Abstract
Insulin-like growth factor binding protein (IGFBP)–1 influences fetal growth by modifying insulin-like growth factor–I (IGF-I) bioavailability. IGFBP-1 phosphorylation, which markedly increases its affinity for IGF-I, is regulated by mechanistic target of rapamycin (mTOR) and casein kinase (CSNK)–2. However, the underlying molecular mechanisms remain unknown. We examined the cellular localization and potential interactions of IGFBP-1, CSNK-2β, and mTOR as a prerequisite for protein–protein interaction. Analysis of dual immunofluorescence images indicated a potential perinuclear co-localization between IGFBP-1 and CSNK-2β and a nuclear co-localization between CSNK-2β and mTOR. Proximity ligation assay (PLA) indicated proximity between IGFBP-1 and CSNK-2β as well as mTOR and CSNK-2β but not between mTOR and IGFBP-1. Three-dimensional rendering of the PLA images validated that IGFBP-1 and CSNK-2β interactions were in the perinuclear region and mTOR and CSNK-2β interactions were also predominantly perinuclear rather than nuclear as indicated by mTOR and CSNK-2β co-localization. Compared with control, hypoxia and rapamycin treatment showed markedly amplified PLA signals for IGFBP-1 and CSNK-2β (approximately 18-fold, P = 0.0002). Stable isotope labeling with multiple reaction monitoring–mass spectrometry demonstrated that hypoxia and rapamycin treatment increased IGFBP-1 phosphorylation at Ser98/Ser101/Ser119/Ser174 but most considerably (106-fold) at Ser169. We report interactions between CSNK-2β and IGFBP-1 as well as mTOR and CSNK-2β, providing strong evidence of a mechanistic link between mTOR and IGF-I signaling, two critical regulators of cell growth via CSNK-2.
During fetal development, insulin-like growth factor (IGF) binding protein (IGFBP)-1, which is primarily secreted by the fetal liver, is the most predominant IGFBP.1 Phosphorylation of IGFBP-1 increases its affinity for IGF-I, resulting in inhibition of IGF-I function.2 Phosphorylation is suggested to play an important role in the development of fetal growth restriction (FGR),3 a condition associated with marked IGFBP-1 hyperphosphorylation that results in reduced bioavailability of IGF-I, a key regulator of fetal growth.4, 5, 6, 7
Casein kinase (CSNK)-2 is a pleiotropic, multifunctional, universal protein kinase crucial in cell differentiation, proliferation, and survival and can phosphorylate multiple proteins.8, 9 CSNK-2 plays a global constitutive role within the cell.10 Two of the three phosphorylated serine residues on IGFBP-1 conform to the general recognition motif for CSNK-2.11 Indeed, we previously reported that inhibition of CSNK-2 activity reduces IGFBP-1 phosphorylation in human hepatocellular carcinoma (HepG2) cells,7, 11 consistent with the possibility that CSNK-2 phosphorylates IGFBP-1 in vivo. CSNK-2 is a ubiquitous serine/threonine kinase localized in the cytoplasm, nuclei, and several other cell organelles.12 It is also bound to the exterior of the cell membrane as an ectokinase, and thus can phosphorylate extracellular proteins.13 Whether IGFBP-1 is phosphorylated by CSNK-2 and at which subcellular location this occurs remain to be fully established.
CSNK-2 is composed of two regulatory α and/or α′ subunits and two noncatalytic β subunits.9 Previous studies have indicated that CSNK-2 is regulated by several protein-protein interactions and that the CSNK-2 subunit β regulates the cellular and biological functions of the kinase.14 CSNK-2 is up-regulated in hypoxia.15 Similarly, IGFBP-1 secretion and phosphorylation are also induced in hypoxia,16, 17 which potently inhibits the action of IGF-I.17 In addition, mechanistic target of rapamycin (mTOR) is a serine/threonine kinase that controls cell growth and metabolism, which is primarily mediated by effects on protein translation.18, 19 Our previous data indicate that inhibition of mTOR stimulates CSNK-2 activity and IGFBP-1 secretion and phosphorylation and decreases IGF-I action, an effect that is prevented by CSNK-2 inhibition.7 Collectively, these data strongly suggest that regulation of IGF-I bioavailability in response to hypoxia is controlled by IGFBP-1 phosphorylation, which is linked to mTOR inhibition17 and activation of protein kinase CSNK-2.7, 11, 17 It is likely that hypoxia can potentiate CSNK-2 activation mediated by mTOR inhibition, resulting in IGFBP-1 hyperphosphorylation.6 Whether the effects of mTOR on IGFBP-1 secretion and phosphorylation may be due to their proximity with CSNK-2 is largely unknown. We speculate that CSNK-2β may be co-localized with IGFBP-1 and mTOR and can mediate IGFBP-1 phosphorylation. This is plausible because CSNK-2β can recognize many biological targets, while retaining specificity.
To address this gap in knowledge, multiple complementary strategies that provide direct visualization of the potential co-localization of CSNK-2β with mTOR and IGFBP-1 in cultured HepG2 cells were used. We applied dual immunofluorescence staining, confocal microscopy, and co-immunoprecipitation under physiologically relevant conditions. Moreover, proximity ligation assay (PLA) was used to determine whether there is proximity between these proteins. Furthermore, three-dimensional rendering of the PLA images was performed to determine localization of the PLA signals for these proteins relative to the nucleus. Subsequently, quantitative proteomics was used by applying stable isotope labeling of amino acids in cell culture (SILAC) combined with multiple reaction monitoring–mass spectrometry (MRM-MS) for mapping targeted sites phosphorylated on IGFBP-1 in response to CSNK-2β/mTOR interactions in hypoxia.
Materials and Methods
Cell Culture
HepG2 cells were purchased from ATCC (Manassas, VA). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/F-12 media supplemented with 10% fetal bovine serum (FBS) (Invitrogen Corp., Carlsbad, CA) at 37°C with atmospheric air (20% oxygen and 5% carbon dioxide). For dual immunofluorescence staining and PLA experiments, HepG2 cells (50,000 per well) were cultured (60% to 70% confluence) on 1:10 poly-l-lysine (Cultrex, Cambridge, MA) coated coverslips and placed in six-well plates.
Hypoxia in Combination with Serum Starvation
On the basis of a previously optimized protocol,17, 20, 21, 22 HepG2 cells were serum starved using DMEM/F-12 supplemented with 2% FBS for 24 hours and then incubated in a hypoxia chamber (Billups-Rothenburg, San Diego, CA). The hypoxia chamber was flushed with a continuous flow of 1% oxygen, 5% carbon dioxide, and balanced nitrogen (BOC Canada Ltd., Mississauga, ON, Canada) for 15 minutes to ensure saturation, which we have shown previously to retain constant oxygen saturation for up to 72 hours.22 The sealed chamber was then placed on an orbital shaker in a tissue culture incubator at 37°C. Cell media and cells in lysis buffer were collected after 24 hours of hypoxic exposure and stored at −80°C.
Inhibition of mTOR Signaling
HepG2 cells were serum starved with 2% FBS for 24 hours similarly as for hypoxia. On the basis of the dose and time dependency treatments, cells were treated with the mTOR inhibitor rapamycin (100 nmol/L) and placed in the hypoxic chamber or cultured normally for 24 hours at 37°C.17 After treatment, cell media and cell lysate were collected and stored at −80°C.
Antibodies
The following primary antibodies were used: monoclonal antibody (mAb) IGFBP-1 (catalog number 0027301; Medix Biochemica, Kauniainen, Finland), mAb mTOR (catalog number sc-136269; Santa Cruz Biotechnology, Dallas, TX), rabbit polyclonal CSNK-2β [gift from Dr. David Litchfield (University of Western Ontario, London, Ontario, Canada)], rabbit polyclonal IGF-I (catalog number PABCa; GroPep, Thebarton, Australia), rabbit polyclonal IGFBP-1 [gift from Dr. Robert Baxter (Kolling Medical Research Institute, Sydney, Australia)], rabbit polyclonal Ki-67 (catalog number ab15580; Abcam, Cambridge, MA), and IgG (L+H) (Bethyl Laboratories, Montgomery, TX). Preimmune serum used as negative controls were Dako mouse IgG1 and Dako rabbit immunoglobulin fraction (normal) (Agilent Technologies, Santa Clara, CA). Secondary antibodies used in immunohistochemistry (IHC) were anti-rabbit Alexa 568 (catalog number A11036) and anti-mouse Alexa 660 (catalog number A21055) (Thermo Fisher, Waltham, MA).
Dual Immunofluorescence
After fixation with 1 mL of 4% paraformaldehyde, the cells in six-well plates (60% to 70% confluency) were washed with phosphate-buffered saline (PBS) (3 times for 5 minutes) and then incubated for 30 minutes with 0.5% Triton X-100 to make them permeable. Cells were encircled with a hydrophobic pen (Dako, Agilent Technologies). All incubations were performed in a humidified chamber at room temperature. After another set of three 5-minute PBS washes, the cells were then treated using Dako Background Sniper (Biocare Medical, Concord, CA) for 10 minutes to reduce nonspecific binding. After a quick PBS wash, the cells were incubated with a combination of primary antibodies. The combinations of antibodies used were as follows: i) mAb IGFBP-1 and rabbit polyclonal CSNK-2β, ii) mAb IGFBP-1 and rabbit polyclonal Ki-67, and iii) mAb mTOR and rabbit polyclonal CSNK-2β antibody. The dilutions for the primary antibodies were IGFBP-1 (1:2500), Ki-67 (1:100), mTOR (1:100), and CSNK-2β (1:1000).
The cells were then washed three times for 5 minutes each with PBS. Combinations of secondary antibodies, anti-rabbit Alexa 568, and anti-mouse Alexa 660 (1:400) were added to the cells for 1 hour and subsequently washed in PBS (3 times for 5 minutes each). The cells were then counterstained with DAPI (Life Technologies, Burlington, ON, Canada) (1:200 dilution) for 2 minutes. Subsequently, the coverslips were mounted on microscope slides (Fisher Scientific, Fairlawn, NJ) with 60 μL of Prolong Diamond Mounting Media (Life Technologies) and dried overnight before imaging.
Confocal Image Acquisition
Images were captured with a 63× Plan-Apochromat oil immersion lens on a Zeiss LSM 510 DUO Vario Confocal, using Zen 2 software version 1.1.1.0 (Carl Zeiss Canada Ltd., North York, ON, Canada). Filters were selected to avoid all crosstalk between channels. Alexa 660 was excited with a 5 MW 633 nm HeNe laser, and the signal was collected through a 650-nm long-pass emission filter. For Alexa 568, a 1 MW 543 nm HeNe laser was used for excitation with a bandpass emission filter of 560 to 615 nm. DAPI signal was excited with a 50 MW 405-50 Diode laser and captured through a 420- to 480-nm bandpass emission filter.
Two- and Three-Dimensional Analyses of Confocal IHC Images
Image rendering in two and three dimensions was performed with Image Pro Premier 3D software version 9.2 (Media Cybernetics, Rockville, MD) using the co-localization module. For representation of nuclear versus cytosolic co-localized signals, the DAPI (nuclear) signal was selected and used to generate a mask of the nuclear area, which was subtracted from the cellular image to generate isolated cytosolic signal.
Iso-surfaces were generated to localize the region of analysis using the red (IGFBP-1), red (mTOR), green (CSNK-2β), and blue (DAPI) channel. A clipping plane was generated to reveal signal localization Iso-surface, and counting thresholds were set using the autobright, or Otsu's thresholding method was used to prevent user bias.
Co-Immunoprecipitation of IGFBP-1, CSNK-2β, and mTOR
Preparation of Protein A Sepharose
Protein A Sepharose (100 μL 50% slurry each; GE Healthcare, Mississauga, ON, Canada) in four different batches was washed in PBS that contained 0.1% Tween-20 at 6500 × g at 4°C. Next, the antibodies IGFBP-1 mAb 6303, CSNK-2β, mTOR, and IgG (L+H) (negative control) were diluted in 5X HEPES buffer (pH 7.4). Each of the diluted antibodies (approximately 200 ng) were incubated separately in tubes that contained the prewashed Protein A Sepharose in PBS (total of 1 mL) that contained Tween-20 and rotated overnight at 4°C. The antibody-coated Protein A Sepharose samples were washed in PBS and Tween-20 and used for immunoprecipitation.
Preparation of Cell Lysate and Immunoblot Analysis
To determine whether i) IGFBP-1 and CSNK-2β, ii) mTOR and CSNK-2β, and iii) IGFBP-1 and mTOR interact under physiologic conditions in vitro, a traditional co-immunoprecipitation approach was used. HepG2 cells were cultured in 25-cm2 flasks for 24 hours, the cell media was aspirated, and the cells were washed using cold PBS. The cells were lyzed using lysis buffer (Cell Signaling Technologies, Beverly, MA) containing protease and phosphatase inhibitor cocktails (Sigma-Aldrich, St. Louis, MO) as described previously.17 The cell lysates were sonicated and cleared by centrifugations at 17,000 × g for 30 minutes at 4°C. Total protein contents were determined by Bradford reagent (BioRad Laboratories, Burlington, ON, Canada). Each of the four aliquots of the cell lysates (500 μg of total protein each) were buffer exchanged against PBS that contained Tween-20 using 10 K Ultracel centrifugal filter units (Millipore, Carrigtwohill, Ireland). The samples were reconstituted to 1 mL in PBS and Tween-20 and were loaded onto precoated Protein A Sepharose beads with IGFBP-1 mAb 6303, CSNK-2β, mTOR, and IgG antibodies separately and incubated rotating overnight at 4°C. The unbound proteins were cleared by centrifugation and the Protein A Sepharose beads bound with the target proteins were washed in PBS and Tween-20 followed by PBS alone. The target proteins were then extracted using 100 μL of the sample buffer [containing 6 mol/L urea, 2% 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate and 20 mmol/L dithiothreitol]. The samples were stored in −80°C for further analysis by immunoblot.
Immunoblot analysis was performed as previously described.7 Briefly, aliquots of the immunoprecipitated samples (10 μL) were probed using immunoblotting and anti-human IGFBP-1 polyclonal antibody to test for the presence of IGFBP-1 in the samples immunoprecipitated using IGFBP-1 mAb 6303 and CSNK-2β antibodies. Polyclonal CSNK-2β antibody was used to test the presence of CSNK-2β. Furthermore, mAb mTOR was used to test the presence of mTOR in the samples immunoprecipitated using IGFBP-1 mAb 6303 or CSNK-2β antibody. In each case, IgG antibody was used as a negative control. After SDS-PAGE, the immunoblots were blocked in 5% milk or BSA for 1 hour and incubated with the corresponding primary antibodies overnight at 4°C. Secondary antibodies were goat anti-mouse or goat anti-rabbit horseradish peroxidase conjugates. The images were developed using Clarity ECL Western blotting substrate (BioRad Laboratories) and captured using the Versa Doc imaging system and Image Lab software version 6.0 (BioRad Laboratories).
PLA
HepG2 cells on coated coverslips (in six-well plates) were grown until 60% to 70% confluent. Cells in culture were treated with rapamycin, hypoxia, or combined hypoxia and rapamycin and in control condition (normoxia) for 24 hours as described above. Subsequently, cells were fixed, encircled with hydrophobic pen, permeabilized, and incubated at room temperature in a humidified chamber, similarly as for immunofluorescent staining. After three 5-minute PBS washes, cells were treated with blocking solution (Sigma-Aldrich) for 30 minutes at 37°C and then probed with specific combination of primary antibodies for 1 hour at room temperature. The combinations were as follows: i) IGFBP-1 mAb 6303 (1:2500) and rabbit polyclonal CSNK-2β (1:1000), ii) mAb mTOR (1:100) and rabbit polyclonal CSNK-2β (1:1000), iii) IGFBP-1 mAb 6303 and rabbit polyclonal IGF-I (1:25), and iv) rabbit polyclonal IGFBP-1 (1:10,000) and mAb mTOR (1:100). The PLA reaction was performed using PLA secondary probes diluted 1:5 in Duolink antibody diluent (Sigma-Aldrich) and incubated for 1 hour. The PLA secondary probes used were Duolink PLA Anti-Rabbit Plus and Anti-Mouse Minus. Subsequent ligation and amplification were performed using reagents and according to the manufacturer's instructions.
Coverslips were mounted on slides with mounting media (Sigma-Aldrich) that contained DAPI counterstain. Image acquisition was performed using fluorescent images acquired on an AxioImager Z1 Epifluorescent Microscope (Carl Zeiss Canada Ltd.). At 20× magnification, 10 regions were randomly selected per slide, and Z-stack images were captured and converted to extended depth of focus TIFF images. PLA quantification was conducted using Image Pro Premier 3D software. PLA spots were identified and cells were manually counted based on positive DAPI staining and quantified using the software based on red signal intensity and spot size. PLA interactions per cell were calculated by dividing the PLA spots by the total number of cells in each region of interest. PLA interactions per cell were plotted, and a t-test was performed using GraphPad Prism software version 6 (GraphPad Software Inc., La Jolla, CA).
Three-Dimensional Image Analysis for PLA Signals
Three-dimensional analysis was performed with Imaris software version 8.0.2 using the iso-surfacing and spots functions. Nuclear (DAPI) signals were iso-surfaced using default thresholds. The PLA signals were iso-surfaced using the automated spots function, which localizes the signal to its origin for accurate spatial representation. Cutaway views were generated with the clipping plane function to reveal localization of the signals relative to the nucleus.
Quantitative Proteomics
SILAC and Sample Preparation
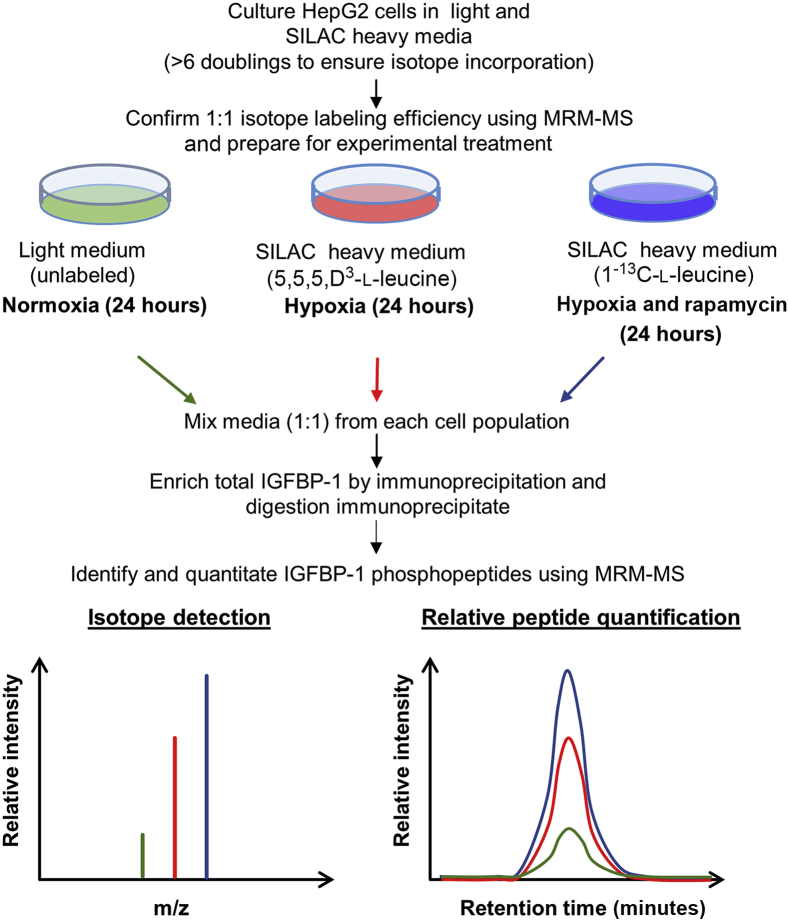
We used quantitative MRM-MS in combination with immunoprecipitation and SILAC23 to quantify the site-specific changes in phosphorylation of IGFBP-1 in HepG2 cells in hypoxia or hypoxia and rapamycin treatments versus controls (normoxia). Three batches of HepG2 cells were grown in custom-made DMEM/F12 supplemented with 10% dialyzed FBS as follows: The first batch was grown in regular DMEM/F12, the second batch was grown in DMEM/F12-SILAC media containing the stable isotope 5,5,5,D3-l-leucine (+3 Da), and the third batch was grown in DMEM/F12-SILAC media containing the stable isotope 1-13C-l-leucine (+1 Da). Cells from each batch were then passaged at least six times to ensure efficient incorporation of the heavy labeled amino acid. Successful 1:1:1 (unlabeled control:5,5,5,D3- l-leucine:1-13C-l-leucine) incorporation of isotope labels was determined by IGFBP-1 immunoprecipitated from cell media, followed by in-solution digestion by both AspN and trypsin and MRM-MS detection. Equal detection of the unlabeled and heavy labeled peptides was obtained by similar peak area, automatically determined by Skyline software version 3.7 (MacCoss Lab Software, Seattle, WA). Once a 1:1:1 incorporation of heavy isotope labels to the control unlabeled samples was determined, cells were then plated in 12-well culture dishes in triplicates for 24 hours and then starved in 2% FBS. After 24 hours, the cells from the second batch were grown in 2% dialyzed FBS DMEM/F12–labeled SILAC media (5,5,5,D3-l-leucine) and were treated in a hypoxia chamber as described earlier (1% oxygen), whereas cells from the third batch were grown in 2% dialyzed FBS DMEM/F12–labeled SILAC media (1-13C- l-leucine) and were treated with 100 nmol/L rapamycin and hypoxia combined. The untreated normoxic cells were grown in 2% dialyzed FBS DMEM/F12. After 24 hours of treatment, the cell media was collected, and 1 mL of each experimental condition (normoxia, hypoxia, and hypoxia and rapamycin) were pooled and buffer exchanged against PBS and concentrated (10×). IGFBP-1 was then immunoprecipitated from the concentrated cell media samples using IGFBP-1 mAb 6303 as described above.
SILAC-Labeled IGFBP-1 Phosphopeptide Identification and Quantitation Using MRM
After immunoprecipitation, the enriched IGFBP-1 samples were subjected to in-solution digestion using both AspN and trypsin endoproteinases as described above. Samples were then cleaned by C18 resin (catalog number Z720038; Sigma-Aldrich) and analyzed by MRM-MS as specified previously.11, 17 In summary, digested peptides were electrosprayed into the mass spectrometer, which monitored a total of 98 peptide-specific transitions with a dwelling time of 50 ms per transition (Supplemental Table S1). Relative changes in IGFBP-1 phosphorylation between the light and two heavy labeled samples were determined by the total peak area of the peptide-specific transitions using Skyline software and normalized by the intensity of an IGFBP-1 internal peptide (NH2-ALPGEQQPLHALTR-COOH). For peptides with multiple phosphorylation sites, specific transitions were monitored that were specific to the presence or absence of a single phosphorylation site (specifically, Y14, b6, and b9 ions for pSer169 and pSer174; y12 and b15 ions for pSer98 and pSer101).
Results
Co-Localization of IGFBP-1 and CSNK-2β
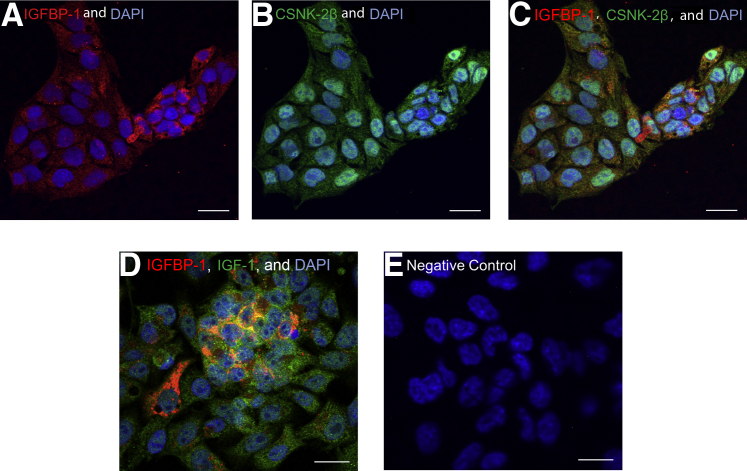
With IGFBP-1 mAb 6303 (Alexa Red) and rabbit polyclonal CSNK-2β (Alexa Green) antibodies and dual immunofluorescent Z-stack confocal images (Figure 1, A–E), IGFBP-1 was found to be present as clusters around the nucleus, suggesting a perinuclear localization (Figure 1A), whereas CSNK-2β appeared to be localized throughout the cell (Figure 1B). Importantly, IGFBP-1 and CSNK-2β were co-localized (Figure 1C). As a positive control, HepG2 cells were dual-stained with IGFBP-1 and its ligand IGF-I, which also depicted a clear yellow overlap (Figure 1D). The negative control with preimmune serum (not included) or in the absence of primary antibodies showed no nonspecific binding (Figure 1E). Together, these findings demonstrate extensive co-localization of IGFBP-1 and CSNK-2β, and strongly suggest that the β regulatory subunit of CSNK-2 likely interacts with IGFBP-1 to regulate IGFBP-1 phosphorylation in HepG2 cells.
Figure 1.
Dual immunofluorescence staining for the co-localization of insulin-like growth factor (IGF) binding protein (IGFBP)-1 and casein kinase (CSNK)-2β. Human hepatocellular carcinoma (HepG2) cells were stained with anti-mouse IGFBP-1 (monoclonal antibody 6303), anti-rabbit IGF-I, and anti-rabbit CSNK-2β antibodies. Corresponding secondary antibodies were Alexa anti-mouse 660 and anti-rabbit 568. Images were captured via confocal microscopy. A and B: IGFBP-1 (red) is predominantly localized in the perinuclear region of the cells (A), whereas CSNK-2β (green) is detected throughout the cell (B). C: Merged channel image shows co-localization predominantly in the perinuclear region (yellow). D: Co-localization of IGFBP-1 (red) and IGF-I (green) in HepG2 cells, indicating a positive control. E: Dual immunofluorescence with no primary antibodies depicting a negative control where no staining was visualized. Scale bars: 20 μm.
Perinuclear Co-Localization of IGFBP-1 and CSNK-2β
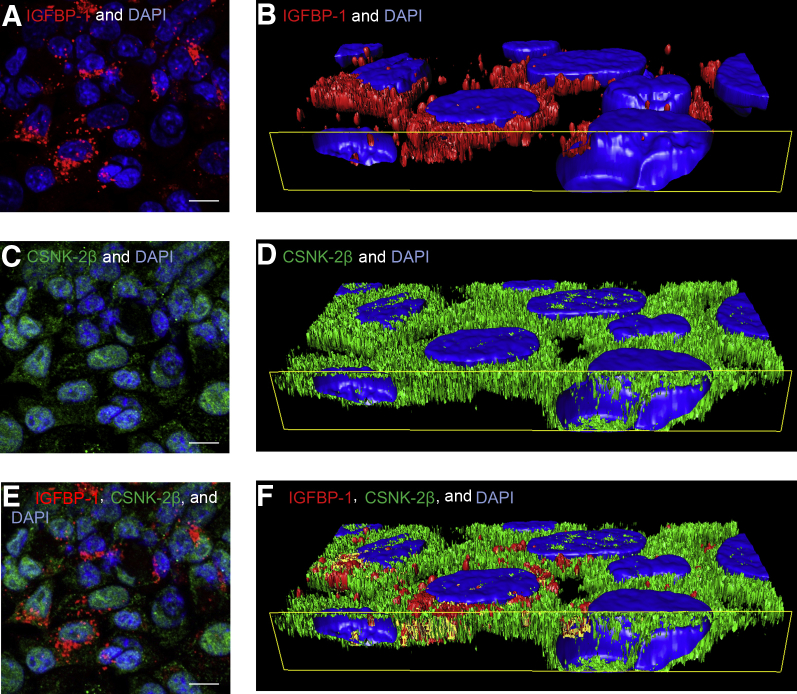
To confirm IGFBP-1/CSNK-2β co-localization, 63× Z-stack images were captured with confocal microscopy (Figure 2, A, C and E). Using imaging software (Image Pro Premier 3D), iso-surfaces were generated for each individual channel (red, green, and blue) through which IGFBP-1 staining was found to be present surrounding the nucleus and varied from cell to cell. These observations suggest a perinuclear localization of IGFBP-1 in a select subpopulation of cells (Figure 2A). Side plane images (Figure 2, B, D, and F) enabled us to examine the cross section of HepG2 cells. CSNK-2β was visualized across the cell and also within the nucleus (Figure 2, C and D). Image Pro Premier 3D software allowed us to merge the red and green iso-surfaces and detect any channel overlap, which is depicted as yellow (Figure 2, E and F). These data suggest that IGFBP-1 and CSNK-2β co-localize predominantly in the perinuclear region, albeit with significant variability among cells, which may be attributable to variations in IGFBP-1 concentrations.
Figure 2.
Localization of insulin-like growth factor binding protein (IGFBP)-1 and casein kinase (CSNK)-2β using Image Pro-Premier 3D software version 9.2 (Media Cybernetics). A, C, and E: After performing dual immunofluorescence using anti-mouse IGFBP-1 (red) and anti-rabbit CSNK-2β (green) antibodies, we captured 63× magnified images on a confocal microscope. B, D, and F: To examine the localization of IGFBP-1 and CSNK-2β and their potential co-localization in HepG2 cells in more detail, iso-surface images were generated with Image Pro-Premier 3D software. Yellow rectangle outlines the cross section of the Z-stack–captured dual-immunofluorescence image. IGFBP-1 is present in the perinuclear region but not inside the nucleus; in addition, the abundance of IGFBP-1 appears to vary from cell to cell (B). CSNK-2β is clearly visualized throughout the cytosol and nucleus of HepG2 cells (D). Merged iso-surface image with the addition of a co-localization channel depicted as yellow, which is present in the perinuclear region of HepG2 cells (F). Scale bars: 10 μm.
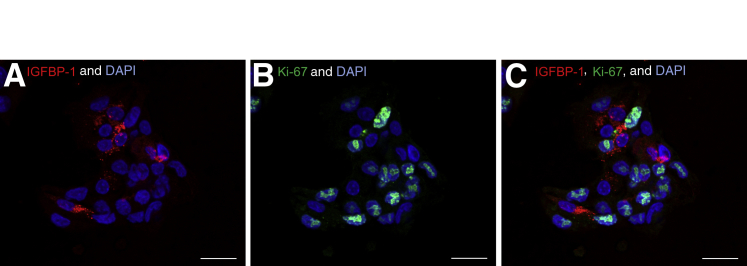
To study the cell-to-cell variation in IGFBP-1 abundance, HepG2 cells were dually stained with IGFBP-1 and Ki-67, a known cell proliferation marker. Ki-67 identified the nucleus of proliferating cells, and proliferating HepG2 cells were deduced to express low levels of IGFBP-1 compared with nonproliferating cells (Figure 3, A–C). These data suggest that nonproliferating HepG2 cells have a higher IGFBP-1 abundance than proliferating HepG2 cells.
Figure 3.
Dual immunofluorescence staining for insulin-like growth factor binding protein (IGFBP)-1 expression in proliferating human hepatocellular carcinoma (HepG2) cells. HepG2 cells were stained with anti-mouse IGFBP-1 (monoclonal antibody 6303) and anti-rabbit Ki-67 antibodies. An antibody targeting Ki-67, a proliferation marker, was used to detect proliferating HepG2 cells. A: IGFBP-1 (red) abundance shows significant cell-to-cell variability. B: Ki-67 (green) identifies growing cells and is clearly present in the nucleus. C: Merged channel image clearly depicts that Ki-67–positive HepG2 cells do not express high amounts of IGFBP-1. Scale bars: 20 μm.
Two-Dimensional Rendering Reveals mTOR and CSNK-2β Co-Localize in the Nucleus
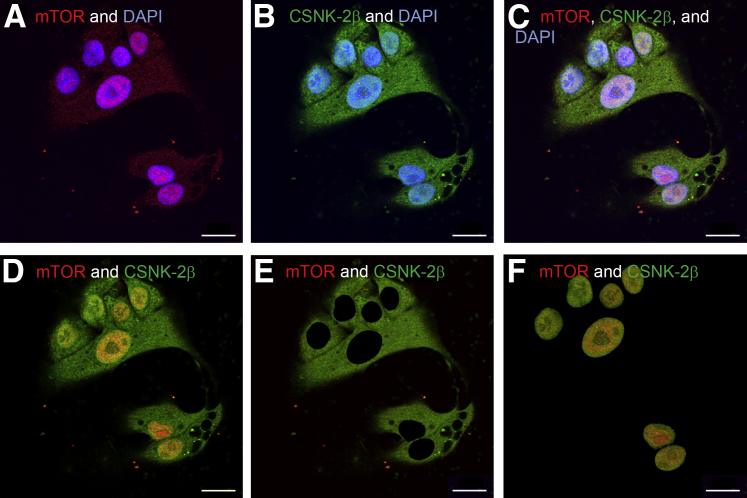
We have previously reported that inhibition of CSNK-2 reduces IGFBP-1 phosphorylation and that IGFBP-1 phosphorylation is increased after mTOR inhibition.7 Together these observations indirectly link mTOR with CSNK-2, suggesting that mTOR may regulate CSNK-2 to modulate IGFBP-1 phosphorylation. To determine the suggested link, it was first determined whether CSNK-2β specifically co-localizes with mTOR. CSNK-2β was present throughout the cell (Figure 1). To determine the subcellular localization of mTOR, dual-immunofluorescence staining of mTOR with CSNK-2β was performed. With the use of anti-mouse mTOR and anti-rabbit CSNK-2β antibodies (Figure 4A), mTOR was localized predominantly in the nucleus, as indicated by the DAPI-stained area. CSNK-2β was present across the cell (Figure 4B). Visualizing the dual immunofluorescent staining overlap suggested a strong co-localization of mTOR and CSNK-2β within the nucleus (Figure 4C), which is best observed when the DAPI signal was removed from the multichannel image (Figure 4, D–F) because it obscures visualization of the nuclear mTOR and CSNK-2β signals. Using Image Pro Premier 3D software, cytosolic and nuclear images depicting mTOR and CSNK-2β staining were extracted. With this method, there was almost no evidence for cytosolic co-localization of mTOR and CSNK-2β (Figure 4E), whereas there was strong co-localization of mTOR and CSNK-2β visualized by yellow in the nucleus (Figure 4F). These data indicate that mTOR and CSNK-2β co-localize mainly in the nucleus in HepG2 cells.
Figure 4.
Dual immunofluorescence staining for the co-localization of mechanistic target of rapamycin (mTOR) and casein kinase (CSNK)-2β. Human hepatocellular carcinoma cells were stained with anti-mouse mTOR and anti-rabbit CSNK-2β antibodies. Corresponding secondary antibodies were Alexa anti-mouse 568 and Alexa anti-rabbit 660, respectively. Images were captured via confocal microscopy. A: mTOR (red) is present throughout the cell but most abundant in the nucleus. B: CSNK-2β (green) is found throughout the cell. C: Merged channel image shows co-localization (yellow). D: Because DAPI staining obscured the depiction of yellow co-localization, dual-immunofluorescence staining is visualized without DAPI. E: To determine the cytosolic co-localization of mTOR and CSNK-2β, we subtracted the positive DAPI staining area from the whole cell image. F: With the use of the positive DAPI stain, nuclear co-localization of mTOR and CSNK-2β is observed. Scale bars: 20 μm.
Co-Localization of IGFBP-1 and CSNK-2β and of CSNK-2β and mTOR as Determined by Immunoprecipitation
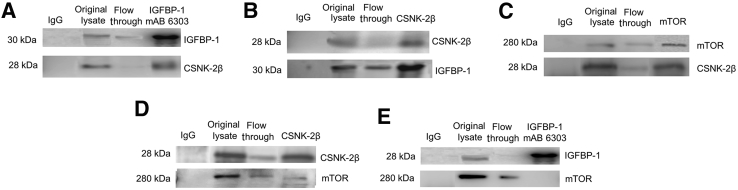
To confirm that interactions between CSNK-2 and IGFBP-1 occur in HepG2 cells under physiologic conditions, immunoprecipitation of IGFBP-1 and the regulatory (β) subunit of CSNK-2 was performed using conventional co-immunoprecipitation. Immunoprecipitation of HepG2 cell lysates using IGFBP-1 mAb 6303 showed that IGFBP-1 immunoprecipitated CSNK-2β (Figure 5A). Furthermore, CSNK-2β antibody also co-immunoprecipitated IGFBP-1 (Figure 5B), corroborating the merged immunofluorescence IHC data with overlapping signals demonstrated in yellow (Figure 1C). Using preimmune IgG, neither IGFBP-1 nor CSNK-2β was detected in the immunoprecipitates. These data indicate that the regulatory subunit CSNK-2β is associated with IGFBP-1.
Figure 5.
Co-immunoprecipitation suggests interactions between insulin-like growth factor binding protein (IGFBP)-1 and casein kinase (CSNK)-2β as well as mechanistic target of rapamycin (mTOR) and CSNK-2 in human hepatocellular carcinoma (HepG2) cell lysate. A: HepG2 cell lysates were immunoprecipitated using IGFBP-1 [monoclonal antibody (mAb) 6303] antibody. The immunoprecipitated proteins were tested on Western blot for IGFBP-1 using IGFBP-1 polyclonal antibody and CSNK-2β antibody. Immunoprecipitation with mAb 6303 co-immunoprecipitates CSNK-2β. B: HepG2 cell lysates were immunoprecipitated using CSNK-2β antibody. The immunoprecipitated proteins were tested on Western blots for IGFBP-1 using IGFBP-1 polyclonal antibody or CSNK-2β. Immunoprecipitation with CSNK-2β co-immunoprecipitates IGFBP-1. C: HepG2 cell lysates were immunoprecipitated using anti-human mTOR monoclonal antibody. The immunoprecipitated proteins were tested on Western blots for mTOR and CSNK-2β. Immunoprecipitation with mTOR antibody co-immunoprecipitates CSNK-2β. D: HepG2 cell lysates were immunoprecipitated using CSNK-2β antibody. The immunoprecipitated proteins were tested on Western blot for mTOR and CSNK-2β. Immunoprecipitation with CSNK-2β co-immunoprecipitates mTOR. E: HepG2 cell lysates were immunoprecipitated using IGFBP-1 (mAb 6303) antibody. The immunoprecipitated proteins were tested on Western blot for mTOR and IGFBP-1. Immunoprecipitation with IGFBP-1 mAb 6303 does not co-immunoprecipitate mTOR.
Whether mTOR immunoprecipitated the CSNK-2β subunit was further examined using co-immunoprecipitation. Immunoprecipitation of HepG2 cell lysates using the mTOR antibody showed that mTOR immunoprecipitated CSNK-2β (Figure 5C). Similarly, CSNK-2β antibody co-immunoprecipitated mTOR (Figure 5D). The relatively lower intensity of the mTOR band in reverse immunoprecipitation suggests a possibly lower capacity of the CSNK-2β subunit to interact with mTOR. In control immunoprecipitation, neither mTOR nor CSNK-2β was detected. Furthermore, there was no co-immunoprecipitation between IGFBP-1 and mTOR (Figure 5E). Together these data provide evidence that the regulatory subunit CSNK-2β is associated with IGFBP-1 and with mTOR, whereas mTOR is not associated with IGFBP-1.
PLA Demonstrates Proximity of IGFBP-1 with CSNK-2β and mTOR with CSNK-2β but Not of mTOR with IGFBP-1
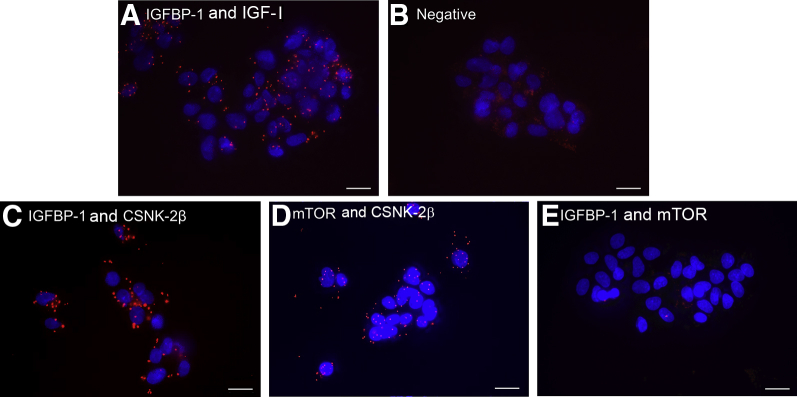
Next, PLA was used as a complementary approach to demonstrate proximity (<40 nm) among IGFBP-1, CSNK-2β, and mTOR in HepG2 cells. As a positive control for the PLA, IGFBP-1 and its ligand IGF-I were used (Figure 6A). No positive PLA interaction was detected in the absence of any primary antibodies (Figure 6B). The proximity between IGFBP-1 and CSNK-2β was then examined with mAb IGFBP-1 and rabbit polyclonal CSNK-2β antibody. Consistent with IHC data (Figure 1), IGFBP-1 and CSNK-2β interaction was observed (Figure 6C). PLA signal was also detected for mTOR and CSNK-2β (Figure 6D). On the other hand, no PLA signals for IGFBP-1 and mTOR were observed (Figure 6E). These data are consistent with the interpretation that there is a direct interaction between IGFBP-1 and CSNK-2β and between CSNK-2β and mTOR, whereas IGFBP-1 does not interact with mTOR directly.
Figure 6.
Interactions of insulin-like growth factor (IGF) binding protein (IGFBP)-1 with mechanistic target of rapamycin (mTOR) and casein kinase (CSNK)-2β visualized by proximity ligation assay (PLA). Representative PLA images where the PLA signal (red) represents close proximity (<40 nm) between two proteins. A: IGFBP-1 interaction with its ligand IGF-I serving as a positive control. B: Without primary antibodies, no PLA signal is observed. C and D: Representative PLA images of IGFBP-1 and CSNK-2β (C) and mTOR and CSNK-2β (D) showing a distinct PLA signal, suggesting interaction among these proteins. E: No signal is visible when PLA targeted IGFBP-1 and mTOR, suggesting minimal interaction between these two proteins. Scale bars: 20 μm.
Three-Dimensional Rendering Reveals Perinuclear and/or Cytosolic PLA Signals for CSNK-2β and IGFBP-1 as Well as for CSNK-2β and mTOR
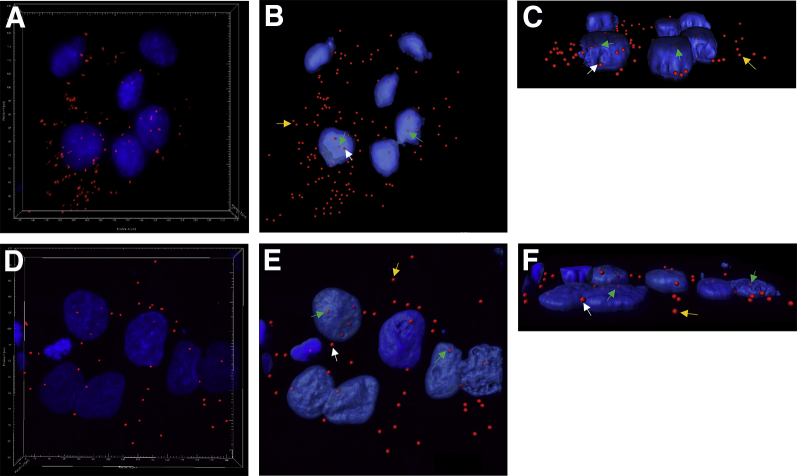
Because dual immunofluorescence co-localization of mTOR and CSNK-2β was detected predominantly in the nucleus (Figure 4), three-dimensional rendering of PLA images was used to clearly determine the subcellular location of the PLA signals relative to the nuclear membrane. Figure 7 shows the rendered three-dimensional views of PLA images of CSNK-2β and IGFBP-1 (Figure 7A), with transparent (Figure 7B) and three-dimensional rendered views (Figure 7C), compared with CSNK-2β and mTOR (Figure 7D) with transparent (Figure 7E) and three-dimensional rotation (Figure 7F). The three-dimensional rotation and clipping plane demonstrates that the PLA signal is located within the nucleus, in the perinuclear region, and in the cytosol. These images demonstrate that there is an interaction between CSNK-2β and IGFBP-1 outside the nucleus. Similarly, CSNK-2β and mTOR signals were predominantly observed in the extranuclear compartment (Figure 7, D–F). Notably, although most of the PLA signals for CSNK-2β and mTOR were extranuclear (Figure 7, E and F), a smaller population of PLA interactions were demonstrated in the intranuclear compartment as revealed in both the transparent (Figure 7E) and rotated (Figure 7F) views. Detection of PLA signals within the nucleus and in the extranuclear region for CSNK-2β and mTOR suggests that CSNK-2β and mTOR may have multiple roles in the cell, and it is likely that their action may be occurring in a variety of locations.
Figure 7.
Three-dimensional rendering of proximity ligation assay (PLA) signals. Representative images of PLA targeting casein kinase (CSNK)-2β and insulin-like growth factor binding protein (IGFBP)-1 in human hepatocellular carcinoma (HepG2) cells demonstrate that the signal is predominantly extranuclear: A: Confocal image of PLA signal from CSNK-2β and IGFBP-1. B: Transparent iso-surface of nuclear outline reveals PLA signals in cytoplasmic area. C: Rotated IGFBP-1/CSNK-2β. Representative images of CSNK-2β and mechanistic target of rapamycin (mTOR) interaction in HepG2 cells demonstrates that the PLA signal is predominantly extranuclear. D: Confocal image of PLA signals in CSNK-2β and mTOR. E: Transparent iso-surface of nuclear outline reveals most PLA spots in extra nuclear location relative to the nucleus. F: Rotated mTOR and CSNK-2β. White arrows indicate perinuclear PLA interactions; gold arrows, cytosolic interactions; and green arrows, nuclear interactions. Original magnification, ×40.
Hypoxia and Rapamycin Treatments Enhance PLA Signals for mTOR and CSNK-2β
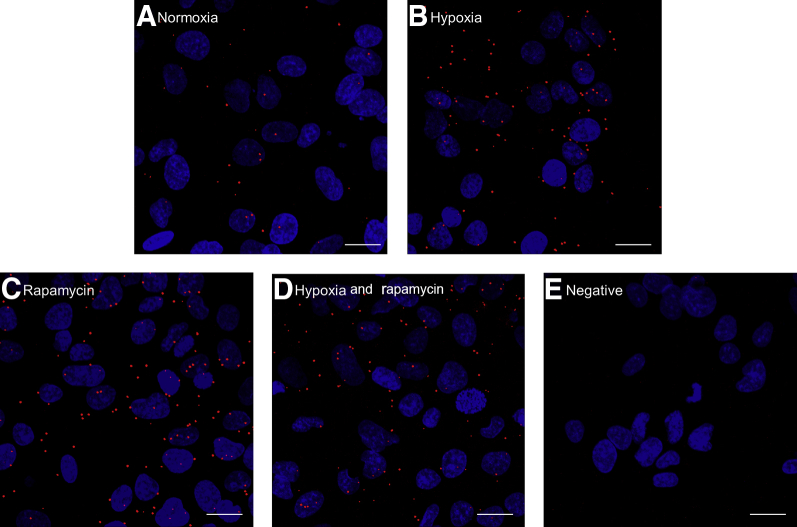
To determine the mechanistic relevance of PLA data, the PLA signals for mTOR with CSNK-2β were tested in the presence of rapamycin and hypoxia compared with normoxic conditions. In normoxia, only a weak signal for CSNK-2β and mTOR was detected (Figure 8A). Compared with normoxia (Figure 8A), PLA signals were increased in response to hypoxia and rapamycin treatment, respectively, as well as when hypoxia and rapamycin were combined (Figure 8, B–D). The PLA signals were specific because no signal was obtained when preimmune serum was used (Figure 8E) or after the omission of one or both primary antibodies (not shown). The PLA application used here thus supported the biological and mechanistic significance of CSNK-2β and mTOR interactions, suggesting that hypoxia promotes contact between mTOR and CSNK-2β in intact HepG2 cells.
Figure 8.
Effects of hypoxia, rapamycin, and hypoxia and rapamycin on the interaction between casein kinase (CSNK)-2β and mechanistic target of rapamycin (mTOR) as visualized by proximity ligation assay (PLA). Representative images of CSNK-2β and mTOR interaction in human hepatocellular carcinoma cells under normoxia (A), hypoxia (B), rapamycin (C), hypoxia and rapamycin (D), and negative control depicting no positive PLA interaction (E). Compared with control (normoxia), hypoxia, rapamycin, or combined hypoxia and rapamycin results in a stronger PLA signal, consistent with an increased interaction between these two proteins in these conditions. Scale bars: 20 μm.
PLA Shows Dynamic Changes in IGFBP-1 and CSNK-2β Proximity After mTOR Inhibition by Hypoxia and Rapamycin
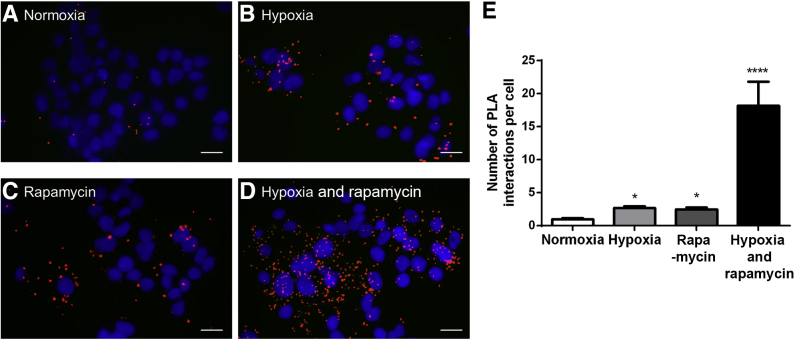
The effects of inhibition of mTOR by rapamycin in hypoxic and normoxic conditions on IGFBP-1 and CSNK-2β PLA interactions were further examined In normoxic conditions, only a weak PLA signal for IGFBP-1 and CSNK-2β was detected (Figure 9). On the other hand, PLA signal was increased in hypoxic and rapamycin treatments individually [approximately 2.5-fold (hypoxia P = 0.0001, rapamycin P = 0.0002)]. Significantly, the PLA signal increased markedly in cells grown under hypoxia and rapamycin combined compared with normoxia [approximately 18.0-fold (P = 0.0002)] (Figure 9, A–E). The PLA signals were specific for the IGFBP-1 and CSNK-2β complex because no signal was obtained when one or both primary antibodies were omitted (not shown). Therefore, we conclude that hypoxia promotes proximity between IGFBP-1 and CSNK-2β in intact cells and that mTOR inhibition and hypoxia act synergistically to markedly enhance the interaction between the two proteins.
Figure 9.
The change in casein kinase (CSNK)-2β and insulin-like growth factor binding protein (IGFBP)-1 interaction when exposed to hypoxia, rapamycin, and hypoxia and rapamycin treatments visualized by proximity ligation assay (PLA). A–D: Representative images of CSNK-2β and IGFBP-1 interaction in human hepatocellular carcinoma cells under normoxia (A), hypoxia (B), rapamycin (C), and hypoxia and rapamycin (D). E: Hypoxia combined with rapamycin treatment markedly increases CSNK-2β and IGFBP-1 interaction. Data are expressed as means ± SEM (E). ∗P < 0.05, ∗∗∗∗P < 0.0001. Scale bars: 20 μm.
SILAC/MRM-MS Shows Notable Enhancement of IGFBP-1 Phosphorylation in Hypoxia Combined with Rapamycin
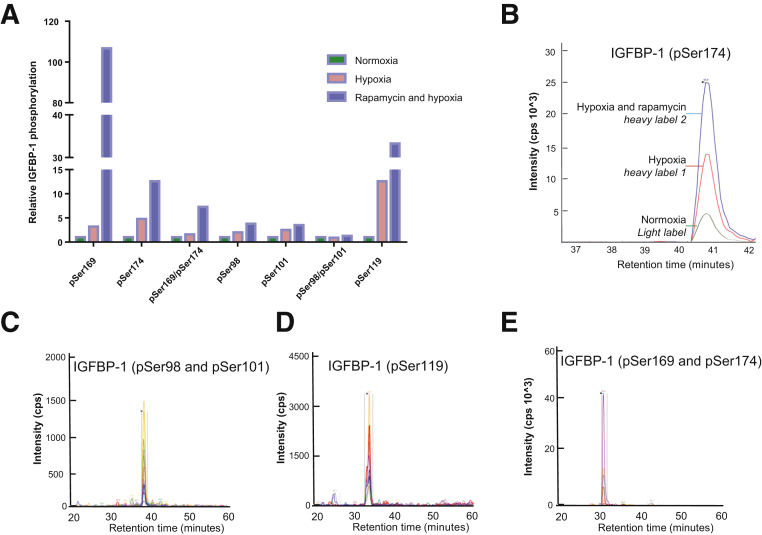
Considering that PLA showed a markedly increased signal for IGFBP-1 and CSNK-2β after hypoxia and rapamycin treatments, SILAC/MRM-MS (Figure 10) was used to confirm that hypoxia and rapamycin treatment also increases IGFBP-1 phosphorylation. SILAC/MRM-MS data confirmed that IGFBP-1 phosphorylation at multiple phosphorylation sites was increased under these conditions (Figure 11). In response to hypoxia, phosphorylation at the Ser169 and Ser174 sites increased by 3.2- and 4.8-fold, respectively, whereas the doubly phosphorylated peptide that contained pSer169 and pSer174 increased by 1.6-fold. Similarly, phosphorylation at the Ser98 and Ser101 sites increased by 2.0- and 2.5-fold, respectively, whereas phosphorylation at Ser119 increased 12.6-fold in hypoxia alone.
Figure 10.
Schematic of the stable isotope labeling with amino acids in cell culture (SILAC) experiment. The light and two heavy cell populations were treated with hypoxia or hypoxia and rapamycin combined and processed together to measure the heavy to light peptide signal ratios. HepG2, human hepatocellular carcinoma; IGFBP-1, insulin-like growth factor binding protein 1; m/z, mass to charge ratio; MRM-MS, multiple reaction monitoring–mass spectrometry.
Figure 11.
Stable isotope labeling with amino acids in cell culture (SILAC) combined with quantitative multiple reaction monitoring–mass spectrometry of insulin-like growth factor binding protein (IGFBP)-1 phosphorylation sites under hypoxia and rapamycin treatments. Human hepatocellular carcinoma cells were grown in regular Dulbecco's modified Eagle's medium/F12 media supplemented with 10% dialyzed fetal bovine serum (control) or grown in SILAC media that contained 5,5,5,D3-l-leucine or 1-1-13C-l-leucine stabile isotope labels. A: Histogram comparing the phosphopeptide intensity of each single or multiple IGFBP-1 phosphorylation sites among experimental conditions. The data are representative of one experiment from pooled 3 mL of media from the three treatments (unlabeled control, 5,5,5,D3-l-leucine–labeled hypoxia, and 1-13C-l-leucine–labeled hypoxia and rapamycin). B: Representative histogram of overlaid total peptide transitions from the three isotope labels and corresponding treatments for the novel IGFBP-1 Ser174 phosphorylated peptide. C–E: Examples of the extracted ion chromatographs showing the optimized transitions used to validate the phosphorylation at Ser98/Ser101 (C), Ser119 (D), and Ser169/Ser174 (E) in the singly or doubly phosphorylated peptides. All mass spectrometry was performed using QTRAP 4000 (AB Sciex, Concord, ON, Canada) with Q3 used as a linear ion trap. p-, phosphorylated.
SILAC/MRM-MS analysis showed that hypoxia and mTOR inhibition in combination much more notably increased IGFBP-1 phosphorylation at Ser169 (106-fold) (Figure 11A). In addition, phosphorylation at Ser174 was increased by 12.6-fold, whereas the doubly phosphorylated pSer169 and pSer174 peptide was enhanced by 7.3-fold. Similar to hypoxia, individual phosphorylation at Ser98 or Ser101 increased by 3.8- and 3.5-fold in response to combined hypoxia and rapamycin, respectively. Phosphorylation levels at Ser119 increased by 33.3-fold in response to hypoxia and rapamycin in combination.
Total peptide transitions from the three isotope labels (normoxia, hypoxia, and hypoxia and rapamycin) for the novel Ser174 phosphorylated IGFBP-1 peptide are depicted as a histogram (Figure 11B). The extracted ion chromatographs showing the optimized transitions used to validate the phosphorylation of IGFBP-1 at Ser98/Ser101, Ser119, and Ser169/Ser174 in the singly or doubly phosphorylated peptides are shown in Figure 11, C–E. Overall, these findings are consistent with PLA data showing that hypoxia and rapamycin promote contact between IGFBP-1 and CSNK-2β, suggesting that these conditions also increase the phosphorylation of IGFBP-1 at discrete sites. On the basis of these findings, we speculate that enhanced CSNK-2β recruitment to IGFBP-1 may result in increased site-specific phosphorylation of IGFBP-1.
Discussion
We provide novel evidence that IGFBP-1 and protein kinase CSNK-2β co-localize, adding to previous functional data that CSNK-2 is the key kinase responsible for the phosphorylation of IGFBP-1 in HepG2 cells.7 Dual-immunofluorescence staining shows that IGFBP-1 and CSNK-2β co-localize in the perinuclear region. With the use of Ki-67 cell proliferation marker, nonproliferating cells were found to express higher levels of IGFBP-1. Furthermore, we report for the first time that CSNK-2β co-localizes with mTOR in the nucleus, whereas IGFBP-1 staining does not coincide with mTOR. Interestingly, using PLA, the proximity amid CSNK-2β and IGFBP-1 was more prominent in the cytosolic compartment, which was consistent with mTOR and CSNK-2β, suggesting that mTOR-mediated CSNK-2 action (ie, phosphorylation of IGFBP-1) may be in the extranuclear region, which has not been previously reported. Importantly, a combination of mTOR inhibition and hypoxia resulted in a marked increase in PLA signals for IGFBP-1 and CSNK-2β, and these treatments also led to greater IGFBP-1 phosphorylation. Because it is believed that phosphorylation of IGFBP-1 in the fetal liver is critical in regulating IGF-I bioavailability and therefore modulating fetal growth, our findings are consistent with the model that CSNK-2 constitutes an important mechanistic link among mTOR, IGFBP-1 phosphorylation, and decreased IGF-I bioavailability. This mechanism may play a critical role in the development of FGR.
We previously reported that fetal liver mTOR signaling is inhibited, protein kinase CSNK-2 is activated, and IGFBP-1 phosphorylation is increased in a baboon model of FGR.7 To provide additional evidence to support a mechanistic link among mTOR signaling, CSNK-2, and IGFBP-1 phosphorylation, in the current report we investigated the subcellular localization of IGFBP-1, CSNK-2β, and mTOR in HepG2 cells. Although dual immunofluorescence imaging demonstrated significant cell-to-cell variation in the localization of IGFBP-1, possibly because of the variability in cell cycle stage, IGFBP-1 is predominantly localized in the perinuclear region in HepG2 cells. CSNK-2β was found to be ubiquitously present throughout the cell, in agreement with studies in other cell types.10, 24 We propose that CSNK-2β interacts with IGFBP-1 and plays a key role in the regulation of IGFBP-1 phosphorylation in HepG2 cells. Moreover, we speculate that CSNK-2 mediates the increased IGFBP-1 phosphorylation in the liver of FGR fetuses.7
Protein kinase CSNK-2 is regulated by several protein-protein interactions in different subcellular compartments, and the interaction of the CSNK-2 subunits can be controlled by close contact among their interacting surfaces.8, 25, 9 The subunit CSNK-2β can participate in the transient formation of distinct complexes.10 Consistent with these findings, in our study, CSNK-2β co-localized with IGFBP-1 and mTOR in HepG2 cells. Because IHC demonstrates co-localization rather than interaction, in situ PLA was used as a complementary strategy to establish a close proximity of proteins. PLA confirmed the close contact between IGFBP-1 and CSNK-2β and mTOR, with CSNK-2β corroborating the dual immunofluorescence findings. However, dual immunofluorescence staining for CSNK-2β and mTOR was most prominent in the nucleus. Using the rendered three-dimensional rotated views of the PLA images of CSNK-2β and mTOR (Figure 7F), we found that only a smaller population of PLA interactions were in the intranuclear compartment (Figure 7, B, C, E, and F). Most of the PLA signals for CSNK-2β and mTOR were present in the extranuclear compartment (Figure 7, B, C, E, and F). These data indicate that the interactions of CSNK-2β and mTOR likely occur predominantly in the extranuclear region, where CSNK-2β and IGFBP-1 PLA signals were detected. On the basis of these findings, we propose that although mTOR and CSNK-2β co-localize in the nucleus, the interaction largely occurs in the cytoplasm, providing plausibility for the specific role of CSNK-2β in mTOR-associated IGFBP-1 phosphorylation.
With IGFBP-1 being a secretory protein, without additional detailed studies it is difficult to determine the precise location where CSNK-2 interacts and phosphorylates IGFBP-1. Nonetheless, the PLA approach confirmed that IGFBP-1 and CSNK-2β are within close proximity (<40 nm), strongly supporting functional interaction between these two proteins. In addition, combined hypoxia and rapamycin treatments resulted in markedly amplified PLA signals for IGFBP-1 and CSNK-2β. Together these findings suggest that the CSNK-2β subunit and the nutrient-sensor mTOR physically interact, consistent with the hypothesis that CSNK-2β–mediated phosphorylation of IGFBP-1 represents a mechanistic link between nutrient sensing and IGF-I signaling.
Using quantitative proteomics, we have previously found that hypoxia and rapamycin increased phosphorylation at distinct IGFBP-1 serine residues.17 Using a MRM-MS strategy, we have also shown that CSNK-2 can directly phosphorylate these three serine residues on IGFBP-1.11 Here, using highly specific and sensitive quantitative SILAC/MRM-MS, we found an increase in IGFBP-1 phosphorylation at the same residues but with a >100-fold increase observed at Ser169 in response to hypoxia and rapamycin. Although our current data are consistent with the possibility that CSNK-2 activity is induced under these conditions, further investigation is necessary to confirm this assumption. These findings also provide strong basis to determine whether IGFBP-1 is a bona fide physiologic CSNK-2 substrate,26 which remains to be established. In addition, the regulation of CSNK-2 is enigmatic, and how its activity or function is controlled via mTOR needs detailed study. The interaction of mTOR and CSNK-2 in the fetal liver may be crucial in controlling fetal growth but also be involved in many other cellular processes, such as in cancer, obesity, diabetes, and autophagy.27, 28
In summary, using complementary methods, this study uncovered close contact between IGFBP-1 and CSNK-2β. These data also suggest that there is a direct interaction between the nutrient-sensing kinase mTOR and CSNK-2β. SILAC/MRM-MS–based quantitative mapping confirmed that the most distinct increase in IGFBP-1 phosphorylation occurred at Ser169, in response to combined hypoxia and mTOR inhibition by rapamycin. Hypoxia inhibits mTOR signaling29 and stimulates CSNK-2 activity.15 The synergistic effects of hypoxia and mTOR inhibition resulted in markedly increased CSNK-2β and IGFBP-1 interaction and IGFBP-1 phosphorylation, which reflects that hypoxia has effects on IGFBP-1 phosphorylation that are independent of mTOR. The phosphorylation of IGFBP-1 in the fetal liver is thought to be critical in regulating IGF-I bioavailability and therefore modulating fetal growth. Our findings are consistent with the model that CSNK-2 constitutes an important mechanistic link among mTOR, IGFBP-1 phosphorylation, and decreased IGF-I bioavailability, which may play a critical role in the development of FGR.
Acknowledgments
We thank Prof. David W. Litchfield (Department of Biochemistry, University of Western Ontario, London, ON, Canada) for his generous support in providing the necessary reagents and the facility for conducting studies with protein kinase CSNK-2; Dr. Robert Baxter (Kolling Institute of Medical Research, University of Sydney, Sydney, Australia) for his kind gift of IGFBP-1 polyclonal antibody; and Biotron Integrated Microscopy, University of Western Ontario, in aiding dual immunofluorescent and PLA image acquisition and analyses.
Footnotes
Supported by the Natural Science and Engineering Council of Canada Discovery grants RGPIN-2016-04752 and RGPIN-2015-05000 and Lawson Health Research Institute grant HRF0512 (M.B.G.) and, in part, NIH grant HD 078313 (M.B.G. and T.J.).
K.B. and M.A.S. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2017.09.009.
Supplemental Data
References
- 1.Han V.K., Matsell D.G., Delhanty P.J., Hill D.J., Shimasaki S., Nygard K. IGF-binding protein mRNAs in the human fetus: tissue and cellular distribution of developmental expression. Horm Res. 1996;45:160–166. doi: 10.1159/000184780. [DOI] [PubMed] [Google Scholar]
- 2.Jones J.I., Busby W.H., Jr., Wright G., Clemmons D.R. Human IGFBP-1 is phosphorylated on 3 serine residues: effects of site-directed mutagenesis of the major phosphoserine. Growth Regul. 1993;3:37–40. [Google Scholar]
- 3.Martina N.A., Kim E., Chitkara U., Wathen N.C., Chard T., Giudice L.C. Gestational age-dependent expression of insulin-like growth factor-binding protein-1 (IGFBP-1) phosphoisoforms in human extraembryonic cavities, maternal serum, and decidua suggests decidua as the primary source of IGFBP-1 in these fluids during early pregnancy. J Clin Endocrinol Metab. 1997;82:1894–1898. doi: 10.1210/jcem.82.6.3974. [DOI] [PubMed] [Google Scholar]
- 4.Abu Shehab M., Khosravi J., Han V.K., Shilton B.H., Gupta M.B. Site-specific IGFBP-1 hyper-phosphorylation in fetal growth restriction: clinical and functional relevance. J Proteome Res. 2010;9:1873–1881. doi: 10.1021/pr900987n. [DOI] [PubMed] [Google Scholar]
- 5.Nissum M., Abu Shehab M., Sukop U., Khosravi J.M., Wildgruber R., Eckerskorn C., Han V.K., Gupta M.B. Functional and complementary phosphorylation state attributes of human insulin-like growth factor-binding protein-1 (IGFBP-1) isoforms resolved by free flow electrophoresis. Mol Cell Proteomics. 2009;8:1424–1435. doi: 10.1074/mcp.M800571-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta M.B. The role and regulation of IGFBP-1 phosphorylation in fetal growth restriction. J Cell Commun Signal. 2015;9:111–123. doi: 10.1007/s12079-015-0266-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abu Shehab M., Damerill I., Shen T., Rosario F.J., Nijland M., Nathanielsz P.W., Kamat A., Jansson T., Gupta M.B. Liver mTOR controls IGF-I bioavailability by regulation of protein kinase CK2 and IGFBP-1 phosphorylation in fetal growth restriction. Endocrinology. 2014;155:1327–1339. doi: 10.1210/en.2013-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litchfield D.W. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J. 2003;369:1–15. doi: 10.1042/BJ20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meggio F., Pinna L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 10.Pinna L.A. Protein kinase CK2: a challenge to canons. J Cell Sci. 2002;115:3873–3878. doi: 10.1242/jcs.00074. [DOI] [PubMed] [Google Scholar]
- 11.Malkani N., Biggar K., Shehab M.A., Li S.S., Jansson T., Gupta M.B. Increased IGFBP-1 phosphorylation in response to leucine deprivation is mediated by CK2 and PKC. Mol Cell Endocrinol. 2016;425:48–60. doi: 10.1016/j.mce.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faust M., Montenarh M. Subcellular localization of protein kinase CK2: a key to its function? Cell Tissue Res. 2000;301:329–340. doi: 10.1007/s004410000256. [DOI] [PubMed] [Google Scholar]
- 13.Veis A., Sfeir C., Wu C.B. Phosphorylation of the proteins of the extracellular matrix of mineralized tissues by casein kinase-like activity. Crit Rev Oral Biol Med. 1997;8:360–379. doi: 10.1177/10454411970080040101. [DOI] [PubMed] [Google Scholar]
- 14.Cao J.Y., Shire K., Landry C., Gish G.D., Pawson T., Frappier L. Identification of a novel protein interaction motif in the regulatory subunit of casein kinase 2. Mol Cell Biol. 2014;34:246–258. doi: 10.1128/MCB.00968-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mottet D., Ruys S.P., Demazy C., Raes M., Michiels C. Role for casein kinase 2 in the regulation of HIF-1 activity. Int J Cancer. 2005;117:764–774. doi: 10.1002/ijc.21268. [DOI] [PubMed] [Google Scholar]
- 16.Popovici R.M., Lu M., Bhatia S., Faessen G.H., Giaccia A.J., Giudice L.C. Hypoxia regulates insulin-like growth factor-binding protein 1 in human fetal hepatocytes in primary culture: suggestive molecular mechanisms for in utero fetal growth restriction caused by uteroplacental insufficiency. J Clin Endocrinol Metab. 2001;86:2653–2659. doi: 10.1210/jcem.86.6.7526. [DOI] [PubMed] [Google Scholar]
- 17.Damerill I., Biggar K.K., Abu Shehab M., Li S.S., Jansson T., Gupta M.B. Hypoxia increases IGFBP-1 phosphorylation mediated by mTOR inhibition. Mol Endocrinol. 2016;30:201–216. doi: 10.1210/me.2015-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mounier C., Dumas V., Posner B.I. Regulation of hepatic insulin-like growth factor-binding protein-1 gene expression by insulin: central role for mammalian target of rapamycin independent of forkhead box O proteins. Endocrinology. 2006;147:2383–2391. doi: 10.1210/en.2005-0902. [DOI] [PubMed] [Google Scholar]
- 19.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seferovic M.D., Gupta M.B. Increased umbilical cord PAI-1 levels in placental insufficiency are associated with fetal hypoxia and angiogenesis. Dis Markers. 2016;2016:7124186. doi: 10.1155/2016/7124186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seferovic M.D., Chen S., Pinto D.M., Gupta M.B. Altered liver secretion of vascular regulatory proteins in hypoxic pregnancies stimulate angiogenesis in vitro. J Proteome Res. 2011;10:1495–1504. doi: 10.1021/pr100879z. [DOI] [PubMed] [Google Scholar]
- 22.Seferovic M.D., Ali R., Kamei H., Liu S., Khosravi J.M., Nazarian S., Han V.K., Duan C., Gupta M.B. Hypoxia and leucine deprivation induce human insulin-like growth factor binding protein-1 hyperphosphorylation and increase its biological activity. Endocrinology. 2009;150:220–231. doi: 10.1210/en.2008-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 24.Peracchia G., Jensen A.B., Culianez-Macia F.A., Grosset J., Goday A., Issinger O.G., Pages M. Characterization, subcellular localization and nuclear targeting of casein kinase 2 from Zea mays. Plant Mol Biol. 1999;40:199–211. doi: 10.1023/a:1006196530079. [DOI] [PubMed] [Google Scholar]
- 25.Olsten M.E., Weber J.E., Litchfield D.W. CK2 interacting proteins: emerging paradigms for CK2 regulation? Mol Cell Biochem. 2005;274:115–124. doi: 10.1007/s11010-005-3072-6. [DOI] [PubMed] [Google Scholar]
- 26.Turowec J.P., Duncan J.S., French A.C., Gyenis L., St Denis N.A., Vilk G., Litchfield D.W. Protein kinase CK2 is a constitutively active enzyme that promotes cell survival: strategies to identify CK2 substrates and manipulate its activity in mammalian cells. Methods Enzymol. 2010;484:471–493. doi: 10.1016/B978-0-12-381298-8.00023-X. [DOI] [PubMed] [Google Scholar]
- 27.Jung C.H., Ro S.H., Cao J., Otto N.M., Kim D.H. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montenarh M. Protein kinase CK2 and angiogenesis. Adv Clin Exp Med. 2014;23:153–158. doi: 10.17219/acem/37040. [DOI] [PubMed] [Google Scholar]
- 29.Foster K.G., Fingar D.C. Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J Biol Chem. 2010;285:14071–14077. doi: 10.1074/jbc.R109.094003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.