Abstract
Bovine mastitis causes significant economic losses in the dairy industry. Effective prevention of bovine mastitis requires an understanding of the infection status of a pathogenic microorganism in a herd that has not yet shown clinical signs of mastitis and appropriate treatment specific for the pathogenic microorganism. However, bacterial identification by culture has drawbacks in that the sensitivity may be low and the procedure can be complex. In this study, we developed a genetic detection method to identify mastitis pathogens using a simple and highly sensitive electrochemical DNA chip which can specifically detect bacterial DNA in milk specimens. First, we selected microorganisms belonging to 12 families and/or genera associated with mastitis for which testing should be performed. Next, we optimized the conditions for amplifying microorganism DNA by loop-mediated isothermal amplification (LAMP) using 32 primers and the use of a DNA chip capable of measuring all pathogens simultaneously. Sample detection could be completed in just a few hours using this method. Comparison of the results obtained with our DNA chip method and those obtained by bacterial culture verified that when the culture method was set to 100%, the total positive concordance rate of the DNA chip was 85.0% and the total negative concordance rate was 86.9%. Furthermore, the proposed method allows both rapid and highly sensitive detection of mastitis pathogens. We believe that this method will contribute to the development of an effective mastitis control program.
Keywords: bovine mastitis, DNA chip, LAMP, milk, pathogen
Bovine mastitis is a disease which occurs when invasive pathogenic microorganisms cause inflammation of the mammary gland. Because the disease results in substantial economic losses, countermeasures for mastitis prevention and management are desirable [1]. The current method for detecting bovine mastitis pathogens is mainly bacterial culture, which has several drawbacks. First, samples with low microbial counts may be unable to colonize the culture plates. Secondly, the viability of the microorganisms present in milk may be lost depending on how the milk is stored prior to the start of culture [6]. Thirdly, optimal culture conditions for each microorganism can be different depending on the bacterial nature, e.g. mycoplasma spp. which may not be easy to culture in the medium. Furthermore, even if colonies are obtained, skilled techniques and experience are required for correct identification. Fourth, the culture process up to bacterial identification is complex and time-consuming.
In this study, we report a detection method for bovine mastitis pathogen genomes which is composed of a process to purify nucleic acid from milk and another to detect the pathogens on a DNA chip as an alternative technique to bacterial culture. The DNA chip detection method using an electrochemically active intercalator of the Hoechst 33258 and DNA probe immobilized on a gold electrode have been reported by the Toshiba Corporation. The DNA chip used to test for mastitis pathogens in this study required DNA amplification by the loop-mediated isothermal amplification (LAMP) method for each pathogen. Several nucleic acid amplification methods, including the PCR method, have been developed so far [2, 14, 17]. Among them, the LAMP method is very promising because DNA can be amplified with high specificity and rapidity under isothermal condition than PCR [14]. Recently, many reports that have detected bacteria or virus by the LAMP method have been published [9, 10]. The identification processes were carried out using one DNA chip per milk sample so that all types of mastitis pathogen were detectable at once [13]. The use of electrochemical DNA chips developed by Toshiba Corporation did not require modification of the target nucleotides by fluorescent or electrical labeling, and therefore accurate detection with high sensitivity and low-cost could be realized. In addition, we compared the results of microorganism culture with the data obtained using a combination of LAMP and DNA chip. We demonstrated that this new detection method is a more sensitive and practical method to identify microorganisms associated with mastitis.
Milk samples
For this study, 81 quarter milk samples were collected from Holstein dairy cows with clinical mastitis in Tokachi sub prefecture, Hokkaido, Japan. To avoid contamination between milk samples through mixing with bacteria normally present in the environment, we used cotton wool dampened with 70% ethanol to wipe the teat end after milking. The milk samples were stored in a refrigerator at 4°C until required.
Pathogenic microorganism types examined
The major pathogenic microorganisms associated with bovine mastitis have been reported by many researchers [3, 5, 7, 12, 18, 19]. In this study we referred to many previous reports, including those involving large-scale analysis, and selected the most common bovine mastitis pathogens. Table 1 shows the selected microorganisms belonging to 12 families and/or genera in this study and the target genes with its GenBank accession numbers used on the electrochemical DNA chip detection. Some pathogens were detected genus-specifically, others species-specifically.
Table 1. Pathogens and target genes used in DNA chip detection.
| Pathogens | Target genes (GenBank accession number) | |
|---|---|---|
| Staphylococcus | Staphylococcus spp. | 16S rRNA (CP017466, KY008762, CP020773) |
| Staphylococcus aureus | Nuc (CP020741) | |
| Streptococcus | Enterococcus spp. | 16S rRNA (CP018128, LC322250) |
| Streptococcus spp. | 16S rRNA (AP018391, MF582800) | |
| Streptococcus agalactiae | recN (CP022537) | |
| Streptococcus uberis | recN (EU917309) | |
| Coliforms | Enterobacteriaceae | rpoA (CP023541) |
| Escherichia coli | uvrC (CP023644) | |
| Klebsiella spp. | metG (CP013985) | |
| Gram positive short bacillus | Corynebacterium bovis | rpoB (JX456369) |
| Trueperella pyogenes | 16S rRNA (KX592207) | |
| Mycoplasma | Mycoplasma (group A)a) | 16S rRNA (MG015946, KX649989) |
| Mycoplasma (group B)a) | 16S rRNA (MF769011) | |
| Prototheca | Prototheca zopfii | 18S-26S rRNA (KX353638) |
| Pseudomonas | Pseudomonas aeruginosa | 16S rRNA (MF687734) |
| Yeast | Candida spp. | 18S-26S rRNA (LC326063, MF927629) |
Some pathogens were detected genus-specifically, others species-specifically. a) Group A is composed of the arginine-undegradable strains, and group B is composed of the arginine-degradable strains in the genus Mycoplasma.
Bacterial culture
Culture of pathogens in milk samples was performed according to the standard identification method of the National Mastitis Council [8]. For mycoplasma, samples were cultured using Hayflik’s liquid medium and then Mycoplasma agar at 37°C and 10% CO2.
Nucleic acid extraction
Nucleic acid was extracted from the milk samples using a Fast ID Genomic DNA Extraction Kit (Genetic ID Inc., Fairfield, IA, U.S.A.). A bead beating process was added to the DNA extraction procedure to break down the varying thicknesses of the pathogen cell walls. Moreover, the separation of protein and fat in the milk samples was improved by adding ammonium sulfate and acetic acid [16].
LAMP reaction
LAMP reactions were performed as described previously [14]. We designed specific LAMP primers capable of stationary amplification of suitable target genes. Supplementary Table 1 shows the individual primer sets designed for all 16 pathogen groups. The reactions were performed at 61°C for 60 min using Loopamp Real-time Turbidimeter LA-200 (Teramecs Co., Ltd., Fushimi, Kyoto, Japan).
DNA chip
DNA samples amplified by the LAMP reaction were stored at −20°C until the detection procedure with the DNA chip. The samples were denatured at 95°C for 5 min before the DNA chip test. We used an electrochemical DNA chip for detection of amplified DNA. In this study, an electrochemical DNA chip mounted with 23 probes was used for detection [4, 15]. To detect the LAMP products on the same DNA chip substrate simultaneously, the optimum 23 probes were selected under the condition that the hybridization was carried out at 55°C for 40 min and washing was carried out at 45°C for 40 min. The LAMP probes were designed specifically for the pathogen strains (supplementary Table 2) and were tested against the target electrode DNA sequences for each pathogen genome. The hybridization reactions between the pathogen-specific probe and the LAMP-amplified DNA products on the DNA chips were measured using probe-specific electrochemically active currents generated by the Toshiba GenelyzerTM (Toshiba Medical Systems Corp., Otawara, Japan). After setting the cassette in the GenelyzerTM system, hybridization reaction, washing and electrochemical detection are performed automatically. Procedures from hybridization to electrochemical detection have been previously reported [17]. The reference value (nA) of each microorganism was determined as the eigenvalue of the mastitis DNA chip and was used to divide the positive/negative. Positive judgment criteria were calculated from the following formula. Positive judgment criteria=(Current anodic peak (ΔIpa) value-Negative control (Ipa) value) >Criterion value (nA) of each bacterial species
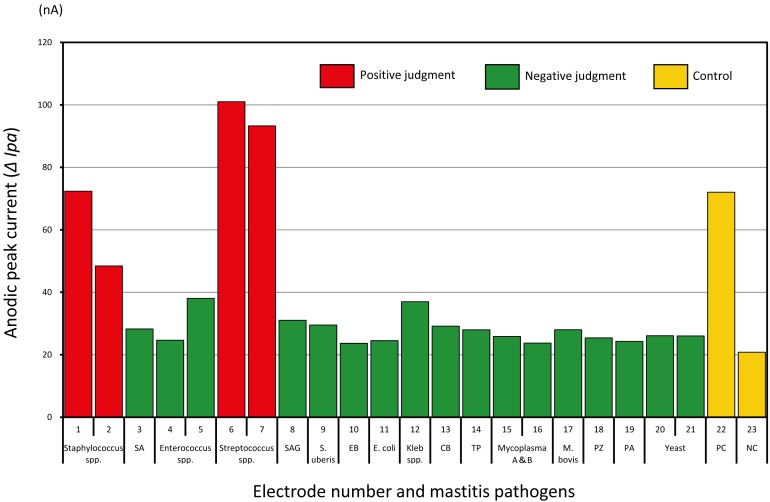
We determined how many different types of pathogens were present in the milk samples from cows with symptoms of clinical mastitis using the LAMP conditions and DNA chip described above. Figure 1 shows an example of the microorganism detection result in a milk sample where nA was detected to be greater than the reference nA value (red bars) for the LAMP products both Staphylococcus spp. and Streptococcus spp. The LAMP products specifically amplified for every pathogen group did not cross-react with the probes for other groups, and the increase in electric current value was only observed with the right probe. It also became clear that the specificity of the probes was not compromised even if several pathogens in the same group were present in the milk sample. Additionally, we confirmed the sensitivity and accuracy of the DNA chip method for detection of pathogens by comparison with the culture method using 81 milk samples obtained from clinical mastitis cases (Table 2). The results obtained with the DNA chip and the bacterial culture procedure were compared according to the type of pathogenic microorganisms identified. The orange grid indicates the number of DNA chip detections for which positive results were obtained although the results of bacterial culture were negative, and the green grids show the number of DNA chip detections for which negative results were obtained, although the results of bacterial culture results were positive. When the culture method was set to 100%, the total positive concordance rate of the DNA chip was 85.0% and the total negative concordance rate was 86.9%. Enterobacteria, Klebsiella, Streptococcus, and Enterococcus were identified as the major groups that were positive using the DNA chip. That is, we were able to identify many environmental pathogens.
Fig. 1.
Mastitis pathogens corresponding to DNA probes and measured anodic peak current (ΔIpa). The reference value (nA) of each microorganism was determined as the eigenvalue of the mastitis DNA chip and was used to divide the positive/negative. Positive judgment criteria were calculated from the following formula. Positive judgment criteria=(Current anodic peak (ΔIpa) value−Negative control (Ipa) value) > Criterion value (nA) of each bacterial species. SA: Staphylococcus aureus, SAG: Streptococcus agalactiae, EB: Enterobacteriea, Kleb spp.: Klebsiella spp., Coryne.: Corynebacterium bovis, TP: Trueperella pyogenes, Myco: Mycoplasma, PZ: Prototheca zopfii, PA: Pseudomonas aeruginosa, PC: Positive control, NC: Negative control. Criterion value (nA) of each bacterial species: Staphylococcus spp. (18 nA), Streptococcus spp. (18 nA), Enterococcus spp. (18 nA), EB (18 nA), E. coli (20 nA), Klebsiella spp. (25 nA), Coryne. (23 nA), TP (12 nA), Mycoplasma spp. (15 nA), M. bovis (18 nA), PZ (23 nA), PA (20 nA), Yeast (23 nA).
Table 2. Comparative results of bacterial culture and DNA chip detection methods.
| Coagulase-negative Staphylococci(CNS) culture | S. aureusculture | Enterococcusculture | Streptococcusculture | S. agalactiaeculture | S. uberisculture | Enterobacteriaeculture | E. coliculture | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | ||
| DNA chip | + | 6 | 14 | 3 | 7 | 9 | 17 | 10 | 19 | 0 | 0 | 2 | 0 | 5 | 56 | 6 | 3 |
| − | 6 | 55 | 1 | 70 | 0 | 55 | 1 | 51 | 0 | 81 | 0 | 79 | 0 | 20 | 1 | 71 | |
| Klebsiella culture | C. bovisculture | T. pyogenesculture | Mycoplasma A+Bculture | Mycoplasma Aculture | Protothecaculture | P. aeruginosaculture | Candidaculture | ||||||||||
| + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | ||
| DNA chip | + | 9 | 25 | 0 | 1 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | 14 | 0 | 1 |
| − | 0 | 47 | 0 | 80 | 0 | 79 | 0 | 79 | 0 | 80 | 0 | 81 | 0 | 66 | 0 | 80 | |
However, there were several differences between the bacterial culture and DNA chip results. The most common difference was that a positive result was obtained with the DNA chip detection method, whereas a negative result was obtained with bacterial culture. These differences were evident in some of the pathogens indicated in bold in Table 2. The discrepancy between the results of culture and genetic-screening methods is a recognized phenomenon which has been reported previously [11]. These differences are considered to be attributable to the high sensitivity of the DNA chip detection method compared to the bacterial culture method. The use of genetic screening as a fast, highly sensitive detection method for pathogenic organisms is a promising technology. However, there is also a risk of detecting non-pathogenic organisms in the sample that are not the cause of disease. The development of technology capable of distinguishing between microbes that are associated with disease symptoms and those that are not is desirable. The presence of non-viable bacteria in a sample also poses a problem. Using bacterial culture methods pathogenicity is only demonstrated by the detection of viable microorganisms whereas, in the DNA chip, dead bacteria may also be counted, introducing confusion into the results. Dead bacteria can be distinguished from viable bacteria by the difference in cell membrane permeability. It should be possible to introduce a pretreatment method before nucleic acid extraction for use with the DNA chip technology to address this issue. This is a priority for future work. There are situations where pathogenic organisms are not present in sufficient numbers to cause overt disease, but when their numbers increase they may require therapeutic interventions. Therefore, bacterial quantification is indispensable for mastitis detection. It will be necessary to improve the system such that hybridization of the target nucleic acid is carried out by a probe, without passing through an amplification process, and also the contaminated microorganism quantity is measured dynamically in the system. A potentially simple solution would be a quasi-quantitative method such as a competition method using DNA or RNA fragments as a concentration standard.
The detection of mastitis-causing bacteria using a combination of LAMP and DNA chip has never been reported, and it is a cutting-edge and epoch-making method applicable worldwide. The advantages of this method include significantly reducing the detection time from 4 days to 2–3 hr when processing less than 10 samples at the same time. It is also possible to easily detect pathogenic microorganisms such as mycoplasma which are difficult to detect using the bacterial culture method. The introduction of this procedure for mastitis detection enables early identification of a pathogenic organism, thus allowing the application of suitable treatment before the microorganism spreads within a herd. Furthermore, the high sensitivity of the DNA chip method can also detect trace amounts of pathogenic microorganisms that may be present before the symptoms of mastitis become evident in dairy cows. We consider that this new detection method using DNA chip technology will be useful as a highly sensitive genetic-screening method for the control of bovine mastitis.
Supplementary Material
Acknowledgments
This work was supported by the Project for Developing Practical Technologies to Promote New Policies in Agriculture (22019) from the Ministry of Agriculture, Forestry and Fisheries, Japan.
REFERENCES
- 1.Blowey R., Edmondson P.2010. The Mastitis Organisms, pp. 33–59. In: Mastitis control in dairy herds, 2nd ed., CAB International, Oxfordshire, U.K. [Google Scholar]
- 2.Compton J.1991. Nucleic acid sequence-based amplification. Nature 350: 91–92. doi: 10.1038/350091a0 [DOI] [PubMed] [Google Scholar]
- 3.Fox L. K.2012. Mycoplasma mastitis: causes, transmission, and control. Vet. Clin. North Am. Food Anim. Pract. 28: 225–237. doi: 10.1016/j.cvfa.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 4.Gao H., Lei Z., Jia J., Wang S., Chen Y., Sun M., Liang C.2009. Application of loop-mediated isothermal amplification for detection of Yersinia enterocolitica in pork meat. J. Microbiol. Methods 77: 198–201. doi: 10.1016/j.mimet.2009.02.001 [DOI] [PubMed] [Google Scholar]
- 5.Hayashi T., Sugita T., Hata E., Katsuda K., Zhang E., Kiku Y., Sugawara K., Ozawa T., Matsubara T., Ando T., Obayashi T., Ito T., Yabusaki T., Kudo K., Yamamoto H., Koiwa M., Oshida T., Tagawa Y., Kawai K.2013. Molecular-based identification of yeasts isolated from bovine clinical mastitis in Japan. J. Vet. Med. Sci. 75: 387–390. doi: 10.1292/jvms.12-0362 [DOI] [PubMed] [Google Scholar]
- 6.Hisaeda K., Koshiishi T., Watanabe M., Miyake H., Yoshimura Y., Isobe N.2016. Change in viable bacterial count during preservation of milk derived from dairy cows with subclinical mastitis and its relationship with antimicrobial components in milk. J. Vet. Med. Sci. 78: 1245–1250. doi: 10.1292/jvms.16-0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogan J., Larry Smith K.2003. Coliform mastitis. Vet. Res. 34: 507–519. doi: 10.1051/vetres:2003022 [DOI] [PubMed] [Google Scholar]
- 8.Hogan J. S., Gonzalez R. N., Harmon R. J., Nickerson S. C., Oliver S. P. 6. Pankey, J. W. and Smith, K. L. 1999. Laboratory Handbook on Bovine Mastitis, Revised ed. National Mastitis Council, Madison. [Google Scholar]
- 9.Ihira M., Yoshikawa T., Enomoto Y., Akimoto S., Ohashi M., Suga S., Nishimura N., Ozaki T., Nishiyama Y., Notomi T., Ohta Y., Asano Y.2004. Rapid diagnosis of human herpesvirus 6 infection by a novel DNA amplification method, loop-mediated isothermal amplification. J. Clin. Microbiol. 42: 140–145. doi: 10.1128/JCM.42.1.140-145.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwamoto T., Sonobe T., Hayashi K.2003. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 41: 2616–2622. doi: 10.1128/JCM.41.6.2616-2622.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keane O. M., Budd K. E., Flynn J., McCoy F.2013. Increased detection of mastitis pathogens by real-time PCR compared to bacterial culture. Vet. Rec. 173: 268. doi: 10.1136/vr.101598 [DOI] [PubMed] [Google Scholar]
- 12.Keefe G. P.1997. Streptococcus agalactiae mastitis: a review. Can. Vet. J. 38: 429–437. [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura N., Ito K., Takahashi M., Hashimoto K., Kawamoto M., Yamanaka M., Taniguchi A., Kamatani N., Gemma N.2007. Detection of six single-nucleotide polymorphisms associated with rheumatoid arthritis by a loop-mediated isothermal amplification method and an electrochemical DNA chip. Anal. Chem. 79: 9484–9493. doi: 10.1021/ac0715468 [DOI] [PubMed] [Google Scholar]
- 14.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T.2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28: E63. doi: 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okada J., Horiuchi H., Hashimoto K., Hirosawa D., Kurosaki Y., Kawamoto K., Yasuda J., Makino S., Gemma N., Nikaido M.2012. Mobile Automatic Detection System for Bacillus anthracis using Electrochemical DNA Chip. J Biosens. Bioelectron. 3: 4. doi: 10.4172/2155-6210.1000126 [DOI] [Google Scholar]
- 16.Unno H., Inada M., Nakamura A., Hashimoto M., Ito K., Hashimoto K., Nikaido M., Hayashi T., Hata E., Katsuda K., Kiku Y., Tagawa Y., Kawai K.2015. Improved rapid and efficient method for Staphylococcus aureus DNA extraction from milk for identification of mastitis pathogens. J. Vet. Med. Sci. 77: 1007–1009. doi: 10.1292/jvms.14-0159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker G. T., Fraiser M. S., Schram J. L., Little M. C., Nadeau J. G., Malinowski D. P.1992. Strand displacement amplification--an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 20: 1691–1696. doi: 10.1093/nar/20.7.1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watts J. L., Lowery D. E., Teel J. F., Rossbach S.2000. Identification of corynebacterium bovis and other coryneforms isolated from bovine mammary glands. J. Dairy Sci. 83: 2373–2379. doi: 10.3168/jds.S0022-0302(00)75126-5 [DOI] [PubMed] [Google Scholar]
- 19.Zaini F., Kanani A., Falahati M., Fateh R., Salimi-Asl M., Saemi N., Farahyar S., Kheirabad A. K., Nazeri M.2012. Identification of Prototheca zopfii from Bovine Mastitis. Iran. J. Public Health 41: 84–88. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.