Abstract
Sulforaphane (SFN) is an isothiocyanate derived from cruciferous vegetables. SFN’s cytoprotective properties have been demonstrated in several models associated with a variety of disorders. Our recent studies have shown that SFN protects against ethanol-induced oxidative stress and apoptosis in neural crest cells (NCCs), an ethanol-sensitive cell population implicated in Fetal Alcohol Spectrum Disorders (FASD). This study is designed to test the hypothesis that SFN can prevent ethanol-induced apoptosis in NCCs by inhibiting HDAC and increasing histone acetylation at the Bcl-2 promoter. We found that exposure to 50 mM ethanol resulted in a significant increase in HDAC activities in NCCs. Treatment with SFN decreased the activities of HDAC in ethanol-exposed NCCs. We also found that SFN treatment significantly increased the expression of acetyl-histone H3 in NCCs treated with ethanol. ChIP-qPCR assay revealed that ethanol exposure significantly decreased acetyl-histone H3 binding to the Bcl-2 promoter while supplementing with SFN reversed the ethanol-induced reduction in acetyl-histone H3 binding to the Bcl-2 promoter. In addition, SFN treatment restored the expression of Bcl-2 in ethanol-exposed NCCs and diminished ethanol-induced apoptosis in NCCs. Treatment with SFN also significantly diminished apoptosis in mouse embryos exposed to ethanol in vivo. These results demonstrate that SFN can epigenetically restore the expression of Bcl-2 and attenuate ethanol-induced apoptosis by increasing histone acetylation at the Bcl-2 promoter and suggest that SFN may prevent FASD through epigenetic regulation of the expression of anti-apoptotic genes.
Keywords: Sulforaphane, ethanol, apoptosis, histone deacetylase, Bcl-2
1. Introduction
Prenatal alcohol exposure can cause a range of structural and functional birth defects, which are defined as Fetal Alcohol Spectrum Disorders (FASD) [1, 2]. It has been estimated that the prevalence of FASD in the US general population range from 0.2 to 7 per 1000 children [3]. Studies have demonstrated that one of the major mechanisms underlying ethanol-induced teratogenesis is excessive cell death in the selective cell population, including neural crest cells (NCCs), which are progenitor cells that can give rise to various cell types [4–7]. Excessive apoptosis in NCCs has been demonstrated to significantly contribute to ethanol-induced embryo abnormalities that are phenotypic characteristics of FASD [8, 9].
Studies from our laboratory and others have shown that multiple signaling pathways are involved in ethanol-induced apoptosis, including Seven in absentia homolog1 (Siah1) [10, 11]; p53 pathways [11, 12], MAPK signaling [13] and Bcl-2 family [14]. It has been reported that ethanol exposure reduced cortical Bcl-2 expression [15] and that the induction of Bcl-2 by elevated interleukin-6 (IL-6) or over-expression of Bcl-2 by using Bcl-2 fusion protein can prevent ethanol-induced apoptosis or toxicity [14, 16]. However, the molecular mechanisms underlying ethanol-induced apoptosis in NCCs remain to be defined. Recent studies from our laboratory have demonstrated that Nrf2 signaling is involved in ethanol-induced apoptosis in NCCs and that ethanol-induced apoptosis in NCCs can be attenuated by sulforaphane (SFN) [7, 17].
SFN is a chemical that is abundant in cruciferous vegetables, including broccoli, cabbage, and cauliflower [18]. SFN shows diverse therapeutic actions in cancer [19, 20] neurodegenerative disorders [21], autism spectrum disorders [22], leukemia [23], and disorders of the immune system [24]. Its cytoprotective properties have also been demonstrated in several models associated with a variety of disorders, including focal cerebral ischemia [25], brain inflammation [26], nephrotoxicity [27] and hepatotoxicity [28]. It has been reported that SFN can activate Nrf2 pathway [29], suppress the activation of NF-kappaB [30], and enhance the proteasome activities [31, 32]. More recently, it was discovered that SFN can regulate gene expression through epigenetic mechanisms, specifically by inhibiting the activities of histone deacetylase (HDAC) [33, 34].
Epigenetic regulation, including DNA methylation and histone modifications, play a critical role in the regulation of gene expression [35]. HDACs are a class of enzymes that remove acetyl groups on a histone and regulate the acetylation of histone together with histone acetyltransferases (HAT) [36]. Recent studies have shown that the inhibition of HDAC activity has been implicated in the treatment of cancer and other disorders [37, 38]. Histone acetylation affects the expression of the genes involved in different pathways, including apoptosis [36, 39, 40]. Acetylation of histone H2B and hypoacetylation of histone H4 are all involved in apoptosis [41, 42]. Given the established importance of apoptosis in NCCs in the pathogenesis of FASD, the role of epigenetic regulation of apoptosis in FASD and the potential of SFN in preventing ethanol-induced apoptosis and teratogenesis deserves to be explored.
The current study, using an NCC cell line, JoMa 1.3 cells, and a mouse FASD model, was designed to test the hypothesis that SFN can prevent ethanol-induced apoptosis in NCCs and mouse embryos by inhibiting HDAC and increasing histone acetylation at the Bcl-2 promoter. For this study, we first determined the effects of ethanol and SFN on the activity of HDAC in NCCs. We next tested whether the increase in the activity of HDAC in ethanol-exposed NCCs will result in reduced acetylation of histone H3 and whether treatment with SFN can increase the levels of acetylated histone H3 in NCCs. We have also tested whether SFN can reverse the ethanol-induced reduction in acetyl-histone H3 binding to the Bcl-2 promoter, restore the expression of Bcl-2 and attenuate ethanol-induced apoptosis in NCCs and mouse embryos. We found that SFN can epigenetically restore the expression of Bcl-2 and attenuate ethanol-induced apoptosis in NCCs by increasing histone acetylation at the Bcl-2 promoter. Treatment with SFN also significantly diminished apoptosis in mouse embryos exposed to ethanol in vivo. These results suggest that SFN may prevent FASD through epigenetic regulation of the expression of anti-apoptotic genes.
2. Materials and Methods
2.1 Cell culture and treatment
NCCs (JoMa1.3 cells) were cultured on cell culture dishes coated with fibronectin as previously described [10]. NCCs were pre-treated with or without 1 µM SFN (LKT Laboratories, St. Paul, MN) for 24 hours. The cells pre-treated without SFN were exposed to 50 mM ethanol alone for 24 hours. The cells pre-treated with SFN were followed by concurrent exposure to SFN and ethanol for additional 24 hours. The stable ethanol level was kept by placing the cell culture dishes in a plastic container containing 50 mM ethanol in distilled water [7].
2.2 Animal care and treatment
C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) were mated for 2 hours early in the light cycle. The time of vaginal plug detection was considered 0 days, 0 h of gestation (GD0:0). Pregnant mice in the experimental groups were given an intraperitoneal (i.p.) injection of SFN at a dosage of 40 mg/kg maternal body weight or PBS on GD 8:12 and then were administered two i.p. doses of ethanol at a dosage of 1.9 g/kg maternal body weight on GD 9:0 and GD 9:4. Control mice were injected with PBS according to the above regimen. Pregnant females were killed by cervical dislocation on GD 9:12 and the embryos were collected for protein preparation. All protocols used in this study were approved by the University of Louisville Institutional Animal Care and Use Committee.
2.3 Nuclear Extraction and HDAC activity assay
Nuclear extracts were isolated from control and treated NCCs by using the EpiQuik Nuclear Extraction Kit (Epigentek, Brooklyn, NY), following the manufacturer’s instructions. The protein concentration of nuclear extracts was measured by BCA Protein Assay Reagent (Pierce, Rockford, IL). Total HDAC activity was measured using the Fluor-de-Lys® HDAC Fluorometric Activity Assay Kit (Enzo Life Sciences, Farmingdale, NY). In brief, 10 µg (3–5 µl) of nuclear extracts and 20–22 µl of assay buffer were added to each well. Then the diluted substrates (25 µl) were added to each well and incubated for 30 min at room temperature. The reaction was stopped by the addition of the assay developer. After incubation with the assay developer for 15 min at room temperature, the activity of HDAC were determined using a multi-mode microplate reader (Molecular Devices, Sunnyvale, CA) with an excitation wavelength of 360 nm and an emission wavelength of 460 nm.
2.4 Quantitative real-time PCR analysis
Total RNAs were isolated from NCCs treated with SFN or ethanol using the QIAGEN RNeasy Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The quantitative real-time PCR was performed on a Rotor-Gene Q real-time thermal cycler (Qiagen, Hilden, Germany) with the FastStart Universal SYBR Green Master qPCR kit (Roche, Mannheim, Germany). The following primer pairs were used: Bcl-2: forward: 5′-GAACTGGGGGAGGATTGTGG-3′; reverse: 5′-GCATGCTGGGGCCATATAGT -3′; β-actin: forward: 5′-AGCCTTCCTTCTTGGGTATGGAATC-3′; reverse: 5′-GGAGCAATGATCTTGATCTTCATGG-3′. The relative changes in gene expression were analyzed by comparing threshold cycle number of Bcl-2 and a reference β-actin mRNA as described previously [7].
2.5 Western blotting
Western blotting was performed as described previously [7]. The protein levels of Acetyl-Histone H3, Histone H3, Histone 2B, Bcl-2, and cleaved caspase-3 were analyzed with the following antibodies, respectively: Acetyl-Histone H3 (Millipore, Temecula, CA), Histone H3 (Millipore, Temecula, CA), Histone 2B (Santa Cruz, Santa Cruz, CA), Bcl-2 (Cell Signaling, Beverly, MA), and Cleaved caspase-3 (Asp175) Rabbit pAb (Cell Signaling, Beverly, MA). The membranes were developed on a Molecular Imager ChemiDoc XRS+ System (Bio-Rad, Hercules, CA) and the intensity of the protein band was analyzed by ImageJ software (1.46b, National Institutes of Health, USA). All Western blot analyses were performed in triplicate.
2.6 ChIP-qPCR analysis of acetyl-histone H3 binding to Bcl-2 promoter
Chromatin preparation and immunoprecipitation from NCCs were carried out with Acetyl-Histone H3 Immunoprecipitation (ChIP) Assay Kit (Millipore, Temecula, CA), according to the manufacturer's protocol. In brief, NCCs were collected from different groups and cross-linked with 1% formaldehyde for 10 min at 37°C. Cross-linking was blocked, and the cells were washed, scraped into conical tubes and lysed in SDS lysis buffer. The lysates were sonicated to shear DNA to the length between 200 and 1000 base pairs with 5 sets of 10-second pulses using a QSonica Q125 sonicator (QSonica, Newtown, CT). This standardized sonicated condition was used in all samples. After centrifugation and dilution with ChIP dilution buffer, 1% of the diluted cell supernatant (around 20 µL) was reserved as an input sample. The supernatant was pre-cleared with Salmon Sperm DNA/protein A agarose beads for 1 hour at 4°C with agitation. The antibody against acetyl-Histone H3 (1µg) (Millipore, Temecula, CA) was then added to the supernatant and incubated overnight at 4°C with rotation, and then incubated with 60 µl Salmon Sperm DNA/protein A agarose beads for 1 hour at 4°C. The Immunoprecipitated complex was then washed and eluted. The histone-DNA crosslinks were reversed, and DNA was purified for Real-time PCR assay. Quantitative real-time PCR was performed on bound and input DNAs with the following primers for Bcl-2: forward: 5′-AAAGAGCTGGATTATAACTA-3′; reverse: 5′-CTTGCGCCATCCTTCCCCGAAAA-3′;
2.7 Analysis of apoptosis
Apoptosis was determined by the analysis of cleavage of caspase-3 and its activity, and TUNEL assay as previously described [10]. Caspase-3 cleavage was determined by Western blot. Caspase activity was determined by using Caspase-Glo® 3/7 Assay Systems (Promega, Madison, WI) according to the manufacturer's protocol. TUNEL assay was performed by using a TiterTACS In Situ Detection Kit (Trevigen, MD, USA), following the manufacturer’s protocols.
2.8 Statistical analysis
Statistical analyses were performed as previously described [7] by using GraphPad Prism software (GraphPad Software, San Diego, CA, USA). All data were expressed as means ± SD of three separate experiments. The difference between groups was compared by One-way ANOVA. Differences between groups were considered significant at p < 0.05.
3. Results
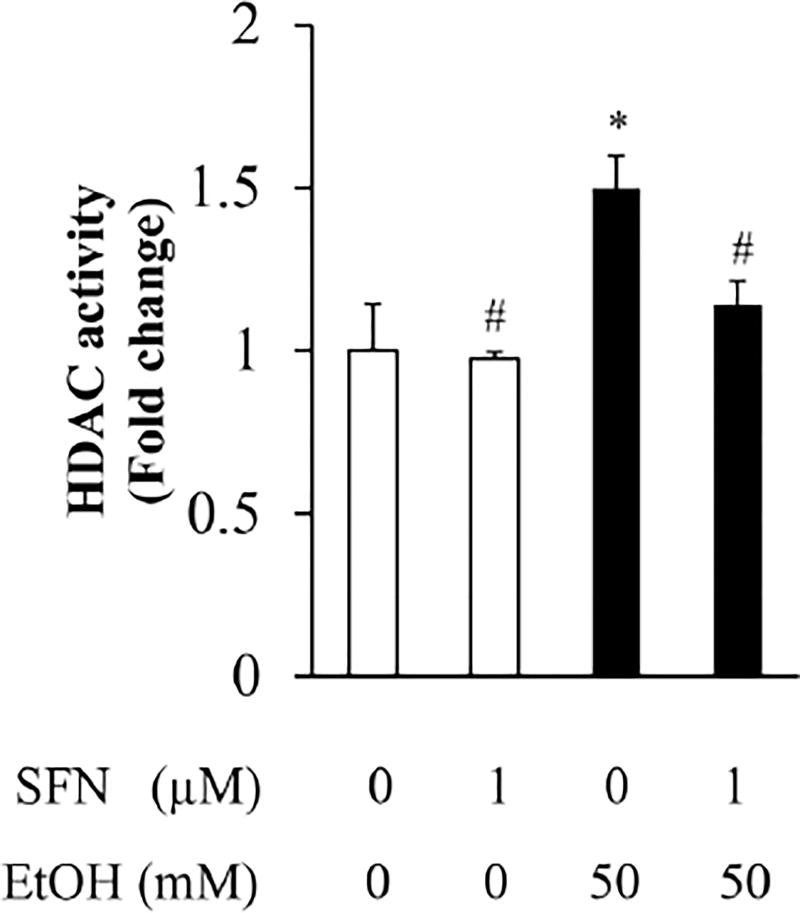
3.1 SFN treatment significantly decreased the activity of HDAC in ethanol-exposed NCCs
To determine the effects of ethanol and SFN on the activity of HDAC in NCCs, cells were treated with 1 µM SFN alone for 24 hours, followed by 24 hours of concurrent exposure to SFN and 50 mM ethanol. As shown in Figure 1, ethanol exposure resulted in a significant increase in the activity of HDAC in NCCs. Treatment with SFN significantly reduced the ethanol-induced increase in the activity of HDAC in NCCs, demonstrating that SFN can reverse the effects of ethanol on the activity of HDAC.
Figure 1. Effects of SFN on HDAC activity in ethanol-exposed neural crest cells.
NCCs were exposed to 50 mM ethanol alone or pretreated with 1 µM SFN for 24 hours, followed by 24 hours of concurrent exposure to SFN and 50 mM ethanol. The nuclear was extracted, and HDAC activity was measured with Fluor-de-Lys® HDAC fluorometric activity assay kit as described in Methods. Data are expressed as fold change over control and represent the mean ± SD of three separate experiments. * p < 0.05 vs. control; # p < 0.05 vs. ethanol.
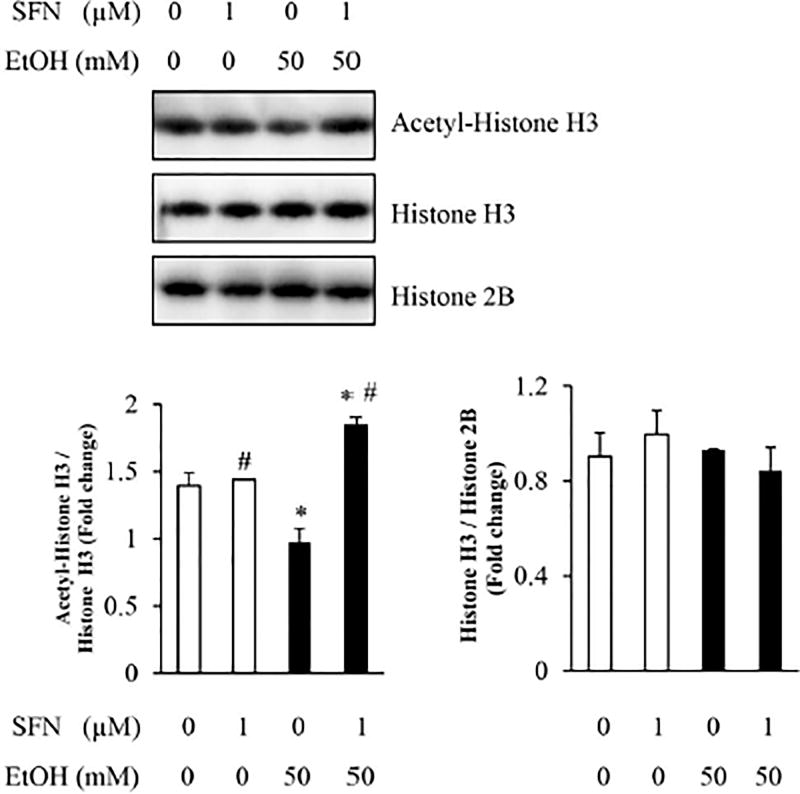
3.2 Treatment with SFN resulted in a significant increase in the levels of acetylated histone H3 in NCCs exposed to ethanol
The acetylation of histone H3, which is tightly controlled by HDAC and HAT, plays a critical role in regulating gene expression [43]. We next determine whether the increase in the activity of HDAC in ethanol-exposed NCCs will result in a reduced acetylation of histone H3 and whether treatment with SFN can increase the levels of acetylated histone H3 in NCCs exposed to ethanol. Western blot analysis revealed that exposure to ethanol or SFN alone or in combination did not result in a significant change in the total protein expression of histone H3. However, ethanol exposure significantly decreased the levels of acetylated histone H3 in NCCs. Co-treatment with SFN significantly increased the acetylation of histone H3 in ethanol-exposed NCCs (Fig. 2). These results indicate that the inhibition of HDAC activity by SFN can reverse the ethanol-induced reduction in the levels of acetylated histone H3 in NCCs.
Figure 2. The expression of acetyl-Histone H3 in NCCs treated with SFN and ethanol.
NCCs were exposed to 50 mM ethanol alone or pretreated with 1 µM SFN for 24 hours, followed by 24 hours of concurrent exposure to SFN and 50 mM ethanol. The expression of acetyl-histone H3, histone H3, and histone 2B was determined by western blot. Data are expressed as fold change over control and represent the mean ± SD of three separate experiments. * p < 0.05 vs. control; # p < 0.05 vs. ethanol.
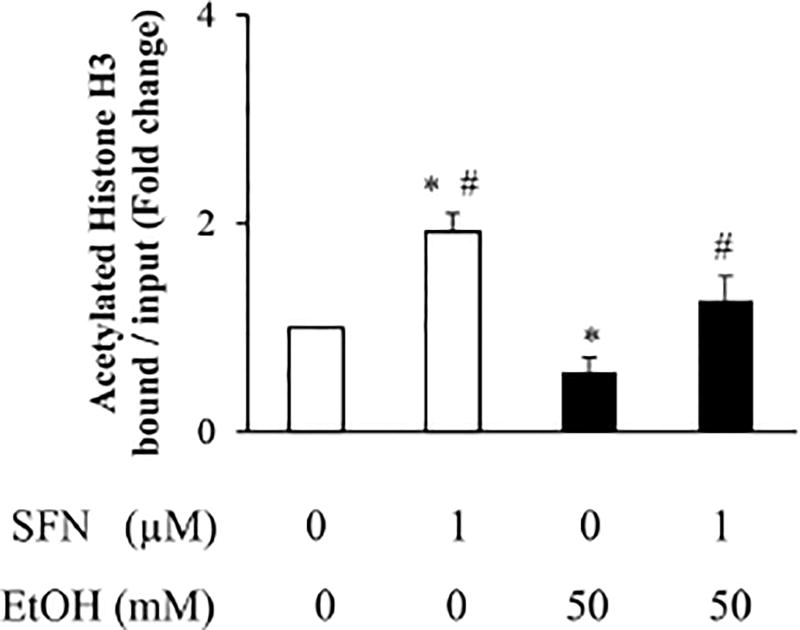
3.3 Supplement with SFN reversed the ethanol-induced reduction in acetyl-histone H3 binding to the Bcl-2 promoter
We next determine whether ethanol exposure can decrease the acetyl-histone H3 binding to the promoter of the anti-apoptotic gene, Bcl-2 and whether SFN treatment can increase the binding of acetylated histone H3 to the promoter of Bcl-2. Using ChIP-qPCR assay, we found that ethanol exposure significantly decreased acetyl-histone H3 binding to the Bcl-2 promoter, while supplementing with SFN greatly reversed the ethanol-induced reduction in acetyl-histone H3 binding to the Bcl-2 promoter (Fig. 3), suggesting that SFN treatment can increase the promoter-associated histone acetylation in Bcl-2.
Figure 3. Effects of SFN on acetylated histone H3 binding to the Bcl-2 promoter in ethanol-exposed NCCs.
NCCs were exposed to 50 mM ethanol alone or pretreated with 1 µM SFN for 24 hours, followed by 24 hours of concurrent exposure to SFN and 50 mM ethanol. The binding of acetyl-histone H3 to the Bcl-2 promoter was measured by quantitative ChIP-qPCR assay with Acetyl-Histone H3 Immunoprecipitation Assay Kit as described in Method. Data are expressed as fold change over control and represent the mean ± SD of three separate experiments. * p < 0.05 vs. control; # p < 0.05 vs. ethanol.
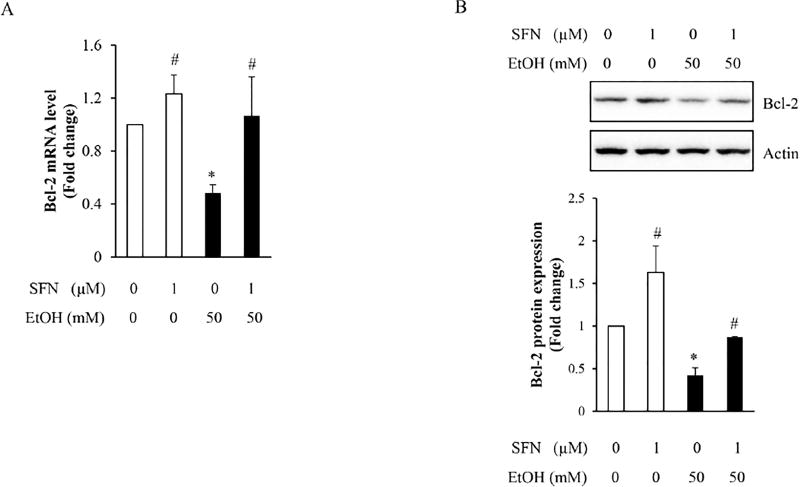
3.4 SFN treatment restored the mRNA and protein expression of Bcl-2 in NCCs treated with ethanol
To determine whether the ethanol-induced reduction in acetyl-histone H3 binding to the promoter of Bcl-2 resulted in a decrease in Bcl-2 transcriptional activation and whether SFN can induce a robust transcriptional activation of Bcl-2 by reversing the ethanol-induced reduction in acetyl-histone H3 binding to Bcl-2 promoter, the mRNA and protein expressions of Bcl-2 were determined in NCCs exposed to ethanol alone or in combination with SFN. We found that ethanol exposure significantly decreased the mRNA expression of Bcl-2 in NCCs. Co-treatment with SFN significantly attenuated the ethanol-induced reduction in Bcl-2 mRNA expression (Fig. 4A). SFN treatment also significantly increased the protein expression of Bcl-2 in ethanol-exposed NCCs, demonstrating that the decrease in Bcl-2 expression can be restored by SFN in ethanol-exposed NCCs (Fig. 4B).
Figure 4. Effects of SFN on the mRNA and protein expression of Bcl-2 in NCCs exposed to ethanol.
NCCs were exposed to 50 mM ethanol alone or pretreated with 1 µM SFN for 24 hours, followed by 24 hours of concurrent exposure to SFN and 50 mM ethanol. The expression of Bcl-2 was determined by real-time PCR (A) or Western blotting (B), respectively. Data are expressed as fold change over control and represent the mean ± SD of three separate experiments. * p < 0.05 vs. control; # p < 0.05 vs. ethanol.
3.5 Epigenetically restoring the expression of Bcl-2 by SFN significantly diminished ethanol-induced apoptosis in NCCs
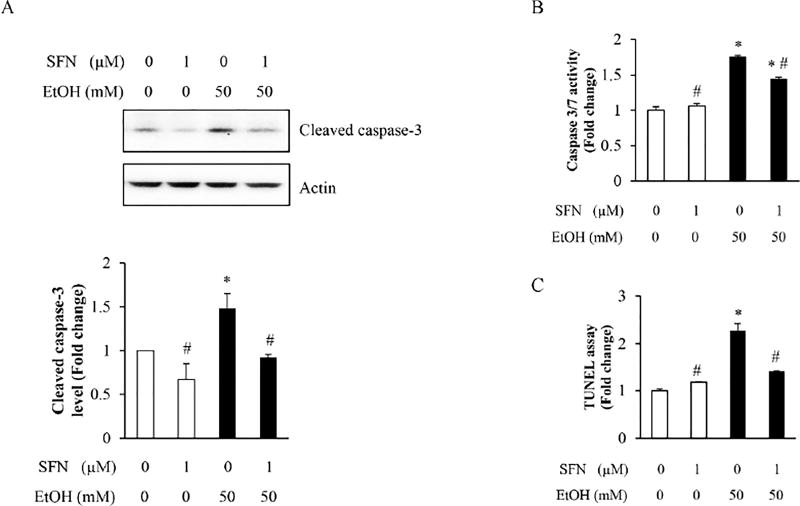
To determine whether epigenetically restoring the expression of Bcl-2 by SFN in NCCs can diminish ethanol-induced apoptosis, apoptosis was determined in NCCs exposed to ethanol or co-treated with ethanol and SFN. We found that ethanol treatment resulted in a significant increase in the cleavage and activity of caspase-3 in NCCs. SFN treatment significantly decreased the cleavage and activity of caspase-3 in ethanol-exposed NCCs, indicating that SFN can diminish ethanol-induced apoptosis in NCCs (Fig. 5A, B). These results were further confirmed by the TUNEL assay, which showed that SFN can significantly reduce ethanol-induced apoptosis in NCCs (Fig. 5C). These results demonstrate that ethanol can induce apoptosis in NCCs by increasing the HDAC activity, reducing the acetyl-histone H3 binding to Bcl-2 promoter and repressing Bcl-2 expression. SFN treatment can prevent ethanol-induced apoptosis in NCCs by epigenetically restoring the expression of the anti-apoptotic protein Bcl-2.
Figure 5. Treatment with SFN attenuated ethanol-induced apoptosis in NCCs.
NCCs were exposed to 50 mM ethanol alone or pretreated with 1 µM SFN for 24 hours, followed by 24 hours of concurrent exposure to SFN and 50 mM ethanol. Apoptosis was determined by the analysis of caspase-3 cleavage (A) and caspase activity assay (B) as well as the TUNEL assay (C). Data are expressed as fold change over control and represent the mean ± SD of three separate experiments. * p < 0.05 vs. control; # p < 0.05 vs. ethanol.
3.6 Treatment with SFN significantly diminished ethanol-induced apoptosis in mouse embryos
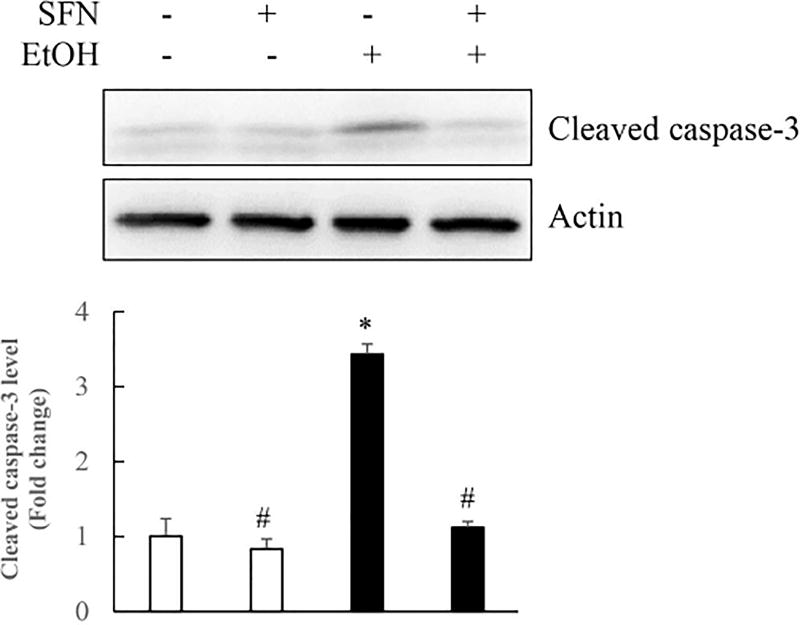
To further confirm that treatment with SFN can provide in vivo protection against ethanol-induced apoptosis in embryos, pregnant mice were given an i.p. injection of SFN at a dosage of 40 mg/kg body weight on GD 8:12, 12 hours before the ethanol treatment. The mice were then administrated two i.p. doses of ethanol at a dosage of 1.9 g/kg on GD 9:0 and GD 9:4. As expected, in vivo ethanol exposure induced the activation of caspase-3, a marker for active apoptosis, in mouse embryos, as evidenced by a three-fold increase in the expression of cleaved caspase-3. Treatment with SFN resulted in a significant reduction in the activation of caspase-3 in ethanol-exposed mouse embryos, as compared with the group treated with ethanol alone (Fig. 6). These results indicate that treatment with SFN is, indeed, effective in preventing apoptosis in mouse embryos exposed to ethanol in vivo.
Figure 6. Co-treatment with SFN significantly diminished ethanol-induced apoptosis in mouse embryos.
Pregnant mice were given an i.p. injection of SFN at a dosage of 40 mg/kg maternal body weight or PBS on GD 8:12 and then were given two i.p. doses of ethanol at a dosage of 1.9 g/kg maternal body weight on GD 9:0 and 9:4. Pregnant mice were killed by cervical dislocation on GD 9:12 and the embryos were collected for protein preparation. Apoptosis was determined by analysis of caspase-3 activation by Western blot. Data are expressed as fold change over control and represent the mean ± SD of three separate experiments. * p < 0.05 vs. control.
4. Discussion
Recent studies have shown that Nrf2-mediated antioxidant response is involved in the susceptibility of NCCs to ethanol-induced apoptosis and that SFN can prevent ethanol-induced apoptosis in NCCs by mediating the induction of Nrf2 signaling [7]. In this study, we found that exposure to ethanol resulted in a significant increase in HDAC activities in NCCs. Treatment with SFN decreased the activities of HDAC in ethanol-exposed NCCs. We also found that SFN treatment significantly increased the expression of acetyl-histone H3 in NCCs treated with ethanol. ChIP-qPCR assay revealed that ethanol exposure decreased acetyl-histone H3 binding to the Bcl-2 promoter while supplementing with SFN reversed the ethanol-induced reduction in acetyl-histone H3 binding to the Bcl-2 promoter. In addition, SFN treatment restored the expression of Bcl-2 in ethanol-exposed NCCs and diminished ethanol-induced apoptosis. These results demonstrate that ethanol can induce apoptosis in NCCs by increasing the HDAC activity, reducing the acetyl-histone H3 binding to Bcl-2 promoter and repressing Bcl-2 expression. SFN can epigenetically restore the expression of Bcl-2 and attenuate ethanol-induced apoptosis by inhibiting HDAC activity and increasing histone acetylation at the Bcl-2 promoter. Treatment with SFN also significantly diminished apoptosis in mouse embryos exposed to ethanol in vivo.
HDACs play a crucial role in regulating the acetylation of histone and other proteins. Acetylation of histone can neutralize the positive charges on the histone complex and decrease its binding ability to DNA, which allows chromatin expansion and facilitates transcription [44]. HDACs prevents transcription by removing the acetyl groups and increase the binding between histone complex and DNA [45]. The effects of ethanol treatment on HDAC activity have been investigated by a number of studies. While it has been reported that global HDAC activity was reduced after an acute ethanol injection [46] or binge ethanol administration [47], and that ethanol treatment has no obvious effect on HDAC activity [48, 49], it has been demonstrated that ethanol exposure can result in a dose-dependent increase in HDAC2 expression in a human neuronal cell line [50]. Ethanol exposure also increased HDAC activity in neurons [51, 52]. It is noteworthy that prenatal ethanol exposure has been found to be able to increase the HDAC activity in adult rat offspring [52, 53]. This is consistent with our results that have shown that ethanol exposure resulted in a significant increase in HDAC activity in NCCs.
Recently, HDACs have been implicated in cancer or other disorders and have become promising therapeutic targets with the potential to reverse or prevent the aberrant epigenetic states associated with a variety of disorders [37, 38] [54, 55]. With an increased appreciation of the importance of dietary factors in health and diseases, in addition to synthetic HDAC inhibitors, a growing number of dietary HDAC inhibitors have been identified, which include EGCG, curcumin, quercetin, butyrate and SFN [56]. SFN is found at high levels in broccoli and other cruciferous vegetables and is metabolized to generate a number of intermediates with HDAC inhibitor activity. It has been reported that SFN inhibited HDAC activity and resulted in both global and localized histone hyperacetylation in human colon cancer cells [56]. Supplementation of broccoli sprouts equated to 105 mg of SFN by human has also been shown to increase the acetylation of histones H3 and H4 in blood cells [57]. In this study, we found that treatment with 1 µM SFN can effectively inhibit the ethanol-induced increase in HDAC activity in NCCs. Inhibition of HDAC by SFN also reversed the ethanol-induced decrease in the levels of acetylated histone H3 in NCCs.
The reversal of acetylation by HDACs associates with transcriptional repression, and the inhibition of HDACs results in hyperacetylation and upregulation of genes. In this study, we have demonstrated that ethanol-induced increased HDAC activity and decreased acetylation of histone H3 resulted in a reduction in the acetyl-histone H3 binding to the promoter of the antiapoptotic gene, Bcl-2 in NCCs. This is consistent with the results from a recent study that has shown that ethanol exposure decreased the acetylation level of histone H3K9 in the promoter region of Bcl-2 [58]. We also found that supplementing with SFN significantly reduced the ethanol-induced reduction in acetyl-histone H3 binding to the Bcl-2 promoter and restored the mRNA and protein expression of Bcl-2 in NCCs treated with ethanol.
The Bcl-2 family proteins, including death antagonists (Bcl-2, Bcl-XL, etc.) and agonists (Bax, Bak, etc.), play a vital role in regulating apoptosis [59]. It has been reported that prenatal ethanol exposure reduced cortical Bcl-2 expression [15]. Studies have also shown that overexpression of Bcl-2 protects the neonatal cerebellum from ethanol-induced neurotoxicity [60]. In this study, we have shown that ethanol exposure significantly decreased the mRNA and protein expression of Bcl-2, indicating that the ethanol-induced reduction in Bcl-2 expression is, at least in part, through increasing HDAC activity and reducing the acetyl-histone H3 binding to the Bcl-2 promoter. We have also shown that SFN co-treatment can restore the expression of Bcl-2 by increasing the acetyl-histone H3 binding to the Bcl-2 promoter and prevent ethanol-induced apoptosis in NCCs and mouse embryos. These results suggest that SFN can prevent ethanol-induced apoptosis through epigenetic regulation of the expression of anti-apoptotic genes.
In conclusion, the results of the current study demonstrate that ethanol exposure can induce apoptosis in NCCs through increasing HDAC activity, reducing the acetyl-histoneH3 binding to the Bcl-2 promoter and repressing Bcl-2 expression. SFN, a potent dietary HDAC inhibitor, can epigenetically restore the expression of Bcl-2 and attenuate ethanol-induced apoptosis by inhibiting HDAC activity and increasing histone acetylation at the Bcl-2 promoter. These findings provide insights into the epigenetic mechanisms involved in the induction of apoptosis in ethanol-exposed embryonic cells. In addition, the potency of SFN in restoring anti-apoptotic gene and preventing ethanol-induced apoptosis in NCCs and mouse embryos, along with the fact that SFN is a bioactive compound derived from broccoli and other cruciferous vegetables, illustrates the potential of a practical and promising nutraceutical-based therapeutic strategy for human FASD.
Highlights.
SFN decreased the activities of HDAC in ethanol-exposed NCCs.
SFN increased the levels of acetylated histone H3 in NCCs exposed to ethanol.
SFN reversed the reduction in acetyl-histone H3 binding to the Bcl-2 promoter.
SFN treatment restored the expression of Bcl-2 in ethanol-exposed NCCs.
SFN significantly diminished ethanol-induced apoptosis in NCCs and mouse embryos.
Acknowledgments
This work was supported by the National Institute of Health Grants AA020265, AA021434, AA024337 (S.-Y.C.), AA032190, and AA022416 (W.F.) from the National Institute on Alcohol Abuse and Alcoholism, AR063630 (X.W.) from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and 1-15-BS-018 (L.C.) from the American Diabetes Association.
List of Abbreviations
- SFN
sulforaphane
- NCCs
neural crest cells
- FASD
Fetal Alcohol Spectrum Disorders
- HDAC
histone deacetylase
- ChIP-qPCR
chromatin immunoprecipitation-quantitative real-time PCR
- Bcl-2
B-cell leukemia/lymphoma-2
- TUNEL
Terminal deoxynucleotidyl transferase dUTP nick end labeling
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Sokol RJ, Delaney-Black V, Nordstrom B. Fetal alcohol spectrum disorder. Jama. 2003;290:2996–9. doi: 10.1001/jama.290.22.2996. [DOI] [PubMed] [Google Scholar]
- 2.Kodituwakku PW. Defining the behavioral phenotype in children with fetal alcohol spectrum disorders: a review. Neuroscience and biobehavioral reviews. 2007;31:192–201. doi: 10.1016/j.neubiorev.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 3.May PA, Gossage JP, Kalberg WO, Robinson LK, Buckley D, Manning M, et al. Prevalence and epidemiologic characteristics of FASD from various research methods with an emphasis on recent in-school studies. Developmental disabilities research reviews. 2009;15:176–92. doi: 10.1002/ddrr.68. [DOI] [PubMed] [Google Scholar]
- 4.Dunty WC, Jr, Chen SY, Zucker RM, Dehart DB, Sulik KK. Selective vulnerability of embryonic cell populations to ethanol-induced apoptosis: implications for alcohol-related birth defects and neurodevelopmental disorder. Alcoholism, clinical and experimental research. 2001;25:1523–35. [PubMed] [Google Scholar]
- 5.Hall BK. The neural crest and neural crest cells: discovery and significance for theories of embryonic organization. Journal of biosciences. 2008;33:781–93. doi: 10.1007/s12038-008-0098-4. [DOI] [PubMed] [Google Scholar]
- 6.Sun H, Chen X, Yuan F, Liu J, Zhao Y, Chen SY. Involvement of seven in absentia homolog-1 in ethanol-induced apoptosis in neural crest cells. Neurotoxicology and teratology. 2014;46:26–31. doi: 10.1016/j.ntt.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Liu J, Chen SY. Sulforaphane protects against ethanol-induced oxidative stress and apoptosis in neural crest cells by the induction of Nrf2-mediated antioxidant response. British journal of pharmacology. 2013;169:437–48. doi: 10.1111/bph.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotch LE, Sulik KK. Experimental fetal alcohol syndrome: proposed pathogenic basis for a variety of associated facial and brain anomalies. American journal of medical genetics. 1992;44:168–76. doi: 10.1002/ajmg.1320440210. [DOI] [PubMed] [Google Scholar]
- 9.Cartwright MM, Smith SM. Increased cell death and reduced neural crest cell numbers in ethanol-exposed embryos: partial basis for the fetal alcohol syndrome phenotype. Alcoholism, clinical and experimental research. 1995;19:378–86. doi: 10.1111/j.1530-0277.1995.tb01519.x. [DOI] [PubMed] [Google Scholar]
- 10.Sun H, Chen X, Yuan F, Liu J, Zhao Y, Chen SY. Involvement of seven in absentia homolog-1 in ethanol-induced apoptosis in neural crest cells. Neurotoxicology and teratology. 2014;46C:26–31. doi: 10.1016/j.ntt.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan F, Chen X, Liu J, Feng W, Wu X, Chen SY. Up-regulation of Siah1 by ethanol triggers apoptosis in neural crest cells through p38 MAPK-mediated activation of p53 signaling pathway. Archives of toxicology. 2016 doi: 10.1007/s00204-016-1746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jana K, Jana N, De DK, Guha SK. Ethanol induces mouse spermatogenic cell apoptosis in vivo through over-expression of Fas/Fas-L, p53, and caspase-3 along with cytochrome c translocation and glutathione depletion. Molecular reproduction and development. 2010;77:820–33. doi: 10.1002/mrd.21227. [DOI] [PubMed] [Google Scholar]
- 13.Morio Y, Tsuji M, Inagaki M, Nakagawa M, Asaka Y, Oyamada H, et al. Ethanol-induced apoptosis in human liver adenocarcinoma cells (SK-Hep1): Fas- and mitochondria-mediated pathways and interaction with MAPK signaling system. Toxicology in vitro : an international journal published in association with BIBRA. 2013;27:1820–9. doi: 10.1016/j.tiv.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Hong F, Kim WH, Tian Z, Jaruga B, Ishac E, Shen X, et al. Elevated interleukin-6 during ethanol consumption acts as a potential endogenous protective cytokine against ethanol-induced apoptosis in the liver: involvement of induction of Bcl-2 and Bcl-x(L) proteins. Oncogene. 2002;21:32–43. doi: 10.1038/sj.onc.1205016. [DOI] [PubMed] [Google Scholar]
- 15.Mooney SM, Miller MW. Effects of prenatal exposure to ethanol on the expression of bcl-2, bax and caspase 3 in the developing rat cerebral cortex and thalamus. Brain research. 2001;911:71–81. doi: 10.1016/s0006-8993(01)02718-4. [DOI] [PubMed] [Google Scholar]
- 16.Balan AG, Myers BJ, Maganti JL, Moore DB. ER-targeted Bcl-2 and inhibition of ER-associated caspase-12 rescue cultured immortalized cells from ethanol toxicity. Alcohol. 2010;44:553–63. doi: 10.1016/j.alcohol.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Dong J, Sulik KK, Chen SY. Nrf2-mediated transcriptional induction of antioxidant response in mouse embryos exposed to ethanol in vivo: implications for the prevention of fetal alcohol spectrum disorders. Antioxidants & redox signaling. 2008;10:2023–33. doi: 10.1089/ars.2007.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keum YS, Jeong WS, Kong AN. Chemopreventive functions of isothiocyanates. Drug news & perspectives. 2005;18:445–51. doi: 10.1358/dnp.2005.18.7.939350. [DOI] [PubMed] [Google Scholar]
- 19.Clarke JD, Dashwood RH, Ho E. Multi-targeted prevention of cancer by sulforaphane. Cancer letters. 2008;269:291–304. doi: 10.1016/j.canlet.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenzi M, Fimognari C, Hrelia P. Sulforaphane as a promising molecule for fighting cancer. Cancer treatment and research. 2014;159:207–23. doi: 10.1007/978-3-642-38007-5_12. [DOI] [PubMed] [Google Scholar]
- 21.Tarozzi A, Angeloni C, Malaguti M, Morroni F, Hrelia S, Hrelia P. Sulforaphane as a potential protective phytochemical against neurodegenerative diseases. Oxidative medicine and cellular longevity. 2013;2013:415078. doi: 10.1155/2013/415078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh K, Connors SL, Macklin EA, Smith KD, Fahey JW, Talalay P, et al. Sulforaphane treatment of autism spectrum disorder (ASD) Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15550–5. doi: 10.1073/pnas.1416940111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fimognari C, Turrini E, Sestili P, Calcabrini C, Carulli G, Fontanelli G, et al. Antileukemic activity of sulforaphane in primary blasts from patients affected by myelo- and lympho-proliferative disorders and in hypoxic conditions. PloS one. 2014;9:e101991. doi: 10.1371/journal.pone.0101991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geisel J, Bruck J, Glocova I, Dengler K, Sinnberg T, Rothfuss O, et al. Sulforaphane protects from T cell-mediated autoimmune disease by inhibition of IL-23 and IL-12 in dendritic cells. Journal of immunology. 2014;192:3530–9. doi: 10.4049/jimmunol.1300556. [DOI] [PubMed] [Google Scholar]
- 25.Zhao J, Kobori N, Aronowski J, Dash PK. Sulforaphane reduces infarct volume following focal cerebral ischemia in rodents. Neuroscience letters. 2006;393:108–12. doi: 10.1016/j.neulet.2005.09.065. [DOI] [PubMed] [Google Scholar]
- 26.Innamorato NG, Rojo AI, Garcia-Yague AJ, Yamamoto M, de Ceballos ML, Cuadrado A. The transcription factor Nrf2 is a therapeutic target against brain inflammation. Journal of immunology. 2008;181:680–9. doi: 10.4049/jimmunol.181.1.680. [DOI] [PubMed] [Google Scholar]
- 27.Yoon HY, Kang NI, Lee HK, Jang KY, Park JW, Park BH. Sulforaphane protects kidneys against ischemia-reperfusion injury through induction of the Nrf2-dependent phase 2 enzyme. Biochemical pharmacology. 2008;75:2214–23. doi: 10.1016/j.bcp.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 28.Baek SH, Park M, Suh JH, Choi HS. Protective effects of an extract of young radish (Raphanus sativus L) cultivated with sulfur (sulfur-radish extract) and of sulforaphane on carbon tetrachloride-induced hepatotoxicity. Bioscience, biotechnology, and biochemistry. 2008;72:1176–82. doi: 10.1271/bbb.70545. [DOI] [PubMed] [Google Scholar]
- 29.Kim HJ, Barajas B, Wang M, Nel AE. Nrf2 activation by sulforaphane restores the age-related decrease of T(H)1 immunity: role of dendritic cells. The Journal of allergy and clinical immunology. 2008;121:1255–61. e7. doi: 10.1016/j.jaci.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song MY, Kim EK, Moon WS, Park JW, Kim HJ, So HS, et al. Sulforaphane protects against cytokine- and streptozotocin-induced beta-cell damage by suppressing the NF-kappaB pathway. Toxicology and applied pharmacology. 2009;235:57–67. doi: 10.1016/j.taap.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Kwak MK, Cho JM, Huang B, Shin S, Kensler TW. Role of increased expression of the proteasome in the protective effects of sulforaphane against hydrogen peroxide-mediated cytotoxicity in murine neuroblastoma cells. Free radical biology & medicine. 2007;43:809–17. doi: 10.1016/j.freeradbiomed.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 32.Gan N, Wu YC, Brunet M, Garrido C, Chung FL, Dai C, et al. Sulforaphane activates heat shock response and enhances proteasome activity through up-regulation of Hsp27. The Journal of biological chemistry. 2010;285:35528–36. doi: 10.1074/jbc.M110.152686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dashwood RH, Ho E. Dietary agents as histone deacetylase inhibitors: sulforaphane and structurally related isothiocyanates. Nutrition reviews. 2008;66(Suppl 1):S36–8. doi: 10.1111/j.1753-4887.2008.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho E, Clarke JD, Dashwood RH. Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. The Journal of nutrition. 2009;139:2393–6. doi: 10.3945/jn.109.113332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature reviews Genetics. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 36.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–52. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 37.Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nature reviews Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 38.Bonfils C, Walkinshaw DR, Besterman JM, Yang XJ, Li Z. Pharmacological inhibition of histone deacetylases for the treatment of cancer, neurodegenerative disorders and inflammatory diseases. Expert opinion on drug discovery. 2008;3:1041–65. doi: 10.1517/17460441.3.9.1041. [DOI] [PubMed] [Google Scholar]
- 39.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 40.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & development. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 41.Fullgrabe J, Hajji N, Joseph B. Cracking the death code: apoptosis-related histone modifications. Cell death and differentiation. 2010;17:1238–43. doi: 10.1038/cdd.2010.58. [DOI] [PubMed] [Google Scholar]
- 42.van Bavel CC, Dieker J, Muller S, Briand JP, Monestier M, Berden JH, et al. Apoptosis-associated acetylation on histone H2B is an epitope for lupus autoantibodies. Molecular immunology. 2009;47:511–6. doi: 10.1016/j.molimm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Bartova E, Pachernik J, Harnicarova A, Kovarik A, Kovarikova M, Hofmanova J, et al. Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. Journal of cell science. 2005;118:5035–46. doi: 10.1242/jcs.02621. [DOI] [PubMed] [Google Scholar]
- 44.Zhu WG, Lakshmanan RR, Beal MD, Otterson GA. DNA methyltransferase inhibition enhances apoptosis induced by histone deacetylase inhibitors. Cancer research. 2001;61:1327–33. [PubMed] [Google Scholar]
- 45.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. Journal of the National Cancer Institute. 2000;92:1210–6. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 46.Botia B, Legastelois R, Alaux-Cantin S, Naassila M. Expression of ethanol-induced behavioral sensitization is associated with alteration of chromatin remodeling in mice. PloS one. 2012;7:e47527. doi: 10.1371/journal.pone.0047527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirpich I, Ghare S, Zhang J, Gobejishvili L, Kharebava G, Barve SJ, et al. Binge alcohol-induced microvesicular liver steatosis and injury are associated with down-regulation of hepatic Hdac 1, 7, 9, 10, 11 and up-regulation of Hdac 3. Alcoholism, clinical and experimental research. 2012;36:1578–86. doi: 10.1111/j.1530-0277.2012.01751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiang M, Denny A, Lieu M, Carreon S, Li J. Histone H3K9 modifications are a local chromatin event involved in ethanol-induced neuroadaptation of the NR2B gene. Epigenetics : official journal of the DNA Methylation Society. 2011;6:1095–104. doi: 10.4161/epi.6.9.16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perkins A, Lehmann C, Lawrence RC, Kelly SJ. Alcohol exposure during development: Impact on the epigenome. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience. 2013;31:391–7. doi: 10.1016/j.ijdevneu.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agudelo M, Gandhi N, Saiyed Z, Pichili V, Thangavel S, Khatavkar P, et al. Effects of alcohol on histone deacetylase 2 (HDAC2) and the neuroprotective role of trichostatin A (TSA) Alcoholism, clinical and experimental research. 2011;35:1550–6. doi: 10.1111/j.1530-0277.2011.01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agudelo M, Yoo C, Nair MP. Alcohol-induced serotonergic modulation: the role of histone deacetylases. Alcohol. 2012;46:635–42. doi: 10.1016/j.alcohol.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao XH, Nguyen HK, Nyomba BL. Prenatal ethanol exposure causes glucose intolerance with increased hepatic gluconeogenesis and histone deacetylases in adult rat offspring: reversal by tauroursodeoxycholic acid. PloS one. 2013;8:e59680. doi: 10.1371/journal.pone.0059680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao XH, Nyomba BL. Hepatic insulin resistance induced by prenatal alcohol exposure is associated with reduced PTEN and TRB3 acetylation in adult rat offspring. American journal of physiology Regulatory, integrative and comparative physiology. 2008;294:R1797–806. doi: 10.1152/ajpregu.00804.2007. [DOI] [PubMed] [Google Scholar]
- 54.Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends in neurosciences. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature reviews Genetics. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myzak MC, Dashwood RH. Histone deacetylases as targets for dietary cancer preventive agents: lessons learned with butyrate, diallyl disulfide, and sulforaphane. Current drug targets. 2006;7:443–52. doi: 10.2174/138945006776359467. [DOI] [PubMed] [Google Scholar]
- 57.Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Experimental biology and medicine. 2007;232:227–34. [PMC free article] [PubMed] [Google Scholar]
- 58.Yan X, Pan B, Lv T, Liu L, Zhu J, Shen W, et al. Inhibition of histone acetylation by curcumin reduces alcohol-induced fetal cardiac apoptosis. Journal of biomedical science. 2017;24:1. doi: 10.1186/s12929-016-0310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 60.Heaton MB, Moore DB, Paiva M, Gibbs T, Bernard O. Bcl-2 overexpression protects the neonatal cerebellum from ethanol neurotoxicity. Brain research. 1999;817:13–8. doi: 10.1016/s0006-8993(98)01173-1. [DOI] [PubMed] [Google Scholar]