Summary
The degree of somatic hypermutation, determined as percent deviation of immunoglobulin heavy chain gene variable region sequence from the germline (IGHV%), is an important prognostic factor in chronic lymphocytic leukaemia (CLL). Currently, a cut-off of 2% deviation or 98% sequence identity to germline in IGHV sequence is routinely used to dichotomize CLL patients into mutated and unmutated groups. Because dissimilar IGHV% cut-offs of 1% to 5% were identified in different studies, we wondered whether no cut-off should be applied and IGHV% treated as a continuous variable. We analysed the significance of IGHV% in 203 CLL patients enrolled on the original frontline fludarabine, cyclophosphamide and rituximab (FCR) trial with a median of 10 years follow-up. Using the Cox Proportional Hazard model, IGHV% was identified as a continuous variable that is significantly associated with progression-free (PFS) and overall survival (OS) (P<0.001). Furthermore, we validated this finding in 323 patients treated with FCR off-protocol and in the total cohort (n=535). Multivariate analysis revealed a continuous trend. Higher IGHV% levels were incrementally associated with favorable PFS and OS in both FCR-treated cohorts (P<0.001, both cohorts). Taken together, our data suggest that IGHV% is a continuous variable in CLL patients treated with FCR.
Keywords: CLL, FCR, IGHV gene, immunoglobulin heavy chain gene
Introduction
The mutation status of the immunoglobulin heavy chain variable region gene (IGHV) is an established prognostic factor in patients with chronic lymphocytic leukaemia (CLL). Fais et al (1998) identified the presence of somatic hypermutation (SHM) in CLL cells and the prognostic significance of the percent of SHM deviation from the germline sequence (IGHV%) in patients with CLL was described the following year (Damle, et al 1999, Hamblin, et al 1999). In recent years a cut-off value of 2% deviation from the germline sequence of IGHV (i.e. 98% identity) has been commonly used in clinical practice and, according to the current dogma, CLL patients with ≥ 2% deviation, or otherwise ≤ 98% identity, are considered ‘mutated’ (M-CLL), whereas CLL patients with <2% IGHV% deviation or >98% identity are considered ‘unmutated’ (U-CLL).(Chiorazzi 2012) The use of a 2% cut-off to classify patients into M-CLL and U-CLL was based on a handful of studies.(Hamblin, et al 2008, Oscier, et al 2010, Tobin, et al 2005) Although other IGHV% cut-offs, such as <1% vs 1–3% vs >3%(Davis, et al 2016), 3%(Krober, et al 2002) and 5%(Lin, et al 2002), have been studied, a 2% cut-off has been regularly used in clinical practice. All of the above studies that aimed to identify an IGHV% cut-off, except one (Davis, et al 2016), analysed a relatively small number of patients who were not uniformly treated and/or had a relatively short follow-up. Using a maximally selected log-rank statistical analysis, Krober et al (2002) another study demonstrated a 3% cut-off, and Tobin, et al (2005) used the Youden index method to identify a similar cut-off. Conversely, Lin et al (2002) suggested that a >5% cut-off is most appropriate because normal CD5+ B cells have <5% IGHV% deviation compared to the germline IGHV sequence.(Fais, et al 1998) Other reports pointed out that assessment of IGHV% levels could be affected by polymorphisms that mimic SHM(Davis, et al 2003, Matsuda, et al 1993), the choice of SHM assessment technique and the choice polymerase chain reaction (PCR) primers(Ghia, et al 2007, Xochelli, et al 2015).
Because different groups established a variety of IGHV% cut-off points, ranging from 1% to 5%, we wondered whether there is no universal cut-off and the IGHV% is a continuous prognostic variable. Furthermore, it is important that in a chronic disease, such as CLL, the prognostic impact of any marker should be based on an analysis with a very long follow-up and large number of patients rather than a short follow-up and limited number of patients. Therefore, to test our hypothesis, we analysed a large cohort of patients with a long follow-up who were treated with the same regimen and whose IGHV mutation status was assessed using the same methodology. We analysed the prognostic impact of IGHV% in 203 patients treated on the original frontline fludarabine, cyclophosphamide and rituximab (FCR) protocol (the FCR-300 trial)(Thompson, et al 2016) in whom the median follow-up was 10 years, and in 332 patients treated with FCR off protocol(Chihara, et al 2016).
Patients and Methods
We analysed the data of 203 patients enrolled on the FCR-300 clinical trial (n=300). Only patients who underwent SHM testing at initial diagnosis were included in this analysis. In addition, we analysed data of 332 off-protocol patients who were treated with FCR-based regimens(Chihara, et al 2016). Treatments administered in the off-protocol group were FCR (n=213), FCR with high dose rituximab (n=47), FCR with granulocyte-macrophage colony-stimulating factor (GM-CSF; n=50) and FCR with granulocyte colony-stimulating factor (n=22). Although IGHV mutation status is now considered relevant in patients with IGHV3-21 mutation with stereotypy subset 2 (Baliakas, et al 2015), we did not have information on the stereotypy status for our cohort; therefore all patients with IGHV3-21 were excluded (Tobin, et al 2003). The standard laboratory workup of the patients enrolled in these clinical trials preceded the currently accepted routine karyotype analysis, fluorescence in situ hybridisation (FISH) analysis, CD38 assessment, ZAP70 protein expression, next generation sequencing and stereotypy status. Therefore, these important data could not be included in our analysis. All patients provided written informed consent, all of which have been approved by the institutional review board.
The SHM testing method was as follows: stored bone marrow or peripheral blood samples from patients which contained at least 10% CD19+/CD5+ CLL cells, as estimated by flow cytometry, were analysed. RNA extraction was followed by cDNA synthesis using random hexamers. Multiplex PCR amplification was performed using the VH leader forward primers and two sets of reverse primers: CH and JH. The primer sequences were:
Forward Primers (VH Leader Primers)
VH1 leader (VH1-F) 5′ CCA TGG ACT GSA YYT GGA GVR TC 3′
VH2 leader (VH2-F) 5′ ATG GAC ACA CTT TGC TMC ACR CTC 3′
VH3 leader (VH3-F) 5′ CCA TGG AGT TKG GRC TBH GCT GG 3′
VH4 leader (VH4-F) 5′ ACA TGA AAC AYC TGT GGT TCT TCC 3′
VH5 leader (VH5-F) 5′ ATG GGG TCA ACC GCC ATC CT 3′
VH6 leader (VH6-F) 5′ ATG TCT GTC TCC TTC CTC ATC 3′
Reverse Primers
| CH | 5′ - TTG GGG CGG ATG CAC T- 3′ |
| JH | 5′ -AAC TGA GGA GAC GGT GAC C- 3′ |
PCR amplification conditions were as follows: incubation at 96°C for 1 min, followed by cDNA amplification for 25 cycles at 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. PCR products were purified and sequenced. Given that our assay is RNA-based, only productive rearrangements were expected to be amplified. The degree of SHM was quantified by comparing the resulting IGHV sequence with the consensus germline sequence in the IMGT/V-QUEST database (http://www.imgt.org) and the percent change of base pairs was calculated. For the calculation, the entire IGHV sequence that includes the complementarity determining region 3 (CDR3) was compared with germline sequence. Sequences less than 150 base pairs in length or without the authentic CDR3 sequence were regarded inadequate for assessment. Ideally, ~300 b.p. sequence is expected, and was achieved in majority of the cases. As these cases were tested as a part of clinical assay, we have minimum reporting criteria set, which included 150 b.p.
Overall survival (OS) was calculated as the number of years from the date of diagnosis to the date of death or last follow-up, and progression-free survival (PFS) was calculated as the number of years from the date of start of treatment to date of last follow-up or disease progression. The Kaplan-Meier product limit method was used to estimate the median survival (OS and PFS). Cox proportional hazard regression models were used to identify any association between covariates and survival (OS and PFS). Classification and regression tree (CART) analysis using the martingale residuals of the Cox models were used to determine the functional form of IGHV% that was included in the multivariate analyses. To validate the significance of IGHV% as a continuous variable, we also analysed the IGHV% after combining the two groups (combined group; n=535). Statistical analysis was performed using the Stata/SE version 14.2 statistical software (Stata Corp. LP, College Station, TX).
Results
The clinical characteristics of 203 patients treated with the original FCR-300 protocol and of 332 patients treated with FCR off protocol (FCR-control group) are summarized in Table I. The median follow-up of patients was 10.7 years (0.1–15.5) in the original FCR-300 trial and 5.6 years (0.2–12.1) in the FCR control group. There were no significant differences in the baseline clinical features of the patients in the two cohorts. Mean number of treatment cycles was 6 (range 1–6) in both cohorts.
Table I.
Baseline characteristics of patients with CLL treated with frontline FCR FCR-300 clinical trial and FCR-based therapies (off-protocol)
| Characteristic | Measure/Category | FCR-300 clinical trial (n=203) | FCR-based therapy (control group) (n=332) |
|---|---|---|---|
| Age, years | Median (range) | 57 (17–85) | 58 (28–84) |
| Age (≥65 years) | n (%) | 49 (24) | 68 (21) |
| Gender | Male/Female (n) | 151/52 | 220/112 |
| Rai stage | (0, 1, 2/3, 4) n (%) | 134 (66)/69 (34) | 217 (65)/115 (35) |
| WBC, ×109/l | Median (range) | 81.5 (2.1–407.8) | 84.2 (1.4–541) |
| Hb, g/l | Median (range) | 125 (61–187) | 125 (23–165) |
| Platelet count, ×109/l | Median (range) | 156 (34–406) | 147 (10–420) |
| β2M (mg/l) | (</≥ 4 mg/l) n (%) | 116 (57)/85 (42) | 220 (67)/111 (33) |
| Number of cycles |
1–3 4–5 6 |
22 30 159 |
77 84 232 |
| Progressed | n (%) | 132 (65) | 158 (48) |
| Died | n (%) | 105 (52) | 64 (20) |
| Follow-up, years | Median (range) | 10.7 (0.1–15.5) | 5.6 (0.2–12.1) |
β2M; β2 microglobulin; CLL: chronic lymphocytic leukaemia; FCR: fludarabine, cyclophosphamide and rituximab; Hb: haemoglobin concentration; WBC: white blood cell count.
Cox proportional hazard models were used to determine the association between IGHV%, as a continuous variable, and survival (PFS and OS) in the FCR-300 trial cohort (Figure 1A, B). The IGHV% deviation was significantly associated with PFS (hazard ratio (HR) 95%; confidence interval (CI): 0.81 (0.76–0.87); P<0.001) and with OS (HR: 95%; CI: 0.84 (0.78–0.91); P<0.001), suggesting that, as the percent deviation of IGHV from the germline sequence increases, the risk of disease progression or death significantly decreases. The CI of 0.81 represents a decrease in the rate of progression of 19% for every 1% increase in the IGHV% (Figure 1A, B). In order to further validate this observation, we analysed the survival of patients treated with FCR off-protocol (n=332) (Figure 1C, D). Similar to patients treated on the FCR-300 study, there was a significant association between PFS (HR: 0.84; CI: (0.79–0.90); P<0.001) and OS (HR: 95%; CI: 0.90 (0.82–0.99); P<0.039) and IGHV% deviation in the FCR control group. Similarly, when we evaluated all 535 patients of the two groups and performed the same analysis (Figure 1E, F) we found that a sustained continuum increase in IGHV% deviation significantly correlated with PFS and OS.
Figure 1. A sustained increase in the percentage deviation of IGHV mutations is significantly associated with a lower risk of PFS and OS.
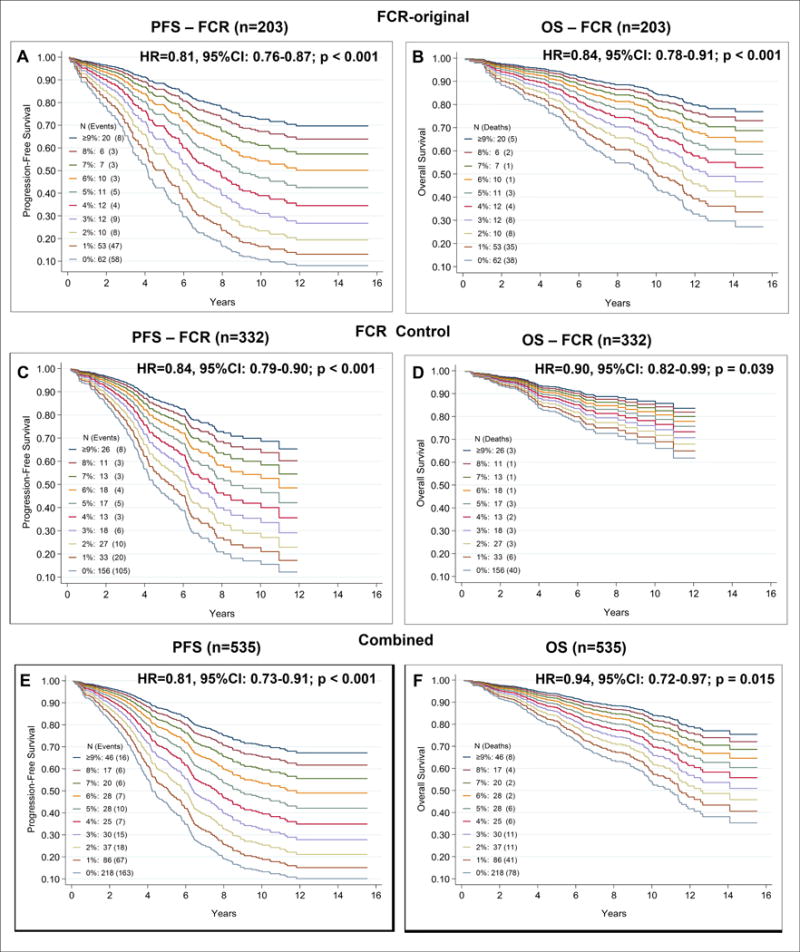
The number of patients and the events in each cohort are depicted in the figure. Univariate analysis identified a significant statistically association between IGHV mutation from the germline sequence and survival (progression-free and overall survival) (A–B) Survival functions were determined after the univariate Cox proportional hazard model was fitted using the continuous IGHV% deviation as a continuous variable A) PFS in patients treated on the original FCR-300 clinical trial (FCR-original, n=203; median follow-up 10.7 years). The estimated five-year survival was 44%, 45%, 40%, 50%, 83%, 82%, 90%, 86%, 83% and 84% at 0, 1, 2, 3, 4, 5, 6, 7, 8 and ≥ 9% of IGHV% deviation, corresponding to various cutoff points for IGHV mutation % deviation (0-0.9, 1-1.9, 2-2.9, 3-3.9 and ≥4% and so on) or otherwise called as (100-99.1, 99-98.1, 98-97.1, 97-96.1 and ≤ 96%) percentage identity, respectively (HR: 95%; CI: 0.81 (0.76–0.87); P <0.001). (B) OS in patients treated on the original FCR-300 clinical trial. The estimated five-year survival was 87%, 70%, 60%, 67%, 92%, 100%, 90%, 100%, 83% and 80% at 0, 1, 2, 3, 4, 5, 6, 7, 8 and ≥ 9% of IGHV%, respectively (HR: 95%; CI: 0.84 (0.78–0.91); P < 0.001). Similar analyses of PFS and OS in patients treated with FCR off-protocol (FCR Control, n=332; median follow up of 5.6 years) is depicted in C and D. (C) PFS in patients treated with FCR-based regimens off-protocol. The estimated five-year survival was 43%, 42%, 84%, 77%, 80%, 78%, 88%, 83%, 71% and 70% at 0, 1, 2, 3, 4, 5, 6, 7, 8 and ≥ 9% of IGHV%, respectively (HR: 95%; CI: 0.84 (0.79–0.90); P<0.001). (D) OS in patients treated with FCR-based regimens off-protocol. Estimated five-year survival was 81%, 83%, 96%, 85%, 80%, 93%, 100%, 93%, 82% and 87% at 0, 1, 2, 3, 4, 5, 6, 7, 8 and ≥ 9% of IGHV%, respectively (HR: 95%; CI: 0.90 (0.82–0.99); P=0.039. (E) PFS in the combined cohorts (i.e., patients treated on the FCR-300 clinical trial and patients treated with FCR-based regimens off-protocol). The estimated five-year survival was 43%, 44%, 71%, 65%, 82%, 79%, 89%, 84%, 76% and 74% at 0, 1, 2, 3, 4, 5, 6, 7, 8 and ≥ 9% of IGHV%, respectively HR: 95%; CI 0.81 (0.73–0.91); P<0.001). F) OS in the combined cohorts (i.e., patients treated on the FCR-300 clinical trial and patients treated with FCR-based regimens off protocol). The estimated five-year survival was 82%, 75%, 86%, 76%, 86%, 96%, 96%, 95%, 82% and 88% at 0, 1, 2, 3, 4, 5, 6, 7, 8 and ≥ 9% of IGHV%, respectively (HR: 95%; CI: 0.84 (0.72–0.97); P =0.015).
95% CI: 95% confidence interval; FCR: fludarabine, cyclophosphamide and rituximab; HR: Hazard ratio; OS: overall survival; PFS: progression-free survival
We then performed univariate and multivariate analyses for PFS and OS using the IGHV% deviation value as a continuous variable rather than the conventionally used cut-off value of 2% (Tables II and III, respectively). The variables that were significantly associated with a better PFS and OS in the univariate analysis were lower β2 microglobulin levels, younger age, lower lactate dehydrogenase levels, undetectable minimal residual disease as assessed by flow cytometry, a higher number of FCR cycles, and a lower white blood cell count (WBC). Multivariate analysis identified IGHV% as being significantly associated with PFS. Other factors significantly associated with longer PFS were number of treatment cycles (4–6) and WBC <200 × 109/l (Table II). Similarly, multivariate analysis identified IGHV% as being significantly associated with OS. Other factors significantly associated with longer OS were the number of treatment cycles (4–6) and age <65 years (Table III).
Table II.
Univariate and multivariate analyses of PFS of CLL patients treated with frontline FCR
| FCR-300 Original | Univariate | Multivariate (continuous) | ||||||
|---|---|---|---|---|---|---|---|---|
| N | Events | HR | 95% CI | P-value | HR | 95% CI | P-value | |
| IGHV% | 203 | 148 | 0.81 | (0.76–0.87) | <0.001 | 0.80 | (0.75–0.86) | <0.001 |
| β2M (mg/l) | 201 | 146 | 1.17 | (1.09–1.27) | <0.001 | |||
| WBC ×109/l | 203 | 148 | 1.00 | (1.00–1.01) | 0.002 | |||
| Cycles (n) | 203 | 148 | 0.71 | (0.62–0.80) | <0.001 | |||
| Age ≥65 years | ||||||||
| No | 154 | 106 | ||||||
| Yes | 49 | 42 | 1.69 | (1.18–2.42) | 0.004 | |||
| β2M ≥ 4 mg/l | ||||||||
| No | 116 | 74 | ||||||
| Yes | 85 | 72 | 2.02 | (1.46–2.80) | <0.001 | |||
| WBC ≥ 200 ×109/l | ||||||||
| No | 190 | 132 | ||||||
| Yes | 21 | 19 | 2.45 | (1.50–3.98) | <0.001 | 2.30 | (1.40–3.77) | 0.001 |
| Cycles | ||||||||
| 1–3 | 22 | 21 | ||||||
| 4–5 | 27 | 19 | 0.36 | (0.19–0.68) | <0.001 | 0.28 | (0.15–0.52) | <0.001 |
| 6 | 154 | 108 | 0.29 | (0.18–0.47) | <0.001 | 0.24 | (0.15–0.39) | <0.001 |
Other variables that were significantly associated with increased risk of progression events in the univariate analysis but not in the multivariate models (and therefore not included in this table) were increased age, high β2 microglobin, elevated lactate dehydrogenase levels, and detectable minimal residual disease (not shown).
95% CI: 95% confidence interval; β2M; β2 microglobulin; CLL: chronic lymphocytic leukaemia; FCR: fludarabine, cyclophosphamide and rituximab; HR: Hazard ratio; PFS: progression-free survival; WBC: white blood cell count.
Table III.
Univariate and multivariate analyses of OS of CLL patients treated with frontline FCR
| FCR on clinical trial – FCR-300 | Univariate | Multivariate (continuous) | ||||||
|---|---|---|---|---|---|---|---|---|
| N | Events | HR | 95% CI | P-value | HR | 95% CI | P-value | |
| IGHV% | 203 | 105 | 0.84 | (0.78–0.91) | <0.001 | 0.82 | (0.76–0.90) | <0.001 |
| β2M (mg/l) | 201 | 103 | 1.24 | (1.15–1.34) | <0.001 | |||
| Cycles (n) | 203 | 105 | 0.66 | (0.58–0.75) | <0.001 | |||
| Ag e≥65 years | ||||||||
| No | 154 | 66 | ||||||
| Yes | 49 | 39 | 2.92 | (1.96–4.35) | <0.001 | 1.92 | (1.23–3.01) | 0.004 |
| β2M ≥ 4 mg/l | ||||||||
| No | 116 | 45 | ||||||
| Yes | 85 | 58 | 2.51 | (1.70–3.71) | <0.001 | |||
| Cycles | ||||||||
| 1–3 | 22 | 19 | ||||||
| 4–5 | 27 | 18 | 0.52 | (0. 27–0.99) | 0.04 | 0.48 | (0.25–0.94) | 0.03 |
| 6 | 154 | 68 | 0.23 | (0.14–0.38) | <0.001 | 0.27 | (0.15–0.48) | <0.001 |
Other variables that were significantly associated with increased risk of death in univariate analysis but not in multivariate models were age >65 years, high β2M, elevated lactate dehydrogenase levels, ZAP70 positivity and undetectable minimal residual disease (not shown)
95% CI: 95% confidence interval; β2M; β2 microglobulin; CLL: chronic lymphocytic leukaemia; FCR: fludarabine, cyclophosphamide and rituximab; HR: Hazard ratio; OS: overall survival.
Because there was a significant difference in the median follow-up and the overall survival curves in the original FCR and the off-protocol FCR groups (Figure 1 B, D), we assessed the causes of death and the first salvage treatments administered to the patients of the two groups. One hundred and four deaths were recorded among the 203 patients of the original FCR-300 cohort: 46 patients died of disease progression, 14 of transformation, 13 from complications of stem cell transplantation (SCT), 11 from solid tumours, 9 from an infection while in remission, 8 from infection during disease progression and 3 from haematological neoplasms. Among the 332 patients of the FCR control group, which had a median follow-up of 5.6 years, 64 deaths were recorded: 21 patients died from other neoplasms (9 solid tumours and 12 haematological malignancies), 15 from disease progression, 8 from disease transformation, 7 from various infections, 4 from pre-existing comorbidities, 4 from complications of SCT and 5 patients died of unknown causes.
In the original FCR-300 trial, 124 relapsed patients received salvage treatment, of which 32 received FCR, 23 miscellaneous chemotherapy regimens, 19 alemtuzumab-based regimens, 16 rituximab with GM-CSF, 10 ibrutinib-based regimens, 10 lenalidomide based regimens, 4 received R-CHOP (rituximab + cyclophosphamide, doxorubicin, vincristine and prednisolone), 4 hyper-CVAD (hyperfractionated cyclophosphamide, doxorubicin, vincristine and dexamethasone + methotrexate and cytarabine) and 6 were treated with radioimmunotherapy. In the FCR control group, 123 patients received salvage treatment. Forty-two patients received an ibrutinib-based regimen, 15 ofatumumab-based regimens, 14 lenalidomide with rituximab, 10 with bendamustine with rituximab and 10 with rituximab and high dose steroids, 8 received FCR and 8 hyper CVAD, 7 were treated with OFAR (oxaliplatin, fludarabine, cytarabine and rituximab), 3 with alemtuzumab, 3 received FBR (fludarabine, bendamustine, rituximab), 2 venetoclax, and one patient was treated with idelalisib. Overall, the FCR off-protocol patients had access to newer, more effective salvage regimens.
Discussion
In this analysis, we assessed the prognostic significance of IGHV mutation status by analysing the absolute value of IGHV% on PFS and OS in a large cohort of patients treated with frontline FCR and followed for up to 16 years. Our analysis revealed that a sustained increase in absolute IGHV% deviation is significantly associated with longer PFS and OS. The higher the IGHV% deviation, the better the PFS and OS, whereas the lower the IGHV%, the higher the risk of disease progression and death. Similar results were obtained in a separate cohort of 323 CLL patients treated with FCR or FCR-based regimens. However, the overall survival rates of patients with lower IGHV% in this cohort were greater (Figure 1D) than those of patients enrolled in the original FCR trial (Figure 1B). These differences could be attributed to a shorter follow-up in the off-protocol cohort and - that a higher proportion of the off-protocol relapsed patients were treated with more effective novel salvage regimens. These salvage therapies were not available at that time for most relapsed patients enrolled in the original FCR-300 trial, most of which were treated with less effective salvage regimens.
Our data suggest that the absolute level of IGHV% deviation rather than a 2% cut-off or 98% cut off for sequence identity is a better and more reliable predictor of treatment outcome. Given that the PFS of patients with high IGHV% deviation followed for up to 16 years might be as high as 70%, the absolute IGHV% rather than the 2% cut-off should be taken into consideration when considering frontline FCR treatment. Nevertheless, although analyses of the two different cohorts yielded similar results, confirmation from large cohorts of FCR-treated CLL patients from other institutions is needed. Furthermore, because of a lack of data on routine karyotype analysis, FISH analysis, next-generation sequencing studies and stereotypy status for these patients, we could not perform a correlative analysis to compare our data with other studies showing that cytogenetic aberrations (del11q and del17p) significantly correlated with IGHV mutation status in CLL patients (Gladstone, et al 2012). On the same note, one study reported that patients with unmutated IGHV and sole del13q abnormality (n=13) had significantly shorter cumulative probability of treatment as compared to mutated IGHV and sole del13q abnormality (n=34), however the OS was not significantly different (Gladstone, et al 2011).
One should keep in mind that the 2% IGHV% deviation cut-off dichotomizing CLL patients into M-CLL (good) and U-CLL (bad), previously reported in patients receiving chemotherapy, might fail to predict survival outcomes in patients treated with novel, non-haemotoxic agents. For example, initial results from the ibrutinib trial(O’Brien, et al 2016) showed that dichotomizing patients into M-CLL and U-CLL is probably irrelevant in patients treated with this agent. Whether assessment of IGHV% as a continuous variable might become a useful prognostic indicator in ibrutinib- or in venetoclax-treated patients remains to be determined. Lin et al (2002) suggested that CLL should be regarded as a disease with a continuous variable phenotype. Our findings support this hypothesis because they suggest that as the percent deviation of IGHV% increases, CLL cells become more susceptible to DNA damaging agents.
Another potential explanation for our results could be derived from previous reports demonstrating a robust correlation between the birth rates of CLL cells, IGHV% and disease course (Murphy, et al 2017, van Gent, et al 2008). In addition, antigen-driven CLLs, i.e., those with significantly higher mutation % deviation, had a better survival then antigen-naïve non-significantly mutated IGHV and unmutated IGHV% (Degan, et al 2004). Furthermore, because the telomere length is longer in patients with higher IGHV% deviation from the germline sequence, these patients have lower proliferation rates and hence a longer survival. In one study (Grabowski, et al 2005), patients with mutated IGHV with shorter telomere lengths were found to have a survival similar to patients with unmutated IGHV. Within the mutated subgroup, patients who responded to CpG-oligodeoxynucleotide (CpG-ODN) stimulation had an inferior outcome compared to those who did not proliferate, i.e., those with a higher IGHV% (Tarnani, et al 2010). Overall, these findings suggest that the higher the IGHV% deviation, the more the cells are anergic to the B-cell receptor (BCR) trigger, have longer telomere lengths, are less responsive to CpG-ODN stimulation, have lower proliferative rates and therefore exhibit longer patient survival.
In summary, we demonstrated that CLL patients’ survival is impacted by a continuous increment of IGHV% deviation. The higher the percent deviation, the better is treatment outcome in patients treated with FCR. Therefore, the prognostic relevance of IGHV% as a continuous, rather than a dichotomized, variable should be further investigated.
Acknowledgments
This study was supported in part by the NIH/NCI award number P30CA016672 and by NCI award number P01CA049639. We thank Chalaire Dawn from scientific publications department of MDACC, for editing the manuscript.
Footnotes
DR. PREETESH JAIN (Orcid ID: 0000-0003-2735-168X)
DR. ANDRÉS E. QUESADA (Orcid ID: 0000-0002-5304-7828)
DR. ZEEV ESTROV (Orcid ID: 0000-0002-1623-3613)
Authorship Contributions:
P.J. and Z.E. designed the study.
P.J., G.N.G., P.T., C.T. and Z.E. analysed results.
P.J., P.T., and Z.E. wrote the paper.
P.J., R.K-S, N.S., R.L. and A.Q. collected data and analysed samples for somatic hypermutation.
M.K., J.B., W.W., J.C., P.T., A.F., Z.E., H.K. and S.O.B. contributed patient samples.
All authors reviewed and gave the final approval for the paper.
Conflicts of Interest Disclosures: The authors declare no competing financial interests for this study.
References
- Baliakas P, Agathangelidis A, Hadzidimitriou A, Sutton LA, Minga E, Tsanousa A, Scarfo L, Davis Z, Yan XJ, Shanafelt T, Plevova K, Sandberg Y, Vojdeman FJ, Boudjogra M, Tzenou T, Chatzouli M, Chu CC, Veronese S, Gardiner A, Mansouri L, Smedby KE, Pedersen LB, Moreno D, Van Lom K, Giudicelli V, Francova HS, Nguyen-Khac F, Panagiotidis P, Juliusson G, Angelis L, Anagnostopoulos A, Lefranc MP, Facco M, Trentin L, Catherwood M, Montillo M, Geisler CH, Langerak AW, Pospisilova S, Chiorazzi N, Oscier D, Jelinek DF, Darzentas N, Belessi C, Davi F, Ghia P, Rosenquist R, Stamatopoulos K. Not all IGHV3-21 chronic lymphocytic leukemias are equal: prognostic considerations. Blood. 2015;125:856–859. doi: 10.1182/blood-2014-09-600874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara D, Thompson PA, Kantarjian HM, O’Brien SM, Ferrajoli A, Jain N, Burger JA, Estrov Z, Faderl S, Wierda WG, Keating MJ. Chemoimmunotherapy for Patients with Chronic Lymphocytic Leukemia: MD Anderson Experience Beyond FCR300. Blood. 2016;128:2047–2047. [Google Scholar]
- Chiorazzi N. Implications of new prognostic markers in chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2012;2012:76–87. doi: 10.1182/asheducation-2012.1.76. [DOI] [PubMed] [Google Scholar]
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- Davis Z, Forconi F, Parker A, Gardiner A, Thomas P, Catovsky D, Rose-Zerilli M, Strefford JC, Oscier D. The outcome of Chronic lymphocytic leukaemia patients with 97% IGHV gene identity to germline is distinct from cases with <97% identity and similar to those with 98% identity. Br J Haematol. 2016;173:127–136. doi: 10.1111/bjh.13940. [DOI] [PubMed] [Google Scholar]
- Davis ZA, Orchard JA, Corcoran MM, Oscier DG. Divergence from the germ-line sequence in unmutated chronic lymphocytic leukemia is due to somatic mutation rather than polymorphisms. Blood. 2003;102:3075. doi: 10.1182/blood-2003-08-2696. [DOI] [PubMed] [Google Scholar]
- Degan M, Bomben R, Bo MD, Zucchetto A, Nanni P, Rupolo M, Steffan A, Attadia V, Ballerini PF, Damiani D, Pucillo C, Poeta GD, Colombatti A, Gattei V. Analysis of IgV gene mutations in B cell chronic lymphocytic leukaemia according to antigen-driven selection identifies subgroups with different prognosis and usage of the canonical somatic hypermutation machinery. Br J Haematol. 2004;126:29–42. doi: 10.1111/j.1365-2141.2004.04985.x. [DOI] [PubMed] [Google Scholar]
- Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, Schulman P, Vinciguerra VP, Rai K, Rassenti LZ, Kipps TJ, Dighiero G, Schroeder HW, Jr, Ferrarini M, Chiorazzi N. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102:1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stilgenbauer S, Stevenson F, Davi F, Rosenquist R, European Research Initiative on CLL. ERIC recommendations on IGHV gene mutational status analysis in chronic lymphocytic leukemia. Leukemia. 2007;21:1–3. doi: 10.1038/sj.leu.2404457. [DOI] [PubMed] [Google Scholar]
- Gladstone DE, Swinnen L, Kasamon Y, Blackford A, Gocke CD, Griffin CA, Meade JB, Jones RJ. Importance of immunoglobulin heavy chain variable region mutational status in del(13q) chronic lymphocytic leukemia. Leuk Lymphoma. 2011;52:1873–1881. doi: 10.3109/10428194.2011.585529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladstone DE, Blackford A, Cho E, Swinnen L, Kasamon Y, Gocke CD, Griffin CA, Bolanos-Meade J, Jones RJ. The importance of IGHV mutational status in del(11q) and del(17p) chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 2012;12:132–137. doi: 10.1016/j.clml.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski P, Hultdin M, Karlsson K, Tobin G, Aleskog A, Thunberg U, Laurell A, Sundstrom C, Rosenquist R, Roos G. Telomere length as a prognostic parameter in chronic lymphocytic leukemia with special reference to VH gene mutation status. Blood. 2005;105:4807–4812. doi: 10.1182/blood-2004-11-4394. [DOI] [PubMed] [Google Scholar]
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- Hamblin TJ, Davis ZA, Oscier DG. Determination of how many immunoglobulin variable region heavy chain mutations are allowable in unmutated chronic lymphocytic leukaemia - long-term follow up of patients with different percentages of mutations. Br J Haematol. 2008;140:320–323. doi: 10.1111/j.1365-2141.2007.06928.x. [DOI] [PubMed] [Google Scholar]
- Krober A, Seiler T, Benner A, Bullinger L, Bruckle E, Lichter P, Dohner H, Stilgenbauer S. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416. [PubMed] [Google Scholar]
- Lin K, Sherrington PD, Dennis M, Matrai Z, Cawley JC, Pettitt AR. Relationship between p53 dysfunction, CD38 expression, and IgV(H) mutation in chronic lymphocytic leukemia. Blood. 2002;100:1404–1409. doi: 10.1182/blood-2001-11-0066. [DOI] [PubMed] [Google Scholar]
- Matsuda F, Shin EK, Nagaoka H, Matsumura R, Haino M, Fukita Y, Taka-ishi S, Imai T, Riley JH, Anand R, Soeda E, Honjo T. Structure and physical map of 64 variable segments in the 3′0.8-megabase region of the human immunoglobulin heavy-chain locus. Nat Genet. 1993;3:88–94. doi: 10.1038/ng0193-88. [DOI] [PubMed] [Google Scholar]
- Murphy EJ, Neuberg DS, Rassenti LZ, Hayes G, Redd R, Emson C, Li K, Brown JR, Wierda WG, Turner S, Greaves AW, Zent CS, Byrd JC, McConnel C, Barrientos J, Kay N, Hellerstein MK, Chiorazzi N, Kipps TJ, Rai KR. Leukemia-cell proliferation and disease progression in patients with early stage chronic lymphocytic leukemia. Leukemia. 2017;31:1348–1354. doi: 10.1038/leu.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien SM, Furman RR, Coutre SE, Flinn IW, Burger J, Blum K, Sharman J, Wierda WG, Jones J, Zhao W, Heerema NA, Johnson AJ, Luan Y, James DF, Chu AD, Byrd JC. Five-Year Experience with Single-Agent Ibrutinib in Patients with Previously Untreated and Relapsed/Refractory Chronic Lymphocytic Leukemia/Small Lymphocytic Leukemia. Blood. 2016;128:233–233. [Google Scholar]
- Oscier D, Wade R, Davis Z, Morilla A, Best G, Richards S, Else M, Matutes E, Catovsky D, Chronic Lymphocytic Leukaemia Working Group UKNCRI. Prognostic factors identified three risk groups in the LRF CLL4 trial, independent of treatment allocation. Haematologica. 2010;95:1705–1712. doi: 10.3324/haematol.2010.025338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnani M, Laurenti L, Longo PG, Piccirillo N, Gobessi S, Mannocci A, Marietti S, Sica S, Leone G, Efremov DG. The proliferative response to CpG-ODN stimulation predicts PFS, TTT and OS in patients with chronic lymphocytic leukemia. Leuk Res. 2010;34:1189–1194. doi: 10.1016/j.leukres.2009.12.020. [DOI] [PubMed] [Google Scholar]
- Thompson PA, Tam CS, O’Brien SM, Wierda WG, Stingo F, Plunkett W, Smith SC, Kantarjian HM, Freireich EJ, Keating MJ. Fludarabine, cyclophosphamide and rituximab achieves long-term disease-free survival in IGHV-mutated chronic lymphocytic leukemia. Blood. 2016;127:303–309. doi: 10.1182/blood-2015-09-667675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin G, Thunberg U, Johnson A, Eriksson I, Soderberg O, Karlsson K, Merup M, Juliusson G, Vilpo J, Enblad G, Sundstrom C, Roos G, Rosenquist R. Chronic lymphocytic leukemias utilizing the VH3-21 gene display highly restricted Vlambda2-14 gene use and homologous CDR3s: implicating recognition of a common antigen epitope. Blood. 2003;101:4952–4957. doi: 10.1182/blood-2002-11-3485. [DOI] [PubMed] [Google Scholar]
- Tobin G, Thunberg U, Laurell A, Karlsson K, Aleskog A, Willander K, Soderberg O, Merup M, Vilpo J, Hultdin M, Sundstrom C, Roos G, Rosenquist R. Patients with chronic lymphocytic leukemia with mutated VH genes presenting with Binet stage B or C form a subgroup with a poor outcome. Haematologica. 2005;90:465–469. [PubMed] [Google Scholar]
- van Gent R, Kater AP, Otto SA, Jaspers A, Borghans JA, Vrisekoop N, Ackermans MA, Ruiter AF, Wittebol S, Eldering E, van Oers MH, Tesselaar K, Kersten MJ, Miedema F. In vivo dynamics of stable chronic lymphocytic leukemia inversely correlate with somatic hypermutation levels and suggest no major leukemic turnover in bone marrow. Cancer Res. 2008;68:10137–10144. doi: 10.1158/0008-5472.CAN-08-2325. [DOI] [PubMed] [Google Scholar]
- Xochelli A, Agathangelidis A, Kavakiotis I, Minga E, Sutton LA, Baliakas P, Chouvarda I, Giudicelli V, Vlahavas I, Maglaveras N, Bonello L, Trentin L, Tedeschi A, Panagiotidis P, Geisler C, Langerak AW, Pospisilova S, Jelinek DF, Oscier D, Chiorazzi N, Darzentas N, Davi F, Ghia P, Rosenquist R, Hadzidimitriou A, Belessi C, Lefranc MP, Stamatopoulos K. Immunoglobulin heavy variable (IGHV) genes and alleles: new entities, new names and implications for research and prognostication in chronic lymphocytic leukaemia. Immunogenetics. 2015;67:61–66. doi: 10.1007/s00251-014-0812-3. [DOI] [PubMed] [Google Scholar]


