Abstract
The activity of protein kinases is often regulated in an intramolecular fashion by signaling domains, which feature several phosphorylation or protein-docking sites. How kinases integrate such distinct binding and signaling events to regulate their activities is unclear, especially in quantitative terms. We have used NMR spectroscopy to show how structural elements within the Abl regulatory module (RM) form synergistically a multilayered allosteric mechanism that enables Abl kinase to function as a finely-tuned switch. We dissected the structure and energetics of the regulatory mechanism to precisely measure the effect of various stimuli, activating or inhibiting, on the Abl kinase activity. The data provide the mechanistic basis for explaining genetic observations and reveal a novel activator region within Abl. Our findings show that drug-resistant mutations in the Abl RM exert their allosteric effect by promoting the activated state of Abl and not by decreasing the drug affinity for the kinase.
INTRODUCTION
The Abl kinase is involved in a number of signaling pathways that control cell growth, survival, invasion, adhesion and migration1–5. In Abl6–8, as in the other human cytoplasmic tyrosine kinases9,10, inhibition and activation is regulated allosterically through intramolecular interactions involving modular domains such as the SH3, SH2 and the kinase domain (KD) (Fig. 1a). Mutations or deletions in the Abl regulatory module (RM) give rise to aberrant forms of Abl, such as the Bcr-Abl, that cause leukemia and other cancers11. Even relatively small increases in activity are sufficient for Abl to cause or mediate tumorigenesis12. Abl RM provides binding sites for the anchoring of adaptor proteins as well as sites that are phosphorylated by other kinases, processes that further regulate Abl activity2,5 (Fig. 1a). Several mutations that confer resistance to the drug imatinib, a KD inhibitor, are located in the RM13,14, but the mechanisms of action remain elusive.
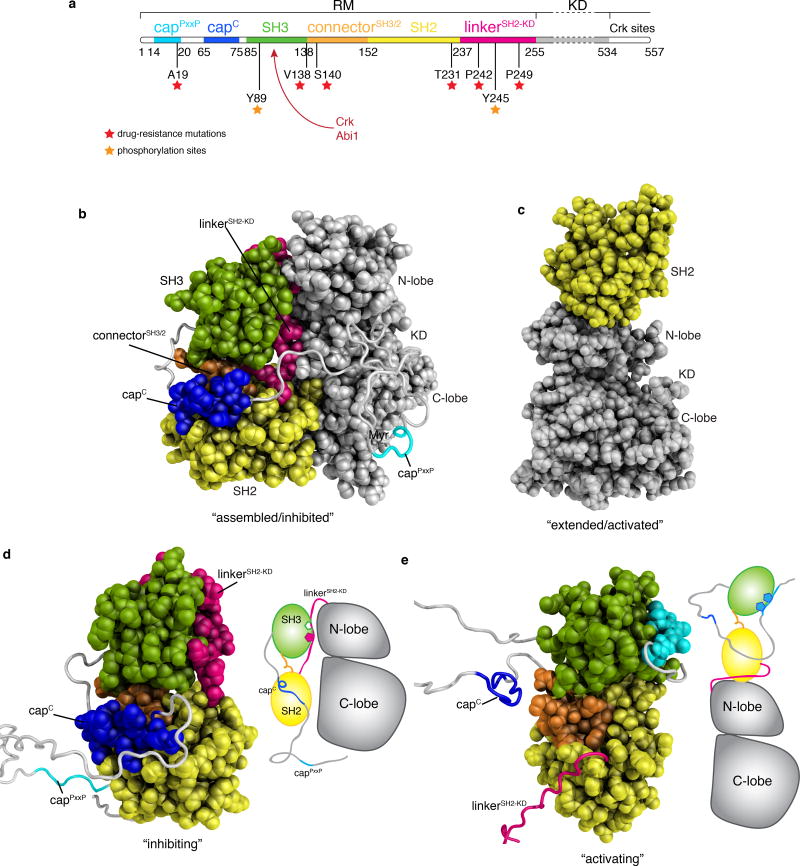
Figure 1.
Structures of Abl. (a) Domain organization of the first N-terminal 557 residues of Abl. The regulatory module (RM) consists of the first 255 residues and its domains and motifs are indicated. The kinase (catalytic) domain (KD) encompasses residues 255–534, whereas the region following (residues 534–557) is disordered and includes a binding site for Crk. Position of imatinib-resistant mutation sites are denoted by red asterisks. Tyrosine phosphorylation sites by Hck and Src kinases are denoted by orange asterisks. CrkSH2 and Abi1 binding sites are indicated by a red arrow. (b) Crystal structure (PDB ID 2FO0) of the myristoylated form of Abl in the assembled state8. The first ~56 N-terminal residues were not visible in the crystal structure and were modeled here as a disordered segment (grey). The various domains and motifs are colored per the color code in panel a. Myr indicates the myristate moiety. (c) Crystal structure (PDB ID 4XEY) of the Abl (SH2-KD) in the extended state12. (d) The lowest-energy solution structure of the isolated Abl RM in its inhibiting state is shown as a space-filling model for the structured regions and as ribbon for the disordered regions. A cartoon of Abl is shown on the right indicating that the isolated Abl RM in its inhibiting state is compatible with the formation of the assembled state of Abl. (e) The lowest-energy solution structure of the isolated Abl RM in its activating state is shown as a space-filling model for the structured regions and as ribbon for the disordered regions. A cartoon of Abl is shown on the right indicating that the isolated Abl RM in its inhibiting state is compatible with the formation of the extended state of Abl. The structures of the two states are compared in Figure 2.
The Abl RM has been observed to dock onto the KD in two distinct modes: in the assembled state6,8,15 the N-terminal myristoyl group is inserted into a pocket in the C-lobe and stabilizes the docking of the RM at the back of the KD giving rise to an autoinhibited form (Fig. 1b); in the extended state the SH2 domain binds to the top of the N-lobe of the KD yielding an activated kinase form (Fig. 1c). The extended Abl conformation15–17 is the most active form of Abl and has been associated with a drastically increased leukemogenic activity14,16,18. Transitions between the inhibited and activated states are central in Abl regulation but are poorly understood. Abl, like many other kinases, is known to function as a graded rather than simply an “on” and “off” switch19,20. Given the dynamic nature of protein kinases21–23 insight into their conformational and energetic landscape24–26 is required to fully understand the underlying mechanisms of regulation and how kinases respond to diverse signals.
RESULTS
Structures of the Abl RM activating and inhibiting states
The Abl RM consists of five distinct regions (Fig. 1a): an unstructured N-terminal region referred to as the cap (residues 1–80), the SH3 domain (residues 85–138), a short linker referred to as the connectorSH3/2 (residues 139–152) that connects the SH3 to the SH2 domain (residues 153–237), and a linker (linkerSH2-KD; residues 238–250) that connects the SH2 to KD (residues 255–534)8. To delineate the conformational landscape of Abl RM, we used NMR27,28 to determine the intrinsic structural and dynamic properties of the isolated Abl RM (residues 1–255) (Supplementary Fig. 1a,b). NMR structure determination (Table 1) showed that the isolated Abl RM adopts two distinct conformations in solution (Fig. 1d,e). In one of the conformations (Fig. 1d), Abl RM adopts a structure very similar to the one seen in the assembled Abl kinase8 (Fig. 1b). In this conformation, the linkerSH2-KD is bound to the polyproline II (PPII)-binding site of the SH3 domain, the C-terminal region of the cap (hereafter capC; residues 65–75) interacts with the SH2 domain, whereas the cap region consisting of residues 1 through 63 is disordered and flexible (Fig. 1d). This finding reveals that the isolated Abl RM intrinsically adopts the conformation that is conducive to the inhibited, assembled form of Abl29 (Fig. 1b). We will refer to this conformation of Abl RM as the “inhibiting” state.
Table 1.
NMR and refinement statistics for protein structures
| Protein | ||
|---|---|---|
|
| ||
| NMR distance and dihedral constraints | Abl (activating) | Abl (inhibiting) |
| Distance constraints | ||
| Total NOE | 3776 | 1360 |
| Intraresidue | 666 | 134 |
| Inter-residue | ||
| Sequential (|i − j| = 1) | 968 | 293 |
| Medium range (2 ≤ |i − j| ≤ 4) | 461 | 187 |
| Long range (|i − j| ≥ 5) | 1681 | 746 |
| Hydrogen bonds | 124 | 124 |
| Total dihedral-angle restraints | ||
| ϕ | 153 | 169 |
| ψ | 146 | 146 |
| Structure statistics | ||
| Violations (mean ± s.d.) | ||
| Distance constraints (Å) | 0.00234 ± 0.016 | 0.272±0.511 |
| Dihedral-angle constraints (°) | 0.39872 ± 1.09 | 0.767±2.57 |
| Max. distance-constraint violation (Å) | 0.40 | 0.34 |
| Max. dihedral-angle violation (°) | 10.30 | 23.52 |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.018 | 0.019 |
| Bond angles (°) | 1.2 | 1.3 |
| Impropers (°) | 0 | 0 |
| Average pairwise r.m.s. deviation (Å)a | ||
| Heavy | 10.0 | 9.9 |
| Backbone | 9.8 | 9.8 |
Pairwise r.m.s.d. was calculated among 20 refined structures.
The second conformation that the isolated Abl RM adopts in solution (Fig. 1e) is drastically different from the inhibiting one. Most notably, the crucial interaction between the linkerSH2-KD and SH3 is disrupted and the canonical PPII-binding site of the SH3 domain is instead engaged by the cap region comprising residues 14–20 (hereafter capPxxP) that carries a PxxP sequence motif, a preferred binding site of Abl SH3 domain30 (Fig. 1e and 2a). Compared to the inhibiting state (Fig. 1d), the SH3 domain in the second conformation undergoes a ~25 Å translation and a ~45° rotation relative to the SH2 domain (Fig. 2a) As a result of this reorientation, SH3 clamps down on SH2 and capC cannot engage the SH2 domain (Fig. 1e and 2b). Notably, in this conformation Abl RM cannot interact optimally with the back of the KD as seen in the assembled structure (Supplementary Fig. 2a–2c). Instead, it is compatible with the activated state of Abl in the extended conformation (Fig. 1e and Supplementary Fig. 2d–2f). Thus, we will refer to the second conformation of Abl RM as the “activating” state (Fig. 1e). It should be noted that formation of the extended Abl conformation necessitates the displacement of the linkerSH2-KD from the SH3 domain and thus only the RM in the activating state and not in the inhibiting state is compatible with the extended form of Abl (Supplementary Fig. 2d–2f).
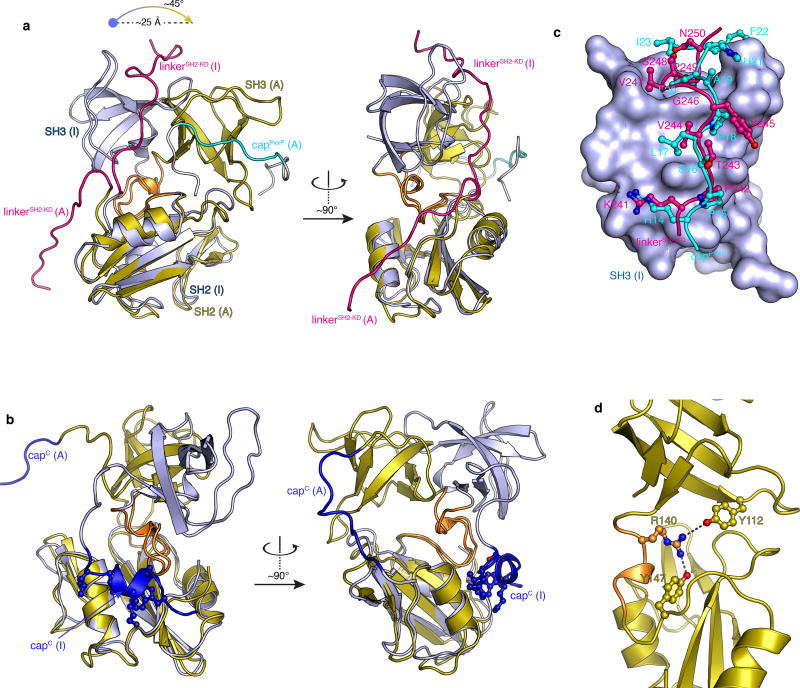
Figure 2.
Structure analysis of the Abl RM activating and inhibiting states. (a) Superposition of the Abl RM activating (A) and inhibiting (I) states highlighting the orientation difference of the SH3-SH2 domains and the conformational changes of the linkerSH2-KD and capPxxP. The two structures are superimposed onto the SH2 domain. (b) Superposition of the Abl RM activating (A) and inhibiting (I) states highlighting the conformational change in capC in the two states. CapC is disordered in the activating state whereas it forms a short helix that docks on a hydrophobic track on the SH2 domain in the inhibiting state. The four capC residues that make the contacts with SH2 are Trp67, Lys70, Leu73 and Leu74. (c) Superposition of the structure of SH3 bound to the linkerSH2-KD (inhibiting state) and bound to the capPxxP motif (activating state). The SH3 domain is shown as a solvent-accessible surface in blue, the linkerSH2-KD in red ball-and-stick and the capPxxP in cyan ball-and-stick. The first few contacts to SH3 are very similar. Lys241, Pro242 and Val244 residues of the linkerSH2-KD are substituted by Arg14, Pro15 and Leu17 in capPxxP thus forming very similar contacts with SH3. However, whereas Tyr245 side chain in linkerSH2-KD is pointing towards the solvent, the topologically equivalent Pro18 in capPxxP forms intimate contacts with SH3. The contacts between SH3 and the linkerSH2-KD residues Val247, Pro249 and Asn250 are substituted by contacts with residues Ala19, Leu20 and Ile23 in capPxxP. Moreover, His21 and Phe22 provide additional contacts to SH3. (d) Structural basis for the stabilization of the activating state by the S140R mutation. Arg140 forms a bifurcated H-bond with Tyr112 (in the SH3) and Tyr147 (in the SH2) in the activating state.
Of note, our data show that capPxxP is an intramolecular SH3-binding site that can compete with and displace the linkerSH2-KD from the SH3 domain thereby destabilizing the inhibiting state (Fig. 2c). CapPxxP appears to form a larger number of contacts to SH3 than the linkerSH2-KD explaining its ability to compete with the linkerSH2-KD for SH3, despite the proximity of the latter to the SH3 binding site (Fig. 2b). Thus, capPxxP constitutes a hitherto unknown activating region within Abl.
Dissection of the multilayered regulatory mechanism
The transition of Abl RM between the inhibiting and activating states is clearly a central process in the activity regulation of Abl kinase. The drastically different arrangement of key structural elements (linkerSH2-KD, capPxxP, capC and the connectorSH3/2) in the inhibiting and activating Abl RM states (Fig. 1d,e and 2) suggests that they influence the relative stability of the two functional states of Abl RM. The NMR data show that the two states are in equilibrium in solution and interconvert rapidly (see Methods and below). We thus sought to dissect the contribution to structure and energetics of each one of the structural elements by characterizing a number of Abl variants. The variants were designed on the basis of the structures of the two states to disrupt or enhance key intramolecular interactions within Abl RM (Fig. 3a). A detailed NMR analysis (Supplementary Fig. 1) of these variants showed that a large number of residues, and especially those located in the connectorSH3/2, are particularly sensitive to even slight changes in the overall structure of Abl RM and are thus excellent probes of the Abl RM conformational ensemble (see Methods). The chemical shift response of all these residues in the variants is very similar and is exemplified by Lys143, whose resonance in the 1H-15N HSQC spectra is located in a relatively uncrowded region of the spectrum and is easy to monitor (Fig. 3a).
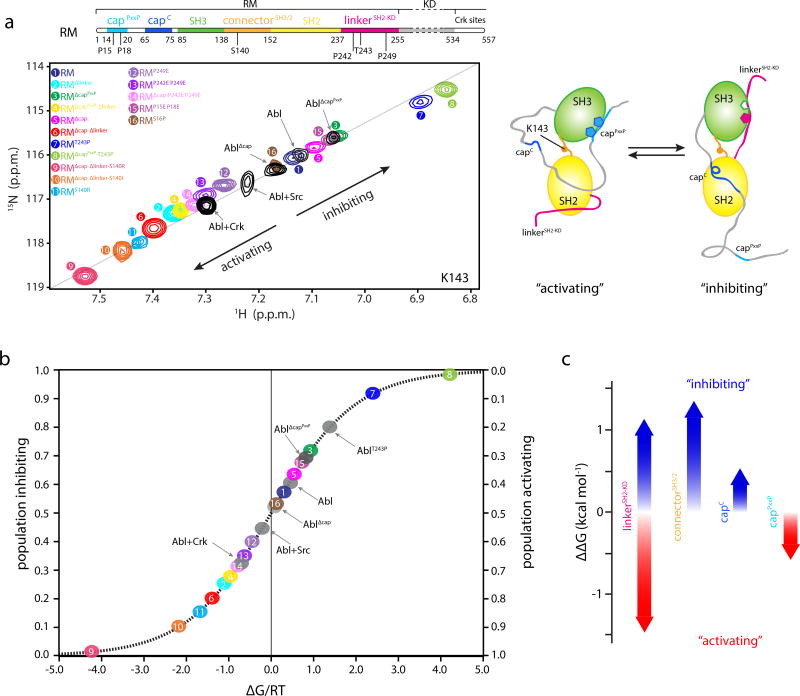
Figure 3.
Populations and energetics of Abl RM inhibiting and activating states. (a) Overlaid 1H-15N heteronuclear single quantum coherence (HSQC) spectra of the indicated Abl RM and Abl variants, showing the residue of K143. K143 is located in the connectorSH3/2 and displays the largest chemical shift range, thus providing the most sensitive probe for determining the populations of the two states in the variants. All of the other residues that have different chemical shifts in the two states show similar linear trend (Supplementary Fig. 3e–3f). Each Abl RM variant is denoted by a number and color. Important amino acid positions that were substituted are indicated on the top panel showing the domain organization of Abl. A schematic highlighting the structural changes in Abl RM as it transitions between the activating and the inhibiting states is shown on the right. (b) Populations of the inhibiting and activating states for the Abl RM and Abl variants determined by NMR from the data on panel A. The populations are plotted as a function of the associated free energy, ΔG/RT, where R is the gas constant, T the temperature and ΔG is given as GA-GI. 0.6 kcal mol−1 change in ΔG corresponds to a change by 1 unit in ΔG/RT at room temperature. As variants get closer to the free energy degeneracy (ΔG/RT=0) small changes in energy will result in substantial changes in the populations. (c) Energy contribution to the stability of the inhibiting and activating states by the four most important regions. Red arrows denote stabilization of the activating state whereas blue arrows denote stabilization of the inhibiting state.
The linkerSH2-KD mediates the juxtaposition of the SH3 domain onto the kinase domain and is thus one of the most important structural regions in Abl regulation29,31–34. Optimizing the binding interface between the linkerSH2-KD and the SH3 by mutating the suboptimal Thr243 to Pro29 is expected to favor the inhibiting state. Indeed, NOESY analysis of Abl RMT243P shows the presence solely of the inhibiting state and Lys143 in Abl RMT243P undergoes a large chemical shift change consistent with a pronounced shift of the equilibrium towards the inhibiting state (Fig. 3a, peak 1→7). Conversely, the double P242E P249E mutation, known to activate Abl31, disrupts the binding of the linkerSH2-KD to SH3 and NMR shows a pronounced shift towards the activating state, with the Lys143 resonance in Abl RMP242E P249E shifting in the opposite direction of that observed in Abl RMT243P (Fig. 3a, peak 1→13). Deletion of the linkerSH2-KD altogether (Abl RMΔlinker) shifts the equilibrium further towards the activating state (Fig. 3a, peak 1 → 2).
Our structural data show that the capPxxP displaces the linkerSH2-KD from the SH3 domain thereby favoring the activating state (Fig. 1e, 2a,b). Indeed, NMR analysis shows that deletion of capPxxP (Abl RMΔcapPxxP) shifts the equilibrium towards the inhibiting state (Fig. 3a, peak 1 → 3). In contrast to capPxxP, which favors the activating state, capC is expected to favor the inhibiting state15 wherein it acts as a clamp on the SH2 while being disordered in the activating state (Fig. 1d,e and 2a,b). Consistent with this hypothesis, deletion of capC shifts the equilibrium towards the activating state (Fig. 3a, peak 3→5, and peak 4→6).
The connectorSH2-KD plays a central role in Abl regulation as has been highlighted by a number of previous reports7,26,35,36. The S140R/I substitution in particular causes a pronounced increase in Abl activity and shows increased resistance to the KD inhibitor imatinib7,13. Notably, the NMR data show that the S140R and S140I substitutions in Abl RM favor markedly the activating state (Fig. 3a, peak 1→11), and NOESY experiments show the presence solely of the activating state. Analysis of the Abl RM structural data reveal that the substitutions favor the activating state either by forming hydrogen bonds (Arg140) or by hydrophobic contacts (Ile140) (Fig. 2d).
NMR study of the entire Abl kinase unit (residues 1–534, hereafter Abl) (Fig. 3a and Supplementary Fig. 1a, c) demonstrates that the conformational landscape of the RM in the context of Abl kinase is very similar to that described above for the isolated RM (Fig. 3a). This is further supported by the observation that the Lys143 chemical shift in Abl and its variants are very similar to that in the corresponding variants of the isolated RM (Fig. 3a). We used NMR to directly monitor the assembled and the extended conformations of Abl in solution. The resonances of Met263 and Met515 provide excellent probes since they are located, respectively, at the binding interface between the N-lobe and the SH2 and the interface between the C-lobe and the SH2 (Fig. 4a,b and Supplementary Fig. 3a). The NMR chemical shift data, combined with NOESY data, demonstrate that Abl populates both the assembled (inhibited) and extended (activated) states in solution, which are in equilibrium and interconvert rapidly (Fig. 4a and Supplementary Fig. 3a,b).
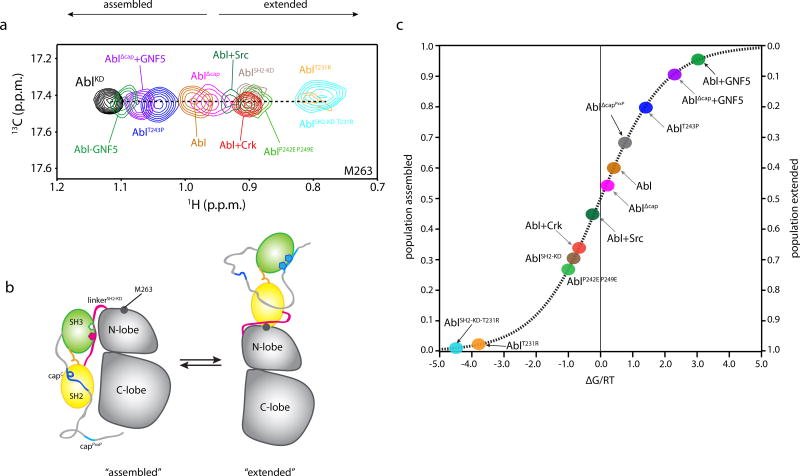
Figure 4.
Populations of Abl assembled and extended states. (a) Overlaid 1H-13C heteronuclear multiple quantum coherence (HMQC) spectra of the indicated Abl variants, showing the residue of M263. M263 is located at the interface between the SH2 and the kinase domain and provides the most sensitive probe for determining the populations of the two states in the variants. (b) Schematic highlighting the structural changes in Abl as it transitions between the assembled and the extended states. The position of M263 is shown as a grey circle. c, Populations of the assembled and extended states for Abl variants determined by NMR from the data on panel a. The populations are plotted as a function of the associated free energy, ΔG/RT, as in Figure 3b.
On the basis of the crystal structure, it was suggested that the T231R substitution stabilizes the extended state via a salt bridge between Arg231 (in SH2) and Glu294 (in the N-lobe) thereby enhancing Abl activity14,16,17,37,38. Our NMR spectra show that the population of the extended state increases substantially in AblT231R, an imatinib-resistance Abl variant, compared to the wild type Abl (Fig. 4a and Supplementary Fig. 3a). Amino acid substitutions in Abl that affect the SH3-linkerSH2-KD binding interface and thus the equilibrium between the inhibiting and the activating states of Abl RM have a direct and pronounced effect on the populations of the assembled and the extended states. For example, the double P242E P249E substitution increases the population of the extended state whereas the T243P substitution increases the population of the assembled state (Fig. 4a and Supplementary Fig. 3a). The present data reveal a direct link between the conformations of the RM (inhibiting vs activating) and the conformations of the entire Abl (assembled vs extended) (Supplementary Fig. 3c).
Energy landscape of the Abl RM
Of note, the NMR resonances of Abl RM residues in all of the characterized variants lie along the linear trajectory between the inhibiting and activating states (Fig. 3a and Supplementary Fig. 3d–3f). This observation provides strong indication that Abl RM populates only these two states in solution. Changes in populations effected by mutations in one of the structural elements, e.g. linker-SH2KD, are reflected in all other structural elements, e.g. capC (Supplementary Fig. 3d–3f) further corroborating a cooperative, two-state transition for Abl RM. Given that the interconversion occurs in the fast exchange regime, the observed chemical shift (δobs) represents population-weighted averaging and thus the population (p) of the two states can be extracted using the equation: δobs=δIpI+δApA (refs 39–41); wherein I and A denote the inhibiting and activating state, respectively (see Methods). In addition, the contribution of each variant to the energetics of the two Abl RM states can be determined using the equation ΔΔGI→A=−RTln(pA/pI) (Fig. 3b and Methods), where ΔΔG is the relative change in the free energy of the two states and pA and pI are the populations of the activating and inhibiting states. Similarly, the NMR resonances of KD residues reporting on the assembled and extended states of Abl (e.g. Met263 in Fig. 4a) can be used to determine the population and energetics of these two functional states in Abl in solution (Fig. 4a,b).
Notably, the relative stability of the inhibiting and activating states in the wild-type Abl RM is roughly equal and thus the two states are almost equally populated (Fig. 3b and 4c). RM lies in the steepest region of the sigmoidal energy curve and this renders RM an ideal switch since even subtle structural alterations that are associated with small energy changes will give rise to a substantial change in the population of the inhibiting/activating states (Fig. 3b and 4c). For example, a change in the free energy of just 0.6 kcal mol−1 will result in a population change of 50% (Fig. 3b and 4c)42,43. Therefore, RM forms a powerful and sensitive switch that respond to even small structural and energetic changes to regulate the Abl kinase activity.
Of note, the intrinsic preference of the simple SH3-SH2 module (RMΔcap Δlinker), that is, in the absence of the flanking cap and linkerSH2-KD regions, is towards the activating state with a population of 81%, versus 19% for the inhibiting state (ΔΔG ~0.8 kcal mol−1 (peak 6 in Fig. 3a,b). The structural element with the strongest effect on shifting the RM towards the inhibiting state is the linkerSH2-KD, which stabilizes the inhibiting state by up to ~2.1 kcal mol−1 (Fig. 3c). Amino acid substitutions at the linkerSH2-KD may stabilize (e.g. T243P) or destabilize (e.g. P242E P249E) the inhibiting state (Fig. 3c, positive or negative ΔΔG) and modulate the equilibrium generating a range of populations between ~20 to 99% for the inhibiting state (Fig. 3b). For example, the P249E substitution decreases the population of the inhibiting state to ~40% and the double P242E P249E substitution even further to ~35%.
The NMR data demonstrate that the capC region stabilizes the inhibiting state by as much as ~0.4 kcal mol−1 (Fig. 3c). This is in agreement with the structural data (Fig. 1b,c) showing that capC acts as a linchpin in the inhibiting state but is disordered and dynamic in the activating state. Deletion of capC favors the activating state, and thereby the extended state in Abl by increasing its population from ~30% to ~50% (Fig. 3b and 4c). In contrast, the capPxxP has the opposite effect to capC as it stabilizes the activating state by as much as ~0.4 kcal mol−1 (Fig. 3b,c and 4c). It does so by displacing the linkerSH2-KD from SH3, which results in the destabilization of the inhibiting state. Optimizing the SH3-capPxxP interface, e.g. by the S16P substitution (Fig. 2b), further favors the activating state (Fig. 3a,b; peak 1→16).
As discussed above, the connectorSH3/2 exerts a major effect by stabilizing the inhibiting state. Indeed, the NMR data show that the S140R substitution in the connectorSH3/2 favors the activating state by up to ~1.1 kcal mol−1 (Fig. 3c). Collectively, our data demonstrate that the linkerSH2-KD and the connectorSH3/2 have the strongest effect on the energetics of the Abl RM inhibiting and activating states, and thus of the Abl assembled and extended states, whereas capC and capPxxP have a significant, albeit weaker effect (Fig. 3c).
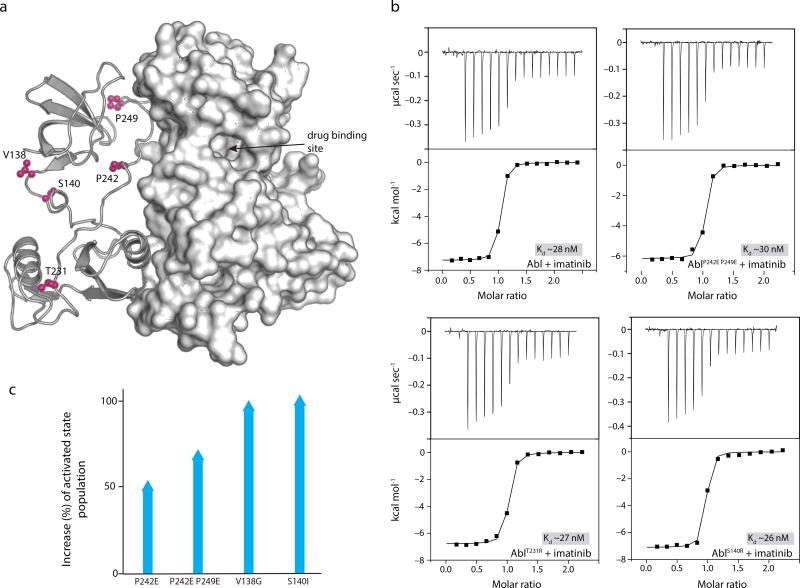
Allosteric drug-resistant mutations
Despite the success in treating chronic myelogenous leukemia by targeting Bcr-Abl with the inhibitor imatinib, a common problem is the development of drug resistance in many patients3. Several of these mutants are found in the Abl RM13,14 and are thus located far away from the drug-binding site (Fig. 1a and 5a). How these mutants exert their allosteric effect cannot be rationalized based on existing structural data and thus the underlying mechanism is not understood13,44. We used isothermal titration calorimetry to measure the thermodynamics of imatinib binding to the Abl variants. The data show that imatinib binds with the same affinity to wild type and the Abl variants (Fig. 5b). Even the individual thermodynamic components (ΔH and ΔS) are essentially identical indicating that, unlike the mutants in the KD drug-binding pocket44, the allosteric drug-resistant mutants in the RM have no effect on imatinib binding. Notably, our NMR data reveal that the allosteric mutants alter the population of inhibited and activated states. Amino acid substitutions in the connector region (V138G, S140R and S140I) and in the linkerSH2-KD (P242E and P249E) give rise to a substantially increased population of the activated state (Fig. 5c). Thus, the allosteric drug-resistant mutants do not interfere directly with the imatinib binding but rather promote the activated state, thereby giving rise to higher overall kinase activity and compromising the inhibitory function of imatinib.
Figure 5.
Mechanistic basis for imatinib resistance of RM mutations. (a) Cartoon of the crystal structure (PDB ID 2FO0) showing the location of the mutations (pink). The drug binding site is located in the KD and far away from the mutation sites. (b) Binding affinity of imatinib for Abl variants. The mutations do not reduce the affinity of imatinib for Abl. (c) Plot shows the increase of the Abl activated sate population as a result of the allosteric mutations in Abl RM.
GNF5 is a potent allosteric inhibitor that binds into the myristic pocket of the C-lobe45 and elicits the same conformational change to the C lobe of KD as the myristic acid8. This conformational change enables the docking of the SH2 to the back of the C lobe. The NMR data show that GNF5 binding to Abl shifts drastically the equilibrium towards the assembled, inhibited state (Fig. 4a). Thus, our findings clearly demonstrate that this allosteric drug45,46 exerts its inhibitory effect by increasing the population of the assembled Abl and thus decreasing the activated state population.
Abl RM functions as a graded switch
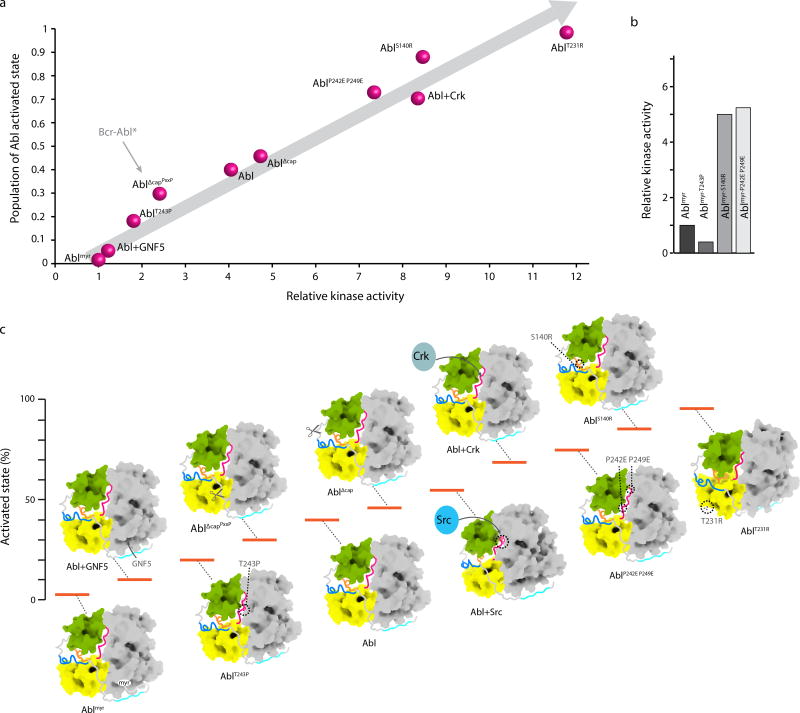
We observed an excellent correlation between the population of the activated state and the relative kinase activity of Abl variants (Fig. 6a). The largest population for the activated state was measured for AblT231R (~99%, Fig. 4c), which exhibits the highest kinase activity, followed by AblS140R and AblP242E/P249E variants (Fig. 6a). On the opposite end, the complex of Abl with the allosteric drug GNF5 exhibits a negligible kinase activity (Fig. 6a), correlated with a low population (~5%) of the activated state (Fig. 4c). The lowest kinase activity, as also reported previously7, is shown by the myristoylated form of Abl (Ablmyr) (Fig. 6a). Although preparation of a highly homogeneous Ablmyr sample for NMR studies has not been possible, the linear correlation produced here between the active state population and the kinase activity (Fig. 6a) provides an excellent tool to estimate the activated state population in Ablmyr. Using this correlation (Fig. 6a) we estimated that the population of the activated state in Ablmyr is ~2–3%. Interestingly, the Ablmyr-T243P mutant further suppresses the kinase activity suggesting that there is an equilibrium between the activated and inhibited states even in Ablmyr (Fig. 6b). We should note that analysis of the Ablmyr structure indicates that the cap linker is sufficiently long to allow formation of the extended structure even when the myristic acid is bound to the C-lobe (Supplementary Fig. 2g). In agreement, the S140R and P242E P249E substitutions increase markedly the kinase activity of Ablmyr (Fig. 6b). This behavior is reminiscent of the activation of Src kinase by substitutions that cause the linkerSH2-KD to be displaced even when the SH2 is still bound to the phosphorylated C-tail47.
Figure 6.
Energy landscape of Abl allosteric regulation. (a) Plot of the activated state population of Abl, as measured by NMR and displayed in Figure 4c, against the measured kinase activity measured at 10 minute time-point shows a linear correlation. The kinase activity is expressed as relative to the activity of Ablmyr. The arrow pointing to AblΔcapPxxP indicates the expected population of the activated state in Bcr-Abl exclusively as a result of the removal of the first 45 residues in the fusion. (b) Kinase activity of Ablmyr variants indicating that further suppression or activation occurs even in the myristoylated form of Abl. (c) Energy landscape of Abl highlighting the changes in the activated state population, and thus of the kinase activity, caused by mutations, deletions, Crk binding and phosphorylation by Src. For clarity only the assembled structure is shown for all states. Uncropped gel images for a and b and quantification are shown in Supplementary Data Set 1.
Response of Abl to physiological activation signals
Abl’s activity is stimulated by the binding of scaffold proteins such as the Crk48, as well as by phosphorylation effected by other kinases (e.g. Src49). Crk, a scaffold protein consisting of a tandem SH2-SH3-SH3, binds through its middle SH3 domain to the proline-rich Crk-binding sites located at the disordered C-terminal tail of Abl50,51 (Fig. 1a) and uses its SH2 domain to bind to the linkerSH2-KD (ref 52). Our NMR data show that disruption of the SH3-linkerSH2-KD interface caused by Crk binding to Abl shifts strongly the equilibrium towards the activated state (population ~70%), and thus gives rise to enhanced kinase activity (Fig. 3a and 4a,b). Src kinase is known to phosphorylate Tyr245 and stimulate Abl activity32,49,53,54. The NMR data show a significant increase in the population of the activated state upon phosphorylation by Src (Fig. 3, 4, and 6a), which is caused by the displacement of the SH3 domain from the back of the kinase due to the steric hindrance induced by Tyr245 phosphorylation (Supplementary Fig. 2h,i).
Discussion
Quantitative description of allosteric mechanisms in multi-domain proteins is generally lacking. The present data provide an atomic view and quantitative description of the energy landscape in the allosteric regulation of the Abl kinase (Fig. 6c). The kinase is inhibited and activated through a fine balance of energetic contributions from several structural elements in the regulatory module that form cooperatively a multilayered regulatory mechanism. The Abl RM makes an ideal switch for several reasons: (i) it is thermodynamically in a regime that allows for efficient suppression or activation even by small changes in energy; (ii) it only adopts two structures (activating and inhibiting) and simple population shift between the two states enables Abl to function as a graded switch; (iii) Perturbations at the individual structural elements making up the multilayered regulatory mechanism act in a synergistic way and can be additive or opposing, giving rise to any possible population in the continuum between full suppression and full activity. (iv) the kinetics of the switch interconversion are fast thereby allowing rapid response to various signals and stimuli; (v) the presence of several structural elements that modulate the conformational equilibrium of the inhibited and activated states provides multiple avenues, via binding or phosphorylation processes, throughout the structure of the RM to affect the equilibrium and thus the kinase activity. Such a regulatory mechanism may be common among the family of cytosolic tyrosine kinases, which share very similar overall structural architectures.
A multilayered regulatory mechanism was also identified to modulate the activity of the proto-oncogene Vav1, a guanine nucleotide exchange factor40,55. This multi-domain protein exists in an equilibrium that is controlled by both thermodynamics and kinetics. The studies revealed a dynamic interplay between the domains of the protein that allows transient exposure and phosphorylation of critical tyrosine residues relieving inhibition.
The kinetics of interconversion between the inhibiting and activating states of Abl RM are very fast with the exchange rate (kex), as estimated by NMR being much larger than 1,500 s−1. Thus, the structural elements that make up the multilayered regulatory mechanism, and serve as binding or phosphorylation sites, are ideally suited to function as regulatory hotspots because they allow quick access to activators or inhibitors enabling Abl to respond rapidly to various signals and stimuli. Rapid phosphorylation of kinases is a common theme (e.g. Btk kinase56) and thus rapid breathing motions within the kinase seem to play an important role to enable such processes.
Removal of the myristoyl group has a pronounced effect on the population of the activated state, which increases to ~40% and thus stimulates the Abl kinase activity (Fig. 6a). Interestingly, our data suggest that the population of the activated state in Bcr-Abl, a fusion that entails the removal of the first 45 residues of Abl (equivalent to the AblΔcapPxxP variant) is only ~30% (considering only the effect caused by the deletion of the cap region) with the majority of the molecules (~70%) being still in the assembled, inhibited state (Fig. 6a). This finding explains previous genetic observations7,13,57 that mutations perturbing intramolecular surfaces can confer imatinib resistance in Bcr-Abl since regulatory constraints are still operational.
The disordered region in the RM of Src, in addition to being myristoylated and phosphorylated, plays a role in interacting with proteins and lipids. Recent reports show that the region is also involved in interactions with the modular domains within the RM to possibly fine tune Src kinase activity58. These findings further highlight the functional similarity between Abl and Src kinases59. The quantitative dissection of the allosteric regulatory mechanism in Abl presented here revealed a hitherto unknown intramolecular activating region (capPxxP) in the cap. It is likely that the recently reported A19V drug-resistant mutation60 located in the capPxxP element can exert its function by promoting the activated state of Abl by means of capPxxP. In this case the underlying mechanism would be similar to the one used by the allosteric drug-resistant mutations located in the connectorSH3/2 and linkerSH2-KD of the Abl RM (Fig. 5).
Online Methods
Protein preparation and purification
Abl fragments used in this work were cloned from full length Abl isoform 1b and have the following boundaries (Supplementary Fig. 1): Abl, residues 1–534; Abl (for studies with Crk) residues 1–557; AblKD, residues 248–534; AblΔcapPxxP, residues 45–534; AblΔcap, residues 76–534; AblSH2-KD, residues 138–534; Abl RM, residues 1–255; Abl RMΔcapPxxP, residues 45–255; Abl RMΔcap, residues 76–255; Abl RMΔlinker, residues 1–239; Abl RMΔcapPxxP Δlinker, residues 45–239; Abl RMΔcap Δlinker, residues 76–239. All constructs were expressed in BL21(DE3) (EMD Chemicals). The Abl RM constructs were cloned into modified pET16b or pET42a vectors (EMD Chemicals) with dual tag of either His10 and maltose binding protein (MBP) or His6 and glutathione S transferase (GST) followed by a TEV protease cleavage site. Abl constructs that include the kinase domain were expressed as described before50. Mutants were generated using site-directed mutagenesis (Agilent). The kinase domain of Src kinase was cloned into pet16b vectors with His10-tag and expressed with similar conditions for Abl kinase domain. CrkII was cloned into the pet42a vector and expression and purification was performed as described61. All constructs were grown at 37 °C. Unlabeled samples were grown in Luria-Bertani (LB) medium and protein synthesis was induced by the addition of 0.2 mM of IPTG at OD600 ~ 0.4 at 18 °C followed by a 12-hr incubation. Cells were lysed by sonication and the cytosolic fraction was separated by centrifugation at 48,000 × g. The lysate was then loaded on Ni-NTA agarose resin (GE) equilibrated with Tris buffer and 1 M NaCl, 3 mM βME at pH 8. Elution was performed with 350 mM imidazole and following TEV protease cleavage the sample was concentrated and loaded on Ni-NTA to remove the protease and tag. The sample was then concentrated and loaded on Superdex 75 or Superdex 200 size-exclusion column (GE). Protein concentration was determined spectrophotometrically at 280 nm using the corresponding extinction coefficient.
Protein isotope labeling for NMR studies
Isotopically labeled samples for NMR studies were prepared by growing the cells in minimal (M9) medium. U-[2H,13C, 15N] labeled samples were prepared for backbone assignment by supplementing the growing medium with 15NH4Cl (1 g l−1) and 2H7,13C6-glucose (2 g l−1) in 99.9% 2H2O (CIL and Isotec). Growing and induction conditions were identical to that used for unlabeled samples. 1H-13C methyl-labeled samples were prepared as described62,63 using 45 mg l−1 of α-ketobutyric acid, 90 mg l−1 of α-ketoisovaleric acid, 50 mg l−1 of 13CH3-Met, 50 mg l−1 of [2H2-13CH3]-Ala. For selective labeling of Phe and Tyr residues, 180 mg l−1 of [13C,15N]-Phe and [13C,15N]-Tyr were added. For selective labeling of Lys, 80 mg l−1 of [15N-2H]-Lys was used. The precursors and amino acids were added 30 minutes before addition of IPTG.
Kinase assay
The kinase assay was carried out in Tris buffer with 100 mM NaCl, 5 mM MgCl2 at pH 7.5. ATP was added to a final concentration of 2 mM to start the reaction. The kinase reaction was started by addition of enzymatic quantity of recombinant Abl kinase and was stopped using 2× Laemmli buffer. Reactions were carried out at room temperature using Crk as substrate12,64, which was recombinantly expressed and purified as previously described65. Final reaction volume was 100µL. At the indicated times, samples were taken and the reaction was stopped using 2× Laemmli buffer. Kinase to substrate ratio was 1:100 (0.1 nM:10 nM). Samples were analyzed using western blotting with the Crk-pTyr221 antibody (Cell Signaling). Quantification was performed using imageJ. For assays with full length Ablmyr and variants, 293T cells were cultured in 6-well polystyrene coated plates in Dulbecco’s modified eagle medium (DMEM). The medium was supplemented with 10% fetal bovine serum (FBS), 100 U ml−1 penicillin, 0.1 mg ml−1 of streptomycin and 2 mM of L-glutamine. Cells were transfected at 70% confluency using X-tremeGENE (Roche). After 48 hours, cells were lysed with RIPA buffer (50mM Tris HCl, pH 8, 150 mM NaCl, 1% NP-40). Lysates were then incubated with 0.5 µg of antibody (OP20 Anti-c-Abl, EMD Millipore) and 60 µL of protein A agarose (Pierce) beads at 4 °C for 3 hr. Beads were washed with Tris buffer in 100 mM NaCl and the reaction carried out with addition of ATP. Samples were analyzed using western blotting with the Crk-pTyr221 antibody (Cell Signaling). For assays with the GNF5, the inhibitor was dissolved in DMSO and added to the reaction mixture prior to the addition of kinase.
Abl phosphorylation by Src
Phosphorylation of Abl by Src kinase was achieved by using catalytic amounts of Src kinase added to Abl kinase in the presence of 5 mM ATP and 5 mM MgCl2 in Tris buffer with 100 mM NaCl, pH 7.5. Abl phosphorylation was monitored by analyzing samples using western blotting with Abl pTyr245, pTyr89 and pTyr412 antibody (Cell Signaling). NMR spectra showed one set of resonances confirming a homogeneous sample.
MALS
Multiangle light scattering was measured using a DAWN HELEOS-II (Wyatt Technology Corporation) downstream of a Shimadzu liquid chromatography system connected to a Superdex 200 10/300 GL (GE Healthcare) gel filtration column. The running buffer was 25 mM KPi, 100 mM NaCl, 0.5 mM EDTA, 3 mM βME, pH 6.5. Protein samples at a concentration of 0.05–0.5 mM were used. The flow rate was set to 0.5 ml min−1 with an injection volume of 200 µl and the light scattering signal was collected at room temperature. The data were analyzed with ASTRA (6.1.5, Wyatt Technology Corporation).
ITC experiments
All calorimetric titrations were performed on an iTC200 microcalorimeter (GE). Protein samples were extensively dialyzed against the ITC buffer containing 25 mM KPi, pH 6.8,100 mM NaCl and 1 mM TCEP at 25 °C. The sample cell was typically filled with ~15 µM of protein and the injection syringe with ~200 µM of inhibitor. Inhibitor solutions were prepared by dissolving the inhibitor in the flow through of the last exchange buffer. Each titration typically consisted of a preliminary injection followed by 14 subsequent injections. Data for the preliminary injection, which are affected by diffusion of the solution from and into the injection syringe during the initial equilibration period, were discarded. The data were analyzed with Origin 7.0.
NMR Spectroscopy
NMR data were collected on Bruker AVANCE III 600-, 700-, 850- and 900-MHz spectrometers equipped with 5-mm cryogenic probes. Spectra were processed with NMRPipe65 and analyzed using NMRView (http://www.onemoonscientific.com) and Sparky (UCSF). Transverse relaxation optimized spectroscopy (TROSY)-based triple-resonance experiments were recorded for backbone resonance assignment. 13Cα, 13Cβ, 13C′, and backbone 1H and 15N chemical shifts were used to compute dihedral angle restraints using TALOS-N65. Side chain assignment in U-[13C,15N] samples was achieved by 3D HCCH-TOCSY, 3D HCCH-COSY and 3D 13C,15N-edited NOESY experiments. Assignments for selectively [1H,13C]-labeled methyl-bearing and aromatic (Phe, Trp, and Tyr) residues were obtained using a combination of 3D 13C,15N-NOESY-HMQC and 13C,15N-HMQC-NOESY-HMQC pulse sequences with mixing time of 300 ms65.
Populations and kinetics of the two states
Only one set of resonances is observed in the NMR spectra for the inhibiting and activating states of the Abl RM variants (Fig. 3a and Supplementary Fig. 3d,e) indicative of a rapid rate of interconversion between them. Similarly, Abl interconverts rapidly between the assembled and extended states given that a single resonance is observed for all the variants (e.g. Met263 in Fig. 4a). Since the interconversion between the two states is fast on the NMR chemical shift time scale, the observed chemical shift (δobs) is population-weighted: δobs=δIpI+δApA39,41; wherein I and A denote the inhibiting and activating state, respectively. The Abl RMΔcapPxxP-T243P and Abl RMΔlinker-S140R variants, respectively, provided the highest populations of the inhibiting and activating states and their chemical shifts were thus assigned as the δI and δA. The largest chemical shift difference between the two states is ~600 Hz, thus setting the lower limit for the exchange rate (kex) to ~ 1,500 s−1.
Structure determination
Initial NOESY experiments of the Abl RM showed the presence of two sets of NOEs that were incompatible with a single structure in solution. For example, the same set of residues in the PPII-binding site of the SH3 domain showed NOEs with residues from both the linkerSH2-KD and the capPxxP region. Preliminary structure analysis of the Abl RM using both sets of the measured NOEs suggested that the protein adopts two distinct structures in solution with the major differences located in the region bound to the SH3 domain and the relative orientation of the SH3-SH2 module. These two structures correspond to the inhibiting (Fig. 1d and Supplementary Fig. 4a) and the activating (Fig. 1e and Supplementary Fig. 4b) states. The fractional population of secondary structure in the flexible regions of the Abl RM were computed using the program δ2D66 (Supplementary Fig. 5). To facilitate the determination of the high-resolution structure of the two states we sought to obtain either one of two states in solution as the major form. Towards this goal, we designed, on the basis of the preliminary structural data, several variants predicted to favor one of the two states. For example, the Abl RMΔcapPxxP variant lacks that capPxxP motif, which competes with the linkerSH2-KD for the SH3 domain, and thus in this case the inhibiting state is favored. Indeed, only one set of NOEs were measured for Abl RMΔcapPxxP corresponding to the inhibiting state. Deletion of the linkerSH2-KD (Abl RMΔlinker) favored, as expected, the activating state. These two variants, together with the Abl RMΔlinker-S140R and Abl RMΔcapPxxP-T243P that produce highly skewed populations of the activating and the inhibiting states, respectively, were used to collect a large number of restraints to refine the preliminary structures of the Abl RM and yield high-resolution structural data (Table 1). The structures were calculated using CYANA 3.9767. Resonance assignment, NOESY peaklists from all NOESY experiments and TALOS-derived dihedral angles and hydrogen bonds inferred from HN-HN NOESY connectivities were used as input to CYANA automated NOE assignment protocol. The 20 lowest-energy structures were refined by restrained molecular dynamics in explicit water with CNS65. The percentage of residues falling in favored and disallowed regions, respectively, of the Ramachandran plot is 100% and 0% for the activating state and 99.2% and 0.8% for the inhibiting state.
Data availability
Coordinates and structure factors for the Abl RM in the activating and inhibiting forms have been deposited in the Protein Data Bank under the accession codes PDB ID 6AMW (“activating”) and PDB ID 6AMV (“inhibiting”). All other data are available from the corresponding authors upon reasonable request.
A Life Sciences Reporting Summary for this paper is available
Supplementary Material
Acknowledgments
The work was supported by National Institutes of Health grant GM122462 to C.G.K.
Footnotes
AUTHOR CONTRIBUTION
T.S., P.R., and C.G.K. designed the study. T.S. performed all biochemical experiments. T.S. and P.R. recorded and analyzed the NMR data. P.R. determined the NMR structures. All authors contributed to and approved the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Bradley WD, Koleske AJ. Regulation of cell migration and morphogenesis by Abl-family kinases: emerging mechanisms and physiological contexts. Journal of cell science. 2009;122:3441–3454. doi: 10.1242/jcs.039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colicelli J. ABL tyrosine kinases: evolution of function, regulation, and specificity. Science Signaling. 2010;3:re6. doi: 10.1126/scisignal.3139re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hantschel O. Structure, regulation, signaling, and targeting of abl kinases in cancer. Genes & Cancer. 2012;3:436–446. doi: 10.1177/1947601912458584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greuber EK, Smith-Pearson P, Wang J, Pendergast AM. Role of ABL family kinases in cancer: from leukaemia to solid tumours. Nature reviews Cancer. 2013;13:559–571. doi: 10.1038/nrc3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khatri A, Wang J, Pendergast AM. Multifunctional Abl kinases in health and disease. J Cell Sci. 2016;129:9–16. doi: 10.1242/jcs.175521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pluk H, Dorey K, Superti-Furga G. Autoinhibition of c-Abl. Cell. 2002;108:247–259. doi: 10.1016/s0092-8674(02)00623-2. [DOI] [PubMed] [Google Scholar]
- 7.Hantschel O, et al. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 8.Nagar B, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 9.Harrison SC. Variation on an Src-like theme. Cell. 2003;112:737–740. doi: 10.1016/s0092-8674(03)00196-x. [DOI] [PubMed] [Google Scholar]
- 10.Pawson T, Kofler M. Kinome signaling through regulated protein-protein interactions in normal and cancer cells. Current opinion in cell biology. 2009;21:147–153. doi: 10.1016/j.ceb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Lin J, Arlinghaus R. Activated c-Abl tyrosine kinase in malignant solid tumors. Oncogene. 2008;27:4385–4391. doi: 10.1038/onc.2008.86. [DOI] [PubMed] [Google Scholar]
- 12.Ganguly SS, et al. c-Abl and Arg are activated in human primary melanomas, promote melanoma cell invasion via distinct pathways, and drive metastatic progression. Oncogene. 2012;31:1804–16. doi: 10.1038/onc.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 14.Sherbenou DW, et al. BCR-ABL SH3-SH2 domain mutations in chronic myeloid leukemia patients on imatinib. Blood. 2010;116:3278–3285. doi: 10.1182/blood-2008-10-183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagar B, et al. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Molecular cell. 2006;21:787–798. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 16.Filippakopoulos P, et al. Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell. 2008;134:793–803. doi: 10.1016/j.cell.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenz S, Deng P, Hantschel O, Superti-Furga G, Kuriyan J. Crystal structure of an SH2-kinase construct of c-Abl and effect of the SH2 domain on kinase activity. Biochem J. 2015;468:283–91. doi: 10.1042/BJ20141492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grebien F, et al. Targeting the SH2-Kinase Interface in Bcr-Abl Inhibits Leukemogenesis. Cell. 2011;147:306–319. doi: 10.1016/j.cell.2011.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kain KH, Gooch S, Klemke RL. Cytoplasmic c-Abl provides a molecular 'Rheostat' controlling carcinoma cell survival and invasion. Oncogene. 2003;22:6071–6080. doi: 10.1038/sj.onc.1206930. [DOI] [PubMed] [Google Scholar]
- 20.Bradshaw JM. The Src, Syk, and Tec family kinases: distinct types of molecular switches. Cellular signalling. 2010;22:1175–1184. doi: 10.1016/j.cellsig.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 22.Foda ZH, Shan Y, Kim ET, Shaw DE, Seeliger MA. A dynamically coupled allosteric network underlies binding cooperativity in Src kinase. Nat Commun. 2015;6:5939. doi: 10.1038/ncomms6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornev AP, Taylor SS. Dynamics-Driven Allostery in Protein Kinases. Trends Biochem Sci. 2015;40:628–47. doi: 10.1016/j.tibs.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faraldo-Gómez JD, Roux B. On the importance of a funneled energy landscape for the assembly and regulation of multidomain Src tyrosine kinases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13643–13648. doi: 10.1073/pnas.0704041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang S, Blachowicz L, Makowski L, Roux B. Multidomain assembled states of Hck tyrosine kinase in solution. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15757–15762. doi: 10.1073/pnas.1004569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corbi-Verge C, et al. Two-state dynamics of the SH3-SH2 tandem of Abl kinase and the allosteric role of the N-cap. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E3372–80. doi: 10.1073/pnas.1303966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu R, Liu D, Cowburn D. Abl kinase constructs expressed in bacteria: facilitation of structural and functional studies including segmental labeling by expressed protein ligation. Molecular BioSystems. 2012;8:1878–1885. doi: 10.1039/c2mb25051a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Oliveira GAP, et al. Intramolecular dynamics within the N-Cap-SH3-SH2 regulatory unit of the c-Abl tyrosine kinase reveal targeting to the cellular membrane. The Journal of biological chemistry. 2013;288:28331–28345. doi: 10.1074/jbc.M113.500926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panjarian S, et al. Enhanced SH3/linker interaction overcomes Abl kinase activation by gatekeeper and myristic acid binding pocket mutations and increases sensitivity to small molecule inhibitors. The Journal of biological chemistry. 2013;288:6116–6129. doi: 10.1074/jbc.M112.431312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saksela K, Permi P. SH3 domain ligand binding: What's the consensus and where's the specificity? FEBS Lett. 2012;586:2609–14. doi: 10.1016/j.febslet.2012.04.042. [DOI] [PubMed] [Google Scholar]
- 31.Barila D, Superti-Furga G. An intramolecular SH3-domain interaction regulates c-Abl activity. Nat Genet. 1998;18:280–282. doi: 10.1038/ng0398-280. [DOI] [PubMed] [Google Scholar]
- 32.Brasher BB, van Etten RA. c-Abl has high intrinsic tyrosine kinase activity that is stimulated by mutation of the Src homology 3 domain and by autophosphorylation at two distinct regulatory tyrosines. The Journal of biological chemistry. 2000;275:35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- 33.Chen S, Brier S, Smithgall TE, Engen JR. The Abl SH2-kinase linker naturally adopts a conformation competent for SH3 domain binding. Protein Science. 2007;16:572–581. doi: 10.1110/ps.062631007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panjarian S, Iacob RE, Chen S, Engen JR, Smithgall TE. Structure and dynamic regulation of Abl kinases. The Journal of biological chemistry. 2013;288:5443–5450. doi: 10.1074/jbc.R112.438382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brasher BB, Roumiantsev S, van Etten RA. Mutational analysis of the regulatory function of the c-Abl Src homology 3 domain. Oncogene. 2001;20:7744–7752. doi: 10.1038/sj.onc.1204978. [DOI] [PubMed] [Google Scholar]
- 36.Young MA, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J. Dynamic coupling between the SH2 and SH3 domains of c-Src and Hck underlies their inactivation by C-terminal tyrosine phosphorylation. Cell. 2001;105:115–126. doi: 10.1016/s0092-8674(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 37.Lamontanara AJ, Gencer EB, Kuzyk O, Hantschel O. Mechanisms of resistance to BCR-ABL and other kinase inhibitors. Biochimica et biophysica acta. 2013;1834:1449–1459. doi: 10.1016/j.bbapap.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Dölker N, et al. The SH2 domain regulates c-Abl kinase activation by a cyclin-like mechanism and remodulation of the hinge motion. PLoS computational biology. 2014;10:e1003863. doi: 10.1371/journal.pcbi.1003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–33. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 40.Yu B, et al. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell. 2010;140:246–56. doi: 10.1016/j.cell.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzeng SR, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–40. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- 42.Akimoto M, et al. Signaling through dynamic linkers as revealed by PKA. Proc Natl Acad Sci U S A. 2013;110:14231–6. doi: 10.1073/pnas.1312644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai CJ, Nussinov R. A unified view of "how allostery works". PLoS Comput Biol. 2014;10:e1003394. doi: 10.1371/journal.pcbi.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barouch-Bentov R, Sauer K. Mechanisms of drug resistance in kinases. Expert Opin Investig Drugs. 2011;20:153–208. doi: 10.1517/13543784.2011.546344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463:501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skora L, Mestan J, Fabbro D, Jahnke W, Grzesiek S. NMR reveals the allosteric opening and closing of Abelson tyrosine kinase by ATP-site and myristoyl pocket inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4437–45. doi: 10.1073/pnas.1314712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moroco JA, et al. Differential sensitivity of Src-family kinases to activation by SH3 domain displacement. PLoS One. 2014;9:e105629. doi: 10.1371/journal.pone.0105629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shishido T, et al. Crk family adaptor proteins trans-activate c-Abl kinase. Genes Cells. 2001;6:431–440. doi: 10.1046/j.1365-2443.2001.00431.x. [DOI] [PubMed] [Google Scholar]
- 49.Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM. c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev. 1999;13:2400–2411. doi: 10.1101/gad.13.18.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jankowski W, et al. Domain organization differences explain Bcr-Abl's preference for CrkL over CrkII. Nature Chemical Biology. 2012;8:590–596. doi: 10.1038/nchembio.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhatt VS, Zeng D, Krieger I, Sacchettini JC, Cho JH. Binding Mechanism of the N-Terminal SH3 Domain of CrkII and Proline-Rich Motifs in cAbl. Biophys J. 2016;110:2630–41. doi: 10.1016/j.bpj.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donaldson LW, Gish G, Pawson T, Kay LE, Forman-Kay JD. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and a Crk-derived phosphopeptide. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14053–14058. doi: 10.1073/pnas.212518799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanis KQ, Veach D, Duewel HS, Bornmann WG, Koleske AJ. Two distinct phosphorylation pathways have additive effects on Abl family kinase activation. Molecular and cellular biology. 2003;23:3884–3896. doi: 10.1128/MCB.23.11.3884-3896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sirvent A, Boureux A, Simon V, Leroy C, Roche S. The tyrosine kinase Abl is required for Src-transforming activity in mouse fibroblasts and human breast cancer cells. Oncogene. 2007;26:7313–23. doi: 10.1038/sj.onc.1210543. [DOI] [PubMed] [Google Scholar]
- 55.Li P, Martins IR, Amarasinghe GK, Rosen MK. Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nat Struct Mol Biol. 2008;15:613–8. doi: 10.1038/nsmb.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, et al. Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate. Elife. 2015;4:e06074. doi: 10.7554/eLife.06074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith KM, Yacobi R, van Etten RA. Autoinhibition of Bcr-Abl through its SH3 domain. Molecular cell. 2003;12:27–37. doi: 10.1016/S1097-2765(03)00274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arbesu M, et al. The Unique Domain Forms a Fuzzy Intramolecular Complex in Src Family Kinases. Structure. 2017;25:630–640. e4. doi: 10.1016/j.str.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 59.Maffei M, et al. The SH3 Domain Acts as a Scaffold for the N-Terminal Intrinsically Disordered Regions of c-Src. Structure. 2015;23:893–902. doi: 10.1016/j.str.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 60.Lee BJ, Shah NP. Identification and characterization of activating ABL1 1b kinase mutations: impact on sensitivity to ATP-competitive and allosteric ABL1 inhibitors. Leukemia. 2017;31:1096–1107. doi: 10.1038/leu.2016.353. [DOI] [PubMed] [Google Scholar]
- 61.Kobashigawa Y, et al. Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Nature Structural & Molecular Biology. 2007;14:503–510. doi: 10.1038/nsmb1241. [DOI] [PubMed] [Google Scholar]
- 62.Saio T, Guan X, Rossi P, Economou A, Kalodimos CG. Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science. 2014;344:1250494. doi: 10.1126/science.1250494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang C, Rossi P, Saio T, Kalodimos CG. Structural basis for the antifolding activity of a molecular chaperone. Nature. 2016;537:202–206. doi: 10.1038/nature18965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Plattner R, et al. A new link between the c-Abl tyrosine kinase and phosphoinositide signalling through PLC-gamma1. Nature cell biology. 2003;5:309–319. doi: 10.1038/ncb949. [DOI] [PubMed] [Google Scholar]
- 65.Sarkar P, Saleh T, Tzeng SR, Birge RB, Kalodimos CG. Structural basis for regulation of the Crk signaling protein by a proline switch. Nat Chem Biol. 2011;7:51–7. doi: 10.1038/nchembio.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Camilloni C, De Simone A, Vranken WF, Vendruscolo M. Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry. 2012;51:2224–31. doi: 10.1021/bi3001825. [DOI] [PubMed] [Google Scholar]
- 67.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–78. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for the Abl RM in the activating and inhibiting forms have been deposited in the Protein Data Bank under the accession codes PDB ID 6AMW (“activating”) and PDB ID 6AMV (“inhibiting”). All other data are available from the corresponding authors upon reasonable request.
A Life Sciences Reporting Summary for this paper is available