Introductory Paragraph
Differential gene expression profiling can be used to uncover the mechanisms by which GWAS associations contribute to pathology1,2. Given that most GWAS hits are regulatory and transcript abundance is in a sense closer to the phenotype2, we hypothesized that summation of risk-allele associated gene expression, namely a Transcriptional risk score (TRS), should provide accurate estimates of disease risk. We integrate summary-level GWAS and eQTL data with RNA-Seq from the RISK study, an inception cohort of pediatric Crohn's disease (CD)3,4. We show that TRS based on genes regulated by IBD variants not only outperform Genetic risk scores (GRS) in distinguishing CD from healthy samples, but also serve to identify patients who in time will progress to complicated disease. Furthermore, our dissection of eQTL effects may be used to distinguish genes whose association with disease is either through promotion or protection, thereby linking statistical association to biological mechanism. The TRS approach constitutes a potential strategy for personalized medicine that enhances inference from static genotypic risk assessment
While GWAS have been very successful in identifying thousands of genetic variants associated with disease, the predictive performance of GRS is limited by the amount of heritability they explain, which is usually low5-8. Given that the majority of variants discovered by GWAS are likely to influence gene regulation, risk scores based on gene expression could constitute an alternative to classical genetic risk scores (GRS). We explored the performance of the TRS in the RISK study, which was designed to identify factors that increase risk of a complicated course of disease and included ileal biopsies from 215 complication-free CD patients and 35 controls profiled at diagnosis with RNA-Seq3,4,9. After careful monitoring for 3 years, 27 of the CD patients progressed to stricturing or penetrating disease, allowing us to ask whether genomic profiles could be used to inform mechanisms of pathogenesis and predict disease status.
We started by considering 232 independent SNPs associated with inflammatory bowel disease (IBD) or one of its main forms, Crohn's disease (CD) or Ulcerative colitis (UC)10. Assigning relevant genes at GWAS loci can be challenging, but eQTL studies provide an effective way to uncover which gene is likely to account for the discovered pathogenic effects. We queried the blood eQTL browser (see URLs), a large meta-analysis of eQTL effects on peripheral blood11, to ascertain genes regulated by IBD-predisposing variants. Around half (n=122, 52.6%) of IBD SNPs act as or are in strong LD (r2>0.8) with at least one cis-eQTL in peripheral blood, for a total of 157 independent candidate genes (∼1.3 candidate genes per SNP, Supplementary Table 1).
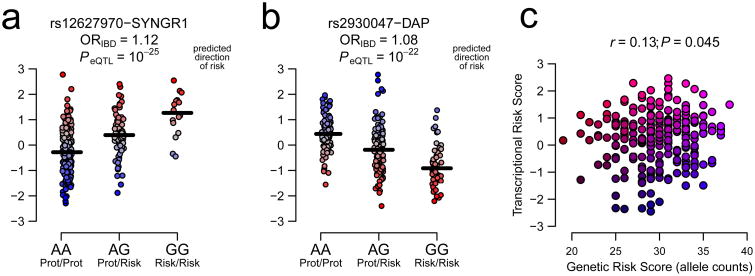
The RNA-Seq samples from the RISK study consist of ileal biopsies, so we next asked whether the aforementioned eQTLs are also active in small intestine (Online Methods). In line with previous studies12-14, we observed considerable sharing of signals (Supplementary Table 2), with strong concordance in direction of effects (70%, P=1.7×10-6, sign test) and including just two cases of reversal of sign between blood and ileum confirmed in GTEx (PNKD and RGS14, Supplementary Fig. 1). This overlap indicates that eQTL effects at IBD-associated SNPs can be used to polarize gene expression relative to risk in order to understand which allele is associated with pathogenesis at each gene. For instance, IBD-risk allele G at rs12627970 increases abundance of SYNGR1 (Fig. 1a), whereas risk allele G at rs2930047 downregulates DAP (Fig. 1b). We can hence polarize transcript abundance, so that in these examples predicted risk of IBD would be highest in individuals with high and low expression of SYNGR1 and DAP, respectively. Summing z-scores over all contributing transcripts identified as eQTL in blood, the TRS is correlated with the GRS, but suggests that different individuals have the highest risk of disease (Fig. 1c).
Figure 1. Transcriptional Risk Scores (TRS) integrate GWAS and eQTL results to measure individual risk of disease based on transcript abundance.
(a) Allele rs12627970-G increases susceptibility to IBD and is associated with elevated expression of SYNGR1. Some individuals with the risk genotype GG show average or even low expression levels, and some individuals with the protective genotype AA have high expression, suggesting that abundance of SYNGR1 provides a different estimate of individual risk of disease than the genotype. (b) By contrast, risk allele rs2930047-G is associated with lower expression of DAP, implying that reduced levels of DAP increase risk of IBD, and hence that inversion of the z-score measures polarized risk of disease. (c) Summation of the polarized transcriptional activity according to eQTL activity (the left-hand y-axis in panels a) and b) summed over all genes, and further standardized, is correlated with an allelic sum GRS plotted on the x-axis, but provides an independent predictor of IBD.
A TRS based on all 157 candidate genes ascertained from the blood eQTL browser distinguishes CD from control individuals (ΔSD=0.51; P=0.0019, Supplementary Fig. 2a), but with just a slight improvement on the performance of a classical weighted allelic sum GRS based on the very same IBD-associated SNPs that also have eQTL activity (ΔSD=0.51; P=0.02). However, this set includes some number of false joint associations due for example to pleiotropy, linkage, or reduced effect sizes in ileum. Several recent methods such as coloc and SMR have been developed to ask in a formal statistical framework whether independent signals are consistent with the same variant producing the signal in both studies15-17. We ran coloc15 for all 157 independent genes (Online Methods), and prioritized 29 genes that have the strongest evidence for colocalization of association signals (H4>80% using GWAS p-values for CD, UC and IBD, Supplementary Table 3 and Supplementary Fig. 3).
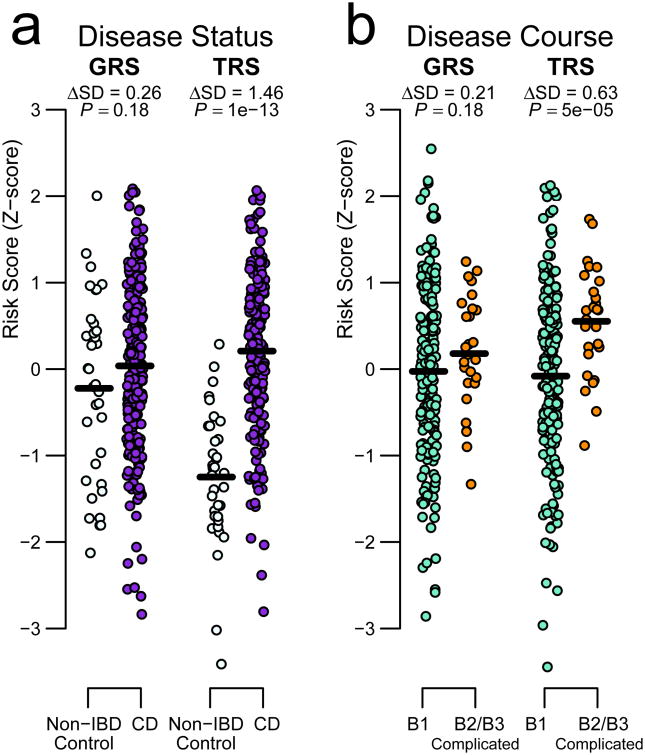
The high confidence set of 29 candidate genes excelled both at distinguishing disease status (Fig. 2a) as well as progression to complicated disease, namely stricturing (B2) or penetrating/fistulizing (B3) disease according to the Montreal classification system (Fig. 2b). The TRS distribution of Crohn's disease samples was highly significantly greater than that of non-IBD individuals, who almost entirely fall below the mean risk score of the cases (ΔSD=1.46; P=1×10-13). Similarly, the small group that progressed to complicated disease showed significantly higher scores than individuals who remained in the milder B1 state (ΔSD=0.63; P=5×10-5). Importantly, this discrimination appears regardless of tissue inflammation, since inflamed and non-inflamed B1 individuals have similar TRS scores (Supplementary Fig. 4). To ensure the robustness of these observations, we repeated the analyses based on a partially overlapping set of 39 genes detected by SMR as targets of IBD-associated variants (PSMR<2.3×10-4, 5% Bonferroni, Online Methods and Supplementary Table 4). This larger list of genes rendered similar results, distinguishing again between B1 and B2/B3 disease behavior (TRS: ΔSD=0.44; P=0.007, Supplementary Fig. 5a,b), confirming the power of TRS.
Figure 2. Transcriptional Risk Scores based on ileal gene expression at diagnosis distinguish status and course of Crohn's disease.
A total of 29 genes were predicted by coloc to be the target of the association with IBD discovered by GWAS. In contrast to classical GRS based on allele counts, risk scores based on summation of standardized expression of these IBD-associated genes (TRS), after polarization according to direction of risk, (a) distinguish between individuals with Crohn's disease (n-210) and (n=35) controls and (b) between CD patients that remain in B1 (n=183) and CD patients that go on to develop complicated disease (B2 and/or B3) within three years after diagnosis (n=27). Differences between groups (in SD units) along with p-values (two-sided t-test) are reported for each comparison.
In contrast, none of the comparisons between Montreal classification groups rendered significant differences when using the corresponding GRS based on GWAS-associated SNPs (e.g. ΔSD=0.26; P=0.18 and ΔSD=0.21; P=0.18, respectively, for the loci ascertained by coloc Fig. 2a,b). Furthermore, genome-wide polygenic risk scores (PRS) assessed using LD pruning8 across the full range of inclusion thresholds failed to approach the TRS performance, peaking at ΔSD=0.69 and P=9×10-4 for 668 SNPs at p<0.001 for the disease-control comparison (Supplementary Fig. 6). Consistent with recent GWAS results indicating independent genetic contributions to susceptibility and prognosis in Crohn's disease18, no PRS approached significance for disease progression, which further highlights the enhanced resolution provided by TRS.
The above results are based on ileal gene expression profiles, but using eQTL that are likely enriched for immune functions, since they were detected in blood from healthy adults. Applying the approach to an independent sample of peripheral blood gene expression, the TRS also distinguished 61 pediatric Crohn's disease cases and 12 controls (ΔSD=1.2; P=4×10-5). We next hypothesized that ileal mucosal samples might include effects that are not observed in peripheral blood, but can be important for IBD pathology, and hence likely to improve the power of TRS. eQTL mapping in 365 RISK samples identified associations at P<10-5 for 40 SNPs with 46 genes that fall in the vicinity (<1Mb) of the 232 SNPs associated with IBD (Online Methods, Supplementary Table 5). These include previously known active associations such as FUT2 and ERAP214,19,20. The list of ileum effects includes 27 genes not described in the blood eQTL browser, 7 of which were selected by coloc as having joint eQTL and GWAS effects consistent with a causal contribution to IBD (H4>80% for the three considered phenotypes, Supplementary Table 5). A TRS based on this short list of 7 loci, using the direction of effect of each eQTL in ileum to polarize risk, failed to separate samples according to disease status (ΔSD=0.17; P=0.32) or course of disease (ΔSD=-0.11; P=0.53). Surprisingly, a 14-gene TRS including 7 more ileum-specific loci exclusively detected by SMR also failed to discriminate cases and controls.
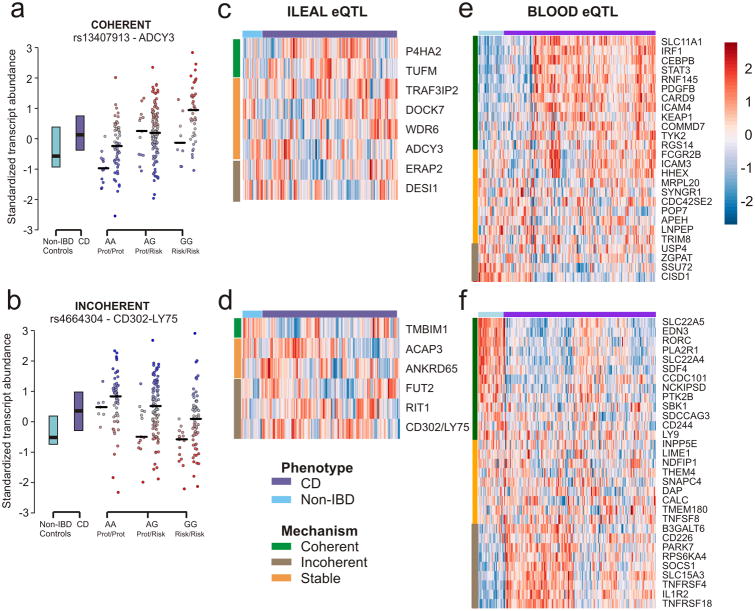
In addition to cis-effects, gene expression is also influenced by a combination of trans-acting genetic effects and environmental effects, both of which tend to produce coordinated patterns of gene expression that may disrupt the expected coherence of the signs of the eQTL and GWAS effects21,22. Specifically, IBD pathology is accompanied by altered expression of many genes as a response to altered intestinal microbiota23,24. For example, Fig. 3a shows how ADCY3 is upregulated in Crohn's disease individuals, consistent with the eQTL direction shown by IBD risk allele rs13407913-G (β=0.14; P=4×10-16), whereas CD302-LY75 is induced in the mucosa of Crohn's patients despite being down-regulated by the GWAS risk allele rs4664304-G (β=-0.065; P=4×10-7, Fig. 3b). Detailed exploration on a gene-by-gene basis shown on Fig. 3c,d suggests that this type of disruption may account for the poor performance of the 14-gene TRS based on ileum eQTL effects. The 3 genes reacting in a coherent fashion (ΔSD>0.3 between cases and controls in the predicted direction) enhance the TRS performance, but they are offset by the 5 genes whose incoherence (ΔSD>0.3 in the opposite direction) diminishes the TRS. The other 6 genes are stable with respect to disease status, not showing a significant difference in expression between cases and controls.
Figure 3. Gene expression polarized according to predicted direction of risk uncovers two divergent mechanisms of association with disease.
For about half of the eQTL, trans- and environmental effects result in coordinated modification of gene expression in cases relative to controls. (a) Example of a coherent association, where individuals with risk genotype GG show increased expression of ADCY3, consistent with the prediction based on the direction of effect of this allele as an eQTL in ileal tissue. Left and right columns of individual points with each genotype correspond to cases and controls respectively. (b) Example of an incoherent association, where in this example individuals with the risk allele have reduced expression in the opposite direction to the overall increased levels of CD302-LY75 in cases. (c,d) Considering eQTL discovered in ileal tissue, 8 genes are controlled by ileal eQTL that increase their expression (c), and 6 that decrease their expression (d). Purple and light blue bars above the heatmaps indicate cases (n=210) and controls (n=35) respectively, and orient how 3 genes are coherent (green bars), 5 are incoherent (red), and 6 stable (orange) with respect to disease. (e,f) Considering eQTL discovered in blood, 26 genes are upregulated by the allele associated with IBD (e), and 31 genes are downregulated (f). In this case, 25 genes are coherent, and just 13 incoherent. The heatmap is color-indexed according to the z-score of each gene from low (blue) to high (red) expression.
By contrast, for the 57 genes detected by either coloc or SMR as target genes based on eQTL effects in blood, there was a clear excess of coherent (n=25) associations over incoherent ones (n=13) (Fig. 3e,f). Clearly, most of the coherent and incoherent genes are strongly co-regulated, implying that environmental or other trans-effects mediate the paradoxical deviation between observed and predicted direction of effect, rather than confounding effects of secondary cis-acting alleles. Examples of incoherence include CD226, an immunoglobulin receptor involved in control of viral infection25 and implicated in several autoimmune diseases26, which is induced in CD patients (ΔSD=1.07) in spite of being downregulated by the GWAS risk allele rs727088-G (P=1×10-46, Supplementary Table 3). Similarly, TNFRSF18 is a receptor of the TNF family with a key role in maintaining self-tolerance27,28 that is also induced in patients (ΔSD=1.49) even though the risk allele decreases its expression (Supplementary Table 3). The functional evidence for both genes suggests a scenario in which induction is protective (e.g. to clear infection in the gastrointestinal tract), and hence individuals with the GWAS risk allele are more prone to developing chronic inflammation because they fail to induce expression sufficiently to fully engage in the defensive response.
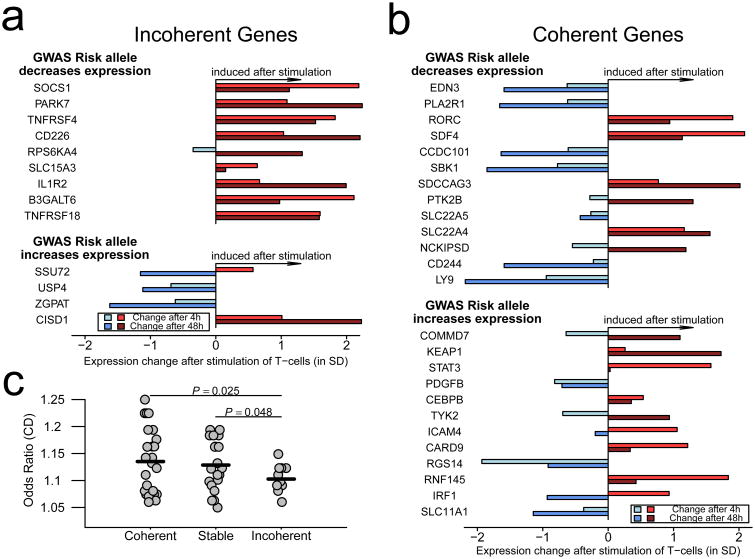
Consistent with this interpretation, analysis of the ImmVar consortium data on 4h and 48h ex vivo response to stimulation29 indicates a common theme for all thirteen incoherent genes. The nine genes that are incoherently upregulated in CD patients are also induced in CD4+ T cells after 48h stimulation with anti-CD3/CD28 beads, whereas three of four genes that are incoherently downregulated in affected individuals are also suppressed after immune stimulation (Fig. 4a). The coherently regulated genes do not show such a consistent pattern (Fig. 4b), suggesting that their disease response may not be due to immune stimulation. This difference between the two sets of genes is significant (p=0.03, Fisher's exact test), and similar results apply to the effects of stimulation with LPS or infection with Influenza virus (not shown).
Figure 4. Incoherent genes show similar patterns in stimulated immune cells and are more weakly associated with IBD according to GWAS.
Changes in gene expression after 4h and 48h in primary T cells stimulated with anti-CD3+CD28 beads as reported by the ImmVar Consortium. (a) All but one of the 13 incoherent genes one show changes in expression at 48h that mimic the inconsistent tendencies observed in CD patients of the RISK cohort. (b) Coherent genes show more diverse changes in patterns of expression. (c) Incoherent genes have significantly lower Odds Ratio of association to IBD by GWAS, than do coherent or stable genes.
Overall, the contrasting behavior of coherent and incoherent genes is consistent with the notion that gene-regulatory IBD risk alleles have detrimental effects through two different mechanisms: some directly promote disease because they regulate gene expression in a manner that is inherently pathogenic, and others fail to safeguard individuals by insufficiently engaging in protective shifts of gene expression. Intriguingly, the latter class generally has odds ratios around 1.1, which is significantly lower than for the remainder (Fig. 4c). Biopsy gene expression profiling of larger cohorts should confirm this inference and further refine our ability to distinguish active and protective risk mechanisms. Other interpretations are also possible, including the possibility that eQTL effects in the ileum are not contributing strongly to pathogenesis, and processes unique to individual genes. An excellent example of the latter is the one incoherent gene which contravenes our model, CISD1, which encodes mitoNEET, an Fe/S-domain protein localized to the mitochondria where it is required for redox sensing30. Mitochondrial function is protective against progression in Crohn's disease4,31, yet transcription of CISD1 is downregulated in patients overall, strongly induced in T-cells by ex vivo stimulation, and the risk allele increases expression.
The existence of incoherent associations highlights the fact that we have much to learn about the relationship between eQTL effects and disease pathogenesis. This phenomenon is likely also to apply to other autoimmune and inflammatory diseases, and further dissection will in turn improve the development of TRS that are predictive of progression to complicated disease, with implications for therapeutic treatment.
URLs
Blood eQTL browser, http://genenetwork.nl/bloodeqtlbrowser/;
GTEx, http://www.gtexportal.org/home/;
IIBDGC trans-ancestry meta-analyses association data, https://www.ibdgenetics.org/downloads.html;
SMR, http://cnsgenomics.com/software/smr/index.html;
coloc R package, https://cran.r-project.org/web/packages/coloc/index.html;
1000 Genomes Project, http://www.internationalgenome.org/1000-genomes-browsers;
Online Methods
Cohort and Outcome Classification
The RISK study is an observational prospective cohort study that aims to develop risk models for predicting complicated course in children with Crohn's disease (CD). From 2008 to 2012, the RISK study recruited more than 1,800 treatment-naive patients with suspected diagnosis of CD at 28 pediatric gastroenterology centers in North America3,4. This disorder is a chronic inflammatory condition of the gastrointestinal tract that results from an inappropriate activation of the immune system thought to be due to a combination of host genetic makeup, enteric flora and microbial or other pathological triggers. A minority of patients progress with time to complicated disease that may require surgery and/or intensive pharmacological therapy. We used the Montreal criteria to classify patients according to disease behavior, distinguishing non-complicated B1 disease (non-stricturing, non-penetrating disease) from complicated disease, composed by B2 (stricturing) and/or B3 (penetrating) behavior32,33.
We ascertained 245 samples from the RISK study that had been profiled with ileal RNA-Seq and genotyped with the IlluminaTM (San Diego, CA, USA) high-density Immunochip array. 35 of the ascertained individuals lacked gut inflammation and were classified as non-IBD controls. The remaining selected individuals showed persisting CD and remained in complication-free B1 status at least through 90 days from initial diagnosis. After three years of follow-up, 17 and 10 patients progressed to B2 and B3 status, respectively. We joined the latter 27 samples to form a group of patients with complicated disease course. The majority of individuals were of European ancestry (n=210, 85.7%), with smaller fractions of samples with African (n=10, 4.1%) and other/mixed (n=25, 10.2%) ethnic origins. More details about outcome classification are available in Kugathasan et al4.
Along with disease behavior, disease location plays a key role in the natural history and clinical course of patients diagnosed with CD. Since a recent study showed that GRS for IBD distinguishes patients with ileal/ileocolonic disease with those with only colonic disease34, we asked whether the TRS also distinguishes these two classes of CD. According to the Paris modification32 of the Montreal classification, pediatric disease is also classified into L1 (ileal only), L2 (colonic only), L3 (ileocolonic) and L4 (upper GI tract). For Supplementary Figure 4 we combined L1 and L3 into inflamed B1 since the biopsies were taken from the ileum, whereas L2 is un-inflamed relative to the site of biopsy. No L4 cases were available. The analysis confirms that the TRS indeed distinguishes L1/L3 from L2 disease.
Processing of RNA-Seq from Ileal biopsies and SNP data from the RISK Cohort
RNA was isolated from ileal biopsies obtained from colonoscopy at diagnosis, and profiles of gene expression were determined using RNA-Seq as previously reported. Reads were mapped to the human genome (hg19) with TopHat 2.0.13 using default parameters35. Aligned reads were transformed with SAMtools36 to quantify the number of reads at the gene level with HTSeq-0.6.137 using default “union” mode. Raw counts were compiled and processed with edgeR38 to obtain normalized counts through trimmed mean of M-values normalization. An in-house R script was then used to inverse rank transform expression estimates for each gene into a standard normal distribution with mean 0 and variance 1. For comparison with GTEx, the data was further transformed into the ‘reads per kilobase per million mapped reads’ metric (RPKM)39 and 13,769 genes with RPKM>1 and >6 read counts in at least 10 individuals were retained. The correlation between the median RPKM per gene in RISK with the median RPKM per gene in 53 tissues available from GTEx (GTEx_Analysis_v6_RNA-seq_RNA-SeQCv1.1.8_gene_median_rpkm.gct, see URLs) had a median Spearman correlation of 0.57 (range 0.39 – 0.88), with the largest correlations corresponding to GTEx “Small_Intestine.Terminal_Ileum” (rs=0.88), “Colon.Transverse” (rs=0.79) and “Stomach” (rs=0.72), confirming similarity of the RISK biopsy data to an external bowel dataset.
The Immunochip was designed to densely genotype 186 distinct loci containing markers associated at genome-wide significance levels (P<5×10-8) with 12 autoimmune and inflammatory diseases, including Crohn's disease and ulcerative colitis. The array was designed to contain all 1000 Genomes pilot phase (September 2009 release) SNPs within 0.1cM recombination blocks (HapMap 3 CEU) around the top associated marker by GWAS40. The initial calling of the Immunochip array before QC contained 192,523 variants. We used the Bioconductor SNPlocs.Hsapiens.dbSNP.20120608 package41 to map autosomal SNPs to GRCh37 and remove i) non-biallelic variants, ii) SNPs not in HW equilibrium (P<10-3) and iii) variants not present in the 1000 Genomes Phase I variant set (March 2012 release). At this point there were 161,540 remaining SNPs. We further removed 49,253 variants with MAF<5% and 10,874 SNPs with >1% missing data rate across all individuals. After QC there were 101,413 genotyped variants available for analysis and all 245 individuals presented <0.1% genotype missing rates. To check relatedness among samples, we calculated pairwise IBD based on 26,233 SNPs obtained after LD pruning using the PLINK routine “—indep 50 5 2”, confirming minimal overall relatedness (PI_HAT<0.05 for 99.3% pairwise comparisons) with just three pairs of first-degree relatives (PI_HAT>0.25).
Selection of SNPs and candidate genes associated to IBD by GWAS
Because our goal was to uncover genes involved in susceptibility to CD, we considered as candidates all genes with a transcription start site (TSS) located ±1 Mb of each of the 232 independent GWAS SNPs previously associated with Inflammatory Bowel Disease10. We examined 7,389 SNP-gene pairs, including 6,180 unique candidate genes (32 genes considered per SNP on average, range: 5 to 620 genes). The eQTL blood browser (see URLs) was queried to ascertain which genes are under control of IBD-associated SNPs. We observed 163 instances in which the GWAS SNP (n=129) or a SNP in LD (n=34, at r2>0.8 in 1KG CEU) act as eQTL (FDR<5%) for the candidate gene located <1Mb from the associated SNP (Supplementary Table 1). In total this resulted in selection of 157 unique genes (six genes were under control of two different IBD SNPs).
Mapping study in RISK cohort to build ileal TRS
A fraction of eQTL variants are known to act in a tissue-specific manner13. We used the RISK ileal biopsies to perform a targeted eQTL study focused on the 7,389 SNP-gene pairs. This analysis aimed to confirm whether eQTL discovered in peripheral blood are also shared in ileal tissue, and to detect ileal-specific eQTLs that can be used to pinpoint at new pathogenic candidate genes.
We applied several QC steps to remove batch effects and normalize the matrix of gene expression in order to carry out the eQTL mapping study. First, we performed a sex incompatibility check comparing the gender recorded for each individual to the expression of XIST and chrY genes EIF1AY, RPS4Y1, DDX3Y and KDM5D. A heatmap based on the expression of the five genes did not show any gender mismatch. Next, we tried to identify outlier samples using D-statistics as done by GTEx13. For each sample, mean correlation of expression with the remaining samples was calculated. All samples showed D>0.9 with no obvious visual outliers from the average correlation of 0.972, and hence all samples were kept for further analysis.
Finally, supervised normalization procedures were used to remove global effects present in the matrix of expression data. The transcriptome shows pervasive co-regulation of transcript abundance that leads to modules of co-regulated genes that share similar biological functions42. Biological variables such as disease can also induce massive changes in gene expression (e.g. thousands of genes are differentially expressed among groups in the RISK study4). Moreover, hidden batch effects and other unknown cofounders can induce spurious correlations at the genome-wide level. All these sources of biological and/or technical variability can hamper the detection of local acting cis-eQTL. We first used unsupervised surrogate variable analysis (SVA)41 to identify hidden confounding factors, deliberately protecting known variables such as gender and disease status (to be included as covariates in the eQTL mapping step). The algorithm detected 14 surrogate variables (sv) that were removed using the supervised normalization of microarray (SNM) procedure42. Specifically, we fit gender and disease status as biological variables and removed the effects of the 14 estimated sv by including these as adjustment variables with the rm=T option.
The eQTL mapping was performed using a linear mixed model implemented in GEMMA43, which allows adjustment for population structure and relatedness among individuals as a random effect through a genetic relationship matrix (GRM) based on the LD-pruned SNP dataset. We tested for associations between genotype and normalized gene expression, including gender and disease status as covariates. Supplementary Table 2 reports association results for 136 available SNP-gene pairs (ileal eQTL association data was not available for the remaining 21 pairs).
Gene selection with SMR and coloc
Detection of nominally significant associations both for eQTL and with IBD at a single SNP does not necessarily imply that the SNP is responsible for both effects. Several recent methods have been designed to increase confidence that colocalization of signals implies that the gene affected by the regulatory SNP is also responsible for the trait association. Coloc uses a Bayesian framework to infer whether the two signals are due to a single site, or two sites in linkage disequilibrium within a genomic region of interest15. It calculates posterior probabilities to quantify the support for five different hypotheses regarding the presence and sharing of causal variants between the two traits under consideration. Similarly, summary-data based Mendelian randomization (SMR) combines GWAS and eQTL summary association data to prioritize target genes with evidence for causal or pleiotropic effects17. We applied both methods to ascertain target genes from the list of 157 aforementioned candidate genes.
We used GWAS summary data for CD, UC and IBD from the publicly available IIBDGC GWAS plus Immunochip trans-ancestry MANTRA meta-analyses (see URLs). For each of the three disease phenotypes, we processed the data considering the sample size indicated in Table 1 in Liu et al.10. For the eQTL effects, we used the cis-eQTL summary data from the largest existing immune-related dataset, namely the eQTL blood browser (see URLs), and converted the reported z-statistics into β and s.e. values following the guidelines from the SMR Supplementary Note by Zhu et al.17. The assigned sample size was 5,311, using Europeans from the 1000 Genomes Project as the reference sample for minor allele frequencies and LD patterns (see URLs). For the coloc analyses, we considered as validated target genes 29 independent loci with 80% or larger posterior probability of the hypothesis of one causal variant common to both traits (H4) for all three of the phenotypes. For the SMR analyses, Supplementary Figure 3a shows the strong relationship between the SMR p-value and highly significant p-values for both the GWAS and eQTL effects. This validates the selection of loci (such as red and brown dots in the figure) that passed Bonferroni for all three of the considered phenotypes (significance threshold P<2.3×10-4 for one phenotype since the p-values are highly correlated). However, it also highlights the likely dependence of the SMR statistic on the significance of the eQTL effects which in turn are strongly influenced by the sample size, as noted by Zhu et al17. In general, inclusion of more high confidence genes would be expected to improve TRS in part by reducing the variance of the score, and it is therefore likely that the small sample size for the ileal eQTL results contributes to the weaker diagnostic performance relative to the larger blood-derived gene set. We also replicated the case-control comparison with an analysis of 13 of the 26 genes recently reported from immune cell-type specific eQTL44 for which replicated directional effects could be inferred (ΔSD=0.73; P=3×10-5), but again larger sample sizes will be needed to establish a high-confidence set of such genes. Due to low density of variants on the Immunochip and the likely presence of multiple causal effects at each locus, computation and interpretation of SMR's HEIDI scores was compromised for half of the loci, and since only four were inferred to be unambiguously causal by this test (P<2.3×10-4 for the three phenotypes), it was deemed not useful for selection of genes for TRS computation. 15 of the loci are common to SMR and coloc, implying that the methods are complementary. Summary results for all considered genomic regions are available in Supplementary Tables 3 and 4.
In addition, we used coloc to select causal genes among the 27 genes controlled by ileum-specific eQTL (P<10-5) discovered in the mapping study described above. To do so, we extended the eQTL mapping study ±500 Kb around the susceptibility SNP for each of the genomic regions and processed the association data in order to run coloc and SMR on these regions. We considered as validated 7 target genes that showed H4>80% for all three of the disease phenotypes. Due to the low number of loci detected through coloc, we complemented the analyses with 7 more loci that passed SMR for all three phenotypes (Bonferroni adjusted P<0.00185 inclusion threshold for one phenotype). Summary results are available in Supplementary Table 5.
Calculation of GRS and TRS
We carried out several comparisons to contrast the predictive power of TRS vs. GRS based on the corresponding GWAS SNPs (those that act as eQTL for the selected genes). For GRS, we used the “score” routine available in PLINK to generate genetic risk scores weighted using the logOR for IBD from GWAS meta-analysis10 (reported in Supplementary Table 1; weighting by the logOR for CD rendered very similar scores at each comparison). In turn, the calculation of the TRS consisted of three steps. First, we used the eQTL activity of GWAS SNPs to infer the direction of risk at each gene selected for the TRS. We used “High Expr.” and “Low Expr.” (available in Supplementary Tables 1 to 5) to denote whether the risk allele associated with disease leads to increased (“High Expr.”) or decreased (“Low Expr.”) gene expression. Next, we polarized expression values so that elevated risk, irrespective of the sign of the effect on expression, adds to the TRS. This was done simply by changing the sign of the z-score for genes labelled as “Low Expr.” (e.g. expression z-scores of -1.5 and +1.3 would transform into +1.5 and -1.3, respectively as indicated on the y-axis of Fig. 1b). Finally, we obtained the TRS for each individual by summing the polarized z-scores over all genes and rank normalizing the distribution. We used t-tests to compare the performance of GRS and TRS between groups.
Calculation of PRS
Polygenic risk scores (PRS) have emerged as the gold standard for overall prediction from GWAS. We used the P+T (pruning+threshold)8 method to build a PRS based on independent SNPs that passed different significance thresholds in GWAS. To avoid loss of power due to the inclusion of correlated SNPs, we first selected 15,135 LD-pruned SNPs from the RISK Immunochip data (by running PLINK's indep-pairwise routine on 5,000 randomly selected individuals from the UK Biobank). Then, we used PLINK's score routine to calculate a battery of PRS based on variants selected across the complete spectrum of significance thresholds for inclusion (from 329 SNPs at P<0.00001 to 9,214 SNPs at P<0.5) in the IIBDGC GWAS plus Immunochip trans-ancestry MANTRA meta-analyses for IBD (see URLs). The performance of the PRS for both the case-control comparison and the indolent-complicated disease comparison at different thresholds is reported in Supplementary Figure 6. The performance of PRS between groups was tested through t-tests.
Coherence and incoherence
For the evaluation of coherence between eQTL and disease effects, we first evaluated whether each transcript is significantly differentially expressed between control and CD samples by at least 0.3 SD units (P∼0.05). Despite the small sample size of controls, clear co-regulation of the up- (Fig. 3c,e) or down- (Fig. 3d,f) regulated genes is clearly visualized. Next, we classified as coherent genes for which the direction of the eQTL effect is the same as the disease (that is, increased expression of the risk allele as well as elevated expression in cases relative to controls; or decreased expression of the risk allele and repressed expression in the cases). Incoherent genes are those with the opposite relationship (that is, either increased expression of the risk allele and repression in cases, or vice-versa). Stable genes are those without clear differential expression between cases and controls.
Whereas our initial proposal for TRS assumed no global impact of disease on gene expression2, the RISK dataset shows that fewer than half of the candidate genes are stable by the above definition. Coherence mathematically tends to enhance the performance of the TRS since it elevates the difference between cases and controls for each gene. By contrast, incoherence diminishes TRS performance since the polarized eQTL effect is counteracted by the influence of disease. Since there is an excess of incoherent associations for the ileal eQTL, the TRS performance is compromised. However, since there is no global differential expression of the GWAS candidate genes between B1 and complicated B2/B3 cases, coherence and incoherence do not affect the ability of the TRS to discriminate these conditions.
Functional evidence from ImmVar project
We used data from the ImmVar project (GEO accession GSE60235) to gain further insight into the coherent and incoherent behaviors detected for some genes included in the TRS. The dataset includes expression profiling with Affymetrix Human Gene 1.0 ST array of resting and activated T-cells from 15 healthy human individuals collected under 5 different conditions29. We downloaded the matrix of normalized gene expression and selected experiments corresponding to three conditions, namely, “Unstimulated 4hr” (n=15), “Activated 4hr” (n=15) and “Activated 48hr” (n=15). For each gene of interest, we transformed expression estimates into a standard normal distribution with mean 0 and variance 1 and performed pairwise comparisons to explore the changes in gene expression at 4h and 48h after stimulation with anti-CD3 and anti-CD28 beads. The changes in average z-score for the selected genes are reported in Fig. 4. We observed similar patterns for both coherent and incoherent genes analyzing a similar ImmVar project that profiled changes in monocyte-derived dendritic cell gene expression after stimulation with LPS or Influenza (GEO accession GSE53166)45.
Data availability
The RNA-Seq data for the 245 individuals included in this study have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE93624 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93624).
Supplementary Material
Acknowledgments
We are grateful to B. Zeng, D. Arafat, H. Somineni, S. Venkateswaran and colleagues from the Gibson and Kugathasan labs for their support and helpful comments. We also would like to thank I. Mendizabal, J. Lachance and K. Jordan for comments on the manuscript. This research was supported by Project 3 (GG, PI) of the NIH program project “Statistical and Quantitative Genetics” grant P01-GM0996568 (B. Weir, University of Washington, Director) as well as research grants from the Crohn's and Colitis Foundation of America (CCFA), New York (New York, USA) to the individual study institutions participating in the RISK study.
Footnotes
Author Contributions: U.M.M and G.G conceived the theoretical framework of the transcriptional risk scores. L.A.D., J.S.H. and S.K. participated in the conception and design of the RISK study. K.M., J.P., T.D.W., A.G., J.D.N., W.V.C., J.R.R., D.R.M., R.K., M.B.H., S.S.B., M.C.S., R.N.B., J.F.M., M.C.D., B.A., M-O. K, and J.C. recruited subjects, collected the data and worked on its curation and analysis. U.M.M. performed the TRS analyses. U.M.M. and G.G. interpreted the results and drafted the manuscript, while L.A.D., J.S.H. and S.K. assisted with interpretation and writing.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Fairfax BP, Knight JC. Genetics of gene expression in immunity to infection. Curr Opin Immunol. 2014;30:63–71. doi: 10.1016/j.coi.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson G, Powell JE, Marigorta UM. Expression quantitative trait locus analysis for translational medicine. Genome Med. 2015;7:60. doi: 10.1186/s13073-015-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haberman Y, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–33. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kugathasan S, et al. Prediction of complicated disease course for children newly diagnosed with Crohn's disease: a multicentre inception cohort study. Lancet. 2017 doi: 10.1016/S0140-6736(17)30317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witte JS, Visscher PM, Wray NR. The contribution of genetic variants to disease depends on the ruler. Nat Rev Genet. 2014;15:765–76. doi: 10.1038/nrg3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–8. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee N, Shi J, Garcia-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walters TD, et al. Increased effectiveness of early therapy with anti-tumor necrosis factor-alpha vs an immunomodulator in children with Crohn's disease. Gastroenterology. 2014;146:383–91. doi: 10.1053/j.gastro.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 10.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabakchiev B, Silverberg MS. Expression quantitative trait loci analysis identifies associations between genotype and gene expression in human intestine. Gastroenterology. 2013;144:1488–96. 1496 e1–3. doi: 10.1053/j.gastro.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Narzo AF, et al. Blood and Intestine eQTLs from an Anti-TNF-Resistant Crohn's Disease Cohort Inform IBD Genetic Association Loci. Clin Transl Gastroenterol. 2016;7:e177. doi: 10.1038/ctg.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hormozdiari F, et al. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am J Hum Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–7. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 18.Lee JC, et al. Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn's disease. Nat Genet. 2017;49:262–268. doi: 10.1038/ng.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ning K, et al. Improved integrative framework combining association data with gene expression features to prioritize Crohn's disease genes. Hum Mol Genet. 2015;24:4147–57. doi: 10.1093/hmg/ddv142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh T, et al. Characterization of expression quantitative trait loci in the human colon. Inflamm Bowel Dis. 2015;21:251–6. doi: 10.1097/MIB.0000000000000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 22.Gibson G, Weir B. The quantitative genetics of transcription. Trends Genet. 2005;21:616–23. doi: 10.1016/j.tig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 23.de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13–27. doi: 10.1038/nrgastro.2015.186. [DOI] [PubMed] [Google Scholar]
- 24.McGovern DP, Kugathasan S, Cho JH. Genetics of Inflammatory Bowel Diseases. Gastroenterology. 2015;149:1163–1176 e2. doi: 10.1053/j.gastro.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nabekura T, et al. Costimulatory molecule DNAM-1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity. 2014;40:225–34. doi: 10.1016/j.immuni.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol. 2015;15:243–54. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- 27.Petrillo MG, et al. GITR+ regulatory T cells in the treatment of autoimmune diseases. Autoimmun Rev. 2015;14:117–26. doi: 10.1016/j.autrev.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Reikvam DH, et al. Increase of regulatory T cells in ileal mucosa of untreated pediatric Crohn's disease patients. Scand J Gastroenterol. 2011;46:550–60. doi: 10.3109/00365521.2011.551887. [DOI] [PubMed] [Google Scholar]
- 29.Ye CJ, et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345:1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiley SE, et al. The outer mitochondrial membrane protein mitoNEET contains a novel redox-active 2Fe-2S cluster. J Biol Chem. 2007;282:23745–9. doi: 10.1074/jbc.C700107200. [DOI] [PubMed] [Google Scholar]
- 31.Novak EA, Mollen KP. Mitochondrial dysfunction in inflammatory bowel disease. Front Cell Dev Biol. 2015;3:62. doi: 10.3389/fcell.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine A, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis. 2011;17:1314–21. doi: 10.1002/ibd.21493. [DOI] [PubMed] [Google Scholar]
- 33.Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–53. doi: 10.1136/gut.2005.082909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cleynen I, et al. Inherited determinants of Crohn's disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2016;387:156–67. doi: 10.1016/S0140-6736(15)00465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 40.Onengut-Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–6. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mecham BH, Nelson PS, Storey JD. Supervised normalization of microarrays. Bioinformatics. 2010;26:1308–15. doi: 10.1093/bioinformatics/btq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–4. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chun S, et al. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat Genet. 2017;49:600–605. doi: 10.1038/ng.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee MN, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data for the 245 individuals included in this study have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE93624 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93624).