Abstract
Purpose
To investigate new genetic risk factors and replicate reported associations with advanced age related macular degeneration (AMD) in a prospective case - control study developed with a Spanish cohort.
Methods
Three hundred and fifty-three unrelated patients with advanced AMD (225 with atrophic AMD, 57 with neovascular AMD, and 71 with mixed AMD) and 282 age-matched controls were included. Functional and tagging SNPs in 55 candidate genes were genotyped using the SNPlex™ genotyping system. Single SNP and haplotype association analysis were performed to determine possible genetic associations; interaction effects between SNPs were also investigated.
Results
In agreement with previous reports, ARMS2 and CFH genes were strongly associated with AMD in the studied Spanish population. Moreover, both loci influenced risk independently giving support to different pathways implicated in AMD pathogenesis. No evidence for association of advanced AMD with other previous reported susceptibility genes, such as CST3, CX3CR1, FBLN5, HMCN1, PON1, SOD2, TLR4, VEGF and VLDLR, was detected. However, two additional genes appear to be candidate markers for the development of advanced AMD. A variant located at the 3´UTR of the FGF2 gene (rs6820411) was highly associated with atrophic AMD, and the functional SNP rs3112831 at ABCA4 showed a marginal association with the disease.
Conclusion
We performed a large gene association study in advanced AMD in a Spanish population. Our findings show that CFH and ARMS2 genes seem to be the principal risk loci contributing independently to AMD in our cohort. We report new significant associations that could also influence the development of advanced AMD. These findings should be confirmed in further studies with larger cohorts.
Keywords: Age related macular degeneration, genetic association, case-control study, CFH, ARMS2, FGF2 and ABCA4
Introduction
Age-related macular degeneration (AMD) is a late-onset, genetically complex disease that causes progressive damage to the macula. Early stages are characterized by the presence of small, intermediate or soft drusen and pigmentary abnormalities in the retinal pigment epithelium (RPE). Progression to advanced stage leads to loss of central vision after the development of two different types of late-stage lesions, choroidal neovascularization (CNV), associated with subretinal haemorrhage and scarring; or geographic atrophy (GA). Advanced AMD is the major cause of untreatable blindness in the Western countries(Friedman et al. 2004). Although neovascular AMD accounts for about 10% of AMD cases, it is responsible for more than 90% of legal blindness due to AMD (Jager et al. 2008).
Several environmental, dietary, and genetic risk factors have been established for AMD development, including age, caucasian race, heredity, and smoking history (Seddon et al. 1997; van Leeuwen et al. 2003). Smoking has been consistently established as a risk factor, resulting in about two- to three-fold increased risk of developing AMD in current-smokers compared with never-smokers (Thornton et al. 2005).
In recent years, major progress has been made in elucidating the AMD genetic basis through the identification of two major risk loci at 1q31 and 10q26, together accounting for above 50% of AMD cases (Edwards et al. 2005; Klein et al. 2005). At 1q31, several risk variants and haplotypes in the complement factor H (CFH) gene have been strongly associated with early and advanced AMD, suggesting an involvement of immune-mediated complement pathway in AMD pathogenesis (Edwards et al. 2005; Hageman et al. 2005; Klein et al. 2005; Hageman et al. 2006; Hughes et al. 2006; Li et al. 2006). In addition, several complement genes have recently been also associated with AMD susceptibility, including complement factor B (CFB), complement 2 (C2), and complement 3 (C3) genes(Gold et al. 2006; Maller et al. 2007; Yates et al. 2007). Besides the complement pathway, the major genetics contributor to AMD risk lies in 10q26 locus at ARMS2/HTRA1 genes. Variants in this region have been consistently reproducible across multiple ethnic groups (Jakobsdottir et al. 2005; Rivera et al. 2005; Dewan et al. 2006; Schmidt et al. 2006; Tanimoto et al. 2007; Weger et al. 2007). To date, many candidate gene association studies have been carried out, describing several other minor susceptibility variants. However, those findings should be considered as inconclusive because of the lack of consistent replication in different populations (Swaroop et al. 2009). Despite this progress in AMD genetic research in the past few years, the total number of loci involved in AMD development and their account for the population attributable risk are far from being fully known. Identification of these genetic and environmental risk factors is the first step towards earlier detection, prevention, and in the future, better treatments.
To further investigate the genetic complexity of advanced AMD in Spain, we performed a large and comprehensive study of candidate genes for advanced AMD, including 350 functional and tagging variants in 55 genes. Our aim was to identify new genetic risk factors, and to replicate the two major and other minor AMD risk loci previously reported. We additionally aimed to explore the combined effects and potential interactions between gene variants.
Materials and Methods
Patient population
A total of 353 case subjects with advanced AMD and 282 age-matched unrelated controls were recruited from ophthalmic clinics in fifteen hospitals from the Spanish multi-centre group of AMD. Subjects were all Caucasian and of Spanish descent.
The diagnosis of AMD was established on the basis of 35° colour pictures obtained of the macular area of each eye, after dilatation of pupils with tropicamide 0.5% and phenylephrine 5%. Fundus photographs were graded according to the Age-Related Eye Disease Study (AREDS) classification for AMD by two trained professionals (2000). AMD patients were categorized into early and advanced AMD, according to status in the more severely affected eye. Briefly, early AMD (grade 2 and 3) was defined as the presence of either soft, distinct drusen with pigmentary irregularities, or soft, indistinct drusen with or without pigmentary irregularities. Advanced AMD (grade 4) was defined as atrophic, neovascular or mixed AMD. As this study focused on end-stage disease, patients with early AMD changes were excluded. Patients were classified in three subgroups: 225 subjects with atrophic AMD, 57 subjects with neovascular AMD, and finally 71 patients with a mixed phenotype, with both geographic atrophy and choroidal neovascularization.
Age-matched controls were recruited from the same hospitals during routine ophthalmic examinations and were above 65 years of age. Control individuals had no evidence of drusen in either eye, macular or retinal disorder after ophthalmic examination, family relationship with the AMD cohort, or family history of maculopathies.
This study was conducted according to the recommendations of the Declaration of Helsinki and approved by the local ethics committees of the participating institutions. Signed informed consent was obtained from all subjects before inclusion in the study. Each participant was given a short questionnaire about sex, smoking, refraction, medical history review, and familial history of AMD. Data on disease status, sex, age, and smoking history of subjects are provided in Table 1.
Table 1.
Baseline characteristics of Age-Related Macular Degeneration patients and controls
| Variable | Controls (N=282) | Cases (N=353) | p-value |
|---|---|---|---|
| Afection status no. (%) | |||
| No AMD | 282 | ||
| Neovascular AMD | 225 (63.7) | ||
| Geographic atrophy | 57 (16.2) | ||
| Mixed AMD | 71 (20.1) | ||
|
| |||
| Sex no. (%) | |||
| Male | 126 (44.7) | 163 (46.2) | 0.707 |
| Female | 156 (55.3) | 190 (53.82) | |
|
| |||
| Mean age (SD) | 75.1 (5.8) | 76.74 (5.82) | 0.003 |
|
| |||
| Smoking history no. (%) | |||
| No. of subjects | 278 | 344 | 0.134 |
| Never smoked | 209 (75.2) | 240 (69.8) | |
| Current or former smoker | 69 (24.8) | 104 (30.2) | |
|
| |||
| HTA no. (%) | |||
| No. of subjects | 280 | 348 | 0.983 |
| no | 133 (47.5) | 165 (47.4) | |
| yes | 147 (52.5) | 183 (52.6) | |
|
| |||
| Diabetes mellitus no. (%) | |||
| No. of subjects | 281 | 348 | 0.874 |
| no | 241 (85.8) | 300 (86.2) | |
| yes | 40 (14.23) | 48 (13.8) | |
|
| |||
| Atheromatous disease no. (%) | |||
| No. of subjects | 280 | 345 | |
| no | 226 (80.7) | 269 (78) | |
| Ischemic cardiopathy | 32 (11.4) | 44 (13) | 0.568 |
| Ischemic stroke | 6 (2.1) | 12 (3.6) | 0.297 |
| Peripheral atherosclerosis | 27 (9.6) | 33 (9.8) | 0.950 |
Candidate genes and SNPs Selection
Fifty-five candidate genes were selected on the basis of biological and genetics knowledge of AMD. We included genes involved in AMD pathogenic mechanisms, such as oxidative damage, chronic inflammation, complement regulation, RPE or photoreceptor death and angiogenesis regulation, by previous expression, knock-out, proteomic or biochemical studies (Mullins et al. 2000; Lambooij et al. 2003; Rakic et al. 2003; Hahn et al. 2004; Martin et al. 2004). We also selected some functional candidate genes located at several loci associated with the disease by previous genome-wide linkage studies, such as 1q32, 3q24-q25, 4q27, 9q33, 12q23.2-24.31, 17q25.1, 19q13.31 (Majewski et al. 2003; Abecasis et al. 2004; Weeks et al. 2004; Fisher et al. 2005; Jun et al. 2005; Barral et al. 2006). Finally, we also studied the two major risk loci for AMD, CFH and ARMS2 genes, and other putative susceptibility genes, such as ABCA4, APOE, CST3, CX3CR1, FBLN5, HMCN1, PON1, SOD2, TLR4, VEGFA and VLDLR. More information about the candidate genes and their selection are given in the Appendix.
Our aim was to examine common variations [Minor Allele Frequency (MAF) > 0.1] in the selected candidate genes for AMD predisposition. Single nucleotide polymorphisms (SNP) selection was based on functional variation and linkage disequilibrium (LD) data from the International HapMap Project [http://www.hapmap.org/] (HapMap 2003). First, we selected all known common non-synonymous coding SNPs deposited in the dbSNP database (Build 126) [http://www.ncbi.nlm.nih.gov/SNP/index.html]. Second, we used FESD, a functional SNP Database [http://variome.kobic.re.kr/FESD/index.php] in order to prioritize putative regulatory SNPs (Kang et al. 2005). Finally, using genotypes and haplotypes from the HapMap Caucasian (CEU) population panel, tagSNPs were selected by using a tagging strategy with the Tagger tool implemented in Haploview (Barrett et al. 2005), using a strong LD tagging criteria of r2> 0.8 and with MAF > 10%. Each candidate gene was covered including an extended region of 10 kb upstream and downstream of the coding region.
Genotyping
Genomic DNA was isolated from peripheral blood using Wizard Genomic DNA Purification Kit (Promega). SNP genotyping was performed by the SNPlex™ genotyping system available in the Santiago node of the National Genotyping Centre of Spain (CEGEN) (Tobler et al. 2005). Genotyping assays were successfully designed for 380 SNPs using the assay design to the SNPlex System Bioinformatics Design Pipeline. SNPlex technology uses oligonucleotide ligation assay (OLA) combined with multiplex PCR technology to achieve allelic discrimination and target amplification. The final products are detected by capillary electrophoresis on 3730xl DNA Analyzer (Applied Biosystems) and analysed with GeneMapper v.4.0 (Applied Biosystems). As a quality control, we tested for any departure from Hardy–Weinberg equilibrium (HWE) in control samples (P>0.001).
Quality measures taken into account for genotyped SNPs to be excluded from the subsequent analysis were: MAF<0.05, genotyping success <80% and failed Hardy-Weinberg equilibrium test in control samples (P>0.001).
Statistical analysis
Analyses of genotyping results were performed using several toolsets implemented in SNPator [http://www.snpator.com/] (Morcillo-Suarez et al. 2008), Haploview 4.0 [http://www.broad.mit.edu/mpg/haploview/] (Barrett et al. 2005) and SNPassoc [http://cran.r-project.org/web/packages/SNPassoc/index.html] software (Rakic et al. 2003). Mann-Whitney U test was used to compare the ages of cases subjects and controls. Chi-square test was used to compare categorical variables and allele or haplotypes frequencies between AMD patients (or in the three AMD subgroups) and controls and to check for Hardy-Weinberg equilibrium (HWE) in control group. Fisher’s exact test was used when allele counts were < 5 by convention. Likelihood ratio test was used to compare genotype frequencies and to investigate interaction effects between SNPs. Dominant, recessive and codominant models were considered and the Akaike information criteria (AIC) was used to choose the genetic model that best fits the data. Adjusted analyses by traditional risk factors of AMD (age, gender and smoking status) were done with logistic regression models. P values, odds ratios (ORs) and 95% confidence intervals are reported. To evaluate the significance of the genetic associations with AMD after adjustment for multiple testing, permutation correction was performed with the association tests of individual SNPs with 10000 simulations (Corrected P<0.05 was considered as significant).
Linkage disequilibrium was assessed using both D´ and r2 measures implemented in Haploview. Haplotype inference was performed by the EM algorithm and haplotype blocks were generated by the algorithm and parameters of Gabriel (Gabriel et al. 2002). Permutation test was used to adjust for multiple testing.
Results
We genotyped 380 SNPs in 55 candidate genes in a Spanish population of 353 patients with advanced AMD and 282 age-matched control subjects. The mean age at examination was 76.2 years for AMD patients (standard deviation [SD], 5.9 years; range, 52–96 years) and 75.1 years for controls (SD, 5.8 years; range, 65-92 years). Although patients were slightly older than controls (P = 0.003), the other factors, such as the gender, smoking status, hypertension, diabetes mellitus, and atheromatous disease, did not differ between cases and controls.
Of the selected SNPs, 27 failed in the SNPlex Genotyping system, 14 had low genotyping call rate and 7 were monomorphic in our population. Therefore, these 48 of the 380 SNPs were not further studied. The success genotyping rate for the remaining SNPs was above 92%. These were tested separately in case and control samples for any departure from the Hardy–Weinberg equilibrium. All the SNP were in Hardy–Weinberg equilibrium in controls samples (P > 0.001) with the exception of two SNPs (rs968451 and rs4858652), which were also excluded from further analyses.
Single-SNP association study
Association analysis was directly assessed with the 330 SNPs (Appendix) and adjusted analyses by age, gender and smoking status were also performed, with similar results (data not shown). After correction for multiple hypothesis testing through permutation analysis (corrected-P < 0.05), SNPs strongly associated with advanced AMD are compiled in Table 2. Additional stratified analyses by AMD subphenotype (neovascular, atrophic or mixed AMD) were also performed and most of the above SNPs remained statistically significant in spite of the smaller number of atrophic and mixed AMD patients.
Table 2.
Single marker analysis. Allele association results
|
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Advanced AMD | Exudative AMD | Athrophic AMD | Mixed AMD | ||||||||
|
| |||||||||||
| Gene | SNP | MAF | Alleles | OR (95% CI) |
P value (Perm. P value)* |
OR (95% CI) |
P value (Perm. P value)* |
OR (95% CI) |
P value (Perm. P value)* |
OR (95% CI) |
P value (Perm. P value)* |
| ABCA4 | rs3112831 | 0,37 | A/G | 1.47 (1.84–1.15) | 0.0015 (0.0369) | 1.01 (1.31–0.79) | 0.027 (0.6537) | 1.84 (2.88–1.19) | 0.006 (0.2428) | 1.58 (2.36–1.06) | 0.0252 (0.8171) |
|
| |||||||||||
| CFH | rs800292 | 0,154 | C/T | 1.68 (2.29–1.24) | 0.0009 (0.0374) | 1.73 (2.47–1.21) | 0.0022 (0.0991) | 1.01 (1.68–0.6) | 0,9758 | 2.83 (5.41–1.47) | 0.0012 (0.0350) |
| CFH | rs3766404 | 0,133 | T/C | 2.21 (3.08–1.58) | 2.17×10−06 (0.0001) | 2.12 (3.1–1.45) | 8.87×10−05 (0.0043) | 2.62 (5.35–1.28) | 0.003 (0.2518) | 2.23 (4.1–1.21) | 0.0084 (0.2695) |
| CFH | rs1329421 | 0,44 | T/A | 2.04 (2.57–1.62) | 8.12×10−10 (< 0.0001) | 1.87 (2.42–1.45) | 1.27×10−06 (< 0.0001) | 2.19 (3.3–1.46) | 0.0001 (0.0076) | 2.54 (3.71–1.74) | 8.15×10−07 (0.0002) |
| CFH | rs1831282 | 0,47 | T/G | 2 (2.52–1.6) | 1.86×10−09 (< 0.0001) | 1.84 (2.37–1.43) | 2.23×10−06 (< 0.0001) | 2.06 (3.1–1.37) | 0.0004 (0.0249) | 2.58 (3.77–1.76) | 6.79×10−07 (0.0002) |
| CFH | rs12144939 | 0,183 | G/T | 2.35 (3.15–1.75) | 6.23×10−09 (< 0.0001) | 2.33 (3.26–1.66) | 5.52×10−7 (< 0.0001) | 2.89 (5.41–1.54) | 0.0006 (0.0323) | 2.07 (3.45–1.24) | 0.0044 (0.1890) |
| CFH | rs2284664 | 0,13 | G/A | 1.71 (2.33–1.21) | 0.002 (0.0395) | 1.83 (2.69–1.24) | 0.002 (0.0905) | 1.03 (1.77–0.6) | 0,9126 | 2.31 (4.45–1.2) | 0.0102 (0.2988) |
| CFH | rs1329428 | 0,31 | G/A | 2.49 (3.19–1.95) | 2.13×10−13 (< 0.0001) | 2.65 (3.52–1.99) | 9.91×10−12 (< 0.0001) | 1.96 (3.08–1.25) | 0.003 (0.1356) | 2.54 (3.94–1.64) | 2.11×10−05 (0.0020) |
|
| |||||||||||
| FGF2 | rs6820411 | 0,122 | A/C | 1.96 (2.83–1.36) | 0.0002 (0.0043) | 1.76 (2.63–1.17) | 0.0057 (0.092) | 2.63 (4.58–1.51) | 0.0004 (0.0144) | 2.13 (3.64–1.24) | 0.0052 (0.0977) |
|
| |||||||||||
| LOC387715 | rs10490924 | 0,324 | T/G | 3.94 (5.14–3.02) | 1.75×10−25 (< 0.0001) | 4.20 (5.61–3.15) | 7.46×10−24 (< 0.0001) | 3.56 (5.49–2.31) | 9.12×10−09 (< 0.0001) | 3.48 (5.19–2.33) | 1.07×10−09 (< 0.0001) |
|
| |||||||||||
| CGREF1 | rs2384571 | 0,453 | C/G | 1.2 (1.51–0.97) | 0,0983 | 1.84 (2.8–1.2) | 0.0045 (0.0484) | ||||
|
| |||||||||||
| APOE | rs7259004 | 0,102 | C/G | 1.08 (1.56–0.75) | 0,6905 | 2.22 (3.68–1.34) | 0.0015 (0.0199) | ||||
OR= Odds ratios
Perm. P value= P value from 10.000 permutations
Significant P values (<0.05) are shown in bold
In agreement with previous reports, both ARMS2 and CFH genes were associated with AMD in Spanish population (de la Fuente et al. 2007; Recalde et al. 2008). Briefly, the T risk allele in the A69S variant (rs10490924) at ARMS2 gene showed the strongest association with late stage AMD (T allele, OR =3.94, P=1.75×10−25; ORhom = 12.64, ORhet = 4.19, P=3.19×10−21). As shown in Table 2, A69S confers similar risks to the three forms of advanced AMD. In the same way, seven alleles at CFH gene were also significantly associated with advanced AMD patients. Between them, two intronic tagging SNPs (rs1329428 and rs1329421) showed the strongest associations (OR=2.49, P=2.13×10−13 and OR= 2.04, P=8.12×10−10, respectively). When neovascular, atrophic and mixed AMD stratified analyses were performed, association with CFH gene remained. No epistatic interactions were detected between rs10490924 at ARMS2 gene and the two most significant SNPs at CFH gene based on the likelihood ratio test (Table 4).
Table 4.
Two loci analysis. CFH and LOC 387715 interactions
| OR (95 % CI)
|
||||
|---|---|---|---|---|
| rs10490924 | ||||
| G/G | G/T | T/T | Interaction P | |
| rs1329421 | ||||
| A/A | 1 | 4.3 (8.2–2.3) | 13.2 (39–4.5) | 0.99871 |
| A/T | 2.3 (4–1.3) | 9.1 (17–4.9) | 33.1 (101.5–10.8) | |
| T/T | 3.9 (7.9–1.9) | 15 (31.7–7.1) | 45.2 (362.8 – 5.6) | |
| rs1329428 | ||||
| G/G | 1 | 4.5 (7.7–2.6) | 12.3 (36.8–4.1) | 0.79878 |
| A/G | 0.6 (1–0.4) | 2.7 (4.6–1.3)5 | 10.7 (37.4–3.1) | |
| A/A | 0.2(0.6–0.1) | 0.5 (1.3–0.2) | 0.8 (5.12–0.13) | |
| rs3766404 | ||||
| T/T | 1 | 3.9 (5.8–2.5) | 13 (31.7–5.4) | 0.64755 |
| C/T | 0.4 (0.7–0.2) | 2.6 (5.4–1.3) | 5.4 (16.7–1.7) | |
| C/C | 0.4 (3.2–0.04) | 0.6 (2.4–0.2) | NA | |
| rs1831282 | ||||
| T/T | 1 | 3.9 (7.5–2) | 10.6 (32–3.5) | 0.9939 |
| C/T | 2.07 (3.7–1.2) | 8.6 (16.3–4.5) | 30.6 (94.8–9.8) | |
| C/C | 3.3 (6.7–1.7) | 14.1 (29.3–6.8) | 52.9 (420.4–6.6) | |
| rs12144939 | ||||
| G/G | 1 | 3.7 (5.8–2.4) | 13.8 (36.1–5.2) | 0.61646 |
| G/T | 0.5 (0.8–0.) | 3 (1.5–5.8) | 6.1 (18.7–2) | |
| T/T | 0.1 (1.1–0.02) | 0.41 (1.5–0.1) | 0 (NA–0) | |
In addition to the previously known risk variants of AMD, a tagging SNP (rs6820411) located 3′ of FGF2 (Fibroblast growth factor 2) was also associated with advanced AMD after permutation correction (allele A OR= 1.96, P=0.0043; OR AA+AC= 2.2, P=0.0076). Also, their effect remained significant after adjusting for other risk factors, including smoking status, age and polymorphisms in CFH and ARMS2 (data not shown). When phenotype subgroups were compared with controls, allele and genotype frequency differences were also found. Although the association remained significant only for atrophic AMD cases after correction for multiple testing, exudative and mixed AMD cases showed a trend toward association (Table 2). Epistatic interactions were not detected between rs6820411 at FGF2 and risk alleles in ARMS2 and CFH based on the likelihood ratio test, as shown in Table 5.
Table 5.
Two locus analysis. FGF2 interactions with CFH and LOC387715
| OR (95 % CI)
|
||||
|---|---|---|---|---|
| rs6820411 | ||||
| C/C | C/A | A/A | Interaction P | |
| CFH | ||||
| rs1329421 | ||||
| A/A | 1 | 2.2 (4.6–1.1) | 0 (0-NA) | 0.500 |
| A/T | 2.1 (3.2–1.4) | 3.7 (6.8–2) | 3.3 (18.6–0.6) | |
| T/T | 3.3 (5.6–2) | 12.5 (37–4.2) | NA | |
| rs1329428 | ||||
| G/G | 1 | 2.3 (4.4–1.2) | 0.6 (9.2–0.04) | 0.373 |
| A/G | 0.5 (0.7–0.3) | 1.1 (2.2–0.6) | 1.1 (6.3–0.2) | |
| A/A | 0.2 (0.34–0.1) | 0.1 (0.5–0.02) | NA | |
| rs3766404 | ||||
| T/T | 1 | 1.9 (3.1–1.2) | 0.7 (3.6–0.1) | 0.124 |
| C/T | 0.4 (0.7–0.3) | 1.1 (2.5–0.4) | NA | |
| C/C | 0.2 (0.7–0.05) | NA | NA | |
| rs1831282 | ||||
| T/T | 1 | 2.1 (4.4–1) | NA | 0.729 |
| C/T | 2 (3–1.33) | 3.8 (7.1–2.1) | 2.2 (10.2–0.5) | |
| C/C | 3.3 (5.5–2) | 10.3 (28–3.8) | NA | |
| rs12144939 | ||||
| G/G | 1 | 2.3 (3.9–1.4) | 1.1 (6.4–.2) | 0.836 |
| G/T | 0.5(0.7–0.3) | 1.1 (2.3–0.5) | 1.4 (15.6–0.1) | |
| T/T | 0.1 (0.5–0.04) | 0.1 (1.2–0.02) | NA | |
|
| ||||
| LOC387715 | ||||
| rs10490924 | ||||
| G/G | 1 | 2.5 (4.5–1.4) | 2.1 (10.9–0.5) | 0.858 |
| G/T | 4.1 (6.2–2.7) | 8.7 (17–4.5) | NA | |
| T/T | 14.5 (31.9–6.6) | 16.1 (72.1–3.6) | NA | |
In order to replicate the putative effect of FGF2, the associated polymorphism in Spanish population was additionally genotyped in a total of 609 AMD cases and 325 healthy Caucasian subjects from US (Swaroop). No evidence of association was found in this cohort (allele association P = 0.17).
Additionally, two variants showed a lesser extent of association with advanced AMD after a multiple testing correction. A functional SNP at ABCA4 gene (rs3112831) had a marginal association (P=0.0015) with advanced AMD. rs2384571, and a tagSNP located in CGREF1 gene, had also shown a significant association but only for atrophic AMD patients (P=0.0045).
Haplotype Association Analysis
To determine whether any of the haplotypes in the candidate genes could be associated with AMD, linkage disequilibrium (LD) analysis and haplotype estimation were performed. Haplotype analysis did not detect any association in the additional candidate genes, only CFH and FGF2 haplotypes showed a strong evidence of association with late AMD after correcting for multiple testing (corrected-P < 0.05). The association between the haplotype carrying risk alleles was either equal or weaker than the association at each individual SNP.
In concordance with previous reports, CFH region showed extensive LD in our population, as shown in Figure 1. Except for rs800292, all SNPs at CFH gene were included in a large LD block. Haplotype estimation in CFH gene in cases and controls identified a common risk haplotype (H1) in 50% of AMD cases versus 35% of controls (OR= 1.99, P<10−9) and three protective haplotypes (H3, H4 and H5), as shown in Figure 1. All of them showed a strong association with AMD in the overall dataset and in the three AMD subgroups (data not shown). The two most associated SNPs at single-marker analysis contained alleles that mainly distinguished between risk and protective haplotypes. The T allele at rs1329421 and the A allele at rs1329428 were exclusively found in the risk and protective haplotypes, respectively.
Figure 1. LD and haplotype maps of the CFH locus in this Spanish population.
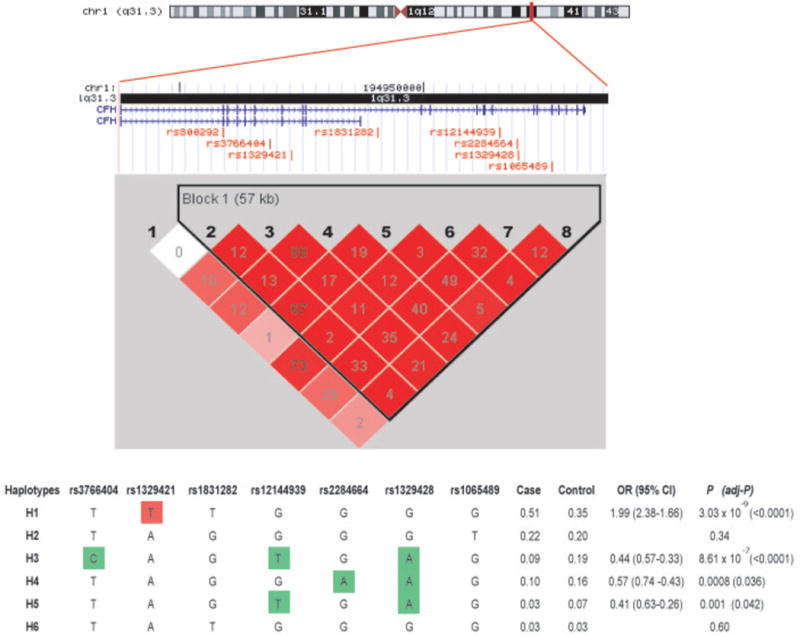
A schematic representation of the intron/exon structure of the CFH gene is indicated above the LD plot. Relative positions of studied SNPs are also indicated. Each box provides r2 values with darker red shades representing stronger LD. Haplotype association analysis in cases and controls were performed for the single haplotype block found at this locus. All of the haplotypes with a frequency of >1% are displayed. The estimated frequencies of the haplotype in cases and controls, ORs, 95% CI and P-values are also shown. The risk haplotype (H1) is shown in red shading, and the protective haplotypes (H3, H4 and H5) are shown in green shading. Alleles exclusively found in these risk and protective haplotypes are boxed.
In the analysis of LD map at FGF2 locus (Figure 2), a small haplotype block of 2 SNPs was found upstream of the gene. Those SNPs were not associated with AMD; however, an extensive LD region was found between the 3′ part of the FGF2 gene and downstream region, comprising NUDT6 gene. Haploview showed six more frequent haplotypes in the analysed population and only one of them (H3) included the risk allele at rs6820411. After multiple testing correction, only this haplotype was significantly associated with AMD risk (OR= 1.83, P<10−4).
Figure 2. LD and haplotype maps of the FGF2 locus in this Spanish population.
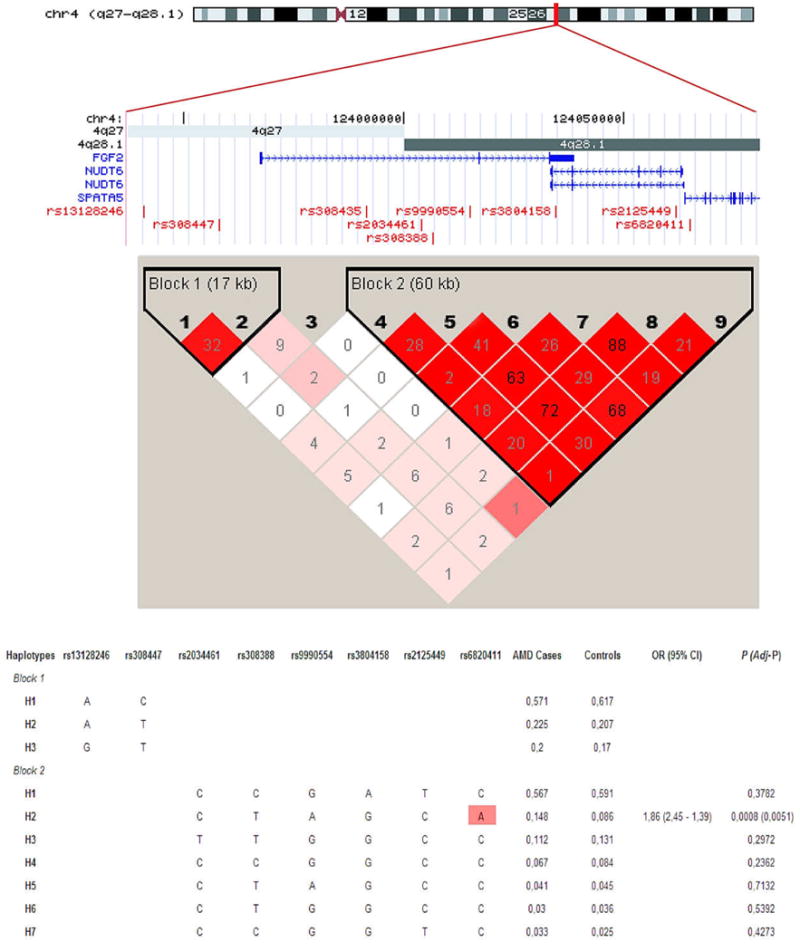
A schematic representation of the intron/exon structure of the FGF2 and NUDT6 genes with the relative positions of tagSNPs, is indicated above the LD plot. Each box provides r2 values with darker red shades representing stronger LD. Haplotype association analysis in cases and controls are also performed on the two haplotype blocks found at this locus. All of the haplotypes with a frequency of >1% are displayed. The estimated haplotypic frequencies in cases and controls, P-values, ORs and 95% CI are also shown. The risk haplotype (H3) is shown in red shading remarking in a box the risk allele at rs6820411.
Discussion
We designed a functional and tagging SNP selection strategy in 55 candidate genes, selected in basis of positional criteria, their contribution to AMD aetiology, and/or their previous implication as risk variant. We determined whether common variations across the candidate genes displayed significant association with advanced AMD or their three phenotypic subgroups (neovascular, atrophic or mixed AMD) in a Spanish population.
Despite using a different set of CFH variants than most of the published studies, our results in Spanish population are in agreement with previous reports in other Caucasian cohorts (Hageman et al. 2005; Klein et al. 2005; Li et al. 2006; Francis et al. 2007). Seven of eight tagSNPs here studied were associated with advanced AMD. Only rs1065489, a missense variant, did not show any evidence of association, as previously reported by Hageman et al (Hageman et al. 2005). rs1329421 and rs1329428, located in the CFH introns 7 and 15, respectively, exhibited the strongest association with AMD in our Spanish cohort. With the exception of rs1329421, all significant variants at CFH gene were previously described as associated to AMD (Hageman et al. 2005; Klein et al. 2005; Li et al. 2006; Francis et al. 2007).
The strong linkage disequilibrium present in this region makes it difficult to distinguish the causal variant of another in linkage disequilibrium with it. Although CFH Y402H could play a causal role in the development of AMD, as postulated by several reports (Skerka et al. 2007; Yu et al. 2007; Ormsby et al. 2008), also other variants could increase the risk of AMD by regulating the expression of CFH or CFH-related genes located within the RCA (Regulation of Complement Activation) locus on chromosome 1. In our Spanish AMD cohort, we could also found a set of common susceptibility and protective haplotypes against AMD, as previously observed in other Caucasian population. Those haplotypes were defined by 7 SNPs, and two of them, T allele at rs1329421 and the A allele at rs1329428 were exclusively found in the risk and protective haplotypes, respectively.
Although we could not asses the AMD risk associated with CFH Y402H due to a unsuccessful genotyping assay with SNPlex platform, we could previously report a risk effect on advanced AMD in a preliminary study with 175 AMD cases and 119 controls (de la Fuente et al. 2007). When we considered only common samples analysed in both cohorts, Y402H variant showed a strong LD with rs1329421 and rs1831282 (r2 = 0.98 and 0.92, respectively). The risk-associated C-allele of the Y402H variant was found to take part of the risk haplotype H1 (data not shown). Therefore, our 7-SNP haplotype seems to fit well with risk haplotypes described in other Caucasian populations.
Our results showed that significant CFH polymorphisms overall associate with a similar frequency with the neovascular, atrophic and mixed AMD subtypes. Neither single variants nor risk haplotype preferentially increased susceptibility to one of these 3 phenotypes. In our report, we did not include early or intermediate stages of AMD; so, whether those polymorphisms also contribute to earlier AMD phenotypes remains to be fully explored.
Our findings also confirm ARMS2 as another principal contributor to advanced AMD risk in Spanish population (Kanda et al. 2007; Fritsche et al. 2008), (de la Fuente et al. 2007; Recalde et al. 2008). In agreement with previous studies, we could not identify gene interaction between CFH and ARMS2. Thus, the A69S polymorphism in ARMS2 is strongly associated with advanced AMD in an independent extent of the CFH polymorphisms.
With regard to the rest of gene variants studied here, most of them did not show allelic or genotype frequency differences between AMD cases and controls. Only rs6820411 at FGF2 gene and rs3112831 at ABCA4 gene maintained statistical significance after multiple testing correction.
Fibroblast growth factor 2 (FGF2) is a widely expressed protein with potent angiogenic activity that promotes growth and differentiation of a broad spectrum of cell types. FGF2 seem to play also an essential role in VEGF-dependent choroidal neovascularisation (Frank 1997; Browning et al. 2008). It was found in high concentration in neovascular tissue in AMD patients and up-regulated in laser-induced choroidal neovascularisation (Ogata et al. 1996; Cameron et al. 2007). In our study, we found a strong association between advanced AMD and rs6820411 even after adjusting for age, sex, smoking, CFH or ARMS2 risk variants. In addition, when endophenotypes were considered, significant association with atrophic AMD was also maintained after multiple testing corrections.
rs6820411 is located downstream of FGF2 in the promoter region of NUDT6 gene, which is transcribed in the opposite direction (Figure 2). NUDT6 transcription generates an overlapping antisense RNA (FGF-AS) implicated in the post-transcriptional regulation of FGF-2 expression and function (Baguma-Nibasheka et al. 2007). HapMap Phase II data shows a large extent of LD as the 3′ FGF2/FGF-AS region, making possible that rs6820411 being in LD with a non-assayed causal variant at FGF-AS. The lack of replication in an admixed US population with patients at early and advanced AMD stages could be reflecting a population and phenotype-dependence of this variant on AMD susceptibility. Epidemiological studies have revealed differences in the prevalence of advanced AMD among different ethnic groups with major rates in Caucasian than African and Asian individuals (2000). As HapMap data showed, rs6820411 is only polymorphic in Caucasian (CEU) population. In addition, despite showing positive association with general AMD in our study, when subclinical forms of the disease were considered, the putative risk allele was only detected in the atrophic forms. Consequently, further resequencing and association analyses of FGF2/FGF-AS region in a larger cohort, phenotypically well characterized in the different clinical AMD phenotypes, are needed to confirm the putative role of FGF-AS in AMD susceptibility.
ABCA4 is the retina-specific ABC transporter gene and responsible for the Stargardt disease, an autosomal recessive form of juvenile macular degeneration. We observed a marginal allelic association with the missense H423R variant (rs3112831) with advanced AMD. Although some authors reported mutations in ABCA4 gene in a small percentage of AMD cases (Allikmets et al. 1997; Shroyer et al. 2001), most of the studies reported no statistical significant association (Rivera et al. 2000; Souied et al. 2000; Guymer et al. 2001; Schmidt et al. 2003). In a similar way, we observed some marginal significant association with two variants at CGREF1 and APOE variants with atrophic and mixed AMD forms. Additionally studies are needed with larger cohorts to confirm those observations.
We could not detect statistically significant association in our population between other minor susceptibility genes and advanced AMD, such as CST3, CX3CR1, FBLN5, HMCN1, PON1, SOD2, TLR4, VEGFA and VLDLR, in agreement with other reports (Schultz et al. 2003; Abecasis et al. 2004; Baird et al. 2004; Hayashi et al. 2004; Bojanowski et al. 2005; Esfandiary et al. 2005; Schmidt et al. 2005; Fuse et al. 2006; Kaur et al. 2006; Seitsonen et al. 2006; Fisher et al. 2007; Richardson et al. 2007; Despriet et al. 2008; Edwards et al. 2008; Utheim et al. 2008). Discrepancies in replication of risk variants in association studies could be caused by population heterogeneity, disease heterogeneity, and/or the use of different diagnostic criteria among cases, but it could also reflect the lack of power to detect modest gene effects with undersized samples. With our sample size, the study reached >80% power at a 5% significance level to detect an odds ratio greater than 1.52 when the allele frequency is 0.05, or an OR > 1.26 for an allele at a frequency of 30%. Since we only examined for association advanced stages of AMD in our study, it could be possible that some of these candidates genes are only associated with the early forms of AMD.
Recently, other risk and protective variants on complement genes have been strongly associated with AMD; however, they could not be assessed here because the study design and genotyping were performed before the variants in these loci were confirmed as risk and protective factors.
In summary, we have replicated the CFH and ARMS2 gene variants association with advanced AMD in the Spanish population. Moreover, as it was previously reported (Deangelis et al. 2008), both loci influence risk independently, giving support to different pathways implicated in the pathogenesis of the disease. No evidence for a role of other previously reported genes in the development of AMD was found. Nevertheless, more extended studies should be performed in order to role out the effect of these genes taking into account different groups of populations and possible interactions with other genetic or environmental factors. Finally, although we have identified a gene variant (rs6820411) within the downstream region of the FGF2 locus, with a novel hypothetical role in the pathogenesis of AMD, we could not replicate our findings in a matched US American set of samples. Validation of the putative effect of this variant deserves further analysis in an extended group of late AMD patients with European descent, well characterized in the different clinical forms of the disease.
Table 3.
Single locus analysis. Genotype association results
|
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMD | Wet AMD | Dry AMD | Mixed | ||||||||
|
|
|||||||||||
| Gene | SNP | Model | Genotype | OR (95% CI) |
P value (Perm. P value)* |
OR (95% CI) |
P value (Perm. P value)* |
OR (95% CI) |
P value (Perm. P value)* |
OR (95% CI) |
P value (Perm. P value)* |
| ABCA4 | rs3112831 | dominant | A/A | 1 | 0.0048 (0.0972) | 1 | 0.0327 (0.58) | 1 | 0.0327 (0.540) | ||
| A/G-G/G | 0.64 (0.88–0.46) | 0.67 (0.97–0.47) | 0.67 (0.97–0.47) | ||||||||
| recessive | A/A-A/G | 1 | 0.0098 (0.224) | ||||||||
| G/G | 0.3 (0.86–0.10) | ||||||||||
|
| |||||||||||
| CFH | rs800292 | recessive | C/C | 1.81 (2.56–1.28) | 0.0008 (0.0368) | 1.88 (2.77–1.26) | 0.0016 (0.08) | 1 | 0.9957 | 2.91 (5.88–1.49) | 0.0008 (0.150) |
| C/T-T/T | 1.85 (2.77–1.23) | ||||||||||
| CFH | rs3766404 | recessive | T/T | 2.32 (3.33–1.59) | 0.00752 (0.0002) | 2.17 (3.33–1.43) | 2.00×10−4 (0.0049) | 2.94 (6.66–1.35) | 0.0028 (0.169) | 2.38 (4.76–1.21) | 0.0065 (0.230) |
| C/T-C/C | |||||||||||
| CFH | rs1329421 | codominant | A/A | 1,00 | 1.61×10−5 (< 0.0001) | 1 | 2.00×10−5 (< 0.0001) | 1 | 0.0015 (0.038) | ||
| A/T | 2.13 (3.06–1.48) | 1.82 (2.73–1.21) | 1.92 (3.88–0.95) | ||||||||
| T/T | 3.75 (5.98–2.36) | 3.18 (5.30–1.91) | 4.23 (9.29–1.92) | ||||||||
| recessive | A/A | 1 | 7.04 E-07 (0.0023) | ||||||||
| A/T-T/T | 4.87 (9.9–2.39) | ||||||||||
| CFH | rs1831282 | codominant | G/G | 1 | 5.71×10−5 (< 0.0001) | 1 | 4.00×10−5 (0.0004) | 1 | 0.0014 (0.078) | 1 | 7.22×10−06 (0.0023) |
| G/T | 2.01 (2.92–1.38) | 1.79 (2.72–1.18) | 1.87 (3.84–0.91) | 3.41 (7.21–1.61) | |||||||
| T/T | 3.57 (5.61–2.28) | 3.04 (5.00–1.85) | 3.7 (8.12–1.69) | 6.33 (14.24–2.82) | |||||||
| CFH | rs12144939 | codominant | G/G | 9.09 (25–2.94) | 3.87×10−5 (< 0.0001) | 11.11 (50–2.56) | 1.30×10−6 (< 0.0001) | 6.66 (50–0.9) | 0.0115 (0.150) | ||
| G/T | 2.08 (2.94–1.44) | 2.00 (2.94–1.35) | 1.81 (3.33–1) | ||||||||
| T/T | 1 | 1,00 | |||||||||
| recessive | G/G | 3.22 (6.25–1.59) | 0.0004 (0.0379) | ||||||||
| G/T-T/T | 1 | ||||||||||
| CFH | rs2284664 | recessive | G/G | 1.75 (2.56–1.21) | 0.002563 (0.0901) | 1.88 (2.85–1.23) | 0.003 (0.082) | 1.07 (2.504–0.57) | 0.8100 | 2.27 (4.54–1.14) | 0.0120 (0.560) |
| A/G-A/A | 1 | 1 | 1 | 1 | |||||||
| CFH | rs1329428 | codominant | G/G | 7.14 (14.28–4) | 2.76×10−09 (< 0.0001) | 8.33(16.66–3.84) | 1.14×10−10 (< 0.0001) | 4 (12.5–1.33) | 0.0130 (0.1672) | 11.11 (50–2.5) | 4.71×10−05 (0.0016) |
| A/G | 2.1 (2.94–1.5) | 2.27 (3.33–1.53) | 1.72 (3.22–1) | 1.96 (3.44–1.11) | |||||||
| A/A | 1 | 1 | 1 | 1 | |||||||
|
| |||||||||||
| FGF2 | rs6820411 | dominant | C/C | 1 | 7.00×10−05 (0.0076) | 1 | 0.0023 (0.18) | 1 | 0.0005 (0.078) | 1 | 0.0095 (0.250) |
| A/C-A/A | 2.20 (3.28–1.48) | 1.97 (3.06–1.27) | 2.20 (3.28–1.48) | 2.28 (4.17–1.24) | |||||||
|
| |||||||||||
| LOC387715 | rs10490924 | codominant | G/G | 1 | 3.19×10−21 (< 0.0001) | 1 | 1.51×10−21 (< 0.0001) | 1 | 6.19×10−07 (< 0.0001) | 1 | 3.9×10−09 (< 0.0001) |
| G/T | 4.19 (6–2.93) | 4.44 (6.6–3) | 2.6 (4.97–1.36) | 5.08 (9.15–2.82) | |||||||
| T/T | 12.64 (25.53–6.25) | 14.4 (30.1.-6.9) | 11.52 (29.35–4.52) | 8.36 (22.87–3.05) | |||||||
|
| |||||||||||
| CGREF1 | rs2384571 | recessive | C/C | 1 | 0.0557 | 2.63 (4.76–1.47) | 0.0013 (0.1672) | ||||
| C/G-G/G | 0.71 (1.01–0.5) | ||||||||||
|
| |||||||||||
| APOE | rs7259004 | dominant | G/G | 1 | 0.7959 | 1 | 0.0019 (0.16) | ||||
| C/G-C/C | 1.05 (1.58–0.7) | 2.54 (4.52–1.43) | |||||||||
OR= Odds ratios
Perm. P value= P value from 10.000 permutations
Significant P values (<0.05) are shown in bold
Table 6.
CFH haplotype analysis using a substracting approach
| Number of SNPs | SNPs in best model | Haplotypes | Case Freq | Control Freq | Chi square | P-value | OR (95 % IC) |
|---|---|---|---|---|---|---|---|
| Protective Haplotypes | |||||||
| 7 | rs3766404 rs1329421 rs1831282 rs12144939 rs2284664 rs1329428 rs1065489 | CAGTGAG | 0.091 | 0.186 | 24215.00 | 8.61×10−07 | 0.44 (0.57 – 0.33) |
| TAGGAAG | 0.100 | 0.164 | 11208.00 | 8×10−04 | 0.57 (0.74 – 0.43) | ||
| TAGTGAG | 0.030 | 0.070 | 10.89 | 0.001 | 0.41 (0.63 – 0.26) | ||
| 6 | rs1329421 rs1831282 rs12144939 rs2284664 rs1329428 rs1065489 | AGTGAG | 0.121 | 0.254 | 37.17 | 1.08×10−09 | 0.4 (0.51 – 0.32) |
| AGGAAG | 0.101 | 0.162 | 10.42 | 0.0012 | 0.58 (0.76 – 0.45) | ||
| 5 | rs1329421 rs1831282 rs2284664 rs1329428 rs1065489 | AGGAG | 0.121 | 0.257 | 39.00 | 4.31×10−10 | 0.39 (0.5 – 0.31) |
| AGAAG | 0.101 | 0.162 | 10.43 | 0.0012 | 0.58 (0.75 – 0.44) | ||
| 4 | rs1329421 rs1831282 rs1329428 rs1065489 | AGAG | 0.222 | 0.419 | 56.84 | 4.74×10−14 | 0.39 (0.48 – 0.32) |
| 3 | rs1329421 rs1831282 rs1065489 | AGG | 0.225 | 0.421 | 56.04 | 7.10×10−14 | 0.4 (0.48 –0.33) |
| 2 | rs1831282 rs1065489 | GG | 0.225 | 0.420 | 55.12 | 1.13×10−13 | 0.4 (0.49–0.33) |
| Risk Haplotypes | |||||||
| 7 | rs3766404 rs1329421 rs1831282 rs12144939 rs2284664 rs1329428 rs1065489 | TTTGGGG | 0.512 | 0.345 | 35165.00 | 3.03×10−09 | 1.99 (2.38 – 1.66) |
| 6 | rs3766404 rs1329421 rs1831282 rs12144939 rs2284664 rs1329428 | TTTGGG | 0.517 | 0.345 | 37.01 | 1.17×10−09 | 2.03 (2.43 – 1.70) |
| 5 | rs3766404 rs1329421 rs1831282 rs2284664 rs1329428 | TTTGG | 0.517 | 0.343 | 38.26 | 6.18×10−10 | 2.05 (2.45 -1.71) |
| 4 | rs3766404 rs1329421 rs2284664 rs1329428 | TTGG | 0.516 | 0.343 | 38.13 | 6.61×10−10 | 2.04 (2.45-1.71) |
| 3 | rs3766404 rs2284664 rs1329428 | TGG | 0.771 | 0.581 | 52.61 | 4.07×10−13 | 2.43 (2.95 – 2) |
| 2 | rs2284664 rs1329428 | GG | 0.773 | 0.581 | 53.73 | 2.31×10−09 | 2.45 (2.98 – 2.02) |
OR= Odds ratios
Acknowledgments
We greatly thanks all the members of the Spanish multi-centre group of AMD: A.García Layana, Clínica Universitaria de Pamplona, Navarra; B. Pazos González, Instituto Gallego de Oftalmologia, Santiago de Compostela; M. Díaz Llopis, Hospital Universitario la Fe, Valencia; C. Torrón, Hospital Universitario Miguel Servet, Zaragoza; R. Coco, IOBA, Valladolid; F. Martínez, Hospital Marques de Valdecilla, Santander; J. Arraiz, Instituto Clínico Quirúrgico de Oftalmología, Bilbao; J. M. Ruiz Moreno, Vissum Instituto Oftalmológico, Alicante; C. Desco Esteban, Fundación Oftalmológica del Mediterráneo, Valencia; E. Esteban, Hospital Virgen de la Macarena, Sevilla; M.S. Figueroa, Hospital Ramón y Cajal, Madrid; F. Gómez-Ulla, Hospital Clínico Universitario de Santiago de Compostela; J. Bañuela Bañuela, Hospital de Alcorcón, Madrid; B. Fernández-Vega Sanz, Instituto Oftalmológico Fernández Vega, Asturias; L. Arias, Hospital Universitario de Bellvitge, Barcelona; and J. Fernández Vigo, Facultad Medicina, Universidad de Extremadura.
The study was supported by grants from the Xunta de Galicia (PGIDIT06PXIB208204PR), the Instituto de Salud Carlos III (EMER07/018), and National Institutes of Health, USA. The sponsor or funding organization had no role in the design or conduct of this research.
References
- Risk factors associated with age-related macular degeneration. A case-control study in the age-related eye disease study: Age-Related Eye Disease Study Report Number 3. Ophthalmology. 2000;107:2224–32. doi: 10.1016/s0161-6420(00)00409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Yashar BM, Zhao Y, Ghiasvand NM, Zareparsi S, Branham KE, Reddick AC, Trager EH, Yoshida S, Bahling J, Filippova E, Elner S, Johnson MW, Vine AK, Sieving PA, Jacobson SG, Richards JE, Swaroop A. Age-related macular degeneration: a high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet. 2004;74:482–94. doi: 10.1086/382786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A, Dean M, Lupski JR, Leppert M. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997;277:1805–7. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- Baguma-Nibasheka M, Li AW, Murphy PR. The fibroblast growth factor-2 antisense gene inhibits nuclear accumulation of FGF-2 and delays cell cycle progression in C6 glioma cells. Mol Cell Endocrinol. 2007;267:127–36. doi: 10.1016/j.mce.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Baird PN, Chu D, Guida E, Vu HT, Guymer R. Association of the M55L and Q192R paraoxonase gene polymorphisms with age-related macular degeneration. Am J Ophthalmol. 2004;138:665–6. doi: 10.1016/j.ajo.2004.04.053. [DOI] [PubMed] [Google Scholar]
- Barral S, Francis PJ, Schultz DW, Schain MB, Haynes C, Majewski J, Ott J, Acott T, Weleber RG, Klein ML. Expanded genome scan in extended families with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47:5453–9. doi: 10.1167/iovs.06-0655. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bojanowski CM, Tuo J, Chew EY, Csaky KG, Chan CC. Analysis of Hemicentin-1, hOgg1, and E-selectin single nucleotide polymorphisms in age-related macular degeneration. Trans Am Ophthalmol Soc. 2005;103:37–44. discussion 44-5. [PMC free article] [PubMed] [Google Scholar]
- Browning AC, Dua HS, Amoaku WM. The effects of growth factors on the proliferation and in vitro angiogenesis of human macular inner choroidal endothelial cells. Br J Ophthalmol. 2008;92:1003–8. doi: 10.1136/bjo.2007.127670. [DOI] [PubMed] [Google Scholar]
- Cameron DJ, Yang Z, Gibbs D, Chen H, Kaminoh Y, Jorgensen A, Zeng J, Luo L, Brinton E, Brinton G, Brand JM, Bernstein PS, Zabriskie NA, Tang S, Constantine R, Tong Z, Zhang K. HTRA1 variant confers similar risks to geographic atrophy and neovascular age-related macular degeneration. Cell Cycle. 2007;6:1122–5. doi: 10.4161/cc.6.9.4157. [DOI] [PubMed] [Google Scholar]
- de la Fuente M, Blanco MJ, Pazos B, Fernandez MI, Carracedo A, Sanchez-Salorio M, Coco RM, Torron C, Gomez AM. Complement factor H. Ophthalmology. 2007;114:193 e1–2. doi: 10.1016/j.ophtha.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Deangelis MM, Ji F, Adams S, Morrison MA, Harring AJ, Sweeney MO, Capone A, Jr, Miller JW, Dryja TP, Ott J, Kim IK. Alleles in the HtrA serine peptidase 1 gene alter the risk of neovascular age-related macular degeneration. Ophthalmology. 2008;115:1209–1215 e7. doi: 10.1016/j.ophtha.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despriet DD, Bergen AA, Merriam JE, Zernant J, Barile GR, Smith RT, Barbazetto IA, van Soest S, Bakker A, de Jong PT, Allikmets R, Klaver CC. Comprehensive analysis of the candidate genes CCL2, CCR2, and TLR4 in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:364–71. doi: 10.1167/iovs.07-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan A, Liu M, Hartman S, Zhang SS, Liu DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, Barnstable C, Pang CP, Hoh J. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–92. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- Edwards AO, Chen D, Fridley BL, James KM, Wu Y, Abecasis G, Swaroop A, Othman M, Branham K, Iyengar SK, Sivakumaran TA, Klein R, Klein BE, Tosakulwong N. Toll-like receptor polymorphisms and age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:1652–9. doi: 10.1167/iovs.07-1378. [DOI] [PubMed] [Google Scholar]
- Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–4. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- Esfandiary H, Chakravarthy U, Patterson C, Young I, Hughes AE. Association study of detoxification genes in age related macular degeneration. Br J Ophthalmol. 2005;89:470–4. doi: 10.1136/bjo.2004.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SA, Abecasis GR, Yashar BM, Zareparsi S, Swaroop A, Iyengar SK, Klein BE, Klein R, Lee KE, Majewski J, Schultz DW, Klein ML, Seddon JM, Santangelo SL, Weeks DE, Conley YP, Mah TS, Schmidt S, Haines JL, Pericak-Vance MA, Gorin MB, Schulz HL, Pardi F, Lewis CM, Weber BH. Meta-analysis of genome scans of age-related macular degeneration. Hum Mol Genet. 2005;14:2257–64. doi: 10.1093/hmg/ddi230. [DOI] [PubMed] [Google Scholar]
- Fisher SA, Rivera A, Fritsche LG, Keilhauer CN, Lichtner P, Meitinger T, Rudolph G, Weber BH. Case-control genetic association study of fibulin-6 (FBLN6 or HMCN1) variants in age-related macular degeneration (AMD) Hum Mutat. 2007;28:406–13. doi: 10.1002/humu.20464. [DOI] [PubMed] [Google Scholar]
- Francis PJ, Schultz DW, Hamon S, Ott J, Weleber RG, Klein ML. Haplotypes in the complement factor H (CFH) gene: associations with drusen and advanced age-related macular degeneration. PLoS ONE. 2007;2:e1197. doi: 10.1371/journal.pone.0001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank RN. Growth factors in age-related macular degeneration: pathogenic and therapeutic implications. Ophthalmic Res. 1997;29:341–53. doi: 10.1159/000268032. [DOI] [PubMed] [Google Scholar]
- Friedman DS, O’Colmain BJ, Munoz B, Tomany SC, McCarty C, de Jong PT, Nemesure B, Mitchell P, Kempen J. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–72. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- Fritsche LG, Loenhardt T, Janssen A, Fisher SA, Rivera A, Keilhauer CN, Weber BH. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40:892–6. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- Fuse N, Miyazawa A, Mengkegale M, Yoshida M, Wakusawa R, Abe T, Tamai M. Polymorphisms in Complement Factor H and Hemicentin-1 genes in a Japanese population with dry-type age-related macular degeneration. Am J Ophthalmol. 2006;142:1074–6. doi: 10.1016/j.ajo.2006.07.030. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, Smith RT, Hageman GS, Dean M, Allikmets R. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–62. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guymer RH, Heon E, Lotery AJ, Munier FL, Schorderet DF, Baird PN, McNeil RJ, Haines H, Sheffield VC, Stone EM. Variation of codons 1961 and 2177 of the Stargardt disease gene is not associated with age-related macular degeneration. Arch Ophthalmol. 2001;119:745–51. doi: 10.1001/archopht.119.5.745. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–32. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Hancox LS, Taiber AJ, Gehrs KM, Anderson DH, Johnson LV, Radeke MJ, Kavanagh D, Richards A, Atkinson J, Meri S, Bergeron J, Zernant J, Merriam J, Gold B, Allikmets R, Dean M. Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: characterization, ethnic distribution and evolutionary implications. Ann Med. 2006;38:592–604. [PMC free article] [PubMed] [Google Scholar]
- Hahn P, Qian Y, Dentchev T, Chen L, Beard J, Harris ZL, Dunaief JL. Disruption of ceruloplasmin and hephaestin in mice causes retinal iron overload and retinal degeneration with features of age-related macular degeneration. Proc Natl Acad Sci U S A. 2004;101:13850–5. doi: 10.1073/pnas.0405146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HapMap CI. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Merriam JE, Klaver CC, Zernant J, Bergen AA, Smith RT, Chang S, Merriam JC, Allikmets R. Evaluation of the ARMD1 locus on 1q25-31 in patients with age-related maculopathy: genetic variation in laminin genes and in exon 104 of HEMICENTIN-1. Ophthalmic Genet. 2004;25:111–9. doi: 10.1080/13816810490514342. [DOI] [PubMed] [Google Scholar]
- Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38:1173–7. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358:2606–17. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Klein BE, Klein R, Fox K, Millard C, Capriotti J, Russo K, Lee KE, Elston RC, Iyengar SK. Genome-wide analyses demonstrate novel loci that predispose to drusen formation. Invest Ophthalmol Vis Sci. 2005;46:3081–8. doi: 10.1167/iovs.04-1360. [DOI] [PubMed] [Google Scholar]
- Kanda A, Chen W, Othman M, Branham KE, Brooks M, Khanna R, He S, Lyons R, Abecasis GR, Swaroop A. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci U S A. 2007;104:16227–32. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Choi KO, Kim BD, Kim S, Kim YJ. FESD: a Functional Element SNPs Database in human. Nucleic Acids Res. 2005;33:D518–22. doi: 10.1093/nar/gki082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur I, Hussain A, Hussain N, Das T, Pathangay A, Mathai A, Hussain A, Nutheti R, Nirmalan PK, Chakrabarti S. Analysis of CFH, TLR4, and APOE polymorphism in India suggests the Tyr402His variant of CFH to be a global marker for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47:3729–35. doi: 10.1167/iovs.05-1430. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambooij AC, van Wely KH, Lindenbergh-Kortleve DJ, Kuijpers RW, Kliffen M, Mooy CM. Insulin-like growth factor-I and its receptor in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2003;44:2192–8. doi: 10.1167/iovs.02-0410. [DOI] [PubMed] [Google Scholar]
- Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, Wade MS, Li Y, Liang L, Zareparsi S, Swaroop A, Abecasis GR. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–54. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Schultz DW, Weleber RG, Schain MB, Edwards AO, Matise TC, Acott TS, Ott J, Klein ML. Age-related macular degeneration–a genome scan in extended families. Am J Hum Genet. 2003;73:540–50. doi: 10.1086/377701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- Martin G, Schlunck G, Hansen LL, Agostini HT. Differential expression of angioregulatory factors in normal and CNV-derived human retinal pigment epithelium. Graefes Arch Clin Exp Ophthalmol. 2004;242:321–6. doi: 10.1007/s00417-003-0838-y. [DOI] [PubMed] [Google Scholar]
- Morcillo-Suarez C, Alegre J, Sangros R, Gazave E, de Cid R, Milne R, Amigo J, Ferrer-Admetlla A, Moreno-Estrada A, Gardner M, Casals F, Perez-Lezaun A, Comas D, Bosch E, Calafell F, Bertranpetit J, Navarro A. SNP analysis to results (SNPator): a web-based environment oriented to statistical genomics analyses upon SNP data. Bioinformatics. 2008;24:1643–4. doi: 10.1093/bioinformatics/btn241. [DOI] [PubMed] [Google Scholar]
- Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. Faseb J. 2000;14:835–46. [PubMed] [Google Scholar]
- Ogata N, Matsushima M, Takada Y, Tobe T, Takahashi K, Yi X, Yamamoto C, Yamada H, Uyama M. Expression of basic fibroblast growth factor mRNA in developing choroidal neovascularization. Curr Eye Res. 1996;15:1008–18. doi: 10.3109/02713689609017649. [DOI] [PubMed] [Google Scholar]
- Ormsby RJ, Ranganathan S, Tong JC, Griggs KM, Dimasi DP, Hewitt AW, Burdon KP, Craig JE, Hoh J, Gordon DL. Functional and structural implications of the complement factor H Y402H polymorphism associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:1763–70. doi: 10.1167/iovs.07-1297. [DOI] [PubMed] [Google Scholar]
- Rakic JM, Lambert V, Munaut C, Bajou K, Peyrollier K, Alvarez-Gonzalez ML, Carmeliet P, Foidart JM, Noel A. Mice without uPA, tPA, or plasminogen genes are resistant to experimental choroidal neovascularization. Invest Ophthalmol Vis Sci. 2003;44:1732–9. doi: 10.1167/iovs.02-0809. [DOI] [PubMed] [Google Scholar]
- Recalde S, Fernandez-Robredo P, Altarriba M, Salinas-Alaman A, Garcia-Layana A. Age-related macular degeneration genetics. Ophthalmology. 2008;115:916–916 e1. doi: 10.1016/j.ophtha.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Richardson AJ, Islam FM, Guymer RH, Cain M, Baird PN. A tag-single nucleotide polymorphisms approach to the vascular endothelial growth factor-A gene in age-related macular degeneration. Mol Vis. 2007;13:2148–52. [PubMed] [Google Scholar]
- Rivera A, Fisher SA, Fritsche LG, Keilhauer CN, Lichtner P, Meitinger T, Weber BH. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–36. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- Rivera A, White K, Stohr H, Steiner K, Hemmrich N, Grimm T, Jurklies B, Lorenz B, Scholl HP, Apfelstedt-Sylla E, Weber BH. A comprehensive survey of sequence variation in the ABCA4 (ABCR) gene in Stargardt disease and age-related macular degeneration. Am J Hum Genet. 2000;67:800–13. doi: 10.1086/303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Haines JL, Postel EA, Agarwal A, Kwan SY, Gilbert JR, Pericak-Vance MA, Scott WK. Joint effects of smoking history and APOE genotypes in age-related macular degeneration. Mol Vis. 2005;11:941–9. [PubMed] [Google Scholar]
- Schmidt S, Hauser MA, Scott WK, Postel EA, Agarwal A, Gallins P, Wong F, Chen YS, Spencer K, Schnetz-Boutaud N, Haines JL, Pericak-Vance MA. Cigarette smoking strongly modifies the association of LOC387715 and age-related macular degeneration. Am J Hum Genet. 2006;78:852–64. doi: 10.1086/503822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Postel EA, Agarwal A, Allen IC, Jr, Walters SN, De la Paz MA, Scott WK, Haines JL, Pericak-Vance MA, Gilbert JR. Detailed analysis of allelic variation in the ABCA4 gene in age-related maculopathy. Invest Ophthalmol Vis Sci. 2003;44:2868–75. doi: 10.1167/iovs.02-0957. [DOI] [PubMed] [Google Scholar]
- Schultz DW, Klein ML, Humpert A, Majewski J, Schain M, Weleber RG, Ott J, Acott TS. Lack of an association of apolipoprotein E gene polymorphisms with familial age-related macular degeneration. Arch Ophthalmol. 2003;121:679–83. doi: 10.1001/archopht.121.5.679. [DOI] [PubMed] [Google Scholar]
- Seddon JM, Ajani UA, Mitchell BD. Familial aggregation of age-related maculopathy. Am J Ophthalmol. 1997;123:199–206. doi: 10.1016/s0002-9394(14)71036-0. [DOI] [PubMed] [Google Scholar]
- Seitsonen S, Lemmela S, Holopainen J, Tommila P, Ranta P, Kotamies A, Moilanen J, Palosaari T, Kaarniranta K, Meri S, Immonen I, Jarvela I. Analysis of variants in the complement factor H, the elongation of very long chain fatty acids-like 4 and the hemicentin 1 genes of age-related macular degeneration in the Finnish population. Mol Vis. 2006;12:796–801. [PubMed] [Google Scholar]
- Shroyer NF, Lewis RA, Yatsenko AN, Wensel TG, Lupski JR. Cosegregation and functional analysis of mutant ABCR (ABCA4) alleles in families that manifest both Stargardt disease and age-related macular degeneration. Hum Mol Genet. 2001;10:2671–8. doi: 10.1093/hmg/10.23.2671. [DOI] [PubMed] [Google Scholar]
- Skerka C, Lauer N, Weinberger AA, Keilhauer CN, Suhnel J, Smith R, Schlotzer-Schrehardt U, Fritsche L, Heinen S, Hartmann A, Weber BH, Zipfel PF. Defective complement control of factor H (Y402H) and FHL-1 in age-related macular degeneration. Mol Immunol. 2007;44:3398–406. doi: 10.1016/j.molimm.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Souied EH, Ducroq D, Rozet JM, Gerber S, Perrault I, Munnich A, Coscas G, Soubrane G, Kaplan J. ABCR gene analysis in familial exudative age-related macular degeneration. Invest Ophthalmol Vis Sci. 2000;41:244–7. [PubMed] [Google Scholar]
- Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto S, Tamura H, Ue T, Yamane K, Maruyama H, Kawakami H, Kiuchi Y. A polymorphism of LOC387715 gene is associated with age-related macular degeneration in the Japanese population. Neurosci Lett. 2007;414:71–4. doi: 10.1016/j.neulet.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Thornton J, Edwards R, Mitchell P, Harrison RA, Buchan I, Kelly SP. Smoking and age-related macular degeneration: a review of association. Eye. 2005;19:935–44. doi: 10.1038/sj.eye.6701978. [DOI] [PubMed] [Google Scholar]
- Tobler AR, Short S, Andersen MR, Paner TM, Briggs JC, Lambert SM, Wu PP, Wang Y, Spoonde AY, Koehler RT, Peyret N, Chen C, Broomer AJ, Ridzon DA, Zhou H, Hoo BS, Hayashibara KC, Leong LN, Ma CN, Rosenblum BB, Day JP, Ziegle JS, De La Vega FM, Rhodes MD, Hennessy KM, Wenz HM. The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. J Biomol Tech. 2005;16:398–406. [PMC free article] [PubMed] [Google Scholar]
- Utheim OA, Ritland JS, Utheim TP, Espeseth T, Lydersen S, Rootwelt H, Semb SO, Elsas T. Apolipoprotein E genotype and risk for development of cataract and age-related macular degeneration. Acta Ophthalmol. 2008;86:401–3. doi: 10.1111/j.1600-0420.2007.01070.x. [DOI] [PubMed] [Google Scholar]
- van Leeuwen R, Klaver CC, Vingerling JR, Hofman A, de Jong PT. The risk and natural course of age-related maculopathy: follow-up at 6 1/2 years in the Rotterdam study. Arch Ophthalmol. 2003;121:519–26. doi: 10.1001/archopht.121.4.519. [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Tsai HJ, Mah TS, Schmidt S, Postel EA, Agarwal A, Haines JL, Pericak-Vance MA, Rosenfeld PJ, Paul TO, Eller AW, Morse LS, Dailey JP, Ferrell RE, Gorin MB. Age-related maculopathy: a genomewide scan with continued evidence of susceptibility loci within the 1q31, 10q26, and 17q25 regions. Am J Hum Genet. 2004;75:174–89. doi: 10.1086/422476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger M, Renner W, Steinbrugger I, Kofer K, Wedrich A, Groselj-Strele A, El-Shabrawi Y, Schmut O, Haas A. Association of the HTRA1 -625G>A promoter gene polymorphism with exudative age-related macular degeneration in a Central European population. Mol Vis. 2007;13:1274–9. [PubMed] [Google Scholar]
- Yates JR, Sepp T, Matharu BK, Khan JC, Thurlby DA, Shahid H, Clayton DG, Hayward C, Morgan J, Wright AF, Armbrecht AM, Dhillon B, Deary IJ, Redmond E, Bird AC, Moore AT. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–61. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- Yu J, Wiita P, Kawaguchi R, Honda J, Jorgensen A, Zhang K, Fischetti VA, Sun H. Biochemical analysis of a common human polymorphism associated with age-related macular degeneration. Biochemistry. 2007;46:8451–61. doi: 10.1021/bi700459a. [DOI] [PubMed] [Google Scholar]


