Tandem mass spectrometry (MS-MS) spectra using CID fragmentation of α- (top) and β-tubulin(bottom) C-terminal tail peptides, each modified with 10 glycine residues. Fragments containing the C-terminus are “y” ions, while those containing the N-terminus are labeled as “b” ions. The fragments are color coded according to how many glycine residues were present on that fragment. For the α-tubulin peptide, most modifications occurred on the final two glutamic acid residues, while the modifications were more dispersed between the final six residues for β-tubulin.
Our mass spectrometry results and NMR results were consistent, showing both mono- and polyglycylation at a variety of modification sites. Interestingly, we observed differences in the localization and extent of modification as measured by NMR and mass spectrometry. For example, we observed a significant fraction of monoglycine on the penultimate amino acid of β-tubulin. This was, at first glance, at odds with the mass spectrometry results, where we did not detect monoglycine modifications of the β-tubulin C-terminal tail. It is possible, though unlikely, that this particular peptide had low sensitivity in the mass spectrometry, because the peptide with three glycine additions was apparent. Our results suggest that particular glutamate is only monoglycated when other residues on the chain are also glycylated. In contrast, we did not detect by NMR a monoglycine addition on the penultimate glutamate of α-tubulin, even though the mass spectrometry strongly suggested that this modification occurred. It could be that this species was not sufficiently abundant in our sample to be detected by NMR, which is not sensitive to low-abundance states. NMR provides valuable complementary information on the degree and position of post-translational modifications, especially in situations where the modification is hard to precisely localize by mass spectrometry.
In contrast to the small effect of glycylation on the conformational ensemble, our NMR results suggested that the ordered tubulin body significantly influences the conformational ensemble of the C-terminal tail. Several residues (B2–5, A3) primarily near the ordered domain of β-tubulin were clearly assignable to two distinct sets of peaks of approximately equal intensity, suggesting that they existed in two distinct conformational ensembles. These groups did not correlate with specific PTM on the tail. Moreover, they were relatively far from the mono- and polyglycine modifications, with intervening residues apparent as only a single peak. This indicates that the two different conformations are more likely to be a result of interactions with the surface of the tubulin body, rather than a result of C-terminal tail glycylation. A lower bound on the exchange rate between the two conformations was determined from the difference in chemical shift between the two peaks. Based on analysis of the B3 threonine (Figure 4), the exchange rate between these two states was slower than 30 Hz, or the lifetime of each state was greater than 30 ms. This is slower than would be expected for any local conformational exchange, but could be consistent with ligand binding or a stable modification.42 It is not expected that these differences were a result of acetylation, as no structural differences have been found using cryo-electron microscopy to compare modified and unmodified tubulins.43 However, this cryo-EM work was of lower resolution than more recent studies that revealed conformational differences in tubulin as a function of nucleotide state.44,45
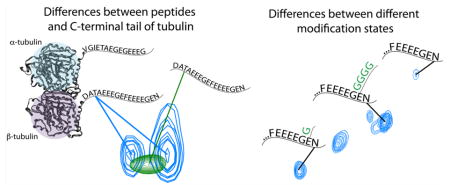
The suggestion that the C-terminal tails might be interacting with the ordered tubulin body led us to compare the C-terminal tails in the context of the dimer to bacterially produced peptides. We hypothesized that the peptides would overlay with unmodified residues of the C-terminal tails, because of the strong dependence of disordered-protein chemical shift on protein amino acid sequence.8 In contrast, transient interactions with the ordered tubulin body would result in differences in chemical shifts between the C-terminal tails and the peptides. Such changes could also arise from the differences in sequence of the peptides, for example in the linker regions used to attach the peptides to the GST tag, though we aimed to design our GST fusion constructs to be sufficiently long to reduce this possibility.
We performed standard 2D and 3D NMR experiments on bacterially produced GST-peptide fusions (Figures 5 and 6). The NMR spectra of the peptides were unchanged when the GST was removed by thrombin cleavage (data not shown). This indicates that the peptides did not have significant interactions with GST. This may also indicate that the C-terminal tails did not have strong self-interactions, as GST forms a dimer. However, the two tails could also be sufficiently far away to prohibit this interaction. In general, the spectra of the C-terminal tails and the peptides were similar; the peak corresponding to a particular residue on the C-terminal tails was near that same residue’s peak on the peptides (Figure 3). This was expected, given the strong effect of amino acid neighbors on disordered protein chemical shift values.8 Among residues represented by multiple peaks in the NMR spectrum due to different modification states, the isolated peptides overlaid most closely with those peaks corresponding to residues with unmodified i − 1 neighbors. This strengthens our supposition that a significant portion of our signal is from the fraction of nonglycylated tubulin.
Figure 6.
Insets of the spectra of Figure 5 with modification states of previous residue annotated, comparing the chemical shifts of C-terminal tails of tubulin with GST-fusion peptides. The peptides overlaid closely with the peaks corresponding to the unmodified form of the C-terminal tail residue. We observed significant chemical shift changes due to the change in the local chemical environment of the residue due to the presence of glycine modifications. Modifications are indicated by stars as in Figure 4.
Several of the residues showed significant differences in chemical shift values of the tubulin dimer as compared to the tubulin peptide. As the C-terminal tails are adjacent in sequence to terminal α-helical segments of the ordered tubulin body, we considered whether the C-terminal tails in the context of the dimer might be biased toward α-helical conformations due to their proximity. However, as discussed above, secondary structural predictions indicate that the C-terminal tails’ conformational ensembles are biased toward β-sheet rather than α-helical conformations. Moreover, there is not a significant shift toward α-helical conformations of the C-terminal tail residues linked to the tubulin dimer as compared to the isolated peptide. For example, the value of δCα – δCβ was not shifted measurably higher, as would be expected for a more α-helical structure.46
The chemical shift changes we observed were small when compared to differences in shift values for ordered proteins undergoing large conformational changes or disordered proteins that become ordered or immobilized upon interaction.47 However, these differences were similar to those seen upon binding in several other disordered systems48–50 and larger than those for disordered protein domains that are nearby, but not interacting with another protein or domain.5,6,9 For example, focusing on the amide proton and nitrogen chemical shifts, our average magnitude of chemical shift change was ΔδHN = 0.02 ppm for the proton and ΔδN = 0.08 ppm for nitrogen chemical shifts. The average of the weighted total shift change (〈Δδtotal〉 = 〈[(WHΔδH)2 + (WNΔδN)2]1/2〉, where WH = 1 and WN = 0.2) was 〈Δδtotal〉 = 0.03 ppm. These shifts were more than twice that seen for either the interaction of PAGE4 with c-Jun48 or the interaction of Pin1 with c-Myc.49 Though their shift was not quantified, it was similar in appearance to that for the tail domain of Nipah virus nucleoprotein interacting with the x-domain.50 In contrast, disordered proteins’ chemical shifts can remain unchanged when the domain is fused to a larger protein or macromolecular complex. For example, all residues that remain visible upon formation of a measles nucleocapsid retain the chemical shifts of the isolated tail domain.9 Additionally, the FG Nups that form the selective barrier to nucleocytoplasmic transport show virtually no shifts aside from the FG repeats that bind to transport factors.5,6 Thus, in comparison to other disordered protein systems, the chemical shift differences we observed when comparing the spectrum of the C-terminal tails to the spectra of the peptides were consistent with a transient protein–protein interaction between the C-terminal tails and the ordered tubulin body.
We propose that transient interactions with the ordered dimer body are responsible for both the slight differences in chemical shift between the C-terminal tails and the peptides as well as for the two conformational ensembles present in residues B2–5, A3 (see Figure 7). These observed chemical shift differences are consistent with simulations showing differences in the conformation ensemble of the C-terminal tails in isolation as compared with the full dimer.51 Direct interactions have also been predicted between the C-terminal tail and charged regions of the tubulin body, because of the high charge of the C-terminal tails.52 Our results indicate that the C-terminal tail do not simply project unhindered from the tubulin body surface, but are in close proximity to the surface to engage in transient interactions with the tubulin body.
Figure 7.
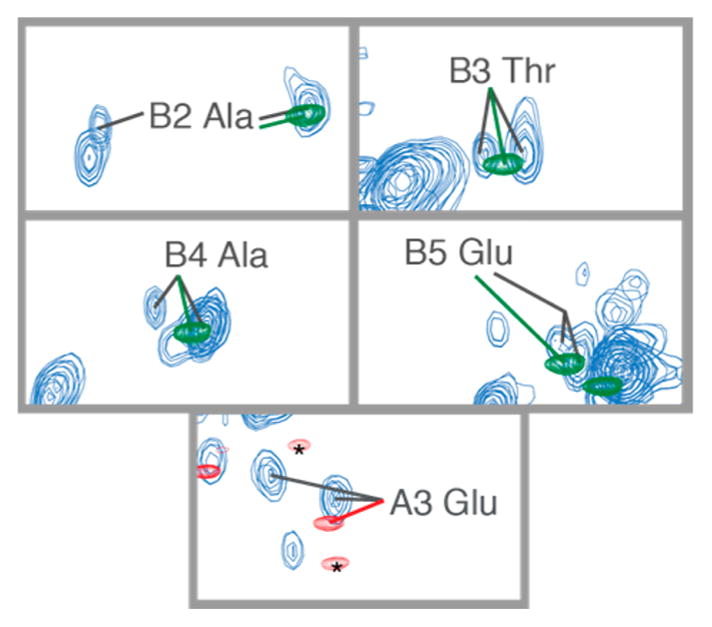
Residues closest to the ordered tubulin body that appeared to be present in two conformational ensembles, presumably due to interaction with the tubulin body surface. These residues appeared in the spectrum as two peaks, neither of which overlayed exactly with the peptide peak.
CONCLUSION
Our experiments demonstrated the successful production of 15N,13C labeled, full length tubulin for analysis by NMR. We focused on the C-terminal tails of α- and β-tubulin, which remained disordered even as part of the full tubulin heterodimer and were not adhered to the dimer surface, as has been proposed.52 The chemical shift values for the C-terminal tails were consistent with other disordered proteins, with a slight propensity for β-sheets (Figure S2). All of our sample was detyrosinated, with additional glycine PTM on the C-terminal tails of α- and β-tubulin (Figures 3 and 4, Table S2). These modifications had large effects on the chemical environment of adjacent residues, but did not appear to cause large conformational changes. Isolated peptides showed chemical shifts significantly different from the C-terminal tails attached to the tubulin body, with differences as large in magnitude as those for binding partner interactions in other disordered protein systems. We observed two distinct conformational ensembles for β-tubulin residues B2–5 and α-tubulin residue A3. This suggests that the C-terminal tails are sensitive to the tubulin body conformation or ligand binding state, and so studies of the tail using only peptides should be interpreted with caution.
The C-terminal tails impact tubulin and MT function, including both intrinsic and extrinsic properties (polymerization rates,53 bending rigidit,y54 and binding of MAP13,55). Several models could explain this behavior. First, charge-based repulsion between the tails of adjacent dimers could alter both polymerization dynamics and bending rigidity. For example, increasing microtubule curvature would bring some tails closer together, which might be energetically unfavorable. However, in physiological buffers, the length scale over which the electric field decays (the Debye screening length) is ~1 nm,56 sufficiently small to raise questions about the potential impact of tail–tail charge–charge interactions for unmodified tails. We hypothesize that heavily polyglutamylated microtubules may exhibit such charge repulsion between tails. Second, changes in the tail could induce conformational changes in the ordered dimer. However, electron microscopy studies have not detected any such changes.17,18 Our data suggest a third possibility; the tails could interact with the tubulin body on adjacent dimers (Figure 8), providing additional stabilizing interactions. This has been proposed to be a mechanism for the C-terminal-tail-dependent variability in bending rigidity at high salt concentrations;54 our observations indicate that this this mechanism is still at play even in low salt. Tail–body interactions could occlude the interface between adjacent tubulin dimers, which would have the most pronounced effect on modulating polymerization rates. We propose that the C-terminal tail modulates intrinsic tubulin behavior through interactions with the dimer surface.
Figure 8.
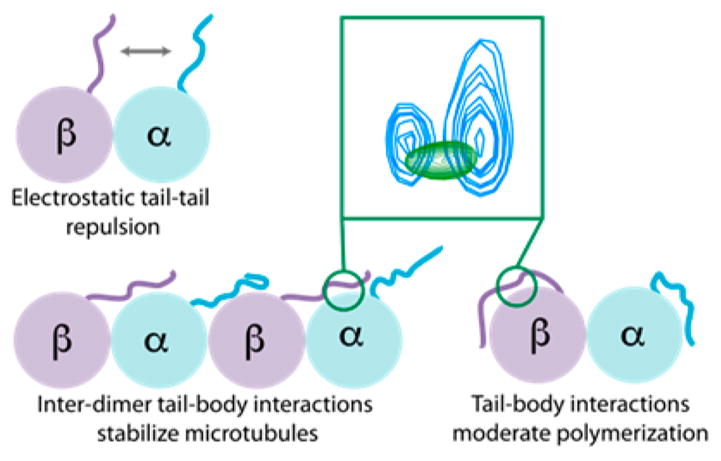
Tubulin C-terminal tails, which can modulate intrinsic properties of microtubules through their intertail electrostatic repulsion and inter- and intradimer tail–body interactions. Our data suggest a significant role for tail–body interactions.
It has been proposed that the conformational ensemble of a disordered protein is important for its cellular function and might be regulated in cells.57 In this work, we found that the conformational ensemble of the disordered tubulin C-terminal tails is relatively insensitive to the level of glycylation but appeared to be sensitive to the presence of the ordered tubulin body. This suggests that C-terminal tail conformation could be regulated by proteins which obscure the sites of interaction on the tubulin body surface. Moreover, the C-terminal tails’ conformational ensembles appear sensitive to either ligand binding or the conformational state of the tubulin body, acting as another mechanism to regulate binding of MAP. More broadly, the cellular regulation of disordered proteins through transient interactions could prove an important mechanism for their regulation.
METHODS
We used progeny from a cross between the B2086 and CU428 strains as the T. thermophila strain (a gift from Mark Winey). T. thermophila were grown initially in SPP (2% protease peptone, 0.2% glucose, 0.1% yeast extracts and 0.003% EDTA ferric salts) then inoculated at a concentration of 103 cells/mL into a variant of CDMA media58 lacking all amino acids, sugars, and nucleotides, but containing the cell pellet from a confluent bacterial culture half of the volume of the T. thermophila culture (grown in M9 complete), as well as penicillin and streptomycin. Cells were grown for 24–48 h, shaking gently at 30 °C until they reached a cell density greater than 105 cells/mL, harvested by centrifugation, and snap frozen and stored at −70 °C until further use.
Tubulin was purified on a TOG affinity column as described previously.26 An NHS-activated column was conjugated with GST-TOG protein. T. thermophila cell pellets were lysed by sonication in BRB80 buffer containing protease inhibitors (PMSF [0.5 mM], benzamadine [1 mM] and leupeptin [25 μg/mL]) and passed over the TOG affinity column. The column was washed with BRB80 containing 1 mM GTP (wash 1), 4.6 mM ATP, 1 mM GTP and 10 mM MgCl2 (wash 2), 1 mM GTP (wash 3), and BRB80 with 10% glycerol and 0.1% Tween 20 (wash 4). The protein was eluted in buffer A (5 mM MES, 0.25 mM EGTA and 0.25 mM MgCl2) with 380 mM ammonium sulfate. The protein was immediately dialyzed into buffer A overnight and concentrated in a spin column. Protease inhibitors and GTP were added to a final concentration of 1 mM GTP. We typically obtained 3 mg of tubulin from 8 L of T. thermophila cell culture (4 L of bacterial culture).
Synthetic tail peptides were produced in BL21 DE3 bacterial cells as GST fusion constructs. The protein sequences after thrombin cleavage are α-peptide, GSEKDYEEVGIETAEGEGEEEG; β-peptide, GSPNSRVDYQDATAEEEGEFEEEGEN. NMR assignment experiments were performed in buffer A on the full GST fusion protein.
All NMR experiments were performed on Agilent 800 MHz VNMRS or 600 MHz INOVA spectrometers using standard BioPac experiments utilizing nonuniform sampling. Nonuniform sampling reconstructions were performed using software from the Wagner lab.59
Mass spectrometry samples were digested with either trypsin (α-tubulin) or chymotrypsin (β-tubulin) and eluted from a Waters nanoAcquity UPLC column directly onto a Thermo Fisher Scientific Orbitrap Velos for MS-MS analysis.
Supplementary Material
Acknowledgments
The authors would like to thank Brian Bayless for the T. thermophila image in Figure 2 and Deborah Wuttke, Art Pardi, and Richard Shoemaker for advice and support. We thank Mark Winey and Alexander Stemm-Wolf for advice and T. thermophila strains, J. Richard McIntosh for advice on tubulin purifications, and William Old for assistance with mass spectrometry. K.P.W. was supported in part by the Interdisciplinary Quantitative Biology (IQ Biology) program at the BioFrontiers Institute, University of Colorado, Boulder (NSF IGERT 1144807). L.E.H. acknowledges the Boettcher Foundation for a Webb-Waring Biomedical Research Award, the University of Colorado Innovative Seed Grant, and the University of Colorado for start-up funds. C.G.P. is funded by NIH-NIGMS GM099820, the Pew Biomedical Scholars Program, and the Boettcher Foundation. The 600 and 800 MHz spectrometers were purchased and supported by the NIH (RR011969, RR16649) and the NSF (DBI-0230966, 960241).
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.6b00507.
Experimental procedures, NMR chemical shift values, secondary structure prediction, and mass spectrometry data (PDF)
References
- 1.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein–protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Borg M, Mittag T, Pawson T, Tyers M, Forman-Kay JD, Chan HS. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci U S A. 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uversky VN. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev. 2011;40:1623. doi: 10.1039/c0cs00057d. [DOI] [PubMed] [Google Scholar]
- 4.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 5.Hough LE, Dutta K, Sparks S, Temel DB, Kamal A, Tetenbaum-Novatt J, Rout MP, Cowburn D. The molecular mechanism of nuclear transport revealed by atomic scale measurements. eLife. 2015;4:e10027. doi: 10.7554/eLife.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milles S, Mercadante D, Aramburu IV, Jensen MR, Banterle N, Koehler C, Tyagi S, Clarke J, Shammas SL, Blackledge M, Gräter F, Lemke EA. Plasticity of an Ultrafast Interaction between Nucleoporins and Nuclear Transport Receptors. Cell. 2015;163:734. doi: 10.1016/j.cell.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gianni S, Morrone A, Giri R, Brunori M. A folding-after-binding mechanism describes the recognition between the transactivation domain of c-Myb and the KIX domain of the CREB-binding protein. Biochem Biophys Res Commun. 2012;428:205–209. doi: 10.1016/j.bbrc.2012.09.112. [DOI] [PubMed] [Google Scholar]
- 8.Tamiola K, Acar B, Mulder FAA. Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J Am Chem Soc. 2010;132:18000–18003. doi: 10.1021/ja105656t. [DOI] [PubMed] [Google Scholar]
- 9.Jensen MR, Communie G, Ribeiro EA, Martinez N, Desfosses A, Salmon L, Mollica L, Gabel F, Jamin M, Longhi S, Ruigrok RWH, Blackledge M. Intrinsic disorder in measles virus nucleocapsids. Proc Natl Acad Sci U S A. 2011;108:9839–9844. doi: 10.1073/pnas.1103270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519:106–109. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- 11.Roll-Mecak A. Intrinsically disordered tubulin tails: complex tuners of microtubule functions? Semin Cell Dev Biol. 2015;37:11–19. doi: 10.1016/j.semcdb.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacroix B, van Dijk J, Gold ND, Guizetti J, Aldrian-Herrada G, Rogowski K, Gerlich DW, Janke C. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J Cell Biol. 2010;189:945–954. doi: 10.1083/jcb.201001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aiken J, Sept D, Costanzo M, Boone C, Cooper JA, Moore JK. Genome-wide Analysis Reveals Novel and Discrete Functions for Tubulin Carboxy-Terminal Tails. Curr Biol. 2014;24:1295–1303. doi: 10.1016/j.cub.2014.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wloga D, Webster DM, Rogowski K, Bré MH, Levilliers N, Jerka-Dziadosz M, Janke C, Dougan ST, Gaertig J. TTLL3 Is a Tubulin Glycine Ligase that Regulates the Assembly of Cilia. Dev Cell. 2009;16:867–876. doi: 10.1016/j.devcel.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Xia L, Hai B, Gao Y, Burnette D, Thazhath R, Duan J, Bré M-H, Levilliers N, Gorovsky MA, Gaertig J. Polyglycylation of Tubulin Is Essential and Affects Cell Motility and Division in Tetrahymena thermophila. J Cell Biol. 2000;149:1097–1106. doi: 10.1083/jcb.149.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alushin GM, Musinipally V, Matson D, Tooley J, Stukenberg PT, Nogales E. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat Struct Mol Biol. 2012;19:1161–1167. doi: 10.1038/nsmb.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garnham CP, Vemu A, Wilson-Kubalek EM, Yu I, Szyk A, Lander GC, Milligan RA, Roll-Mecak A. Multivalent Microtubule Recognition by Tubulin Tyrosine Ligase-like Family Glutamylases. Cell. 2015;161:1112–1123. doi: 10.1016/j.cell.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szyk A, Deaconescu AM, Piszczek G, Roll-Mecak A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat Struct Mol Biol. 2011;18:1250–1258. doi: 10.1038/nsmb.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jimenez MA, Evangelio JA, Aranda C, Lopez-Brauet A, Andreu D, Rico M, Lagos R, Andreu JM, Monasterio O. Helicity of alpha(404–451) and beta(394–445) tubulin C-terminal recombinant peptides. Protein Sci Publ Protein Soc. 1999;8:788–799. doi: 10.1110/ps.8.4.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otter A, Kotovych G. The solution conformation of the synthetic tubulin fragment Ac-tubulin-α(430–441)-amide based on two-dimensional ROESY experiments. Can J Chem. 1988;66:1814–1820. [Google Scholar]
- 22.Sugiura M, Maccioni RB, Cann JR, York EJ, Stewart JM, Kotovych G. A Proton Magnetic Resonance and a Circular Dichroism Study of the Solvent Dependent Conformation of the Synthetic Tubulin Fragment Ac Tubulin, Alpha (430–441) Amide and its Interaction with Substance-P. J Biomol Struct Dyn. 1987;4:1105–1117. doi: 10.1080/07391102.1987.10507701. [DOI] [PubMed] [Google Scholar]
- 23.Mishima M, Maesaki R, Kasa M, Watanabe T, Fukata M, Kaibuchi K, Hakoshima T. Structural basis for tubulin recognition by cytoplasmic linker protein 170 and its autoinhibition. Proc Natl Acad Sci U S A. 2007;104:10346–10351. doi: 10.1073/pnas.0703876104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janke C. The tubulin code: Molecular components, readout mechanisms, and functions. J Cell Biol. 2014;206:461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verhey KJ, Gaertig J. The Tubulin Code. Cell Cycle. 2007;6:2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- 26.Widlund PO, Podolski M, Reber S, Alper J, Storch M, Hyman AA, Howard J, Drechsel DN. One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol Biol Cell. 2012;23:4393–4401. doi: 10.1091/mbc.E12-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pucciarelli S, Ballarini P, Sparvoli D, Barchetta S, Yu T, Detrich I, Miceli C. Distinct Functional Roles of β-Tubulin Isotypes in Microtubule Arrays of Tetrahymena thermophila, a Model Single-Celled Organism. PLoS One. 2012;7:e39694. doi: 10.1371/journal.pone.0039694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vemu A, Garnham CP, Lee D-Y, Roll-Mecak A. Generation of differentially modified microtubules using in vitro enzymatic approaches. Methods Enzymol. 2014;540:149–166. doi: 10.1016/B978-0-12-397924-7.00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redeker V, Levilliers N, Vinolo E, Rossier J, Jaillard D, Burnette D, Gaertig J, Bré M-H. Mutations of Tubulin Glycylation Sites Reveal Cross-talk between the C Termini of α- and β-Tubulin and Affect the Ciliary Matrix in Tetrahymena. J Biol Chem. 2005;280:596–606. doi: 10.1074/jbc.M408324200. [DOI] [PubMed] [Google Scholar]
- 30.Garnham CP, Roll-Mecak A. The chemical complexity of cellular microtubules: Tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton. 2012;69:442–463. doi: 10.1002/cm.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alper JD, Decker F, Agana B, Howard J. The motility of axonemal dynein is regulated by the tubulin code. Biophys J. 2014;107:2872–2880. doi: 10.1016/j.bpj.2014.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi H, Shimada I. Production of isotopically labeled heterologous proteins in non-E. coli prokaryotic and eukaryotic cells. J Biomol NMR. 2010;46:3–10. doi: 10.1007/s10858-009-9377-0. [DOI] [PubMed] [Google Scholar]
- 33.Aldag I, Bockau U, Rossdorf J, Laarmann S, Raaben W, Herrmann L, Weide T, Hartmann MW. Expression, secretion and surface display of a human alkaline phosphatase by the ciliate Tetrahymena thermophila. BMC Biotechnol. 2011;11:11. doi: 10.1186/1472-6750-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boldrin F, Santovito G, Gaertig J, Wloga D, Cassidy-Hanley D, Clark TG, Piccinni E. Metallothionein Gene from Tetrahymena thermophila with a Copper-Inducible-Repressible Promoter. Eukaryotic Cell. 2006;5:422–425. doi: 10.1128/EC.5.2.422-425.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boldrin F, Santovito G, Formigari A, Bisharyan Y, Cassidy-Hanley D, Clark TG, Piccinni E. MTT2, a copper-inducible metallothionein gene from Tetrahymena thermophila. Comp Biochem Physiol, Part C: Toxicol Pharmacol. 2008;147:232–240. doi: 10.1016/j.cbpc.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Sahu D, Bastidas M, Showalter SA. Generating NMR chemical shift assignments of intrinsically disordered proteins using carbon-detected NMR methods. Anal Biochem. 2014;449:17–25. doi: 10.1016/j.ab.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luchko T, Torin Huzil J, Stepanova M, Tuszynski J. Conformational Analysis of the Carboxy-Terminal Tails of Human β-Tubulin Isotypes. Biophys J. 2008;94:1971–1982. doi: 10.1529/biophysj.107.115113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liokatis S, Dose A, Schwarzer D, Selenko P. Simultaneous Detection of Protein Phosphorylation and Acetylation by High-Resolution NMR Spectroscopy. J Am Chem Soc. 2010;132:14704–14705. doi: 10.1021/ja106764y. [DOI] [PubMed] [Google Scholar]
- 39.Okuda M, Nishimura Y. Real-time and simultaneous monitoring of the phosphorylation and enhanced interaction of p53 and XPC acidic domains with the TFIIH p62 subunit. Oncogenesis. 2015;4:e150. doi: 10.1038/oncsis.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wloga D, Rogowski K, Sharma N, van Dijk J, Janke C, Eddé B, Bré MH, Levilliers N, Redeker V, Duan J, Gorovsky MA, Jerka-Dziadosz M, Gaertig J. Glutamylation on α-Tubulin Is Not Essential but Affects the Assembly and Functions of a Subset of Microtubules in Tetrahymena thermophila. Eukaryotic Cell. 2008;7:1362–1372. doi: 10.1128/EC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vinh J, Langridge JI, Bré MH, Levilliers N, Redeker V, Loyaux D, Rossier J. Structural Characterization by Tandem Mass Spectrometry of the Posttranslational Polyglycylation of Tubulin. Biochemistry. 1999;38:3133–3139. doi: 10.1021/bi982304s. [DOI] [PubMed] [Google Scholar]
- 42.Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochim Biophys Acta, Proteins Proteomics. 2011;1814:942–968. doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howes SC, Alushin GM, Shida T, Nachury MV, Nogales E. Effects of tubulin acetylation and tubulin acetyltransferase binding on microtubule structure. Mol Biol Cell. 2014;25:257–266. doi: 10.1091/mbc.E13-07-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nogales E, Zhang R. Visualizing microtubule structural transitions and interactions with associated proteins. Curr Opin Struct Biol. 2016;37:90–96. doi: 10.1016/j.sbi.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue Y, Yuwen T, Zhu F, Skrynnikov NR. Role of Electrostatic Interactions in Binding of Peptides and Intrinsically Disordered Proteins to Their Folded Targets. 1. NMR and MD Characterization of the Complex between the c-Crk N-SH3 Domain and the Peptide Sos. Biochemistry. 2014;53:6473–6495. doi: 10.1021/bi500904f. [DOI] [PubMed] [Google Scholar]
- 48.He Y, Chen Y, Mooney SM, Rajagopalan K, Bhargava A, Sacho E, Weninger K, Bryan PN, Kulkarni P, Orban J. Phosphorylation-induced Conformational Ensemble Switching in an Intrinsically Disordered Cancer/Testis Antigen. J Biol Chem. 2015;290:25090–25102. doi: 10.1074/jbc.M115.658583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helander S, Montecchio M, Pilstål R, Su Y, Kuruvilla J, Elvén M, Ziauddin JME, Anandapadamanaban M, Cristobal S, Lundström P, Sears RC, Wallner B, Sunnerhagen M. Pre-Anchoring of Pin1 to Unphosphorylated c-Myc in a Fuzzy Complex Regulates c-Myc Activity. Structure. 2015;23:2267–2279. doi: 10.1016/j.str.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baronti L, Erales J, Habchi J, Felli IC, Pierattelli R, Longhi S. Dynamics of the Intrinsically Disordered C-Terminal Domain of the Nipah Virus Nucleoprotein and Interaction with the X Domain of the Phosphoprotein as Unveiled by NMR Spectroscopy. ChemBioChem. 2015;16:268–276. doi: 10.1002/cbic.201402534. [DOI] [PubMed] [Google Scholar]
- 51.Freedman H, Luchko T, Luduena RF, Tuszynski JA. Molecular dynamics modeling of tubulin C-terminal tail interactions with the microtubule surface. Proteins: Struct, Funct, Genet. 2011;79:2968–2982. doi: 10.1002/prot.23155. [DOI] [PubMed] [Google Scholar]
- 52.Tuszynski JA, Carpenter EJ, Huzil JT, Malinski W, Luchko T, Luduena RF. The evolution of the structure of tubulin and its potential consequences for the role and function of microtubules in cells and embryos. Int J Dev Biol. 2006;50:341–358. doi: 10.1387/ijdb.052063jt. [DOI] [PubMed] [Google Scholar]
- 53.Sackett DL, Bhattacharyya B, Wolff J. Tubulin subunit carboxyl termini determine polymerization efficiency. J Biol Chem. 1985;260:43–45. [PubMed] [Google Scholar]
- 54.Bouxsein NF, Bachand GD. Single Filament Behavior of Microtubules in the Presence of Added Divalent Counterions. Biomacromolecules. 2014;15:3696–3705. doi: 10.1021/bm500988r. [DOI] [PubMed] [Google Scholar]
- 55.Valenstein ML, Roll-Mecak A. Graded Control of Microtubule Severing by Tubulin Glutamylation. Cell. 2016;164:911–921. doi: 10.1016/j.cell.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phillips R, Kondev J, Theriot J. Physical Biology of the Cell. 1. Garland Science; New York: 2008. [Google Scholar]
- 57.Tompa P. Unstructural biology coming of age. Curr Opin Struct Biol. 2011;21:419–425. doi: 10.1016/j.sbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 58.Cassidy-Hanley DM. Tetrahymena in the laboratory: strain resources, methods for culture, maintenance, and storage. Methods Cell Biol. 2012;109:237–276. doi: 10.1016/B978-0-12-385967-9.00008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyberts SG, Takeuchi K, Wagner G. Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J Am Chem Soc. 2010;132:2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.