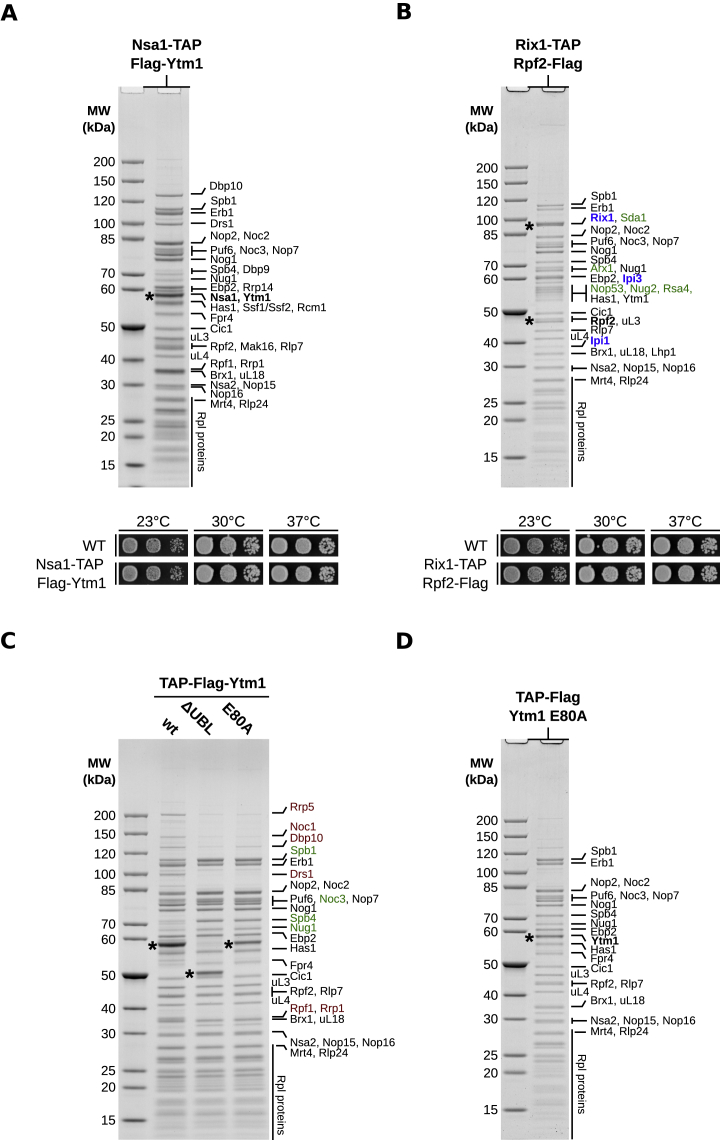
Figure S1.
Affinity Purification of Nucleolar Pre-60S Particles for Cryo-EM Analysis, Related to Figure 1
(A and B) Split affinity purifications of pre-60S intermediates (upper panels) purified through Nsa1-TAP Flag-Ytm1 (A) and Rix1-TAP Rpf2-Flag (B). Final eluates were used for cryo-EM and analyzed by SDS-PAGE and Coomassie staining. Major protein bands were identified by mass-spectrometry and are labeled on the right side of the gels. The bait proteins are shown in bold and indicated by asterisks. The Rix1-TAP Rpf2-Flag purification purifies nucleolar (state E) and nucleoplasmic (state F) pre-60S particles (B). Nucleoplasmic assembly factors are indicated in green and the Rix1 subcomplex members (Rix1, Ipi3, Ipi1) in blue. Growth analysis (lower panels) of the Nsa1-TAP Flag-Ytm1 (A) and the Rix1-TAP Rpf2-Flag (B) strains in comparison to the untagged wild-type strain (DS1-2b). Cells were spotted in 10-fold serial dilution on YPD medium and cell growth at the indicated temperatures was monitored after 2 days.
(C) Affinity purifications of the indicated plasmid-encoded Ytm1 variants fused to an N-terminal TAP-Flag tag and expressed from the endogenous promoter. The plasmids were transformed into a wild-type strain (W303). Final eluates of the purifications were analyzed by SDS-PAGE and Coomassie staining and co-purifying proteins are indicated on the right side of the gel and bands corresponding to the Ytm1 bait proteins are marked with an asterisk. Proteins decreased in the Ytm1 ΔUBL/E80A purifications compared to the Ytm1 wild-type particle are shown in red and factors increased on the mutant particles are indicated in green.
(D) Affinity purification of the Ytm1 E80A particle used for cryo-EM analyzed by SDS-PAGE. Proteins bands were identified by mass-spectrometry and are labeled on the right side of the Coomassie stained gel.