Abstract
Although many studies have been conducted worldwide for Equus caballus papillomavirus (EcPV), limited information is available on the virus in Japan. We recently collected one classical viral papillomatosis sample (E150904) from a racing horse in Japan. Papillomavirus infection was confirmed by histopathology, immunohistochemistry and PCR assays, and the sample was diagnosed as epithelial papilloma. Full-length genome of the virus was cloned and sequenced. It was 7,613 bp in length and had the same genome organization with reported EcPV-1. Moreover, phylogenetic analysis based on L1 gene revealed that the infection was caused by a variant of EcPV-1. This is the first report of EcPV infection in Japan, and would further contribute to the molecular epidemiological and pathological studies for EcPV.
Keywords: EcPV type 1, equine papillomatosis, initial detection, racing horse
Equus caballus papillomavirus (EcPV, also named equine papillomavirus) is associated with diseases in horse including classical viral papillomatosis, genital papillomas/papillomatosis, and aural plaques [9]. Papillomaviruses (PVs) have strict tissue specificity, thus each of these diseases is associated with a different type of PVs. To date 7 types of EcPV have been identified and are classified into 3 genera according to the nucleotide sequence diversity in the open reading frame (ORF) of the L1 gene [5, 9]. EcPV type 1 (EcPV-1) was first isolated from cutaneous papillomas [3, 6]. So far all EcPV-1 have been reported to be associated with the classical papillomatosis which is a typical benign proliferative lesion and usually transient in young horses younger than 3 years [3, 6, 7, 9]. EcPV-2 and EcPV-7 were found in genital papillomas/papillomatosis [4, 5, 8], while EcPV-3, -4, -5 and -6 were detected in aural plaques [2, 4, 5, 7].
Although many studies for EcPV have been conducted worldwide since the first EcPV was identified in 1986 [6], limited information is available on EcPV in Japan. Therefore, this study aimed to investigate the presence of EcPVs in Japan. We recently collected one classical viral papillomatosis sample (E150904) from a racing horse (Thoroughbred, colt, 2 years old) in Japan. E150904 was one of the 10 millet or rice grain sized papillomas located on the muzzle of the animal. DNA was extracted with the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. One pair of oligonucleotide primer EcPVF/EcPVR (5′-CCDGCHHTBGGNGAGTAYTGG-3′/5′-TGCCAGCANARNCCATTGTT-3′) was designed according to the published genome sequences of EcPVs (GenBank access number: AF394740, AF498323, NC003748, HM461973, NC012123, NC017862, NC020085, NC020084, NC020500 and NC020501) to amplify 502 base pair (bp) segment of L1 gene. The amplification of the primer set was carried out using an initial denaturation at 94°C for 5 min followed by 35 cycles of 94°C for 30 sec, 50°C for 30 sec, 72°C for 40 sec, and a final extension of 72°C for 10 min. The sample E150904 show positive by PCR. The 502 bp PCR product was cloned and sequenced. The sequence analysis using BLAST demonstrated 97.4% nucleotide identity with EcPV-1 (NC003748) [3], suggesting that the papillomatosis was associated with EcPV-1 infection.
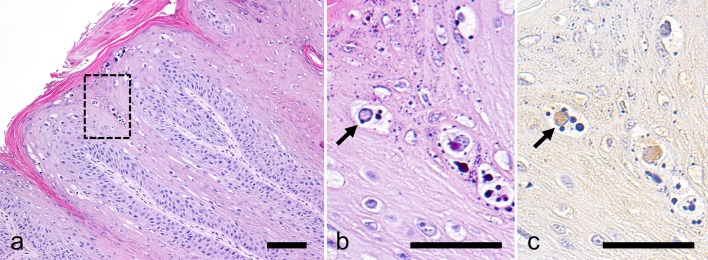
Tissue sample of E150904 was fixed with 10% formalin and routinely embedded in paraffin. Sections were stained with haematoxylin and eosin for histopathological examination. For immunohistochemistry, mouse anti-papillomavirus antibody (clone BPV-1/1H8 + CAMVIR, Abcam, Cambridge, U.K.) was used for primary antibody, and horseradish peroxidase labeled detection kit (Envision+ System, Dako Japan, Kyoto, Japan) was used. Antigen-antibody complex were visualized using 3,3′- diaminobenzidine. Sections were counterstained with haematoxylin. For negative control, primary antibody was substituted with Tris-buffered saline. The neoplasm consisted of hyperkeratotic squamous epithelial cells arranged in papillary pattern. Pale basophilic intranuclear inclusion bodies were observed occasionally in epithelial cells; the inclusion bodies were strongly positive for BPV antigens by immunohistochemistry (Fig. 1). Based on these histopathological characteristics and the presence of PV antigens, the neoplastic mass was diagnosed as epithelial papilloma associated with papillomavirus infection.
Fig. 1.
Histopathological and immunohistochemical findings. (a and b) Haematoxylin and eosin stain. (b) a higher magnification of the boxed part of (a). (c) PV immunostaining. The neoplasm consists of papillary growth of squamous epithelium (a). There are intranuclear inclusion bodies (b) that are immunopositive for PV antigen (arrow). Bar=100 µm (a); 50 µm (b and c).
Full-length genome sequences were amplified from the sample E150904 by using 5 pairs of oligonucleotide primer (Table 1) which designed according to published EcPV-1 genome sequences [3]. Subsequently, PCR products were purified using a QIAquick gel extraction kit (Qiagen). Five overlapping fragments were cloned into pMD20-T (Takara, Otsu, Japan) and then sequenced respectively. To reconstitute the full-length genome sequence, overlapping sequences were assembled into contiguous sequences using Seqman DNASTAR software (Lasergene) (DNASTAR, Madison, WI, U.S.A.). The sequence of the whole genome of EcPV-1 from E150904 had 7,613 bp (GenBank MF288893), shared 99.7% nucleotide identity with 3 PVs of EcPV-1 (AF394740, NC003748 and AF498323) reported so far, and had the same genome organization with them. The whole EcPV-1 genome of previously reported was either 7,610 bp (NC003748 and AF498323) or 7,613 bp (AF394740) in length. The difference of three nucleotides (GCG) exists within coding regions of L2, resulting in one amino acid (alanin) insertion or deletion. In the present study, EcPV1 from E150904 revealed to have the same insertion of alanin in L2 (the insertion located in 5,169–5,171 nt in whole genome and 1,404–1,406 nt in L2 gene). Moreover, L1 gene shared 99.5% (AF394740) and 99.6% (NC003748 and AF498323) nucleotide identity with reported EcPV-1. According to PV taxonomy following the general criteria established by the International Committee on the Taxonomy of Viruses (ICTV), a PV is recognized as a variant if the complete genome has been cloned and the DNA sequence of the L1 ORF differs less than 2% homology with the closest known PV type [1]. Based on this criterion, it is concluded that the PV from E150904 is a variant of EcPV-1.
Table 1. Oligonucleotides for whole genome amplification and cloning.
| Name | Sequence | Location (NC003748) | Predict length |
|---|---|---|---|
| EcPV1clone1F | CCTGGGGACGGTTCGCATTT | 6,550–6,569 | 1,788 |
| EcPV1clone1R | GCATCTCTCACAGGCGTACC | 708–727 | |
| EcPV1clone2F | GCCGTGTGTACCGTCTGCTG | 545–564 | 2,148 |
| EcPV1clone2R | CCTGCTTTGCCTGTGATTGG | 2,673–2,692 | |
| EcPV1clone3F | CGATGGGAGTCCTGTTTACC | 2,375–2,394 | 1,533 |
| EcPV1clone3R | CCACCTTGTTTACAACGTCC | 3,888–3,907 | |
| EcPV1clone4F | CCTTTACCCTAACCCCCTCCT | 3,680–3,700 | 1,721 |
| EcPV1clone4R | GCACCCGCATGGTTTGGTCAC | 5,380–5,400 | |
| EcPV1clone5F | GTTGATAGTCCAGACACCTCG | 5,110–5,130 | 1,660 |
| EcPV1clone5R | CATCAGGCAAGAACGAGCAGG | 6,749–6,769 |
In the present study, the EcPV-1 was isolated from a racing horse whose age is less than 3 years, in accordance with the conclusion that viral classical papillomatosis typically affects horses younger than 3 years [9]. Moreover, the racing horse was born and growing up in Japan without any travel overseas. Therefore, it suggested that the infection was occurred in Japan. Further studies are required to determine whether EcPV-1 exist in local horse herd. This study is the first report of EcPV infection in Japan, and would further contribute to the molecular epidemiological and pathological studies for EcPV.
REFERENCES
- 1.de Villiers E. M., Fauquet C., Broker T. R., Bernard H. U., zur Hausen H.2004. Classification of papillomaviruses. Virology 324: 17–27. doi: 10.1016/j.virol.2004.03.033 [DOI] [PubMed] [Google Scholar]
- 2.Fairley R. A., Haines D. M.1992. The electron microscopic and immunohistochemical demonstration of a papillomavirus in equine aural plaques. Vet. Pathol. 29: 79–81. doi: 10.1177/030098589202900110 [DOI] [PubMed] [Google Scholar]
- 3.Ghim S. J., Rector A., Delius H., Sundberg J. P., Jenson A. B., Van Ranst M.2004. Equine papillomavirus type 1: complete nucleotide sequence and characterization of recombinant virus-like particles composed of the EcPV-1 L1 major capsid protein. Biochem. Biophys. Res. Commun. 324: 1108–1115. doi: 10.1016/j.bbrc.2004.09.154 [DOI] [PubMed] [Google Scholar]
- 4.Lange C. E., Tobler K., Ackermann M., Favrot C.2011. Identification of two novel equine papillomavirus sequences suggests three genera in one cluster. Vet. Microbiol. 149: 85–90. doi: 10.1016/j.vetmic.2010.10.019 [DOI] [PubMed] [Google Scholar]
- 5.Lange C. E., Vetsch E., Ackermann M., Favrot C., Tobler K.2013. Four novel papillomavirus sequences support a broad diversity among equine papillomaviruses. J. Gen. Virol. 94: 1365–1372. doi: 10.1099/vir.0.052092-0 [DOI] [PubMed] [Google Scholar]
- 6.O’Banion M. K., Reichmann M. E., Sundberg J. P.1986. Cloning and characterization of an equine cutaneous papillomavirus. Virology 152: 100–109. doi: 10.1016/0042-6822(86)90375-2 [DOI] [PubMed] [Google Scholar]
- 7.Postey R. C., Appleyard G. D., Kidney B. A.2007. Evaluation of equine papillomas, aural plaques, and sarcoids for the presence of Equine papillomavirus DNA and Papillomavirus antigen. Can. J. Vet. Res. 71: 28–33. [PMC free article] [PubMed] [Google Scholar]
- 8.Scase T., Brandt S., Kainzbauer C., Sykora S., Bijmholt S., Hughes K., Sharpe S., Foote A.2010. Equus caballus papillomavirus-2 (EcPV-2): an infectious cause for equine genital cancer? Equine Vet. J. 42: 738–745. doi: 10.1111/j.2042-3306.2010.00311.x [DOI] [PubMed] [Google Scholar]
- 9.Torres S. M., Koch S. N.2013. Papillomavirus-associated diseases. Vet. Clin. North Am. Equine Pract. 29: 643–655. doi: 10.1016/j.cveq.2013.08.003 [DOI] [PubMed] [Google Scholar]