Abstract
Background
Alloantibodies to RBC antigens can cause significant hemolytic events. Prior studies have demonstrated that inflammatory stimuli in animal models and inflammatory states in humans, including autoimmunity and viremia, promote alloimmunization. However, molecular mechanisms underlying these findings are poorly understood. Given that type 1 interferons (IFNα/β) regulate antiviral immunity and autoimmune pathology, the hypothesis that IFNα/β regulates RBC alloimmunization was tested in a murine model.
Study Design and Methods
Leuko-reduced murine RBCs expressing the human KEL glycoprotein were transfused into control mice (WT), mice lacking the unique IFNα/β receptor (IFNAR1−/−), or bone marrow chimeric mice lacking IFNAR1 on specific cell populations. Anti-KEL IgG production, expressed as mean fluorescence intensity (MFI), and B cell differentiation were examined.
Results
Transfused WT mice produced anti-KEL IgG alloantibodies (peak response MFI=50.4). However, the alloimmune response of IFNAR1−/− mice was almost completely abrogated (MFI=4.2, p<0.05). The response of bone marrow chimeric mice lacking IFNAR1 expression in all hematopoietic cells or specifically in B cells was also diminished (MFI=3.8 and 5.4, respectively, compared to control chimeras, MFI=65.6, p<0.01). Accordingly, transfusion-induced differentiation of IFNAR1−/− B cells into germinal center B cells and plasma cells was significantly reduced, compared to WT B cells.
Conclusions
This study demonstrates that B cells require signaling from IFNα/β to produce alloantibodies to the human KEL glycoprotein in mice. These findings provide a potential mechanistic basis for inflammation-induced alloimmunization. If these findings extend to human studies, patients with IFNα/β-associated conditions may have an elevated risk of alloimmunization and benefit from personalized transfusion protocols.
Keywords: Type I interferon, alloimmunization, KEL, inflammation
Introduction
With approximately 15 million units of red blood cells (RBC) transfused per year, RBC transfusion is the most common procedure performed in United States hospitals 1. Unlike ABO and Rh antigens, other common RBC antigens, including Kell, Duffy, and Kidd, are not routinely matched between donor units and recipients. Exposure to allogeneic RBC antigens during transfusion leads to production of antigen-specific alloantibodies in 3–10% of transfusion recipients and 30–50% of patients with sickle-cell disease 2,3. Subsequent exposure to the RBC antigen may result in potentially fatal hemolytic transfusion reactions 4. Additionally, transfusion-dependent patients with hemoglobinopathies, including sickle cell disease, may have rare or multiple RBC alloantibodies. Thus, compatible RBC units for these patients can be exceedingly limited or unattainable, resulting in anemia-associated morbidity and mortality 5,6. Elucidating factors that influence alloantibody production could improve prediction of these alloimmune responses, and allow for intervention to reduce alloimmunization and hemolytic events.
One such factor is the inflammatory state of the transfusion recipient 7–11. Ramsey and Smietana reported an increased prevalence of autoimmune disease in alloimmunized patients 8. More recent studies have reported an increased incidence of alloimmunization in patients with inflammatory bowel disease, febrile transfusion reactions, and sickle cell-mediated acute chest syndrome 7,10,11. Recently, Evers et al. reported that patients with viremia also have an elevated risk of alloimmunization 12. However, patients with Gram-negative bacteremia had significantly lower rates of alloimmunization. Accordingly, studies in mouse models of alloimmunization have demonstrated that viral mimetics promote RBC alloimmune responses 13–15, while lipopolysaccharide (LPS) inhibits responses 16. These findings suggest that specific inflammatory pathways associated with certain conditions, including some autoimmune diseases and viral infection, promote RBC alloimmune responses. Recently, Arneja et al demonstrated a role for interleukin (IL)-6 in T cell responses to stored RBCs 17. However, other cytokine-mediated mechanisms underlying inflammation-induced alloimmunization have not been investigated.
The inflammatory conditions that promote alloimmunization are associated with the production of numerous inflammatory cytokines, including tumor necrosis factor α (TNFα). IL-12, and IL-6. One class of inflammatory cytokines that regulates both antiviral immunity and autoimmune pathology is the type I Interferon (IFNα/β) family, consisting of 13 IFNα genes and 1 IFNβ gene. In a virally infected cell, recognition of viral nucleic acids by ubiquitously expressed pattern recognition receptors (PRRs), including Toll-like receptors, results in production and secretion of IFNα/β. Binding of IFNα/β to its dimeric receptor, consisting of IFNα Receptor (IFNAR) 1 and IFNAR2, induces expression of interferon stimulated genes (ISGs) that inhibit viral replication and dissemination 18.
Long-term viral protection is dependent on adaptive immune responses that produce neutralizing antibodies. Studies in murine models of viral infection and immunization have shown that IFNα/β can enhance or inhibit antibody responses. Multiple studies have reported that injection of IFNα/β promotes antibody responses to some soluble antigens by increasing activation of dendritic cells and lymphocytes 19–21. However, Muller et al. generated IFNAR1-deficient mice (IFNAR1−/−) and reported that wildtype (WT) and IFNAR1−/− mice produced comparable IgM and IgG responses to immunization with multiple soluble antigens, including ovalbumin and a viral glycoprotein22. More recent studies have demonstrated that IFNAR1−/− mice produce elevated levels of protective antibodies in chronic viral infections 23–25. Thus, IFNAR1−/− mice can mount antibody responses to multiple viral and soluble antigens.
IFNAR signaling has also been implicated in the pathogenesis of multiple autoimmune diseases, including myositis, scleroderma, rheumatoid arthritis, and Sjogren’s syndrome 26–29. Patients with systemic lupus erythematosus (SLE) frequently have elevated serum IFNα/β and ISG expression, which correlate with increased auto-antibody levels and disease severity30–33. This association has led to the use of anti-IFNα/β therapy in clinical trials 34,35.
The role of IFNα/β in RBC alloimmunization has not been previously investigated. In comparison to viral and soluble antigens, RBC-bound antigens differ in their route of administration, quantity of antigen, duration of exposure, and accessibility to niches within secondary lymphoid tissue 14,36,37. Thus, the role of IFNα/β in RBC alloimmunization may differ from their role in autoimmunity, viral infections, or immunization. Given that viral infections and multiple autoimmune diseases induce IFNα/β production and promote RBC alloimmunization, we tested the hypothesis that IFNα/β regulates alloimmune responses to RBC antigens. Given the large number of antigenic differences between human donors and recipients, we utilized a murine transfusion model that allows examination of alloimmune responses to a single donor RBC antigen. Using this model, we assessed the role of IFNα/β in promoting RBC alloimmunization. Further, we assessed the contribution of cell-specific IFNα/β signaling to the RBC alloimmune response.
Materials and Methods
Mice
C57BL/6 and congenic C57BL/6-Ly5.1 WT mice were purchased from Charles River Laboratories (Wilmington, MA). muMT−, TCRα−/−, and Zbtb46-DTR mice were purchased from Jackson Laboratories (Bar Harbor, ME). IFNAR1−/− and KEL RBC transgenic mice (previously published as KEL2B mice) were previously described 22,38. All mice had been backcrossed to the C57BL/6 background for more than 8 generations. All animal protocols were approved by the Yale Institutional Animal Care and Use Committee (IACUC). In accordance to the IACUC protocol, experimental groups consisted of 4–5 mice per independent experiment.
RBC Transfusion
Peripheral blood of KEL RBC mice was collected in 12% Citrate Phosphate Dextrose Adenine (CPDA-1, Jorgensen Labs, Melville, NY) by retro-orbital bleeding and leuko-reduced with a Pall (East Hills, NY) syringe filter. WT, IFNAR1−/−, or chimeric mice were transfused i.v. with 75 μL leuko-reduced packed RBCs, the mouse equivalent of 1 unit of human RBCs. For secondary responses, mice were re-transfused 28 days later. For IFNAR1 blocking experiments, WT mice were i.p. injected with 600 μg anti-mouse IFNAR1 (clone: MAR1-5A3, BioXCell, West Lebanon, NH), isotype control mouse IgG1 (MOPC-21, BioXCell) or PBS 24 hours prior to and 3 and 7 days after transfusion.
Generation of bone marrow chimeras
Recipient WT C57BL/6 (CD45.2+) or IFNAR1−/− (CD45.2+) mice were exposed to 2 rounds of X-ray irradiation (6.35 Gy, 3 hours apart) using an X-RAD 320 irradiator (Precision X-ray Inc., North Branford, CT). Recipients were i.v. injected 2–4 hrs after irradiation with a total of 3×106 bone marrow cells from WT C57BL/6-Ly5.1 (CD45.1+), IFNAR1−/−, muMT− (CD45.2+), TCRα−/− (CD45.2+) and/or Zbtb46-DTR (CD45.1+, CD45.2+) mice. Peripheral blood was analyzed for lymphocyte reconstitution 6 weeks after bone marrow transfer. Mice were transfused 8–9 weeks following bone marrow reconstitution. For chimeras generated with Zbtb46-DTR bone marrow, DTR-expressing cells were depleted with 2 i.p. injections of diphtheria toxin (DT, Sigma-Aldrich, St. Louis, MO) 3 (60 ng/g) and 1 day (40ng/g) prior to transfusion as previously described39,40. Doses of DT were titrated in Zbtb46-DTR chimeras to achieve efficient conventional DC depletion.
Measurement of anti-KEL alloantibodies
Transfusion of murine KEL RBCs induces anti-KEL antibody production in allogeneic mice that do not express the human KEL glycoprotein38. These anti-KEL antibodies formed following transfusion of KEL RBCs are defined as anti-KEL alloantibodies. Serum anti-KEL IgG was measured by flow cytometric crossmatch 7, 14, 21, and 28 days after transfusion as previously described 41. Briefly, serum from transfused mice was incubated with KEL RBCs and subsequently stained for RBC-bound IgG (Goat anti-mouse IgG APC, Jackson ImmunoResearch, West Grove, PA). Undiluted serum was used to maximize the sensitivity of the assay. The adjusted MFI was calculated by subtracting the reactivity of serum with syngeneic C57BL/6 RBCs from the reactivity of serum with KEL RBCs. Figure data illustrates the peak antibody response, 21–28 days following transfusion. Flow cytometry of RBCs was performed using a BD FACS Calibur (San Jose, CA) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Flow cytometric Analysis
Single cell suspensions of peripheral blood leukocytes and splenocytes were analyzed following RBC lysis. Bone marrow was collected by flushing tibias with media. For dendritic cell analysis, spleens were minced with a razor blade prior to cell filtration with a 70 μM nylon mesh. Suspensions were stained with fluorescently conjugated antibodies specific for cell surface proteins, including CD45.2 (Clone: 104), CD138 (Syndecan-1), CD19 (6D5), TCRβ (H57-597), IgD (11-26c.2a), I-A/I-E (MHC II, M5/114.15.2), and GL7 from Biolegend (San Diego, CA); CD45.1 (A20), CD11c (N418), B220 (RA3-6B2), and IFNAR1 (MAR1-5A3) from Ebioscience (San Diego, CA), and Fas/CD95 (Jo2) from BD Biosciences (San Jose, CA). Zombie-NIR (Biolegend) was used to exclude dead cells. Cells were acquired with a Miltenyi MACSQuant flow cytometer and analyzed using FlowJo.
In vitro B cell cultures
B cells were isolated from WT and IFNAR1−/− spleens by negative selection using a magnetic cell isolation kit (StemCell Technologies, Vancouver, BC). B cell purity analyzed by flow cytometry was 90–95%. B cells (1×106/mL) were cultured in complete RPMI previously described 42 containing anti-CD40 (10 μg/mL FGK4.5) with or without recombinant IFNα (100 U/mL, HC1040, Hycult Biotech, Netherlands) for 72 hrs. Development of plasma cells (CD19+IgDloB220loCD138+) was measured by flow cytometry.
Statistics
Statistical analysis was performed using GraphPad Prism software (San Diego, CA). Statistical significance between two groups was determined using an unpaired t-test or Mann Whitney U test for parametric and non-parametric data, respectively. Significance between three groups was determined using a Kruskal-Wallis test with a Dunn’s post-test. All anti-KEL IgG, germinal center B cell, and primary plasma cell data was analyzed using non-parametric tests. B cell culture data was analyzed with an unpaired two-tailed t-test. Bars on graphs indicate the mean of the data, and the circles represent individual data points from one mouse or one culture. Statistical significance was determined for all individually performed experiments containing at least 4 mice per group. With the exception of B cell culture data, Figures in the primary text display results of one experiment of 2–3 completed experiments with 3–5 mice per group. Results of repeated experiments are shown in Supplemental figures.
Results
IFNα/β regulates alloimmunization to KEL-expressing RBCs
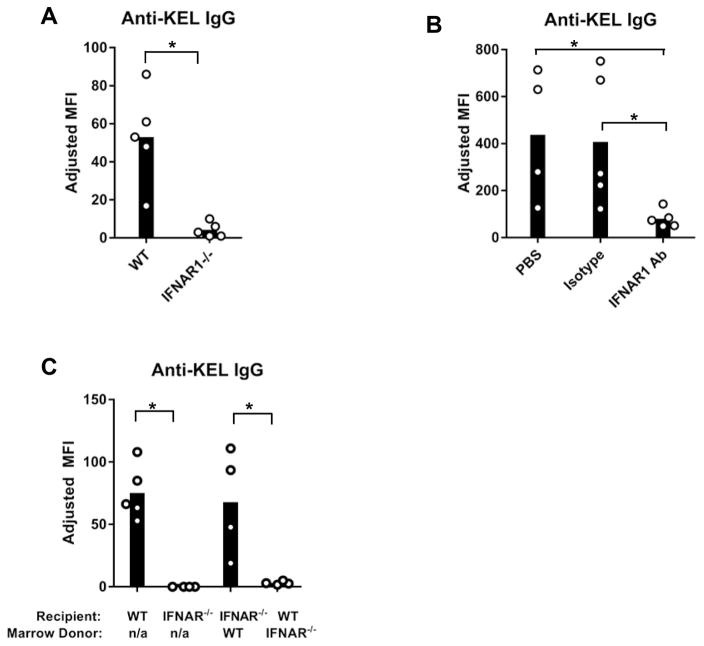
IFNα/β is induced in inflammatory conditions associated with increased alloimmunization. Thus, we examined whether expression of the only receptor for IFNα/β, IFNAR, was required for RBC alloimmunization. We utilized a murine model in which donor murine RBCs express the human KEL glycoprotein in a RBC specific manner (KEL RBCs) 38. As shown in Figure 1A, WT mice transfused with leuko-reduced KEL RBCs produced anti-KEL glycoprotein IgG alloantibodies in the absence of an adjuvant. In contrast, in 3 out of 3 experiments, the anti-KEL antibody response of IFNAR1−/− mice was almost completely abrogated after transfusion of KEL RBCs (Figure 1A and Supplemental Figure 1A). The abrogated response could be due to altered lymphoid structure or leukocyte development in IFNAR1−/− mice. To address this possibility, the anti-KEL IgG response was examined in WT mice treated with an IFNAR1 blocking antibody (MAR1-5A3), an isotype control IgG1 antibody, or PBS. Compared to control groups, MAR1-5A3 treated mice produced significantly lower levels of anti-KEL IgG following transfusion (Figure 1B and Supplemental Figure 1B). Collectively, these results indicate that IFNα/β critically regulates alloimmunization to KEL-expressing RBCs.
Figure 1. IFNα/β promotes alloimmunization to KEL RBCs.
Peripheral blood of KEL-expressing transgenic mice was leuko-reduced and transfused into recipients on Day 0. Serum anti-KEL IgG was measured by flow cytometric crossmatch. The adjusted MFI was calculated by subtracting the reactivity of serum with WT RBCs from serum reactivity with KEL RBCs. (A) Anti-KEL IgG in serum of WT or IFNAR1−/− mice 28 days after transfusion with KEL RBCs. (B) Serum anti-KEL IgG of transfused WT mice injected i.p. with anti-IFNAR1 blocking antibody (MAR1-5A3), an isotype control IgG1 antibody (MOPC-21), or PBS on Day −1, +2, and +7. (C) Anti-KEL IgG of transfused WT, IFNAR1−/−, and bone marrow chimeric mice. Recipients were irradiated and reconstituted with donor bone marrow cells, 8 weeks prior to transfusion. “n/a”; non applicable. One of 2 (B–C) or 3 (A) independent experiements with 4–5 mice/group. Data from repeated experiments are shown in Supplemental Figure 1. Bars on graphs indicate the mean of the data, and the circles represent individual data points from one mouse.*p<0.05 by (A, C) Mann Whitney U test and (B) Kruskal-Wallis test with a Dunn’s post-test.
Hematopoietic cell expression of IFNα/β receptors in transfusion recipients is required for KEL RBC alloimmunization
IFNAR1 is expressed by multiple hematopoietic and non-hematopoietic cells. Additionally, IFNAR signaling has been reported to promote humoral immune responses by activating multiple cell types 18. To determine whether hematopoietic or non-hematopoietic IFNAR1 expression is required for alloimmunization following KEL RBC transfusion, alloimmune responses in bone marrow chimeras were examined. Recipients were irradiated and reconstituted with donor bone marrow 8 weeks prior to transfusion. As shown in Figure 1C, reconstitution of irradiated IFNAR1−/− mice with WT bone marrow rescued the alloimmune response of IFNAR1−/− mice. In contrast, irradiated WT mice reconstituted with IFNAR1−/− bone marrow failed to produce anti-KEL IgG following transfusion (Figure 1C and Supplemental Figure 1C). Thus, IFNAR1 expression by hematopoietic cells is required for KEL-RBC alloimmunization.
IFNAR expression in conventional dendritic cells is dispensable for RBC alloimmunization
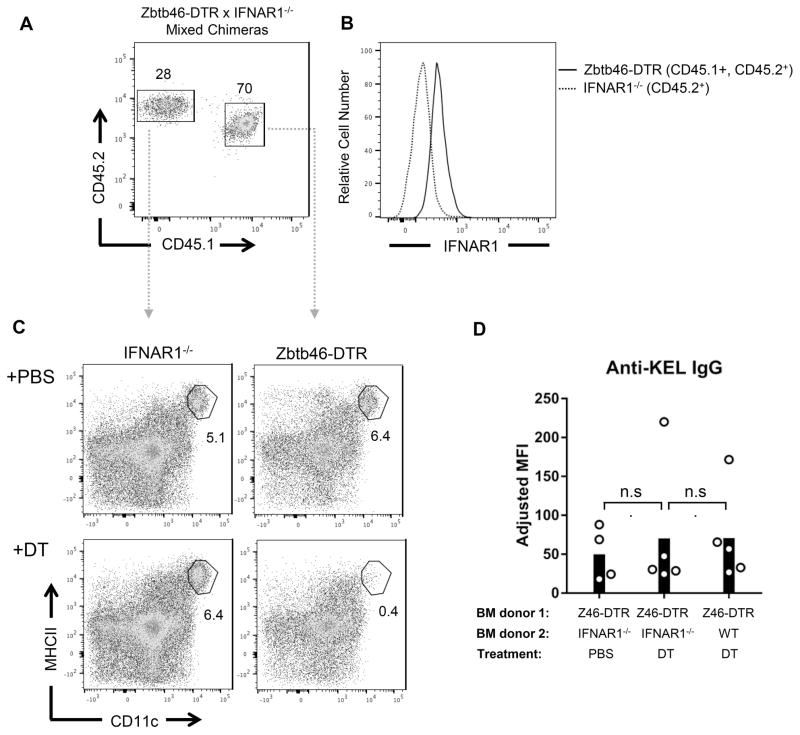
To determine which hematopoietic cell type(s) utilize(s) IFNα/β to promote alloantibody production, responses were assessed in mixed bone marrow chimeras that lack IFNAR1 expression in specific cell types. Given that prior studies have established a role for IFNα/β in promoting dendritic cell maturation and antigen presentation 43,44, we hypothesized that IFNAR signaling in conventional dendritic cells (cDCs) may regulate alloimmunization. To generate chimeras lacking IFNAR1 expression in cDCs, we utilized Zbtb46-DTR mice, which specifically express the diphtheria toxin receptor (DTR) on cDCs 45. WT mice were irradiated and reconstituted with a mixture of Zbtb46-DTR (CD45.1+, CD45.2+) bone marrow and either WT (CD45.1+) or IFNAR1−/− (CD45.2+) bone marrow. CD45 congenic markers were used to identify cells derived from different donors (Figure 2A). After 8 weeks of reconstitution, analysis of IFNAR1 expression on splenocytes from Zbtb46-DTR x IFNAR1−/− chimeras confirmed that CD45.2+ cells were derived from IFNAR1−/− bone marrow (Figure 2B). Zbtb46-DTR-expressing cDCs were then depleted by injecting diphtheria toxin (DT) prior to transfusion. As a result, nearly all remaining cDCs in DT-treated Zbtb46-DTR x IFNAR1−/− chimeras were derived from IFNAR1−/− BM. (Figure 2C). However, following transfusion, DT and PBS-treated chimeras produced comparable levels of anti-KEL IgG. The anti-KEL IgG levels were also comparable to those in DT-treated chimeras containing WT cDCs (Zbtb46-DTR x WT, Figure 2D and Supplemental Figure 2A). These results indicate that IFNAR1 expression on cDCs is not required for alloantibody responses to KEL-expressing RBCs.
Figure 2. IFNAR expression by conventional dendritic cells is dispensable for RBC alloimmunization.
Mixed chimeras were generated by reconstituting irradiated CD45.2+ WT recipients with a 4:1 mixture of Zbtb46-DTR (CD45.1+, CD45.2+) and either IFNAR1−/− (CD45.2+) or WT (CD45.1+) bone marrow. (A) Representative flow cytometric analysis of splenocytes derived from bone marrow donors in Zbtb46-DTR x IFNAR1−/− chimeras. (B) IFNAR1 expression of splenocytes gated in (A). (C) Representative flow cytometric analysis of spleen CD11c+ MHCII+ conventional dendritic cells in Zbtb46-DTR x IFNAR1−/− chimeras treated with PBS or diptheria toxin (DT) 8 weeks after bone marrow transfer. (D) Serum anti-KEL IgG produced by indicated chimeras treated with PBS or DT prior to transfusion with KEL RBCs. Z46-DTR = Zbtb46-DTR. Numbers on plots indicate percent of CD19− TCRβ − cells within the drawn gate. Representative of 3 independent experiments with 4–5 mice/group. n.s., not significant by Kruskal-Wallis test with a Dunn’s post-test. Data from repeated alloimmunization experiments are shown in Supplemental Figure 2A.
T cell IFNAR expression is not required for alloimmunization to KEL RBCs
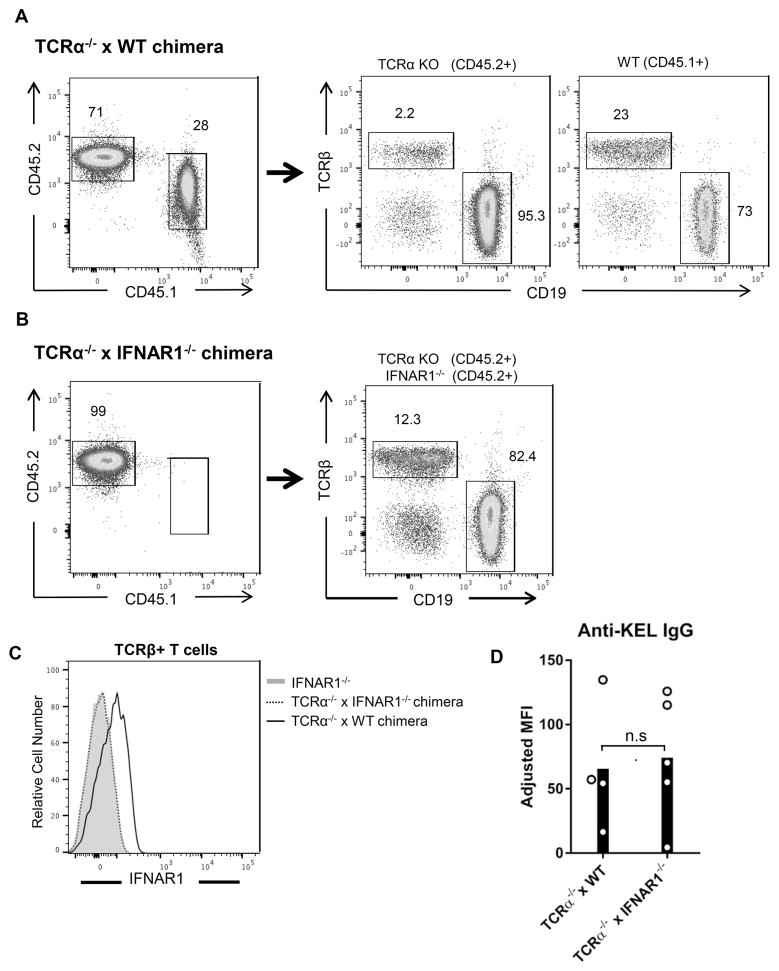
Studies have demonstrated that IFNAR signaling in T cells can regulate humoral immune responses in viral infection and immunization models 21,46. To determine whether T cell IFNAR signaling regulates alloimmunization to KEL RBCs, mixed chimeras that lack IFNAR1 expression specifically on T cells were generated. Irradiated IFNAR1−/− (CD45.2+) recipients were reconstituted with bone marrow from T cell-deficient TCRα−/− (CD45.2+) mice and either WT (CD45.1+) or IFNAR1−/− bone marrow. IFNAR1−/−, rather than WT mice were used as bone marrow recipients to ensure that irradiation-resistant recipient thymocytes in TCRα−/− x IFNAR1−/− chimeras lacked IFNAR1 expression. Following reconstitution, T cells in TCRα−/− x WT chimeras were predominately derived from donor CD45.1+ WT bone marrow. However, recipient CD45.2+ irradiation-resistant T cells were also present (Figure 3A). TCRα−/− x IFNAR1−/− chimeras were only generated with CD45.2+ bone marrow (Figure 3B). However, measurement of IFNAR1 expression in T cells confirmed that TCRα−/− x IFNAR1−/− chimeras contained only IFNAR1-deficient T cells. (Figure 3C). Following transfusion of KEL RBCs, both groups of chimeras produced comparable amounts of anti-KEL IgG (Figure 3D and Supplemental Figure 2B). These results indicate that IFNAR expression by T cells is not required for alloimmunization to KEL RBCs.
Figure 3. T cell IFNAR expression is not required for RBC alloimmunization.
Mixed chimeras were generated by reconstituting irradiated IFNAR1−/− (CD45.2+) recipients with a 4:1 mixture of TCRα−/− (CD45.2+) and either IFNAR1−/− (CD45.2+) or WT (CD45.1+) bone marrow. (A–B) Representative reconstitution analysis of peripheral blood lymphocytes from (A) TCRα−/− x WT and (B) TCRα−/− x IFNAR1−/− chimeras. Numbers on plots indicate percent of live (left), CD45.2+ (middle), or CD45.1+ (right) cells within the drawn gate. (C) Relative expression of IFNAR1 on peripheral blood TCRβ+ T cells from IFNAR1−/− mice and the indicated chimeras. (D) Anti-KEL IgG in serum of indicated chimeras following transfusion with KEL RBCs. Representative of 3 independent experiments with 4–5 mice/group per experiment. n.s., not significant by Mann Whitney U test. Data from repeated alloimmunization experiments are shown in Supplemental Figure 2B.
IFNAR expression by B cells is required for alloimmunization to KEL RBCs
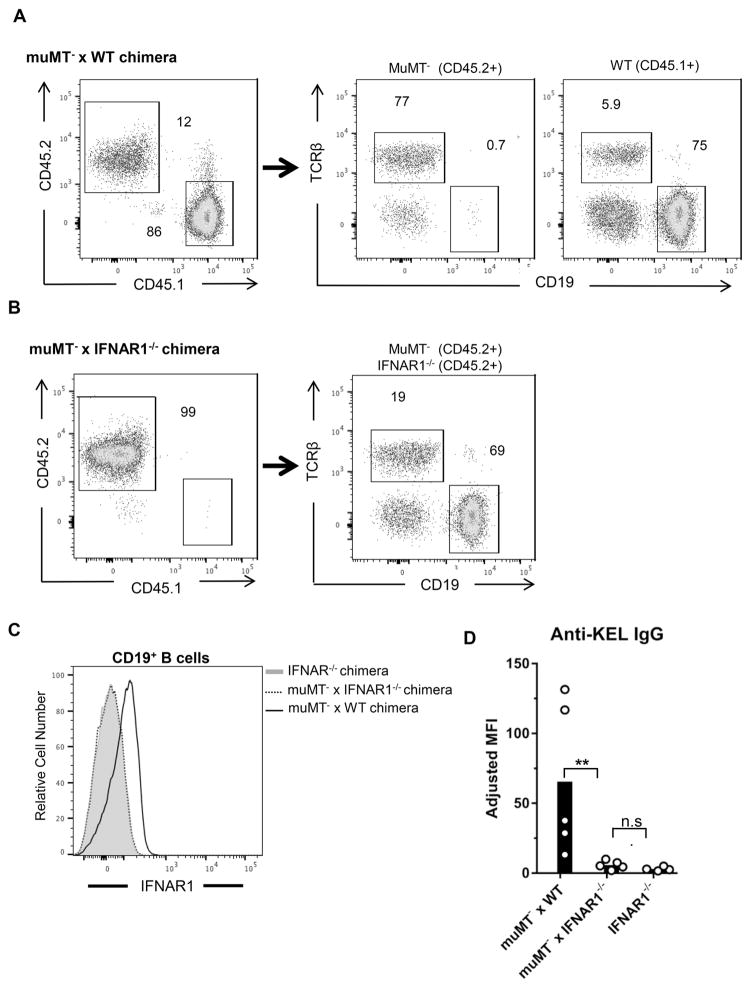
Studies of viral infection and immunization have also shown that IFNα/β can directly promote B cell activation and antibody production 21,47,48. To examine whether IFNAR signaling in B cells regulates RBC alloimmunization, mixed chimeras were generated by reconstituting irradiated CD45.2+ WT mice with bone marrow from B cell-deficient muMT− (CD45.2+) mice and either WT (CD45.1+) or IFNAR1−/− (CD45.2+) mice. Chimeras reconstituted with only IFNAR1−/− bone marrow were also generated to serve as controls. Following reconstitution, nearly all CD19+ B cells in muMT− x WT chimeras were derived from CD45.1+ WT bone marrow (Figure 4A). In muMT− x IFNAR1−/− chimeras, expression of CD45.2 on all cells excluded a similar analysis (Figure 4B). However, analysis of IFNAR1 expression demonstrated that all B cells in muMT− x IFNAR1−/− chimeras were IFNAR1-deficient (Figure 4C). Upon transfusion with KEL RBCs, muMT− x WT chimeras produced alloantibodies. However, like IFNAR1−/− control chimeras, muMT− x IFNAR1−/− chimeras produced significantly lower levels of anti-KEL IgG (Figure 4D and Supplemental Figure 2C). Thus, IFNAR signaling in B cells regulates alloimmunization to KEL RBCs.
Figure 4. IFNAR expression by B cells is required for alloimmunization to KEL RBCs.
Mixed chimeras were generated by reconstituting irradiated WT (CD45.2+) recipients with a 2:1 mixture of muMT− (CD45.2+) and either IFNAR1−/− (CD45.2+) or WT (CD45.1+) bone marrow. Chimeras reconstituted with only IFNAR1−/− bone marrow served as negative controls for IFNAR1 expression and alloimmunization. (A–B) Representative reconstitution analysis of peripheral blood lymphocytes from (A) muMT− x WT and (B) muMT− x IFNAR1−/− chimeras. Numbers on plots indicate percent of live (left), CD45.2+ (middle), or CD45.1+ (right) cells within the drawn gate. (C) IFNAR1 expression on CD19+ B cells from indicated chimeras. (D) Serum anti-KEL IgG of indicated chimeras 28 days following transfusion with KEL RBCs. Representative of 3 independent experiments with 4–5 mice per group. **p<0.01 and n.s., not significant, by Mann Whitney U test. Data from repeated alloimmunization experiments are shown in Supplemental Figure 2C.
IFNα/β promotes germinal center B cell and plasma cell differentiation
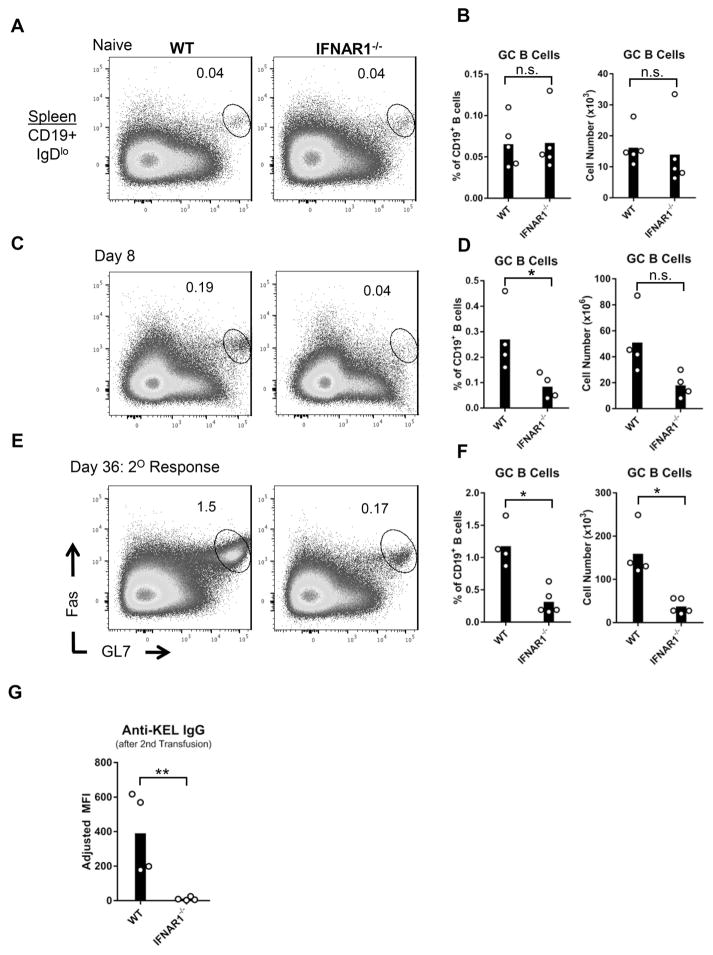
Upon antigen recognition, follicular B cells differentiate into germinal center (GC) B cells that ultimately produce antibody producing plasma cells. To determine whether IFNα/β regulates B cell differentiation, we examined GC B cell and plasma cell formation in IFNAR1−/− and WT mice following KEL RBC transfusion. Prior to transfusion, similar levels of GC B cells (CD19+IgDloFas+GL7hi) were detected in spleens of IFNAR1−/− and WT mice (Figure 5A–B and Supplemental Figure 3A). Compared to WT controls, the percentage of GC B cells in IFNAR1−/− mice was significantly decreased, 8 days following transfusion. The total number of GC B cells was statistically different between the 2 groups in 1 of 2 experiments (Figure 5C–D and Supplemental Figure 3B). To enrich for KEL-specific germinal center B cells and plasma cells, mice were re-transfused with KEL RBCs on Day 28. Eight days following a second transfusion, the percentage and number of GC B cells in IFNAR1−/− mice were reduced compared to WT controls (Figure 5E,F and Supplemental Figure 3C). In addition, despite a potential increase in IFNAR1−/− GC B cells following a second transfusion, anti-KEL IgG responses by IFNAR1−/− mice remained fully abrogated (Figure 5G and Supplemental Figure 3D). Differentiation of plasma cells was also assessed 14 days after the second transfusion. The percent and total number of bone marrow plasma cells (CD19+IgDloB220loCD138+) were significantly reduced in IFNAR1−/− mice, compared to WT controls (Figure 6A–C and Supplemental Figure 4). These results indicate that IFNα/β regulates KEL alloimmune responses by promoting GC B cell and plasma cell differentiation.
Figure 5. IFNα/β promotes germinal center B cell development.
IFNAR1−/− and WT mice were transfused with KEL RBCs. (A, C, E) Representative flow cytometric analysis of spleen GC B cells (CD19+IgDloFas+GL7hi) from (A) naïve or transfused mice (C) 8 or (E) 36 days after transfusion. Plots are gated on live CD19+IgDlo cells. Numbers on plots indicate the percent of CD19+ B cells within the drawn region. (B, D, F) GC B cells quantified as percent of CD19+ B cells (left) and cell number (right). (G) Serum anti-KEL IgG measured by flow cytometric cross-match 36 days after transfusion. (E–G) Mice received a second transfusion 28 days following the first transfusion. Data are from one of two independent experiments with 3–5 mice per group. *p<0.05, **p<0.01, n.s., not significant, by Mann Whitney U test. Data from a repeated independent experiment are shown in Supplemental Figure 3.
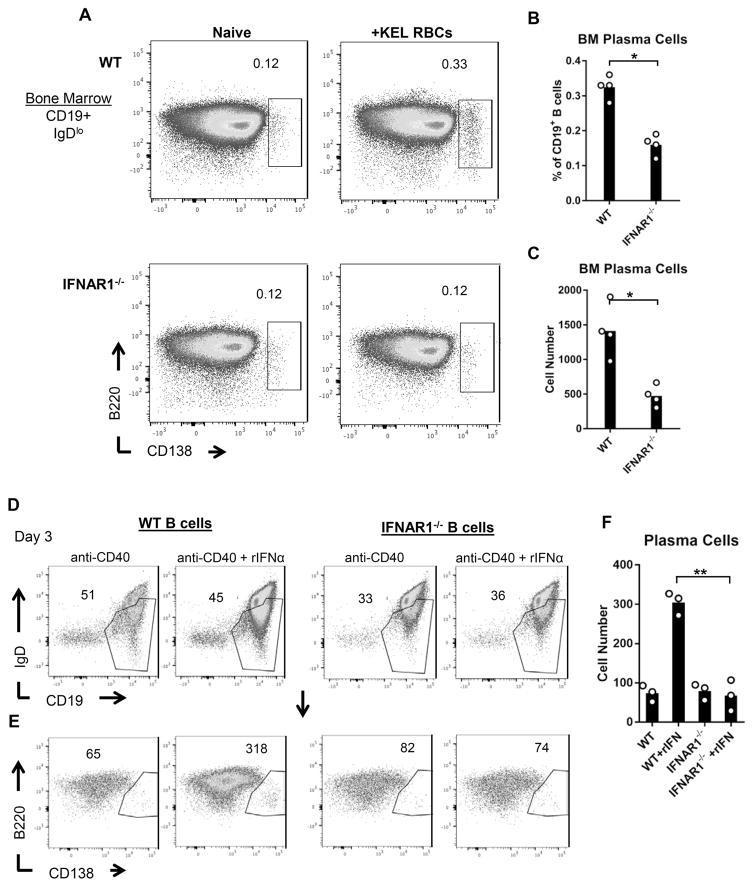
Figure 6. IFNα/β promotes plasma cell differentiation.
(A–C) WT and IFNAR1−/− mice were transfused 28 days following an initial transfusion, and plasma cells were analyzed 14 days later. (A) Representative flow cytometric analysis of plasma cells (CD19+IgDloB220loCD138+) from bone marrow of naïve and transfused mice. Plots are gated on live CD19+IgDlo cells. Numbers on plots indicate the percent of CD19+ B cells within the drawn region. (B–C) Bone marrow plasma cells were quantified as (B) percent of CD19+ B cells and (C) cell number. Data show one of two independent experiments with 4 mice per group. *p<0.05 by Mann Whitney U test. Data from a repeated independent experiment are shown in Supplemental Figure 4. (D–F) B cells were isolated from spleens of WT and IFNAR1−/− mice by magnetic cell separation. Cells were cultured for 72 hrs in the presence of anti-CD40 +/− rIFNα. (D–E) Representative flow cytometric analysis of live (D) CD19+IgDlo B cells and (E) B220loCD138+ plasma cells gated on regions drawn in D). Numbers on plots indicate the (D) percent of live cells or (E) the total number of cells in the drawn regions. Representative of 3 independent experiments. (F) Summary data of the number of plasma cells generated in the indicated cultures. **p<0.01 by unpaired 2-tailed t-test.
Given that IFNAR expression is abrogated in all cell types of IFNAR1−/− mice, it is possible that IFNAR expression in other hematopoietic and non-hematopoietic cells may influence plasma cell differentiation. To address this possibility, we isolated WT and IFNAR1−/− spleen B cells and examined plasma cell differentiation in ex vivo cultures in the presence and absence of recombinant IFNα (rIFNα). Magnetically selected B cells were cultured for 72 hrs in the presence of the anti-CD40 antibody, FGK4.5, to promote cell survival. In accordance with prior studies 48,49, the addition of rIFNα to WT cultures resulted in elevated production of plasma cells (CD19+IgDloB220loCD138+), compared to cultures lacking rIFNα. However, the addition of rIFNa did not increase plasma cell development in IFNAR1−/− B cell cultures (Figure 6D–F). This result demonstrates that IFNα/β directly promotes B cell differentiation into antibody-producing plasma cells.
Discussion
Identifying patients with an elevated risk of transfusion would allow interventions, such as extended antigen matching, to inhibit alloimmunization and hemolytic events. However, diagnostic tests to predict alloimmunization have not been developed. This is in part due to the lack of understanding of cellular and molecular pathways that promote alloimmunization. In this study, we demonstrate that recipient expression of interferon receptors (IFNAR) is required for alloimmunization to the human KEL glycoprotein in a murine transfusion model. Although the receptor for IFNα/β is expressed by many non-hematopoietic and hematopoietic cell types, we demonstrate that IFNAR expression by B cells critically regulates the humoral alloimmune response. We further show that IFNAR promotes germinal center B cell and plasma cell differentiation following transfusion.
IFNα/β has been shown to have diverse effects on humoral immune responses to varying infectious organisms and immunogenic antigens 21–24. Given that IFNAR1−/− and WT mice were reported to produce similar antibody responses in many other models 22, the abrogated RBC alloimmune response of IFNAR1−/− mice was unlikely due to potentially altered lymphoid architecture or hematopoiesis in IFNAR1−/− mice. Rather, our interpretation of these data is that binding of IFNα/β to IFNAR activates downstream signaling that is required for alloimmunization to KEL RBCs. This conclusion is supported by the finding that treatment of WT mice with an IFNAR1 blocking antibody significantly inhibited the anti-KEL IgG response.
These findings provide insight into previously reported studies in mouse transfusion models. Treatment of recipient mice with inflammatory pathogen associated molecular patterns (PAMPs), including poly(I:C) and CpG, promotes alloimmunization to RBCs expressing KEL or other alloantigens 13–15. Poly(I:C) is a mimetic of viral double stranded RNA (dsRNA) that induces robust production of IFNα/β by many cell types. Our demonstration that IFNAR expression is required for KEL RBC alloimmunization raises the possibility that poly(I:C) promotes alloimmunization by inducing IFNα/β. However, this should be formally tested in transfusion models that require the use of poly(I:C) to induce alloimmunization.
Multiple studies have successfully utilized mixed bone marrow chimeras to examine the role of IFNAR signaling in specific cell types 21,50. Using this approach, we found that while IFNAR expression by B cells was critical for anti-KEL alloimmune resonses, IFNα/β-mediated responses by cDCs and T cells were dispensable. In contrast, prior studies utilizing IFNα/β injections to increase antibody responses have reported that IFNα/β-mediated responses by cDCs, T cells, and B cells promoted humoral immune responses to soluble antigens 19,21. This apparent discrepancy might reflect inherent differences between responses to soluble and RBC-bound antigens 36,37. In addition, although RBC alloimmunization to other antigens requires DC presentation to T cells 39,40, it is possible that T cell help is not required for anti-KEL responses in this model. Further, IFNAR signaling in T cells or DCs may be able to promote B cell alloantibody production. In this case, deletion of IFNAR from either cell type would not inhibit alloimmunization. These findings may also indicate that IFNα/β-mediated contributions of T cells and cDCs to the alloimmune response may depend on the level of available IFNα/β, which may be produced and consumed in localized niches accessible to B cells. Unlike IFNα/β treated mice, we observed that the level of serum IFNα in mice transfused with KEL RBCs was nearly undetectable (unpublished results). However, our IFNAR1−/− results indicate that there is a source of IFNα/β that stimulate B cells to produce anti-KEL antibodies. We hypothesize that transfused RBCs might be associated with damage associated molecular patterns (DAMPs) that can bind to PRRs and induce IFNα/β production. Such DAMPs may be present on RBCs, platelets, or remaining leukocytes. Future studies are needed to identify the sensor(s) and ligand(s) that promote IFNα/β production in alloimmunization.
Analysis of B cell differentiation in IFNAR1−/− mice revealed that IFNα/β regulates GC B cell and plasma cell differentiation in response to KEL RBC transfusion. Additionally, in accordance with prior studies of human B cells co-cultured with IFNα/β-producing plasmacytoid DCs 51,52, rIFNα directly induced plasma cell differentiation in cultures of isolated spleen B cells. These findings are consistent with results from viral infection and immunization models 47,48,53,54 and provide insight into the role of IFNα/β in transfusion-induced B cell differentiation. We hypothesize that IFNα/β may regulate the selection, proliferation and/or maintenance of KEL-specific GC B cell clones. However, plasma cell formation is the result of numerous B cell processes including antibody class switching, affinity maturation, and cell migration. Thus, other mechanisms underlying IFNα/β-induced B cell differentiation during alloimmunization are plausible and are the focus of further study.
In summary, we demonstrate that expression of the IFNα/β receptor by B cells is required for alloimmunization to the human KEL glycoprotein on transfused murine RBCs. We further show that IFNα/β promotes GC B cell and plasma cell differentiation following RBC transfusion. These findings provide a potential mechanistic basis for inflammation-induced alloimmunization. If they extend to human studies, patients with IFNα/β-associated conditions, including viral infection and autoimmunity, or those treated with IFN-based therapies, may have an elevated risk of alloimmunization.
Supplementary Material
Acknowledgments
Funding: This work was supported by grants from the National Blood Foundation (R13672) (DRG) and NIH/NHLBI (R01 HL126076) (JEH) and (T32 HL007974-14) (Brian Smith, M.D.).
This work was supported by grants from the National Blood Foundation (R13672) (DRG) and NIH/NHLBI (R01 HL126076) (JEH) and (T32 HL007974-14) (Brian Smith, M.D.).
Footnotes
Conflicts of Interest: The authors declare that they have no conflicts of interest relevant to the manuscript submitted to TRANSFUSION.
Authorship: DRG, AI, and JEH planned the experiments completed by DRG, JL, PN, MS, JM, SP, and JEH. All authors made experimental suggestions. DRG wrote the initial draft of the manuscript, and all authors edited the manuscript and approved the final version of the manuscript.
References
- 1.Pfuntner A, Wier LM, Stocks C. Most Frequent Procedures Performed in U.S. Hospitals, 2010: Statistical Brief #149 Healthcare Cost and Utilization Project (HCUP) Statistical Briefs. Rockville (MD): 2006. [Google Scholar]
- 2.Kormoczi GF, Mayr WR. Responder individuality in red blood cell alloimmunization. Transfus Med Hemother. 2014;41:446–51. doi: 10.1159/000369179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yazdanbakhsh K, Ware RE, Noizat-Pirenne F. Red blood cell alloimmunization in sickle cell disease: pathophysiology, risk factors, and transfusion management. Blood. 2012;120:528–37. doi: 10.1182/blood-2011-11-327361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(FDA) UDoHaHS. Fatalities Reported to the FDA Following Blood Collection and Transfusion: Annual Summary for Fiscal Year 2014. 2014 http://www.fda.gov/downloads/biologicsbloodvaccines/safetyavailability/reportaproblem/transfusiondonationfatalities/ucm459461.pdf.
- 5.Nickel RS, Hendrickson JE, Fasano RM, Meyer EK, Winkler AM, Yee MM, Lane PA, Jones YA, Pashankar FD, New T, Josephson CD, Stowell SR. Impact of red blood cell alloimmunization on sickle cell disease mortality: a case series. Transfusion. 2016;56:107–14. doi: 10.1111/trf.13379. [DOI] [PubMed] [Google Scholar]
- 6.Telen MJ, Afenyi-Annan A, Garrett ME, Combs MR, Orringer EP, Ashley-Koch AE. Alloimmunization in sickle cell disease: changing antibody specificities and association with chronic pain and decreased survival. Transfusion. 2015;55:1378–87. doi: 10.1111/trf.12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fasano RM, Booth GS, Miles M, Du L, Koyama T, Meier ER, Luban NL. Red blood cell alloimmunization is influenced by recipient inflammatory state at time of transfusion in patients with sickle cell disease. Br J Haematol. 2015;168:291–300. doi: 10.1111/bjh.13123. [DOI] [PubMed] [Google Scholar]
- 8.Ramsey G, Smietana SJ. Multiple or uncommon red cell alloantibodies in women: association with autoimmune disease. Transfusion. 1995;35:582–6. doi: 10.1046/j.1537-2995.1995.35795357881.x. [DOI] [PubMed] [Google Scholar]
- 9.Ryder AB, Hendrickson JE, Tormey CA. Chronic inflammatory autoimmune disorders are a risk factor for red blood cell alloimmunization. Br J Haematol. 2015 doi: 10.1111/bjh.13781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papay P, Hackner K, Vogelsang H, Novacek G, Primas C, Reinisch W, Eser A, Mikulits A, Mayr WR, Kormoczi GF. High risk of transfusion-induced alloimmunization of patients with inflammatory bowel disease. Am J Med. 2012;125:717e1–8. doi: 10.1016/j.amjmed.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 11.Yazer MH, Triulzi DJ, Shaz B, Kraus T, Zimring JC. Does a febrile reaction to platelets predispose recipients to red blood cell alloimmunization? Transfusion. 2009;49:1070–5. doi: 10.1111/j.1537-2995.2009.02116.x. [DOI] [PubMed] [Google Scholar]
- 12.Evers D, van der Bom JG, Tijmensen J, Middelburg RA, de Haas M, Zalpuri S, de Vooght KM, van de Kerkhof D, Visser O, Pequeriaux NC, Hudig F, Zwaginga JJ. Red cell alloimmunisation in patients with different types of infections. Br J Haematol. 2016 doi: 10.1111/bjh.14307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao W, Yu J, Heck S, Yazdanbakhsh K. Regulatory T-cell status in red cell alloimmunized responder and nonresponder mice. Blood. 2009;113:5624–7. doi: 10.1182/blood-2008-12-193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryder AB, Zimring JC, Hendrickson JE. Factors Influencing RBC Alloimmunization: Lessons Learned from Murine Models. Transfus Med Hemother. 2014;41:406–19. doi: 10.1159/000368995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, Heck S, Yazdanbakhsh K. Prevention of red cell alloimmunization by CD25 regulatory T cells in mouse models. American Journal of Hematology. 2007;82:691–6. doi: 10.1002/ajh.20959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrickson J, Roback JD, Hillyer CD, Easley KA, Zimring JC. Discrete toll like receptor agonists have differential effects on alloimmunization to red blood cells. Transfusion. 2008:1869–77. doi: 10.1111/j.1537-2995.2008.01801.x. [DOI] [PubMed] [Google Scholar]
- 17.Arneja A, Salazar JE, Jiang W, Hendrickson JE, Zimring JC, Luckey CJ. Interleukin-6 receptor-alpha signaling drives anti-RBC alloantibody production and T-follicular helper cell differentiation in a murine model of red blood cell alloimmunization. Haematologica. 2016;101:e440–e4. doi: 10.3324/haematol.2016.149278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Bon A, Schiavoni G, D’Agostino G, Gresser I, Belardelli F, Tough DF. Type 1 interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 2001;14:461–70. doi: 10.1016/s1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- 20.Proietti E, Bracci L, Puzelli S, Di Pucchio T, Sestili P, De Vincenzi E, Venditti M, Capone I, Seif I, De Maeyer E, Tough D, Donatelli I, Belardelli F. Type I IFN as a natural adjuvant for a protective immune response: lessons from the influenza vaccine model. J Immunol. 2002;169:375–83. doi: 10.4049/jimmunol.169.1.375. [DOI] [PubMed] [Google Scholar]
- 21.Le Bon A, Thompson C, Kamphuis E, Durand V, Rossmann C, Kalinke U, Tough DF. Cutting edge: enhancement of antibody responses through direct stimulation of B and T cells by type I IFN. J Immunol. 2006;176:2074–8. doi: 10.4049/jimmunol.176.4.2074. [DOI] [PubMed] [Google Scholar]
- 22.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–21. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 23.Davidson S, Crotta S, McCabe TM, Wack A. Pathogenic potential of interferon alphabeta in acute influenza infection. Nat Commun. 2014;5:3864. doi: 10.1038/ncomms4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, Schreiber RD, de la Torre JC, Oldstone MB. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–11. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, Aronow BJ, Karp CL, Brooks DG. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340:202–7. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Assassi S, Mayes MD, Arnett FC, Gourh P, Agarwal SK, McNearney TA, Chaussabel D, Oommen N, Fischbach M, Shah KR, Charles J, Pascual V, Reveille JD, Tan FK. Systemic sclerosis and lupus: points in an interferon-mediated continuum. Arthritis Rheum. 2010;62:589–98. doi: 10.1002/art.27224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bave U, Nordmark G, Lovgren T, Ronnelid J, Cajander S, Eloranta ML, Alm GV, Ronnblom L. Activation of the type I interferon system in primary Sjogren’s syndrome: a possible etiopathogenic mechanism. Arthritis Rheum. 2005;52:1185–95. doi: 10.1002/art.20998. [DOI] [PubMed] [Google Scholar]
- 28.Higgs BW, Liu Z, White B, Zhu W, White WI, Morehouse C, Brohawn P, Kiener PA, Richman L, Fiorentino D, Greenberg SA, Jallal B, Yao Y. Patients with systemic lupus erythematosus, myositis, rheumatoid arthritis and scleroderma share activation of a common type I interferon pathway. Ann Rheum Dis. 2011;70:2029–36. doi: 10.1136/ard.2011.150326. [DOI] [PubMed] [Google Scholar]
- 29.Olsen N, Sokka T, Seehorn CL, Kraft B, Maas K, Moore J, Aune TM. A gene expression signature for recent onset rheumatoid arthritis in peripheral blood mononuclear cells. Ann Rheum Dis. 2004;63:1387–92. doi: 10.1136/ard.2003.017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–5. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crow MK, Kirou KA, Wohlgemuth J. Microarray analysis of interferon-regulated genes in SLE. Autoimmunity. 2003;36:481–90. doi: 10.1080/08916930310001625952. [DOI] [PubMed] [Google Scholar]
- 32.Feng X, Wu H, Grossman JM, Hanvivadhanakul P, FitzGerald JD, Park GS, Dong X, Chen W, Kim MH, Weng HH, Furst DE, Gorn A, McMahon M, Taylor M, Brahn E, Hahn BH, Tsao BP. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54:2951–62. doi: 10.1002/art.22044. [DOI] [PubMed] [Google Scholar]
- 33.Ytterberg SR, Schnitzer TJ. Serum interferon levels in patients with systemic lupus erythematosus. Arthritis Rheum. 1982;25:401–6. doi: 10.1002/art.1780250407. [DOI] [PubMed] [Google Scholar]
- 34.Petri M, Wallace DJ, Spindler A, Chindalore V, Kalunian K, Mysler E, Neuwelt CM, Robbie G, White WI, Higgs BW, Yao Y, Wang L, Ethgen D, Greth W. Sifalimumab, a human anti-interferon-alpha monoclonal antibody, in systemic lupus erythematosus: a phase I randomized, controlled, dose-escalation study. Arthritis Rheum. 2013;65:1011–21. doi: 10.1002/art.37824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao Y, Richman L, Higgs BW, Morehouse CA, de los Reyes M, Brohawn P, Zhang J, White B, Coyle AJ, Kiener PA, Jallal B. Neutralization of interferon-alpha/beta-inducible genes and downstream effect in a phase I trial of an anti-interferon-alpha monoclonal antibody in systemic lupus erythematosus. Arthritis Rheum. 2009;60:1785–96. doi: 10.1002/art.24557. [DOI] [PubMed] [Google Scholar]
- 36.Kontos S, Grimm AJ, Hubbell JA. Engineering antigen-specific immunological tolerance. Curr Opin Immunol. 2015;35:80–8. doi: 10.1016/j.coi.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 37.Grimm AJ, Kontos S, Diaceri G, Quaglia-Thermes X, Hubbell JA. Memory of tolerance and induction of regulatory T cells by erythrocyte-targeted antigens. Sci Rep. 2015;5:15907. doi: 10.1038/srep15907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith NH, Henry KL, Cadwell CM, Bennett A, Hendrickson JE, Frame T, Zimring JC. Generation of transgenic mice with antithetical KEL1 and KEL2 human blood group antigens on red blood cells. Transfusion. 2012;52:2620–30. doi: 10.1111/j.1537-2995.2012.03641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calabro S, Gallman A, Gowthaman U, Liu D, Chen P, Liu J, Krishnaswamy J, Nasciemento MS, Xu L, Patel SR, Williams A, Tormey CA, Hod EA, Spitalnik SL, Zimring JC, Hendrickson JE, Stowell SR, Eisenbarth SC. Bridging channel dendritic cells induce immunity to transfused red blood cells. Journal of Experimental Medicine. 2016 doi: 10.1084/jem.20151720. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibb DR, Calabro S, Liu D, Tormey CA, Spitalnik SL, Zimring JC, Hendrickson JE, Hod EA, Eisenbarth SC. The Nlrp3 Inflammasome Does Not Regulate Alloimmunization to Transfused Red Blood Cells in Mice. EBioMedicine. 2016;9:77–86. doi: 10.1016/j.ebiom.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stowell SR, Girard-Pierce KR, Smith NH, Henry KL, Arthur CM, Zimring JC, Hendrickson JE. Transfusion of murine red blood cells expressing the human KEL glycoprotein induces clinically significant alloantibodies. Transfusion. 2014;54:179–89. doi: 10.1111/trf.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibb DR, El Shikh M, Kang DJ, Rowe WJ, El Sayed R, Cichy J, Yagita H, Tew JG, Dempsey PJ, Crawford HC, Conrad DH. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J Exp Med. 2010;207:623–35. doi: 10.1084/jem.20091990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito T, Amakawa R, Inaba M, Ikehara S, Inaba K, Fukuhara S. Differential regulation of human blood dendritic cell subsets by IFNs. J Immunol. 2001;166:2961–9. doi: 10.4049/jimmunol.166.5.2961. [DOI] [PubMed] [Google Scholar]
- 44.Montoya M, Schiavoni G, Mattei F, Gresser I, Belardelli F, Borrow P, Tough DF. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood. 2002;99:3263–71. doi: 10.1182/blood.v99.9.3263. [DOI] [PubMed] [Google Scholar]
- 45.Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, Idoyaga J, Cheong C, Yao KH, Niec RE, Nussenzweig MC. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209:1153–65. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Havenar-Daughton C, Kolumam GA, Murali-Krishna K. Cutting Edge: The direct action of type I IFN on CD4 T cells is critical for sustaining clonal expansion in response to a viral but not a bacterial infection. J Immunol. 2006;176:3315–9. doi: 10.4049/jimmunol.176.6.3315. [DOI] [PubMed] [Google Scholar]
- 47.Coro ES, Chang WL, Baumgarth N. Type I IFN receptor signals directly stimulate local B cells early following influenza virus infection. J Immunol. 2006;176:4343–51. doi: 10.4049/jimmunol.176.7.4343. [DOI] [PubMed] [Google Scholar]
- 48.Fink K, Lang KS, Manjarrez-Orduno N, Junt T, Senn BM, Holdener M, Akira S, Zinkernagel RM, Hengartner H. Early type I interferon-mediated signals on B cells specifically enhance antiviral humoral responses. Eur J Immunol. 2006;36:2094–105. doi: 10.1002/eji.200635993. [DOI] [PubMed] [Google Scholar]
- 49.Poeck H, Wagner M, Battiany J, Rothenfusser S, Wellisch D, Hornung V, Jahrsdorfer B, Giese T, Endres S, Hartmann G. Plasmacytoid dendritic cells, antigen, and CpG-C license human B cells for plasma cell differentiation and immunoglobulin production in the absence of T-cell help. Blood. 2004;103:3058–64. doi: 10.1182/blood-2003-08-2972. [DOI] [PubMed] [Google Scholar]
- 50.Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, Lee H, Arthur CD, White JM, Kalinke U, Murphy KM, Schreiber RD. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Douagi I, Gujer C, Sundling C, Adams WC, Smed-Sorensen A, Seder RA, Karlsson Hedestam GB, Lore K. Human B cell responses to TLR ligands are differentially modulated by myeloid and plasmacytoid dendritic cells. J Immunol. 2009;182:1991–2001. doi: 10.4049/jimmunol.0802257. [DOI] [PubMed] [Google Scholar]
- 52.Jego G, Palucka AK, Blanck JP, Chalouni C, Pascual V, Banchereau J. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity. 2003;19:225–34. doi: 10.1016/s1074-7613(03)00208-5. [DOI] [PubMed] [Google Scholar]
- 53.Zhu J, Huang X, Yang Y. Type I IFN signaling on both B and CD4 T cells is required for protective antibody response to adenovirus. J Immunol. 2007;178:3505–10. doi: 10.4049/jimmunol.178.6.3505. [DOI] [PubMed] [Google Scholar]
- 54.Bach P, Kamphuis E, Odermatt B, Sutter G, Buchholz CJ, Kalinke U. Vesicular stomatitis virus glycoprotein displaying retrovirus-like particles induce a type I IFN receptor-dependent switch to neutralizing IgG antibodies. J Immunol. 2007;178:5839–47. doi: 10.4049/jimmunol.178.9.5839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.