Abstract
Copper nanoparticles (CuNPs) are of great interest due to their extraordinary properties such as high surface-to-volume ratio, high yield strength, ductility, hardness, flexibility, and rigidity. CuNPs show catalytic, antibacterial, antioxidant, and antifungal activities along with cytotoxicity and anticancer properties in many different applications. Many physical and chemical methods have been used to synthesize nanoparticles including laser ablation, microwave-assisted process, sol-gel, co-precipitation, pulsed wire discharge, vacuum vapor deposition, high-energy irradiation, lithography, mechanical milling, photochemical reduction, electrochemistry, electrospray synthesis, hydrothermal reaction, microemulsion, and chemical reduction. Phytosynthesis of nanoparticles has been suggested as a valuable alternative to physical and chemical methods due to low cytotoxicity, economic prospects, environment-friendly, enhanced biocompatibility, and high antioxidant and antimicrobial activities. The review explains characterization techniques, their main role, limitations, and sensitivity used in the preparation of CuNPs. An overview of techniques used in the synthesis of CuNPs, synthesis procedure, reaction parameters which affect the properties of synthesized CuNPs, and a screening analysis which is used to identify phytochemicals in different plants is presented from the recent published literature which has been reviewed and summarized. Hypothetical mechanisms of reduction of the copper ion by quercetin, stabilization of copper nanoparticles by santin, antimicrobial activity, and reduction of 4-nitrophenol with diagrammatic illustrations are given. The main purpose of this review was to summarize the data of plants used for the synthesis of CuNPs and open a new pathway for researchers to investigate those plants which have not been used in the past.
Graphical abstract.
Proposed Mechanism for Antibacterial activity of copper nanoparticles.
Keywords: Phytosynthesis, Copper nanoparticles, Phytochemicals, Cytotoxicity, Catalytic activity, Antibacterial activity
Background
Nanoparticles (NPs) have a number of interesting applications in the industrial field such as space technology, magnetism, optoelectronics and electronics, cosmetics, and catalytic, pharmaceutical, biomedical, environmental, and energy applications [1, 2]. The extraordinary properties of NPs such as ductility, high yield strength, hardness, flexibility, rigidity, high surface-to-volume ratio, macroquantum tunneling effect, and quantum size are attributable as compared to properties of bulk materials having the same chemical composition [3]. Indeed, the properties of NPs, which may considerably differ from those observed for fine particles, are higher specific surface area, specific optical properties, lower melting points, specific magnetizations, mechanical strength, and numerous industrial applications [4]. Copper nanoparticles (CuNPs) are of great interest due to easy availability, low cost, and their similar properties to those of noble metals [5–9]. CuNPs can also be used in sensors, heat transfer systems [10–12], and electronics (fuel cell and solar cell), as catalysts in many reactions and as bactericidal and antimicrobial agents used to coat hospital equipment [13–19].
Many physical and chemical methods including laser ablation [20], microwave-assisted process, sol-gel [21], co-precipitation [22], pulsed wire discharge [23], vacuum vapor deposition [24], high-energy irradiation [25], lithography [26], mechanical milling [27], photochemical reduction, electrochemistry [28–32], electrospray synthesis [33], hydrothermal reaction [34], microemulsion [35], and chemical reduction are used to synthesize nanoparticles. Although physical and chemical methods produce well-defined and pure nanoparticles, these methods are neither cost-effective nor eco-friendly due to the use of toxic chemicals. One of the most important criteria of nanotechnology is the development of eco-friendly, nontoxic, and clean green chemistry procedures [36]. Hence, biosynthesis of nanoparticles contains a green chemistry-based method which employs different biological bodies such as plants [37, 38], actinomycetes [39, 40], fungus [41–44], bacteria [45–49], yeast [50–52], and viruses [53, 54]. Biological entities offer a nontoxic, clean, and environment-friendly approach to synthesize the NPs with a wide range of size, physicochemical properties, shapes, and compositions [55].
Copper nanoparticles were synthesized and stabilized in the literature by using different plants such as Euphorbia esula [56], Punica granatum [57], Ocimum sanctum [58], Ginkgo biloba [59], Calotropis procera [60], Lawsonia inermis [61], Citrus medicalinn [62], Camellia sinensis [63], Datura innoxia [64], Syzygium aromaticum [65], Sesamum indicum [66], Citrus limon, Turmeric curcumin [67], Gloriosa superba L. [68], Ficus carica [69], Aegle marmelos [70], Caesalpinia pulcherrima [71], Cassia fistula [72], Leucas aspera, Leucas chinensis [73], Delonix elata [74], Aloe barbadensis Miller [75], Thymus vulgaris [76], Phyllanthus emblica [77], Magnolia kobus [78], Eucalyptus [79], Artabotrys odoratissimus [80], Capparis zeylanica [81], Vitis vinifera [82], Hibiscus rosa-sinensis [83], Zingiber officinale [84], Datura metel [85], Zea mays [86], Urtica, Matricaria chamomilla, Glycyrrhiza glabra, Schisandra chinensis, Inula helenium, Cinnamomum [87], Dodonaea viscosa [88], Cassia auriculata [89], Azadirachta indica, Lantana camera, Tridax procumbens [90], Allium sativum [91], Asparagus adscendens, Bacopa monnieri, Ocimum bacilicum, Withania somnifera [92], Smithia sensitiva, Colocasia esculenta [93], Nerium oleander [94], and Psidium guajava [95]; by using different algae/fungi such as Phaeophyceae [96], Stereum hirsutum [97], and Hypocrea lixii [98]; and by using some microorganisms such as Pseudomonas fluorescens [99] and Enterococcus faecalis [100] cultures.
Biosynthesis of Copper Nanoparticles
Parts of Plant Used for Extract
Different parts of plants are used for the preparation of plant extracts such as leaves, seeds, barks, fruits, peel, coir, roots, and gum. Leaves and roots are used in two ways. Firstly, fresh leaves and roots are used for the preparation of plant extracts, and secondly, dry leaves and roots in powder form are used.
Procedure for the Synthesis of CuNPs
For the synthesis of CuNPs, plant extract was prepared by using different parts of different plants. For synthesis of the extract part of the plant of interest, leaves are collected and washed with tap water and then with distilled water to remove dust particles. The washed leaves are used further in two ways. First, these leaves are sun dried for 1–2 h to remove the residual moisture. Known weights of these sun-dried leaves are divided into small parts and soaked in deionized water or ethanol solution. This mixture is stirred for 24 h at room temperature by using a magnetic stirrer and then filtered for further use. Second, these leaves are sun dried for 4–7 days or dried in an oven at 50 °C for 1 day and powdered using a domestic blender. Known weight of plant powder is mixed in water or ethanol solution and then stirred and filtered.
For the synthesis of CuNPs, aqueous solution of precursor salts such as copper sulfate, copper chloride, copper acetate, and copper nitrate with different concentrations is mixed with plant extract. Aqueous solution of sodium hydroxide is also prepared and added to the reaction mixture to control the pH medium. The reaction mixture is strongly shaken for different time intervals in an electric shaker and heated in an oven at different time intervals and at different temperatures. The formation of CuNPs can also take place at room temperature and is confirmed by changing the color of the reaction mixture. At the end, nanoparticles were centrifuged and dried at different temperatures. Reaction optimizations take place by changing the pH of the mixture, concentration of precursor salt, heating time, and temperature of reaction mixture. In the literature, different plants have been used for the formation of copper nanoparticles by using different precursor salts with different reaction conditions as shown in Table 1. From the table, it can be seen that the different reaction conditions affect the shape and size of copper nanoparticles.
Table 1.
Data for synthesis of copper nanoparticles under different reaction conditions
| Plants | Part of plant | Active compounds in plant | Precursor salt | Concentration of salt | Reaction conditions | Characterization | Size | Shape | References |
|---|---|---|---|---|---|---|---|---|---|
| Euphorbia esula | Leaves | Flavonoids and phenolic acids | Copper chloride | 5 mM | Temp 120 °C, pH 9, time 20 min | UV, FTIR, XRD, TEM | 20–110 nm | Spherical | [56] |
| Punica granatum | Peels | – | Copper sulfate | 50 mM | Temp 80 °C for 10 min and 40 °C for 4 h | UV, FTIR, PSA, TEM | 15–20 nm | Spherical | [57] |
| Ocimum sanctum | Leaves | Terpenoids, alcohols, ketones, esters, aldehydes, and carboxylic acids | Copper sulfate | 1 mM | Room temp | UV, FTIR, PSA, TEM, MZS | 25 nm | Rod, cylindrical, elliptical | [58] |
| Leaves | Copper sulfate | 1 mM | Room temp | UV, FTIR, EDX, SEM | 150–200 nm | Spherical | [115] | ||
| Ginkgo biloba | Leaves | Polyphenols, quercetin | Copper chloride | 5 mM | Temp 80 °C, pH 9, time 30 min | UV, FTIR, EDS, TEM | 15–20 nm | Spherical | [59] |
| Calotropis procera | Latex | Cysteine proteases | Copper acetate | 3 mM | Room temp | UV, FTIR, XRD, TEM, EDAX | 15 ± 1.7 nm | Spherical | [60] |
| Lawsonia inermis | Leaves | – | Copper sulfate | 10 mM | Temp 100 °C, pH 11, time 30 min | UV, FTIR, HRTEM, SEM, DMOM | – | [61] | |
| Citrus medicalinn | Fruit juice | Ascorbic acid, saponins, and flavonoids | Copper sulfate | 100 mM | Temp 60–100 °C | UV, FTIR, NTA, XRD | 33 nm | – | [62] |
| Camellia sinensis | Leaves | Flavonoids, phenolic acids, terpenoids, and polysaccharides | Copper chloride | 1 mM | Temp 100 °C, time 3 h | UV, FTIR, EDX, TEM, SEM | 15–25 nm | Spherical | [63] |
| Leaves | – | Copper chloride | 10 mM | Temp 90 °C | FTIR, EDX, TEM, SEM, XRD, NTA | 10–40 nm | Spherical | [104] | |
| Datura innoxia | Leaves | – | Copper sulfate | 1 mM | – | UV, FTIR, EDX, FESEM | 90–200 nm | Spherical | [64] |
| Syzygium aromaticum | Flowers | Eugenol | Copper sulfate | 1 mM | Room temp, pH 3.43 | UV, FTIR, XRD, TEM, SEM | 5–40 nm | – | [65] |
| Sesamum indicum | Seeds | – | Copper sulfate | 10 mM | – | UV | – | – | [66] |
| Citrus limon and Turmeric curcumin | Fruit | Curcuminanilineazomethine | Copper chloride | 1 mM | – | UV, FTIR, XRD, HRTEM, SEM | 60–100 nm | Spherical | [67] |
| Gloriosa superba L. | Leaves | – | Copper sulfate | 1 mM | Room temp | UV, FTIR | – | – | [68] |
| Gossypium | Gum | Hydroxyl, acetyl, carbonyl, and carboxylic groups | Copper nitrate | 40 mM | Room temp, pH 12 | TEM, SAXS, UV, XRD | 19 nm | Spherical | [116] |
| Ficus carica | Leaves | – | Copper chloride | 10 mM | Temp 25 °C, pH 8, time 30 min | UV, SEM, XRD | 50–120 nm | – | [69] |
| Aegle marmelos | Leaves | Polyphenols, alkenoids, phenylpropanoid, and terpenoids | Copper chloride | 1 mM | – | UV, FTIR, XRD | 48 nm | Spherical | [70] |
| Caesalpinia pulcherrima | Flowers | – | Copper nitrate | 1 mM | – | UV, FTIR, XRD, SEM, EDAX | 18–20 nm | Spherical | [71] |
| Cassia fistula | Flowers | – | Copper sulfate | 1 mM | Room temp | UV, FTIR, XRD, SEM | 20 | – | [72] |
| Leucas aspera | Leaves | – | Copper sulfate | 1 mM | – | UV | – | – | [73] |
| Leucas chinensis | Leaves | – | Copper sulfate | 1 mM | – | XRD, FESEM, EDX | 60.23 nm | – | [117] |
| Delonix elata | Flowers | – | Copper sulfate | 1 mM | – | UV, FTIR, XRD, SEM | 20 | – | [74] |
| Aloe barbadensis Miller | Flowers | – | Copper acetate | 5 mM | Temp 50 °C, time 30 min | UV, FTIR, FESEM | 40 nm | Spherical | [75] |
| Thymus vulgaris | Leaves | – | Copper sulfate | 0.2 M | Temp 80 °C, time 4 h | BET, TEM, SAED, FTIR, XRD, XRF, FESEM, EDS | – | – | [76] |
| Phyllanthus emblica | Fruit | Tannin, saponin, flavonoid, alkaloid, quinone, anthraquinone, anthocyanosides, phenols | Copper sulfate | 20 mM | Temp 60–80 °C, pH 10 | UV, FTIR, XRD, SEM, EDAX | 15–30 nm | Flakes | [77] |
| Magnolia kobus | Leaves | Copper sulfate | 1 mM | Temp 25–95 °C | ICP, EDS, XPS, SEM, HRTEM | 40–100 nm | Spherical | [78] | |
| Eucalyptus | Leaves | Flavonoids and phenolic acids | Copper sulfate | 1 mM | – | UV, FTIR, XRD | 38.62 nm | – | [79] |
| Artabotrys odoratissimus | Leaves | – | Copper sulfate | 1 mM | Temp 95 °C | PSA | 35 nm | – | [80] |
| Capparis zeylanica | Leaves | – | Copper sulfate | – | UV, FTIR, SEM, EDX, XRD, TEM | 50–100 nm | Cubical | [81] | |
| Vitis vinifera | Leaves | – | Copper acetate | 1% | – | UV, FTIR, XRD | 3–6 nm | – | [82] |
| Hibiscus rosa-sinensis | Leaves | Polyphenols, flavonoids, proteins, lignins, xanthones | Copper nitrate | 50 mM | – | UV, FTIR, TEM | – | – | [83] |
| Zingiber officinale | – | – | – | – | – | FTIR, XRD, EDX, TEM, SAED | 10.13 nm | Cubical | [84] |
| Datura metel | Leaves | Alkaloids, terpenoids, and phenolic groups | – | – | Time 10 min | UV, PSA, TEM, EDX, FTIR | – | – | [85] |
| Zea mays | Leaves | – | Copper sulfate | 10 mM | Room temp, time 1 h | UV, XRD, EDAX, FTIR | 40 nm | Mixed | [86] |
| Urtica | Leaves | Flavonoids, quercetin, rutin, morin | Copper sulfate | – | Temp 70 °C | UV, SEM, XRD | 6.5 nm | – | [87] |
| Matricaria chamomilla | Leaves | Flavonoids | Copper sulfate | – | Temp 70 °C | UV, SEM, XRD | 58.77 nm | – | [87] |
| Glycyrrhiza glabra | Leaves | Flavonoids | Copper sulfate | – | Temp 70 °C | UV, SEM, XRD | 28.21 nm | – | [87] |
| Schisandra chinensis | Leaves | Quercetin, rutin, morin | Copper sulfate | – | Temp 70 °C | UV, SEM, XRD | 32 nm | – | [87] |
| Inula helenium | Leaves | Flavonoids | Copper sulfate | – | Temp 70 °C | UV, SEM, XRD | 32.41 nm | – | [87] |
| Cinnamomum | Leaves | Flavonoids | Copper sulfate | – | Temp 70 °C | UV, SEM, XRD | 48.8 nm | – | [87] |
| Dodonaea viscosa | Leaves | Santin, penduletin, alizarin, pinocembrin, tannins, saponins | Copper chloride | 1 mM | Temp 50 °C, pH 10 | UV, XRD, AFM, HRTEM, SAED | 30–40 nm | Spherical | [88] |
| Cassia auriculata | Leaves | – | Copper sulfate | 1 mM | – | FESEM, XRD, FTIR | 38–43 nm | Spherical | [89] |
| Azadirachta indica | Leaves | – | Fehling solution | – | – | UV | – | – | [90] |
| Lantana camera | Leaves | – | Fehling solution | – | – | UV | – | – | [90] |
| Tridax procumbens | Leaves | – | Fehling solution | – | – | UV | – | – | [90] |
| Allium sativum | – | Copper sulfate | 10 mM | – | UV, FTIR, SEM, XRD, TEM | 100 nm | Spherical | [91] | |
| Asparagus adscendens | Leaves | – | copper sulfate | 1 mM | – | UV, FTIR, TEM, SAED | 10–15 nm | Spherical | [92] |
| Bacopa monnieri | Leaves | – | copper sulfate | 1 mM | – | UV, FTIR, TEM, SAED | 50–60 nm | Spherical | [92] |
| Ocimum bacilicum | Leaves | – | copper sulfate | 1 mM | – | UV, FTIR, TEM, SAED | 40–60 nm | Spherical | [92] |
| Withania somnifera | Leaves | – | copper sulfate | 1 mM | – | UV, FTIR, TEM, SAED | 50–60 nm | Mixed | [92] |
| Smithia sensitiva | Leaves | Tannin, saponin, flavonoid, anthraquinone glycoside, steroids | Copper sulfate | 1 mM | – | UV, FTIR, SEM, NTA | 136 nm | – | [93] |
| Leaves | Tannin, saponin, flavonoid, anthraquinone glycoside, steroids | Copper acetate | 1% | – | UV, FTIR, SEM, NTA | 50 nm | – | [93] | |
| Colocasia esculenta | Leaves | Tannin, flavonoid, alkaloid, cardiac glycoside, terpenoids, phenols | Copper sulfate | 1 mM | – | UV, FTIR, SEM, NTA | 57 nm | – | [93] |
| Leaves | Tannin, flavonoid, alkaloid, cardiac glycoside, terpenoids, phenols | Copper acetate | 1% | – | UV, FTIR, SEM, NTA | 44 nm | – | [93] | |
| Nerium oleander | Leaves | Copper sulfate | 1 mM | – | UV, FTIR | – | – | [94] | |
| Psidium guajava | Fruit | Flavonoid, alkaloid, steroids, glycoside, terpenoids, phenols | Copper sulfate | 20 mM | Room temp, pH 10 | UV, FTIR, XRD, EDAX, TEM, SEM | 15–30 nm | Flakes | [95] |
Effect of Reaction Parameters on Properties of NPs
The concentration of plant extract plays a main role in reducing and stabilizing the CuNPs. It has been reported that by increasing the concentration of plant extract, the number of particles increased [88]. By increasing the concentration of plant extract, the concentration of phytochemicals increased and the reduction of copper salt also increased. Due to the fast reduction of the metal salt, the size of the nanoparticles also decreased [101].
The size and structure of CuNPs are highly affected by the copper salt. The morphology of nanoparticles changes when the salt (e.g., copper chloride, copper acetate, copper nitrate, or copper sulfate) is used in the presence of sodium hydroxide. It was reported that the shape was triangular and tetrahedron in the case of copper chloride, rod-shaped in the case of copper acetate, and spherical in the case of copper sulfate [102]. By increasing the concentration of the precursor salt, the size of the CuNPs also increased.
The synthesis of CuNPs gives best results by varying the pH of the reaction medium within the preferred range. The size of nanoparticles was controlled by changing the pH value of the reaction mixture. At higher pH, smaller-sized nanoparticles were obtained compared to those obtained at low pH value. This difference can be attributed to the difference in reduction rate of the metal salts by plant extract. The inverse relation between the value of pH and the size of nanoparticle showed that an increase in pH value enables us to obtain small-sized spherical nanoparticles while a decrease in pH value gives large-sized (rod-shaped and triangular) nanoparticles. The effect on absorption spectra of different values of pH (4, 6, 8, 10, and 12) is represented in Fig. 1 [36]. It was reported that the addition of plant extract to CuCl2 did not lead to the formation of CuNPs but, instead, the CuNPs were obtained by changing the pH of the reaction mixture to basic medium. The same behavior was observed by Wu and Chen, and it was concluded that pH plays an important role in the synthesis of CuNPs [103].
Fig. 1.
Parts of the plant used for the preparation of plant extract
Mechanism for Phytosynthesis of Copper Nanoparticles
Phytochemical Screening: a Qualitative Analysis
Phytochemical screening analysis is a chemical analysis carried out for the detection of phytochemicals in different plants. Fresh plant extract with chemicals or chemical reagents is used for this analysis [77] as shown in Table 2.
Table 2.
Phytochemical screening analysis
| Test for phytochemicals | Amount of plant extract | Chemicals used | End point for confirmation of phytochemical |
|---|---|---|---|
| Carbohydrate | 2 mL | Few drops of concentrated sulfuric acid and 1 mL of Molisch’s reagent | Reddish or purple color |
| Tannins | 2 mL | 4 mL of 5% ferric chloride | Greenish black or dark blue color |
| Saponins | 2 mL | 2 mL of distilled water and shake for 15 min | Layer of foam on surface |
| Flavonoids | 2 mL | 1 mL of 2 N sodium hydroxide | Yellow color |
| Alkaloids | 2 mL | Few drops of Mayer’s reagent and 2 mL of concentrated HCl | White precipitate or green color |
| Anthraquinone | 1 mL | Few drops of 10% ammonia solution | Pink color precipitates |
| Anthocyanosides | 1 mL of filtrate | 5 mL HCl | Pale pink color |
Phytochemicals for Reduction of Metal and Stabilizing the NPs
Green synthesis of CuNPs by the use of phytochemicals offers more flexible control over the shape and size of the NPs (i.e., by changing reaction temperature, concentration of plant extract, metal salt concentration, reaction time, and pH of reaction mixture). Color change of the reaction medium indicates reduction of the metal ion and formation of NPs. The green reduction of the copper salts starts instantly, and the formation of copper nanoparticles is indicated by the color change of the reaction mixture. Phytochemicals have a main role in first reducing the metal ions and then stabilizing the metal’s nuclei in the form of nanoparticles as shown in Fig. 2. The interaction of phytochemicals with metal ions and the concentration of these phytochemicals control the shape and size of CuNPs.
Fig. 2.
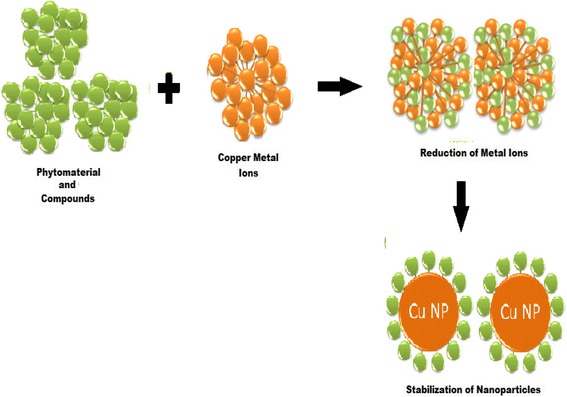
A protocol for reducing the metal ions and then stabilizing the metal’s nuclei
Flavonoids contain polyphenolic compounds, e.g., quercetin, catechins, flavanones, isoflavones, santin, penduletin, alizarin, pinocembrin, anthocyanins, flavones, tannins, and saponins, which are present in different plants such as Ginkgo biloba [59], Citrus medicalinn [62], Phyllanthus emblica [77], Hibiscus rosa-sinensis [83], and Dodonaea viscosa [93]. These compounds play a main role in reducing and chelating the metal. Various functional groups present in the flavonoids are responsible for the reduction of the copper ion. It has been assumed that a reactive hydrogen atom in the flavonoids may be released during the tautomeric alterations of the enol form to the keto form which can reduce copper ions to form copper nuclei or CuNPs. For example, it is assumed that in the case of Ginkgo biloba plant extracts, it is the transformation of quercetin (flavonoid) which plays a main role in the reduction of copper metal ions into copper nuclei or CuNPs due to the change of enol form to keto form as shown in Fig. 3.
Fig. 3.
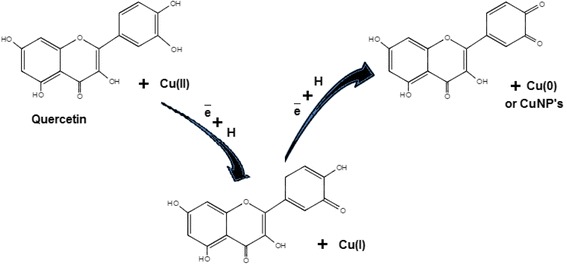
Reduction of copper ions by quercetin
During the synthesis process of CuNPs, metal ions with monovalent or divalent oxidation states are converted into zero-oxidation copper nuclei and these nuclei are merged to obtain different shapes. During the nucleation, nuclei aggregate to form different shapes such as wires, spheres, cubes, rods, triangles, pentagons, and hexagons. Some flavonoids have an ability to chelate the CuNPs with their π electrons and carbonyl groups. Quercetin and santin are flavonoids with strong chelating activity due to the presence of two functional groups involving the hydroxyls and carbonyls. These groups chelate with copper nanoparticles by following the previous mechanism and also explain the ability of adsorption of santin (flavonoid) on the surface of CuNPs as shown in Fig. 4.
Fig. 4.
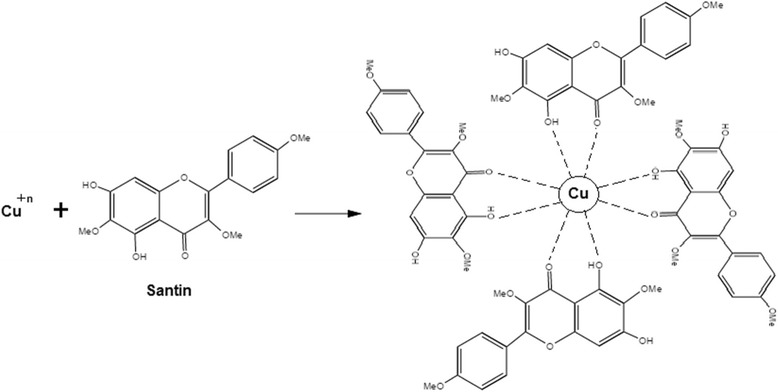
Stabilization of copper nanoparticles by santin
It was assumed that the protein molecules (superoxide dismutase, catalase, glutathione) in different plants such as Hibiscus rosa-sinensis [83] and Camellia sinensis [104] display a high reducing activity for the formation of nanoparticles from metal ions but their chelating activity is not excessive. Sugars such as monosaccharides (glucose), disaccharides (maltose and lactose), and polysaccharides in Camellia sinensis plant [63] can act as reducing agents or antioxidants and have a series of tautomeric transformations from ketone to aldehyde.
Other phytochemicals such as polyphenols (e.g., ellagic acid and gallic acid) which are present in Hibiscus rosa-sinensis [40], phenylpropanoids (phenylalanine, tyrosine) in Aegle marmelos [70], terpenoids in Ocimum sanctum and Asparagus adscendens [58, 92], cysteine proteases in Calotropis procera [60], curcuminanilineazomethine in Turmeric curcumin [67], ascorbic acid in Citrus medicalinn [62], eugenol in Syzygium aromaticum [65], and alkaloids in Aegle marmelos [70] play the same role of reducing the copper ions and stabilizing the copper nanoparticles. Carbohydrates, anthraquinone, quinone, and anthocyanoside in Phyllanthus emblica [77]; lignins and xanthones in Hibiscus rosa-sinensis [83]; and cardiac glycoside, triterponoid, carotenoid glycoside, and anthraquinone glycoside in Colocasia esculenta plant [93] are also phytochemicals which are present in extracts of different plants and act as reducing and stabilizing agents. Examples of certain phytochemicals with structures are shown in Fig. 5.
Fig. 5.
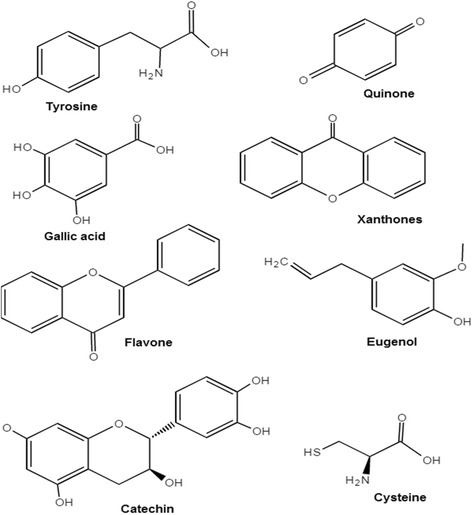
Phytochemicals with their structures
Characterization Techniques
For characterization of synthesized nanoparticles, different techniques were used such as ultraviolet-visible spectroscopy (UV-vis), transmission electron microscopy (TEM), small-angle X-ray scattering (SAXS), Fourier transform infrared spectroscopy (FTIR), X-ray fluorescence spectroscopy (XRF), X-ray diffraction (XRD), X-ray photoelectron spectroscopy (XPS), scanning electron microscopy (SEM), field emission scanning electron microscopy (FESEM), particle size analysis (PSA), Malvern Zetasizer (MZS), energy-dispersive X-ray spectroscopy (EDX/EDS), nanoparticle tracking analysis (NTA), X-ray reflectometry (XRR), Brunauer-Emmett-Teller analysis (BET), selected area electron diffraction (SAED), and atomic force microscopy (AFM) (Table 3).
Table 3.
Characterization techniques and limitations
| Technique | Main role | Limitations | Sensitivity | Ref. |
|---|---|---|---|---|
| Ultraviolet-visible spectroscopy (UV-vis) | Concentration and shape of NPs can be measured | Only for liquid samples | UV-visible regions 200–800 nm | [22] |
| Fourier transform infrared spectroscopy (FTIR) | Nature of bonds and functional groups can be determined | Structure and size of NPs cannot be measured | 20 Å–1 μm | [22] |
| X-ray diffraction (XRD) | Size and crystallinity of nanoparticles can be measured | Composition of NPs and plasmon cannot be found | 1 nm | [36] |
| Scanning electron microscopy (SEM) | Shape and size of nanostructures can be determined | Samples must be solid and cannot detect elements with atomic number < 11 | < 1 nm | [115] |
| Field emission scanning electron microscopy (FESEM) | All structural and morphological investigations are carried out by this technique | Does not give a concentration of NPs | < 1 nm | [117] |
| Transmission electron microscopy (TEM) | Shape and size of nanostructures can be determined | Particles with size < 1.5 nm cannot be determined | < 1.5 nm | [92] |
| Particle size analysis (PSA) | Measured the distribution of size in the sample of solid or liquid particulate materials | – | 1 nm–1 μm | [57, 58] |
| Malvern Zetasizer (MZS) | Measured the size of NPs, zeta potential, and protein mobility | In nanorange | – | [58] |
| Energy-dispersive X-ray spectroscopy (EDX/EDS) | Composition of NPs can be analyzed | Particles with size < 2 nm cannot be analyzed | < 2 nm | [59, 60] |
| Nanoparticle tracking analysis (NTA) | Visualize and measure particle size, concentration, and fluorescent properties of a nanoparticle | – | 30–10 nm | [62] |
| Small-angle X-ray scattering (SAXS) | Size and shape conformation | Lower resolution range | 50–10 Å | [116] |
| X-ray reflectometry (XRR) | Determination of thickness, density, and roughness | Layer thickness 0.1–1000 nm | – | [116] |
| X-ray fluorescence spectroscopy (XRF) | Chemical composition and concentration can be measured | Limited in their ability to measure precisely and accurately | – | [76] |
| X-ray photoelectron spectroscopy (XPS) | Elemental composition of nanoparticles can be analyzed | Decomposition of samples occurred | 3–92 nm | [78] |
| Brunauer-Emmett-Teller analysis (BET) | Specific surface area is measured | 0.35–2 nm | [76] | |
| Selected area electron diffraction (SAED) | Technique that can be performed inside a TEM | Cannot be recommended for quantitative identification techniques | – | [76] |
| Atomic force microscopy (AFM) | Particle size and characterization | For gas and liquid samples | 1 nm–8 μm | [88] |
Applications of Copper Nanoparticles
Due to their outstanding chemical and physical properties, large surface-to-volume ratio, constantly renewable surface, low cost, and nontoxic preparation, CuNPs have been of great interest for applications in different fields. Copper nanoparticles show catalytic activity, antibacterial activity, cytotoxicity or anticancer activity, antioxidant activity, and antifungal activity in different applications. In catalytic activity, copper nanoparticles are used for the Huisgen [3 + 2] cycloaddition of alkynes and azides in many solvents under ligand-free conditions [59], 1-methyl-3-phenoxy benzene, 3,3-oxybis(methylbenzene) [94], synthesis of 1-substituted 1H-1,2,3,4-tetrazole [76], adsorption of nitrogen dioxide, and adsorption of sulfur dioxide [66]. In most of the transition metals catalyzed, Ullmann coupling-reaction ligands, such as phosphines, are reported in the literature and most ligands are expensive, difficult to prepare, and moisture sensitive. For this work, synthesized copper nanoparticles are used for ligand-free Ullmann coupling of diphenyl ether. Different dyes and toxic organic compounds and pesticides present in industrial waste are very harmful for the environment and living organisms. Copper nanoparticles are used for degradation of different dyes such as methylene blue [73], degradation of atrazine [86], and reduction of 4-nitrophenol [76].
Among the antimicrobial agents, copper compounds have been commonly used in agriculture as herbicides [105], algaecides [106], fungicides [107], and pesticides as well as in animal husbandry as a disinfectant [108] (shown in Table 4). The biogenic copper nanoparticles showed powerful antibacterial activity against gram-positive and gram-negative pathogens such as Pseudomonas aeruginosa (MTCC 424), Micrococcus luteus (MTCC 1809), Enterobacter aerogenes (MTCC 2832) [57], Salmonella enterica (MTCC 1253), Rhizoctonia solani, Xanthomonas axonopodis pv. citri, Xanthomonas axonopodis pv. punicea [58], Escherichia coli (ATCC 14948) [62], Staphylococcus aureus (ATCC 25923), Bacillus subtilis (ATCC 6633), Pediococcus acidilactici [69], and Klebsiella pneumoniae (MTCC 4030). In antifungal activity, copper nanoparticles are used against Alterneria carthami, Colletotrichum gloeosporioides, Colletotrichum lindemuthianum, Drechslera sorghicola, Fusarium oxysporum f.sp. carthami, Rhizopus stolonifer, Fusarium oxysporum f.sp. ciceris, Macrophomina phaseolina, Fusarium oxysporum f.sp. udum, Rhizoctonia bataticola [58], Candida albicans, Curvularia, Aspergillus niger, and Trichophyton simii [67]. In cytotoxicity, copper nanoparticles are used for a study on HeLa, A549, MCF7, MOLT4, and BHK21 cell lines (cancer tumors) [60, 104].
Table 4.
Catalytic, antibacterial, cytotoxicity or anticancer, antioxidant, and antifungal activities of copper nanoparticles
| Biological entity | Activity | In/against | Concentration of NPs | References |
|---|---|---|---|---|
| Euphorbia esula | Catalytic | Reduction of 4-nitrophenol | 25 μL | [56] |
| Catalytic | Ligand-free Ullmann coupling of diphenyl ether, 1-methyl-3-phenoxy benzene, and 3,3-oxybis(methylbenzene) | 1 mL | [56] | |
| Punica granatum | Antibacterial | Enterobacter aerogenes, Micrococcus luteus, Salmonella enterica, and Pseudomonas aeruginosa | 100 μg/L | [57] |
| Ocimum sanctum | Antibacterial | Rhizoctonia solani, Xanthomonas axonopodis pv. citri, Xanthomonas axonopodis pv. punicea | – | [58] |
| Antifungal | Alterneria carthami, Colletotrichum gloeosporioides, Colletotrichum lindemuthianum, Drechslera sorghicola, Fusarium oxysporum f.sp. carthami, Rhizopus stolonifer, Fusarium oxysporum f.sp. ciceris, Macrophomina phaseolina, Fusarium oxysporum f.sp. udum, and Rhizoctonia bataticola | – | [58] | |
| Ginkgo biloba | Catalytic | Huisgen [3 + 2] cycloaddition of azides and alkynes | 10 mol% | [59] |
| Calotropis procera | Cytotoxicity | Study on HeLa, A549, and BHK21 cell lines (cancer tumors) | 120 μM | [60] |
| Citrus medicalinn | Antibacterial | Propionibacterium acnes (MTCC 1951), Salmonella typhi (ATCC 51812), K. pneumoniae (MTCC 4030), P. aeruginosa, and Escherichia coli | 20 μL | [62] |
| Antifungal | Fusarium culmorum (MTCC 349) and Fusarium oxysporum (MTCC 1755) | 20 μL | [62] | |
| Camellia sinensis | Antibacterial | Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus, and Bacillus subtilis | 2, 4, 6, and 8 μg/L | [63] |
| Anticancer | HT-29, MCF7, and MOLT4 cell lines | 80 μg/mL | [104] | |
| Datura innoxia | Antibacterial | Xanthomonas oryzae pv. oryzae | [64] | |
| Sesamum indicum | Catalytic | Adsorption of nitrogen dioxide and sulfur dioxide | 0.01–0.06 g | [66] |
| Citrus limon and Turmeric curcumin | Antibacterial | Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus, and Bacillus subtilis | – | [67] |
| Antifungal | Candida albicans, Curvularia, Aspergillus niger, Trichophyton simii | – | [67] | |
| Ficus carica | Antibacterial | Pediococcus acidilactici | 10 μg/mL | [69] |
| Leucas aspera | Catalytic | Degradation of methylene blue | 1 mL | [73] |
| Thymus vulgaris | Catalytic | Reduction of 4-nitrophenol and synthesis of 1-substituted 1H-1,2,3,4-tetrazole | 50 g and 15 mg, respectively | [76] |
| Phyllanthus emblica | Antibacterial | Staphylococcus aureus and Escherichia coli | – | [77] |
| Magnolia kobus | Antibacterial | Escherichia coli (ATCC 25922) | – | [78] |
| Capparis zeylanica | Antibacterial | Gram-positive and gram-negative pathogens | – | [81] |
| Vitis vinifera | Antibacterial | Bacillus subtilis and Escherichia coli (ATCC 25922) | – | [82] |
| Hibiscus rosa-sinensis | Antibacterial | Bacillus subtilis and Escherichia coli (ATCC 25922) | – | [83] |
| Antioxidant | Hydrogen peroxide scavenging assay was assessed | – | [83] | |
| Zingiber officinale | Antibacterial | Staphylococcus aureus (ATCC 25923), Bacillus subtilis, and Escherichia coli | – | [84] |
| Zea mays | Catalytic | Degradation of atrazine | 30 mg | [86] |
| Dodonaea viscosa | Antibacterial | Staphylococcus aureus (ATCC 25923), Bacillus subtilis, Escherichia coli, and K. pneumoniae (MTCC 4030) | – | [88] |
| Azadirachta indica | Antibacterial | Escherichia coli | – | [90] |
| Lantana camera | Antibacterial | Escherichia coli | – | [90] |
| Antifungal | Aspergillus niger | – | [90] | |
| Tridax procumbens | Antibacterial | Escherichia coli | – | [90] |
| Antifungal | Aspergillus niger | – | [90] | |
| Allium sativum | Antibacterial | Escherichia coli, Bacillus subtilis | 75 and 50 μL, respectively | [91] |
| Asparagus adscendens | Antibacterial | Staphylococcus aureus | – | [92] |
| Bacopa monnieri | Antibacterial | Bacillus subtilis, Escherichia coli, Pseudomonas aeruginosa | – | [92] |
| Nerium oleander | Antibacterial | Escherichia coli, Staphylococcus aureus, Bacillus subtilis, K. pneumoniae, Salmonella typhi | 35 μL | [94] |
| Psidium guajava | Antibacterial | Escherichia coli, Staphylococcus aureus | – | [95] |
Hypothetical Mechanism of Antimicrobial Activity
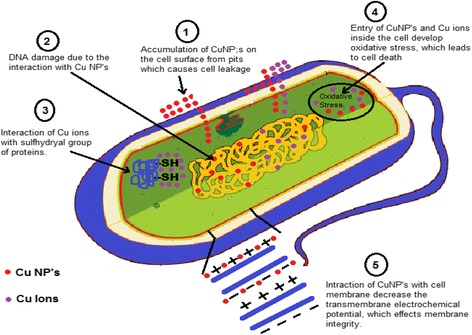
It was observed that CuNPs have an excellent antimicrobial activity and only limited reports presented the mechanism of the antibacterial activity of copper nanoparticles in the literature, but these mechanisms were hypothetical. It was observed that bacteria and enzymes/proteins were destroyed due to the interaction of CuNPs with –SH (sulfhydryl) group [109, 110]. It was also reported that the helical structure of DNA molecules become disturbed by the interaction of CuNPs [111]. The interaction of CuNPs with the cell membrane of bacteria decreased the transmembrane electrochemical potential, and due to the decrease in transmembrane electrochemical potential, it affected the membrane integrity [112]. It was assumed that metal NPs release their respective metal ions. Copper nanoparticles and copper ions accumulate on the cell surface of the bacteria and form pits in the membrane, causing leakage of the cellular component from the cell and inside the cell, causing oxidative stress which leads to cell death [112–114]. A hypothetical mechanism of antibacterial activity representing the above possibilities is shown in Fig. 6.
Fig. 6.
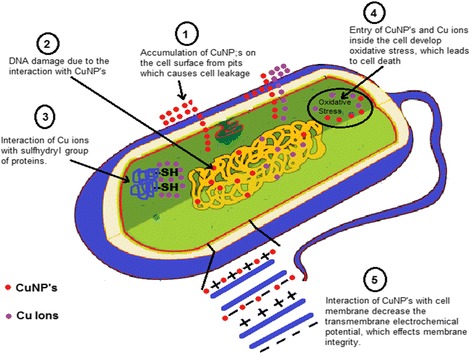
Mechanism for antibacterial activity of copper nanoparticles
Catalytic Activity for Reduction of 4-Nitrophenol
4-Nitrophenol (4-NP) which is usually found in agricultural wastewaters and industrial products is hazardous and not environment-friendly. Hydrogenation or reduction of 4-NP, which is converted into 4-aminophenol (4-AP), takes place in the presence of CuNPs. CuNPs can catalyze the reaction to overcome the kinetic barrier by assisting electron transfer from the donor borohydrate ions to the acceptor 4-NP.
Catalytic activity of the synthesized CuNPs has been studied in the reduction of 4-nitrophenol in aqueous medium at room temperature in the presence of aqueous solution of sodium borohydride [56]. The reduction of 4-NP by using CuNPs is a simple and environment-friendly process. Catalytic efficiency of CuNPs for the reduction of 4-NP was examined by using a UV-vis spectrometer. It was observed that the maximum absorption peak for 4-NP in aqueous medium was at 317 nm and the adsorption peak shifted to 403 nm by adding sodium borohydride due to the formation of 4-nitrophenolate ions. A peak at 403 nm remained unaffected even after 2 days, which indicated that the reduction of 4-NP cannot take place in the absence of a catalyst. After adding the CuNPs, the absorption peak of the solution shifted to 300 nm and the peak at 403 nm completely disappeared which indicated the reduction of 4-NP to 4-AP without any side product. A hypothetical mechanism for the reduction of 4-NP is shown in Fig. 7. In the mechanism, 4-NP and sodium borohydride are present in the solution in the form of ions. The protons of the borohydride ion are adsorbing on the surface of the copper nanoparticles and BO2 produced. 4-Nitrophenolate ions also adsorb on the surface of the CuNPs. Due to the adsorption of both protons and 4-nitrophenolate ion, CuNPs overcome the kinetic barrier of reactants and 4-nitrophenolate ion is converted into 4-aminophenolate ion. After conversion, desorption of the 4-aminophenolate ion takes place and it is converted into 4-aminophenol.
Fig. 7.
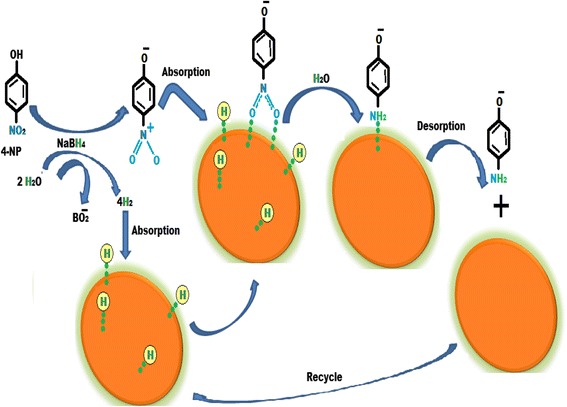
Mechanism for the reduction of 4-nitrophenol
Conclusions
This paper has reviewed and summarized recent information of biological methods used for the synthesis of copper nanoparticles (CuNPs) using different plants. Green synthesis of CuNPs has been proposed as a valuable alternative to physical and chemical methods with low cytotoxicity, economic prospects, environment-friendly, enhanced biocompatibility, feasibility, and high antioxidant activity and high antimicrobial activity of CuNPs. The mechanism of biosynthesis of NPs is still unknown, and more research needs to be focused on the mechanism of formation of nanoparticles and understanding of the role of phytochemicals in the formation of NPs. This review gives data of plants used in the synthesis of copper nanoparticles, synthesis procedure, and the reaction parameters which affect the properties of synthesized CuNPs. A phytochemical screening analysis is a chemical analysis used to identify the phytochemicals such as detection of carbohydrates, tannins, saponins, flavonoids, alkaloids, anthraquinones, and anthocyanosides in different plants. The mechanism of reduction of copper ion by quercetin and stabilization of copper nanoparticles by santin is described in this paper. Characterization techniques used in the literature for copper nanoparticles are UV-vis, FTIR, XRD, SEM, FESEM, TEM, PSA, MZS, EDX, NTA, SAXS, XRR, XRF, XPS, BET, SAED, and AFM. Copper nanoparticles show catalytic activity, antibacterial activity, cytotoxicity or anticancer activity, antioxidant activity, and antifungal activity in different applications. Hypothetical mechanisms of antimicrobial activity and reduction of 4-nitrophenol with diagrams are shown in this paper.
CuNPs with different structural properties and effective biological effects can be fabricated using new green protocols in the coming days. The control over particle size and, in turn, the size-dependent properties of CuNPs will open the new doors of their applications. This study provides an overview of synthesis of CuNP by using plant extract, microbial extract, and naturally occurring biomolecules. Although all these green protocols for CuNP synthesis have their own advantages and limitations, the use of plant extract as a reductant is more beneficial as compared to the use of microbial extract because of the rapid rate of production of nanoparticles with former green reductant.
Acknowledgments
Funding
The authors confirmed that they did not receive any funding for this article.
Availability of Data and Materials
The authors agreed to share data to any recommended repositories.
Authors’ Contributions
MID collected all the data and write the whole manuscript. FA also contributes to this article by studying more than 100 relevant articles and also helped in collecting some data. ZH helped in the writing of this manuscript. She is a native speaker of English from UK. She revised the whole manuscript and improved its English language. MM helped in the writing of the interpretation of antimicrobial effect. She also helped in explaining the green mechanism. All authors read and approved the final manuscript.
Authors’ Information
MID received his PhD in Physical Chemistry from the Islamia University of Bahawalpur in 2013. He joined the Institute of Chemistry, University of the Punjab, Lahore, Pakistan, in November 2009. His field of interest is adsorption by activated carbon, theoretical chemistry, computational chemistry, and material chemistry. Currently, he is working on high-surface-area activated carbon. His research work is published in different international journals and presented at various international conferences held worldwide. He has published over 42 research articles in leading international journals. This review article is a 3-year effort of MID.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Puzyn T, Leszczynska D, Leszczynski J. Toward the development of “nano-QSARs”: advances and challenges. Small. 2009;5:2494–2509. doi: 10.1002/smll.200900179. [DOI] [PubMed] [Google Scholar]
- 2.Leszczynski J. Bionanoscience: nano meets bio at the interface. Nat Nanotechnol. 2010;5:633–634. doi: 10.1038/nnano.2010.182. [DOI] [PubMed] [Google Scholar]
- 3.Puzyn T, Leszczynski J, Cronin MT. Recent Advances in QSAR Studies: Methods and Applications. 2010. [Google Scholar]
- 4.Raigond P, Raigond B, Kaundal B, Singh B, Joshi A, Dutt S. Effect of zinc nanoparticles on antioxidative system of potato plants. J Env. Biol. 2017;38:435. [Google Scholar]
- 5.Ahmadi SJ, Outokesh M, Hosseinpour M, Mousavand T. A simple granulation technique for preparing high-porosity nano copper oxide (II) catalyst beads. Particuology. 2011;9:480–485. doi: 10.1016/j.partic.2011.02.010. [DOI] [Google Scholar]
- 6.Ramgir N, Datta N, Kaur M, Kailasaganapathi S, Debnath AK, Aswal D, Gupta S. Metal oxide nanowires for chemiresistive gas sensors: issues, challenges and prospects. Colloid Surface A. 2013;439:101–116. doi: 10.1016/j.colsurfa.2013.02.029. [DOI] [Google Scholar]
- 7.Ruparelia JP, Chatterjee AK, Duttagupta SP, Mukherji S. Strain specificity in antimicrobial activity of silver and copper nanoparticles. Actabiomaterialia. 2008;4:707–716. doi: 10.1016/j.actbio.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Ren G, Hu D, Cheng EW, Vargas-Reus MA, Reip P, Allaker RP. Characterisation of copper oxide nanoparticles for antimicrobial applications. Int J Antimicrob AG. 2009;33:587–590. doi: 10.1016/j.ijantimicag.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 9.T. Theivasanthi, M. Alagar, arXiv preprint arXiv (2011) Studies of copper nanoparticles effects on microorganisms:1110.1372
- 10.Eranna G, Joshi B, Runthala D, Gupta R. Oxide materials for development of integrated gas sensors—a comprehensive review. Crit Rev Solid State. 2004;29:111–188. doi: 10.1080/10408430490888977. [DOI] [Google Scholar]
- 11.Guo Z, Liang X, Pereira T, Scaffaro R, Hahn HT. CuO nanoparticle filled vinyl-ester resin nanocomposites: Fabrication, characterization and property analysis. Compos Sci Technol. 2007;67:2036–2044. doi: 10.1016/j.compscitech.2006.11.017. [DOI] [Google Scholar]
- 12.Padil VVT, Černík M. Green synthesis of copper oxide nanoparticles using gum karaya as a biotemplate and their antibacterial application. Int J Nanomedicine. 2013; [DOI] [PMC free article] [PubMed]
- 13.Stoimenov PK, Klinger RL, Marchin GL, Klabunde KJ. Metal oxide nanoparticles as bactericidal agents. Langmuir. 2002;18:6679–6686. doi: 10.1021/la0202374. [DOI] [Google Scholar]
- 14.Gabbay J, Borkow G, Mishal J, Magen E, Zatcoff R, Shemer-Avni Y. Copper oxide impregnated textiles with potent biocidal activities. J Ind Textiles. 2006;35:323–335. doi: 10.1177/1528083706060785. [DOI] [Google Scholar]
- 15.Borkow G, Zatcoff RC, Gabbay J. Reducing the risk of skin pathologies in diabetics by using copper impregnated socks. Med Hypotheses. 2009;73:883–886. doi: 10.1016/j.mehy.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 16.Borkow G, Gabbay J, Dardik R, Eidelman AI, Lavie Y, Grunfeld Y, Ikher S, Huszar M, Zatcoff RC, Marikovsky M. Molecular mechanisms of enhanced wound healing by copper oxide-impregnated dressings. Wound Repair Regen. 2010;18:266–275. doi: 10.1111/j.1524-475X.2010.00573.x. [DOI] [PubMed] [Google Scholar]
- 17.Umer A, Naveed S, Ramzan N, Rafique MS. Selection of a suitable method for the synthesis of copper nanoparticles. Nano. 2012;7:1230005. doi: 10.1142/S1793292012300058. [DOI] [Google Scholar]
- 18.Nasirian A. Synthesis and Characterization of Cu Nanoparticles and Studying of Their Catalytic Properties. Dimension. 2012;2:159–164. [Google Scholar]
- 19.Magaye R, Zhao J, Bowman L, Ding M. Genotoxicity and carcinogenicity of cobalt-, nickel- and copper-based nanoparticles. Exp Therapeutic Med. 2012;4:551–561. doi: 10.3892/etm.2012.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bajaj G, Chaudhary A, Naaz H, Kumar B, Soni R. Laser ablation synthesis of Zn/ZnO core-shell nanoparticles. IEEE. 2007:940–2.
- 21.Jia F, Zhang L, Shang X, Yang Y. Non-aqueous sol–gel approach towards the controllable synthesis of nickel nanospheres, nanowires, and nanoflowers. Adv Mater. 2008;20:1050–1054. doi: 10.1002/adma.200702159. [DOI] [Google Scholar]
- 22.Imran Din M, Rani A. Recent Advances in the Synthesis and Stabilization of Nickel and Nickel Oxide Nanoparticles: A Green Adeptness. Int J Ana Chem. 2016;2016 [DOI] [PMC free article] [PubMed]
- 23.Tanori J, Pileni MP. Control of the shape of copper metallic particles by using a colloidal system as template. Langmuir. 1997;13:639. doi: 10.1021/la9606097. [DOI] [Google Scholar]
- 24.Lisiecki I, Pileni MP. Synthesis of copper metallic clusters using reverse micelles as microreactors. J Am Chem Soc. 1993;115:3887–3896. doi: 10.1021/ja00063a006. [DOI] [Google Scholar]
- 25.Treguer M, de Cointet C, Remita H, Khatouri J, Mostafavi M, Amblard J, Belloni J, De Keyzer R. Dose rate effects on radiolytic synthesis of gold− silver bimetallic clusters in solution. J Phy Chem B. 1998;102:4310–4321. doi: 10.1021/jp981467n. [DOI] [Google Scholar]
- 26.Zhang G, Wang D. Fabrication of heterogeneous binary arrays of nanoparticles via colloidal lithography. J Am Chem Soc. 2008;130:5616–5617. doi: 10.1021/ja710771j. [DOI] [PubMed] [Google Scholar]
- 27.S-H W, Chen D-H. Synthesis of high-concentration Cu nanoparticles in aqueous CTAB solutions. J Colloid Interf Sci. 2004;273:165–169. doi: 10.1016/j.jcis.2004.01.071. [DOI] [PubMed] [Google Scholar]
- 28.Chen W, Cai W, Zhang L, Wang G, Zhang L. Sonochemical processes and formation of gold nanoparticles within pores of mesoporous silica. J Colloid Interf Sci. 2001;238:291–295. doi: 10.1006/jcis.2001.7525. [DOI] [PubMed] [Google Scholar]
- 29.Eustis S, Hsu H-Y, El-Sayed MA. Gold Nanoparticle Formation from Photochemical Reduction of Au3+ by Continuous Excitation in Colloidal Solutions. A Proposed Molecular Mechanism. J Phy Chem B. 2005;109:4811–4815. doi: 10.1021/jp0441588. [DOI] [PubMed] [Google Scholar]
- 30.Khaydarov RA, Khaydarov RR, Gapurova O, Estrin Y, Scheper T. Electrochemical method for the synthesis of silver nanoparticles. Nano Res. 2009;11:1193–1200. doi: 10.1007/s11051-008-9513-x. [DOI] [Google Scholar]
- 31.Shanmugavadivu M, Kuppusamy S, Ranjithkumar R. Synthesis of pomegranate peel extract mediated silver nanoparticles and its antibacterial activity. Drug Deliv. 2014;2:174–182. [Google Scholar]
- 32.Frattini A, Pellegri N, Nicastro D, De Sanctis O. Effect of amine groups in the synthesis of Ag nanoparticles using aminosilanes. Mater Chem Phys. 2005;94:148–152. doi: 10.1016/j.matchemphys.2005.04.023. [DOI] [Google Scholar]
- 33.Basak S, Chen D-R, Biswas P. Electrospray of ionic precursor solutions to synthesize iron oxide nanoparticles: modified scaling law. Chem Eng Sci. 2007;62:1263–1268. doi: 10.1016/j.ces.2006.11.029. [DOI] [Google Scholar]
- 34.Chen D, Xu R. hydrothermal synthesis and characterization of nanocrystallineγ-Fe2O3particles. J Solid State Chem. 1998;137:185–190. doi: 10.1006/jssc.1997.7631. [DOI] [Google Scholar]
- 35.Chen D-H, S-H W. Synthesis of nickel nanoparticles in water-in-oil microemulsions. Chem Mater. 2000;12:1354–1360. doi: 10.1021/cm991167y. [DOI] [Google Scholar]
- 36.Din MI, Arshad F, Rani A, Aihetasham A, Mukhtar M, Mehmood H. Single step green synthesis of stable copper oxide nanoparticles as efficient photo catalyst material. Biomed Mater. 2017;9:41–48. [Google Scholar]
- 37.Philip D. Green synthesis of gold and silver nanoparticles using Hibiscus rosa sinensis. Phys E. 2010;42:1417–1424. doi: 10.1016/j.physe.2009.11.081. [DOI] [Google Scholar]
- 38.Kumar P, Singh P, Kumari K, Mozumdar S, Chandra R. a green approach for the synthesis of gold nanotriangles using aqueous leaf extract of Callistemon viminalis. Mater Lett. 2011;65:595–597. doi: 10.1016/j.matlet.2010.11.025. [DOI] [Google Scholar]
- 39.Ahmad A, Senapati S, Khan MI, Kumar R, Sastry M. Extracellular Biosynthesis of Monodisperse Gold Nanoparticles by a Novel Extremophilic Actinomycete, Thermomonospora sp. Langmuir. 2003;19:3550–3553. doi: 10.1021/la026772l. [DOI] [Google Scholar]
- 40.Sastry M, Ahmad A, Khan MI, Kumar R. Biosynthesis of metal nanoparticles using fungi and actinomycete. Current Sci. 2003;85:162–170. [Google Scholar]
- 41.Mukherjee P, Ahmad A, Mandal D, Senapati S, Sainkar SR, Khan MI, Parishcha R, Ajaykumar P, Alam M, Kumar R. Fungus-mediated synthesis of silver nanoparticles and their immobilization in the mycelial matrix: a novel biological approach to nanoparticle synthesis. Nano Lett. 2001;1:515–519. doi: 10.1021/nl0155274. [DOI] [Google Scholar]
- 42.Ahmad A, Senapati S, Khan MI, Kumar R, Ramani R, Srinivas V, Sastry M. Intracellular synthesis of gold nanoparticles by a novel alkalotolerant actinomycete, Rhodococcus species. Nanotechnol. 2003;14:824. doi: 10.1088/0957-4484/14/7/323. [DOI] [Google Scholar]
- 43.Ahmad A, Senapati S, Khan MI, Kumar R, Sastry M. Extra-/intracellular biosynthesis of gold nanoparticles by an alkalotolerant fungus, Trichothecium sp. J Biomed Nanotechnol. 2005;1:47–53. doi: 10.1166/jbn.2005.012. [DOI] [Google Scholar]
- 44.Bhainsa KC, D’Souza S. Extracellular biosynthesis of silver nanoparticles using the fungus Aspergillus fumigatus. Colloid Surface B. 2006;47:160–164. doi: 10.1016/j.colsurfb.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 45.Roh Y, Lauf R, McMillan A, Zhang C, Rawn C, Bai J, Phelps T. Microbial synthesis and the characterization of metal-substituted magnetites. Solid State Commun. 2001;118:529–534. doi: 10.1016/S0038-1098(01)00146-6. [DOI] [Google Scholar]
- 46.Lengke M, Southam G. Bioaccumulation of gold by sulfate-reducing bacteria cultured in the presence of gold (I)-thiosulfate complex. Geochimica Cosmochimica Acta. 2006;70:3646–3661. doi: 10.1016/j.gca.2006.04.018. [DOI] [Google Scholar]
- 47.Nair B, Pradeep T. Coalescence of nano-clusters and formation of submicron crystallites assisted by Lactobacillus strains. Cryst Grow Design. 2002;2:293–298. doi: 10.1021/cg0255164. [DOI] [Google Scholar]
- 48.Klaus-Joerger T, Joerger R, Olsson E, Granqvist C-G. Bacteria as workers in the living factory: metal-accumulating bacteria and their potential for materials science. Trend. Biotechnol. 2001;19:15–20. doi: 10.1016/s0167-7799(00)01514-6. [DOI] [PubMed] [Google Scholar]
- 49.Husseiny M, El-Aziz MA, Badr Y, Mahmoud M. Biosynthesis of gold nanoparticles using Pseudomonas aeruginosa. Spectrochimica Acta Part A. 2007;67:1003–1006. doi: 10.1016/j.saa.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 50.Dameron C, Reese R, Mehra R, Kortan A, Carroll P, Steigerwald M, Brus L, Winge DR. Biosynthesis of cadmium sulphide quantum semiconductor crystallites. Nature. 1989;38:596–597. doi: 10.1038/338596a0. [DOI] [Google Scholar]
- 51.Kowshik M, Ashtaputre S, Kharrazi S, Vogel W, Urban J, Kulkarni SK, Paknikar K. Extracellular synthesis of silver nanoparticles by a silver-tolerant yeast strain MKY3. Nanotechnol. 2002;14:95. doi: 10.1088/0957-4484/14/1/321. [DOI] [Google Scholar]
- 52.Mohanpuria P, Rana NK, Yadav SK. Biosynthesis of nanoparticles: technological concepts and future applications. Nano Res. 2008;10:507–517. doi: 10.1007/s11051-007-9275-x. [DOI] [Google Scholar]
- 53.Lee S-W, Mao C, Flynn CE, Belcher A. Ordering of quantum dots using genetically engineered viruses. Science. 2002;296:892–895. doi: 10.1126/science.1068054. [DOI] [PubMed] [Google Scholar]
- 54.Merzlyak A, Lee S-W. Phage as templates for hybrid materials and mediators for nanomaterial synthesis. Curr Opinion Chem Bio. 2006;10:246–252. doi: 10.1016/j.cbpa.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 55.Husen A, Khwaja SS. Phytosynthesis of nanoparticles: concept, controversy and application. Nanoscale res let. 2014;9:229–252. doi: 10.1186/1556-276X-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nasrollahzadeh M, Sajadi SM, Khalaj M. Green synthesis of copper nanoparticles using aqueous extract of the leaves of Euphorbia esula L and their catalytic activity for ligand-free Ullmann-coupling reaction. RSC Advance. 2014;4:47313–47318. doi: 10.1039/C4RA08863H. [DOI] [Google Scholar]
- 57.Kaur P, Thakur R, Chaudhury A. Calotropis procera: A phytochemical and pharmacological review. Chem Lett Review. 2016;9:33–38. doi: 10.1080/17518253.2016.1141238. [DOI] [Google Scholar]
- 58.Shende S, Gaikwad N, Bansod S. Synthesis and evaluation of antimicrobial potential of copper nanoparticle against agriculturally important Phytopathogens. Synthesis. 2016;1:4. [Google Scholar]
- 59.Nasrollahzadeh M, Sajadi SM. Green synthesis of copper nanoparticles using Ginkgo biloba L. leaf extract and their catalytic activity for the Huisgen [3 + 2] cycloaddition of azides and alkynes at room temperature. J Colloid Interface Sci. 2015;457:141–147. doi: 10.1016/j.jcis.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Harne S, Sharma A, Dhaygude M, Joglekar S, Kodam K, Hudlikar M. Novel route for rapid biosynthesis of copper nanoparticles using aqueous extract of Calotropis procera L. latex and their cytotoxicity on tumor cells. Colloid Surface B. 2012;95:284–288. doi: 10.1016/j.colsurfb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 61.Cheirmadurai K, Biswas S, Murali R, Thanikaivelan P (2014) RSC Adv 4:19507–19511
- 62.Shende S, Ingle AP, Gade A, Rai M. Green synthesis of copper nanoparticles and conducting nanobiocomposites using plant and animal sources. W J Microb Biotechnol. 2015;31:865–873. doi: 10.1007/s11274-015-1840-3. [DOI] [PubMed] [Google Scholar]
- 63.Keihan AH, Veisi H, Veasi H. Green synthesis and characterization of spherical copper nanoparticles as organometallic antibacterial agent. 2017. [Google Scholar]
- 64.Kala A, Soosairaj S, Mathiyazhagan S, Raja P. Green synthesis of copper bionanoparticles to control the bacterial leaf blight disease of rice. Curr Sci. 2016;110:10. doi: 10.18520/cs/v110/i10/2011-2014. [DOI] [Google Scholar]
- 65.Subhankari I, Nayak P. Synthesis of copper nanoparticles using Syzygium aromaticum (Cloves) aqueous extract by using green chemistry. World J Nano. Sci Technol. 2013;2:14–17. [Google Scholar]
- 66.Nenavath G, Sirisha D, Hasheena M, Asthana S (2014) Adsorptive removal of aqueous SO2 by using Orange Peel Powder IJERT 12:39–51.
- 67.Jayandran M, Haneefa MM, Balasubramanian V. Green synthesis of copper nanoparticles using natural reducer and stabilizer and an evaluation of antimicrobial activity. J Chem Pharm Res. 2015;7:251–259. [Google Scholar]
- 68.Rajanaika H, Lingaraju K, Manjunath K, Kumar D, Nagaraju G, Suresh D, Nagabhushana H. Green Nanotechnology: Biomimetic Synthesis of Metal Nanoparticles Using Plants and Their Application in Agriculture and Forestry. JTUSCI. 2015;9:7–12. [Google Scholar]
- 69.Gültekin DD, Güngör AA, Önem H, Babagil A, Nadaroğlu H. Synthesis of Copper Nanoparticles Using a Different Method: Determination of Its Antioxidant and Antimicrobial Activity. J Turkish Chem Soc A. 2016;3:623–636. doi: 10.18596/jotcsa.287299. [DOI] [Google Scholar]
- 70.Kulkarni V, Kulkarni P. Synthesis of copper nanoparticles with aegle marmelos leaf extract. Nanosci Nanotechnol. 2014;8:401–404. [Google Scholar]
- 71.Kurkure RV, Jaybhaye S, Sangle A. Synthesis of Copper/Copper Oxide nanoparticles in ecofriendly and non-toxic manner from floral extract of Caesalpinia pulcherrima. IJRITCC. 2016;4:363–366. [Google Scholar]
- 72.Valli G, Suganya M. GREEN SYNTHESIS OF COPPER NANOPARTICLES USING CASSIA FISTULA FLOWER EXTRACT. J.Bio.Innov. 2015;4:162–170. [Google Scholar]
- 73.Hase J, Bharati G, Deshmukh K, Phatangre K, Rahane N, Dokhe Shital A. Synthesis and characterization of Cu nanoparticles of Leucas chinensis L plant. EJPMR. 2016;3:241–242. [Google Scholar]
- 74.Valli G, Suganya M. Biogenic synthesis of copper nanoparticles using Delonix elata flower extract. J Chem Pharm Research. 2015;7:776–779. [Google Scholar]
- 75.Mohsenzadeh JKS. Rapid, Green, and Eco-Friendly Biosynthesis of Copper Nanoparticles Using Flower Extract of Aloe Vera. Syn React Inorg Met. 2015;45:895–898. doi: 10.1080/15533174.2013.862644. [DOI] [Google Scholar]
- 76.Rostami-Vartooni A, Alizadeh M, Bagherzadeh M. Rapid, Green, and Eco-Friendly Biosynthesis of Copper Nanoparticles Using Flower Extract of Aloe Vera. Beilstein. J Nanotechnol. 2015;6:2300–2309. doi: 10.3762/bjnano.6.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jyothi PG, Aishwarya B, Guruprasad R. GREEN SYNTHESIS OF COPPER NANOPARTICLES FROM PHYLLANTHUS EMBLICA AND TO STUDY ITS UV- PROTECTION PROPERTY BY DANIO RERIO (ZEBRAFISH) EMBRYOS. W. J Pharm Pharmaceutical Sci. 2016;5(2016):1133–1140. [Google Scholar]
- 78.Lee H-J, Lee G, Jang NR, Yun JH, Song JY, Kim BS. Biological synthesis of copper nanoparticles using plant extract. Nanotechnol. 2011;1:371–374. [Google Scholar]
- 79.Kolekar R, Bhade S, Kumar R, Reddy P, Singh R, Pradeepkumar K. Biosynthesis of copper nanoparticles using aqueous extract of Eucalyptus sp. plant leaves. Curr Sci. 2015;109:255. [Google Scholar]
- 80.Kathad U, Gajera H. Synthesis of copper nanoparticles by two different methods and size comparison. Int. J Pharm Bio Sci. 2014;5:533–540. [Google Scholar]
- 81.Saranyaadevi K, Subha V, Ravindran RE, Renganathan S. Synthesis and characterization of copper nanoparticle using Capparis zeylanica leaf extract. Int J Chem Technol Res. 2014;6:4533–4541. [Google Scholar]
- 82.Rozina SSM, Shaikh R, Sawant MR, Sushama B. Biosynthesis of Copper Nanoparticles using Vitis vinifera Leaf Extract and Its Antimicrobial Activity. Der Pharm Lett. 2016;8(2016):265–272. [Google Scholar]
- 83.Subbaiya R, Selvam M. Green synthesis of copper nanoparticles from Hibicus rosa-sinensis and their antimicrobial, antioxidant activities. Res J Pharm Biol. Chem Sci. 2015;6:1183–1190. [Google Scholar]
- 84.Chitra K, Manikandan A, Moortheswaran S, Reena K, Antony SA. Zingiber officinale extracted green synthesis of copper nanoparticles: structural, morphological and antibacterial studies. Adv Sci Eng. Medicine. 2015;7:710–716. [Google Scholar]
- 85.Parikh P, Zala D, Makwana B. Biosynthesis of copper nanoparticles and their antimicrobial activity. Inst Post Studies Res KSV Uni. India. 2014:1–15.
- 86.Sriram T, Pandidurai V, Kuberan T. DEGRADATION OF ATRAZINE USING COPPER NANOPARTICLES SYNTHESISED FROM THE LEAF EXTRACT OF Zea mays. J Sci. 2016;6:232–238. [Google Scholar]
- 87.Shah M, Fawcett D, Sharma S, Tripathy SK, Poinern GEJ. Green synthesis of metallic nanoparticles via biological entities. Material. 2015;11:7278–7308. doi: 10.3390/ma8115377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Daniel SK, Vinothini G, Subramanian N, Nehru K, Sivakumar M. Biosynthesis of Cu, ZVI, and Ag nanoparticles using Dodonaea viscosa extract for antibacterial activity against human pathogens. J. Nano Res. 2013;15:1319. doi: 10.1007/s11051-012-1319-1. [DOI] [Google Scholar]
- 89.Dr A, Chandra M. SYNTHESIS OF COPPER NANOPARTICLES USING BIO METHOD IN CASSIA AURICULATA LEAVES EXTRACT. W. J Pharm Res. 2017:1058–65.
- 90.V. A. Mane, PATIL. N, S. S. Gaikwad (2016) EXTRACELLULAR SYNTHESIS OF COPPER NANOPARTICLES USING DIFFERENT PLANT EXTRACT. Int J Appl Natural Sci (IJANS) 5: 33–38
- 91.Joseph AT, Prakash P, Narvi SS. phytofabrication and characterization of copper nanoparticles using allium sativum and its antibacterial activity. IJSET. 2016;4:463–473. [Google Scholar]
- 92.Thakur S, Rai R, Sharma S. STUDY THE ANTIBACTERIAL ACTIVITY OF COPPER NANOPARTICLES SYNTHESIZED USING HERBAL PLANTS LEAF EXTRACTS. Int J Bio-Technol Res. 2014;4:21–34. [Google Scholar]
- 93.Damle S, Sharma K, Bingi G, Shah H. A Comparative Study of Green Synthesis of Silver and Copper Nanoparticles using Smithia sensitiva (Dabzell), Cassia tora (L.) and Colocasia esculenta (L.). Int J Pure App. Biosci. 2016;4:275–281. [Google Scholar]
- 94.Gopinath M, Subbaiya R, Selvam MM, Suresh D. Synthesis of copper nanoparticles from Nerium oleander leaf aqueous extract and its antibacterial activity. Int J Curr Microbiol. Appl Sci. 2014;3:814–818. [Google Scholar]
- 95.Caroling G, Priyadharshini MN, Vinodhini E, Ranjitham AM, Shanthi P. biosynthesis of copper nanoparticles using aqueous guava extract. Int J Pharm. BioSci. 2015;5:25–43. [Google Scholar]
- 96.Abboud Y, Saffaj T, Chagraoui A, El Bouari A, Brouzi K, Tanane O, Ihssane B. Biosynthesis, characterization and antimicrobial activity of copper oxide nanoparticles (CONPs) produced using brown alga extract (Bifurcaria bifurcata) Appl Nanosci. 2014;4:571–576. doi: 10.1007/s13204-013-0233-x. [DOI] [Google Scholar]
- 97.Cuevas R, Durán N, Diez M, Tortella G, Rubilar O. Extracellular biosynthesis of copper and copper oxide nanoparticles by Stereum hirsutum, a native white-rot fungus from chilean forests. J Nanomater. 2015;16:57. [Google Scholar]
- 98.M.R. Salvadori, L.F. Lepre, R.M.A. Ando, C.A.O Nascimento, B. Correâ (2013) Biosynthesis and Uptake of Copper Nanoparticles by Dead Biomass of Hypocrea lixii Isolated from the Metal Mine in the Brazilian Amazon Region. [DOI] [PMC free article] [PubMed]
- 99.Shantkriti S, Rani P. Biological synthesis of copper nanoparticles using Pseudomonas fluorescens. Int J Curr Microb. Appl Sci. 2014;3:374–383. [Google Scholar]
- 100.Ashajyothi C, Jahanara K, Chandrakanth K. BIOSYNTHESIS AND CHARACTERIZATION OF COPPER NANOPARTICLES FROM ENTEROCOCCUS FAECALIS. Int J PharmaBiosci. 2014;5:204–211. [Google Scholar]
- 101.Din MI, Rehan R. Synthesis, Characterization, and Applications of Copper Nanoparticles. Analytical Lett. 2017;50:50–62. doi: 10.1080/00032719.2016.1172081. [DOI] [Google Scholar]
- 102.Shankar S, Rhim J-W. Effect of copper salts and reducing agents on characteristics and antimicrobial activity of copper nanoparticles. Mater Lett. 2014;132:307–311. doi: 10.1016/j.matlet.2014.06.014. [DOI] [Google Scholar]
- 103.SH W, Chen DH. Synthesis of high-concentration Cu nanoparticles in aqueous CTAB solutions. J Colloid Interface Sci. 2004;273:165–169. doi: 10.1016/j.jcis.2004.01.071. [DOI] [PubMed] [Google Scholar]
- 104.Keihan AH, Veisi H, Veasi H. Green synthesis and characterization of spherical copper nanoparticles as organometallic antibacterial agent. Appl. Organometal Chem. 2017;31
- 105.Mastin B, Rodgers JH., Jr Toxicity and bioavailability of copper herbicides (Clearigate, Cutrine-Plus, and copper sulfate) to freshwater animals. Arch Environ Con Tox. 2000;39:445–451. doi: 10.1007/s002440010126. [DOI] [PubMed] [Google Scholar]
- 106.de Oliveira-Filho EC, Lopes RM, Paumgartten FJR. Comparative study on the susceptibility of freshwater species to copper-based pesticides. Chemosphere. 2004;56:369–374. doi: 10.1016/j.chemosphere.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 107.Garcia PC, Rivero RM, Ruiz JM, Romero L. the role of fungicides in the physiology of higher plants: implications for defense responses. Bot Review. 2003;69:162–172. doi: 10.1663/0006-8101(2003)069[0162:TROFIT]2.0.CO;2. [DOI] [Google Scholar]
- 108.Mortazavi F, An J, Dubinett S. p120-catenin is transcriptionally downregulated by FOXC2 in nonsmall cell lung cancer cells. Molecular Canc Res. 2010;8:762–774. doi: 10.1158/1541-7786.MCR-10-0004. [DOI] [PubMed] [Google Scholar]
- 109.Das R, Gang S, Nath SS, Bhattacharjee R. Linoleic acid capped copper nanoparticles for antibacterial activity. J Bio-nanosci. 2010;4:82–86. [Google Scholar]
- 110.Schrand AM, Rahman MF, Hussain SM, Schlager JJ, Smith DA, Syed AF. Metal-based nanoparticles and their toxicity assessment. Nano-med Nano-biotechnol. 2010;2:544–568. doi: 10.1002/wnan.103. [DOI] [PubMed] [Google Scholar]
- 111.Kim J-H, Cho H, Ryu S-E, Choi M-U. Effects of Metal Ions on the Activity of Protein Tyrosine Phosphatase VHR: Highly Potent and Reversible Oxidative Inactivation by Cu2+ Ion. Arch Biochem Biophys. 2000;382:72–80. doi: 10.1006/abbi.2000.1996. [DOI] [PubMed] [Google Scholar]
- 112.Deryabin D, Aleshina E, Vasilchenko A, Deryabina T, Efremova L, Karimov I, Korolevskaya L. Investigation of Copper Nanoparticles Mechanisms Tested by Luminescent Escherichia coli Strains. Nanotech Russia. 2013;8:402–408. doi: 10.1134/S1995078013030063. [DOI] [Google Scholar]
- 113.Khursheed A, Ahmed B (2015) Microwave Accelerated Green Synthesis of Stable Silver Nanoparticles with Eucalyptus globulus Leaf Extract and Their Antibacterial and Antibiofilm Activity on Clinical Isolates. [DOI] [PMC free article] [PubMed]
- 114.Samia S, Ahmed B, Khan MS, Al-Shaeri Musarrat M. Inhibition of growth and biofilm formation of clinical bacterial isolates by NiO nanoparticles synthesized from Eucalyptus globulus plants. J. Microbial Pathogenesis. 2017;111:375–387. doi: 10.1016/j.micpath.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 115.Mekala J, Rajan M, Ramesh R. Synthesis and characterization of copper nanoparticles using Tridax procumbens and its application in degradation of bismarck brown. Int. J ChemTech Res. 2016;9:498–507. [Google Scholar]
- 116.Suresh Y, Annapurna S, Bhikshamaiah G, Singh A. Green Luminescent Copper Nanoparticles. IJSER. 2014;5:156–160. [Google Scholar]
- 117.Hase GJ, Bharati KT, Deshmukh KK, Phatangre ND, Rahane A, Dokhe S. Synthesis and Characterization of anthracite oxide nanoparticles of anthracite coal. IJSER. 2016;7:638–641. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors agreed to share data to any recommended repositories.


