Abstract
Flap endonuclease 1 (FEN1) plays a crucial role in both DNA replication and damage repair. In this study, FEN1 expression and its clinical-pathologic significance in non–small-cell lung cancer (NSCLC) was investigated. Quantitative RT-PCR and immunohistochemistry analysis identified that both FEN1 mRNA and protein were highly overexpressed in about 36% of 136 cancer tissues compared to adjacent tissues, in which FEN1 was generally undetectable. Notably, patients with FEN1-overexpressed cancers were prone to have poor differentiation and poor prognosis. A strong positive correlation between the levels of FEN1 and Ki-67 staining was identified in these NSCLC tissues (r = 0.485), suggesting overexpressed FEN1 conferred a proliferative advantage to NSCLC. Furthermore, knockdown of FEN1 resulted in G1/S or G2/M phase cell cycle arrest and suppressed in vitro cellular proliferation in NSCLC cancer cells. Consistently, a selective FEN1 inhibitor was shown to effectively inhibit cellular proliferation of NSCLC cells in a dose-dependent manner. Additionally, knockdown of FEN1 significantly attenuated homologous DNA repair efficiency and enhanced cytotoxic effects of cisplatin in NSCLC cells. Taken together, these findings have indicated that overexpressed FEN1 represents a prognostic biomarker and potential therapeutic target for NSCLC treatment, which warrants further study.
Lung cancer is the leading cause of cancer-related death worldwide. Non–small-cell lung cancer (NSCLC) accounts for 85% of lung cancers. Although there have been advances in targeted therapies and even immunotherapy, the 5-year survival rate for NSCLC remains only 15%.1, 2 Clearly, new approaches are required for the development of more effective therapies for this devastating disease. DNA damage has been long recognized as a causal factor for cancer development, and inappropriate DNA repair may lead to malignant transformation of cells through inactivation of tumor suppressors or activation of oncogenes,3 whereas enhanced DNA repair in cancers have been associated with treatment resistance.4, 5
Flap endonuclease-1 (FEN1) is an important member of the structure-specific nuclease family that participates in numerous DNA pathways, including Okazaki fragment maturation, stalled replication fork rescue, telomere maintenance, long-patch base excision repair, and apoptotic DNA fragmentation, in almost all organisms.6, 7, 8 Base excision repair is one of the main methods of DNA damage repair and thus is important in the process of cancer development.9 Due to its pivotal role in DNA repair, FEN1 functions as a key enzyme in maintaining genomic stability and protecting against carcinogenesis.10, 11, 12 In agreement with its essential role in genomic stability, deficiency of FEN1 has been shown to predispose to cancers.13, 14 Zheng et al11, 12 previously identified several loss-of-function FEN1 mutations in human cancer specimens, and exemplified mice with a FEN1 mutant developed spontaneous lung cancer at high frequency at their late life stages. This notion was further supported by the clinical identification of two FEN1 69G>A and 4150G>T single nucleotide polymorphisms that were reproducibly associated with lower FEN1 expression, increased DNA damage, and high risk in hepatocellular carcinoma, esophageal cancer, gastric and colorectal cancer, and lung cancer.15, 16
Several studies have revealed that FEN1 was highly expressed in breast cancer tissues,17 lung, testis, and brain tumors,18 as well as prostate cancer.19 Wang et al20 reported that overexpression of FEN1 in gastric cancer was correlated with tumor size, lymphatic metastasis, and degree of differentiation. Another recent study showed that FEN1 overexpression was associated with high grade, high stage, and poor survival in breast and ovarian epithelial cancer.21 Moreover, Wang et al22 recently observed that the level of FEN1 was inversely correlated with cancer drug and radiation resistance and with survivorship in breast cancer patients. Another study showed that down-regulation of FEN1 expression in glioma cells increased the cells' sensitivity to methyl methane-sulfonate and temozolomide damage.18 Interestingly, FEN1 has recently been reviewed as one of the deregulated DNA damage response proteins in 15 human cancers.23 Thus, FEN1 potentially represents a novel therapeutic target, and targeting FEN1 may benefit broad-spectrum cancer and current chemotherapies.24, 25 Although a FEN1-specific inhibitor is not clinically available, searching for effective FEN1 inhibitors is underway.23, 25 Notably, a small molecular compound, FEN1 inhibitor SC13, was recently developed and has shown cytotoxic and inhibitory activity in human breast cancer in a mouse model.26
Significant roles of FEN1 in human NSCLC have not been well investigated. This study examined FEN1 expression and its correlation with malignant characteristics of NSCLC patients such as grade, stage, survival, and sensitivity to chemotherapy drugs. The findings have indicated that overexpressed FEN1 may represent a prognostic biomarker and potential target in NSCLC treatment, warranting future further study.
Materials and Methods
Patients and Clinical Data Collection
The study was reviewed and approved by the institutional review board of the City of Hope National Medical Center (Duarte, CA). A total of 154 patients with pathologically diagnosed NSCLC, including adenocarcinoma and squamous cell carcinoma lung cancers, who received primary and curative surgical operations without neoadjuvant treatment between 2002 and 2014 were recruited in the study. The details of their demographic and clinicopathologic characteristics are summarized in Table 1.
Table 1.
The Correlation between Clinicopathologic Characteristics and FEN1 IHC Staining in NSCLC
| Variable∗ | FEN1 IHC score, n (%) |
P value | |||
|---|---|---|---|---|---|
| − | + | ++ | +++ | ||
| Age group, years | |||||
| ≤60 | 24 (28) | 4 (29) | 3 (18) | 7 (39) | 0.5777 |
| >60 | 63 (72) | 10 (71) | 14 (82) | 11 (61) | |
| Sex | |||||
| Male | 27 (31) | 8 (57) | 6 (35) | 10 (56) | 0.0939 |
| Female | 60 (69) | 6 (43) | 11 (65) | 8 (44) | |
| Tobacco history | |||||
| Never used | 16 (25) | 0 (0) | 1 (8) | 1 (7) | 0.1603 |
| Previous used | 13 (20) | 0 (0) | 4 (33) | 4 (27) | |
| Current used | 36 (55) | 7 (100) | 7 (58) | 10 (67) | |
| Histology | |||||
| Adenocarcinoma | 81 (94) | 13 (93) | 16 (94) | 16 (89) | 0.6708 |
| SCC | 1 (1) | 1 (7) | 0 (0) | 1 (6) | |
| Adenosquamous | 4 (5) | 0 (0) | 1 (6) | 1 (6) | |
| Differentiation | |||||
| Well differentiated | 22 (27) | 2 (14) | 0 (0) | 0 (0) | 0.0120 |
| Moderately differentiated | 35 (43) | 4 (29) | 8 (47) | 12 (67) | |
| Poorly differentiated | 25 (30) | 8 (57) | 9 (53) | 6 (33) | |
| Node status | |||||
| Node negative | 28 (64) | 3 (75) | 3 (60) | 7 (100) | 0.2686 |
| Node positive | 16 (36) | 1 (25) | 2 (40) | 0 (0) | |
| Recurrence | |||||
| No | 62 (74) | 11 (85) | 10 (59) | 15 (88) | 0.2002 |
| Yes | 22 (26) | 2 (15) | 7 (41) | 2 (12) | |
| Ki-67 average | |||||
| Negative <1% | 49 (59) | 8 (57) | 5 (29) | 4 (22) | 0.0006 |
| Moderate 1%–<5% | 22 (27) | 3 (21) | 5 (29) | 1 (6) | |
| High 5%–25% | 9 (11) | 2 (14) | 5 (29) | 9 (50) | |
| Higher >25% | 3 (4) | 1 (7) | 2 (12) | 4 (22) | |
−, <1%; +, 1% to <5%; ++, 5% to 25%; +++, >25%; IHC, immunohistochemistry; SCC, squamous cell carcinoma.
Pearson χ2 test.
siRNA Transfection, Cell Proliferation, and Cell Cycle Analysis
Human lung cancer cell lines A549, H1299, and H460 were purchased from ATCC (Manassas, VA) in 2014, and authenticated by DNA short tandem repeat profiling analysis in October 2016. The human FEN1 siRNA (sc-36338) and control siRNA (sc-36338) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). For siRNA transfection, a total of 1.5 × 105 cells/well was seeded in 6-well plates, and transfected with 45 pmol of FEN1 siRNA or control siRNA according to the protocol of lipofectamine RNAiMAX (Thermo Fisher Scientific, Waltham, MA). Cell proliferation was measured at 72 hours post-transfection or 96 hours post-transfection if the cells were treated with chemotherapy at 24 hours post-transfection. Cell proliferation was monitored at 72 hours using the colorimetric MTT assay (CellTiter 96 AQueous One Solution Cell Proliferation Assay; Promega Corporation, Madison, WI). For cell cycle analysis, cells transfected with control or FEN1 siRNA 72 hours were stained with propidium iodide staining solution and analyzed by a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Data were acquired with Cell Quest software version 5.1 (BD Biosciences, Franklin Lakes, NJ), and the percentages of G1/G0, S, and G2/M phase cells were calculated with MODFIT LT software version 3.3 (BD Biosciences).
Cisplatin Treatment and Apoptosis Analysis
A549 and H460 cells were transected with siRNA for 24 hours, followed by exposure to various doses of cisplatin (5 to 20 μmol/L) for a defined period. Cytotoxicity of FEN1 inhibitor SC13 (a generous gift from Dr. Zhigang Guo, Nanjing Normal University, Nanjing, China) and cisplatin was determined by the MTT assay. For apoptotic assays, cisplatin-induced apoptosis was measured by flow cytometry analysis of Alexa Fluor 488 Annexin-V and propidium iodide staining according to the manufactory's kit protocol (Thermo Fisher Scientific). Meanwhile, total proteins were also extracted from these cells for Western blot analysis of DNA damage and the apoptotic proteins markers, including p53 and cleaved caspase-3.
Quantitative RT-PCR
Total RNA was extracted from paired normal and malignant lung tissues using TRIzol Reagent (Thermo Fisher Scientific) according to the user protocol. cDNA was synthesized from 0.5 μg of total RNA using the Superscript III First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative RT-PCR was conducted using SYBR Green PCR Master Mix (Thermo Fisher Scientific) to measure the FEN1 mRNA level in paired adjacent normal and tumor tissues, and β-actin was used as an internal control for mRNA expression, respectively. Data were presented as the relative quantity of targets, normalized with respect to the internal control, or relative to a calibrator sample.
Immunohistochemistry and Western Blot Analysis
Mouse monoclonal anti-FEN1 antibody from GeneTex (San Antonio, TX) and rat monoclonal anti–Ki-67 antibody clone MIB-1 (M7240) from Dako (Glostrup, Denmark) were used for immunohistochemistry (IHC) analysis. IHC analyses were performed as described previously.27 Quantitative analysis of IHC staining was used to assess the scanned images on the basis of the percentage of positively stained cells and staining intensity. FEN1 or Ki-67 stained tissues were scored as negative −, +, ++, or +++ if <1%, 1% to <5%, 5% to 25%, or >25%, respectively, of cancer cells in a tissue displayed positive cytoplasmic or nuclear staining.27 Antibodies against p21, p53, total caspase-3, cleaved caspase-3, and phosphorylated histone H2AX at Ser139 (γH2AX) were purchased from Cell Signaling Technology (Danvers, MA). About 20 μg of total protein was used for Western blot analysis.
DNA Repair Assays
Reporter cell lines for green fluorescent protein (GFP)-based DNA damage repair assays were established by stable transfection of A549 cells with the pimEJ5GFP reporter plasmid for non-homologous end joining (NHEJ)28 and the pHPRT-DRGFP reporter plasmid for homologous repair (HR),29 respectively, and selected with 0.3 μg/mL puromycin. The resultant A549/H460-EJ5GFP and A549/H460-DRGFP cells were first transfected with control or FEN1 siRNA for 24 hours, and then transiently transfected with a predetermined mixture of pCBA-Scel plasmid to express I-Scel endonuclease and a plasmid to express DsRed (mCherry-A) protein that served as the control for transfection efficiency. After culturing for another 48 hours, 5 × 105 cells per transfection were analyzed by fluorescence-activated cell sorting to count total GFP and DsRed protein–positive cells. Each assay was performed three times, and data were presented as the ratio of GFP-positive to DsRed-positive cells among whole cells.
Data Management and Statistical Analysis
Group comparisons for continuous data were conducted using t-tests or χ2 tests, and for quantitative variables were analyzed with the Wilcoxon signed-rank test or the Spearman rank correlation test. Kaplan-Meier analysis was used to assess survival. Log-rank tests were used to compare patient overall survival and time to tumor recurrence between subgroups. The results of the cell function experiments were presented as the means ± SEM. Statistical significance was set at P < 0.05.
Results
Overexpression of FEN1 in NSCLC Tissues
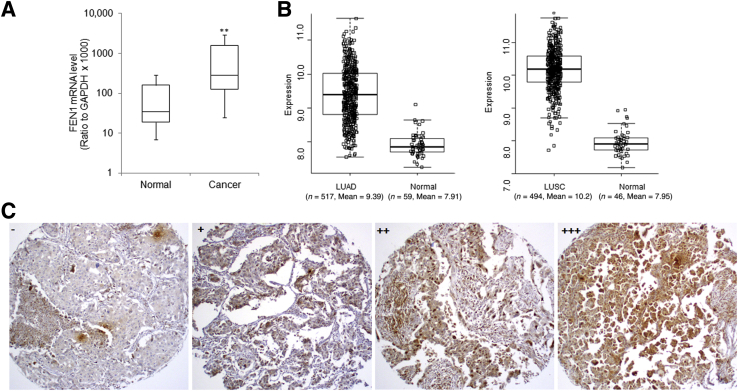
FEN1 mRNA expression in paired normal and NSCLC tissues was examined, and the results of quantitative RT-PCR showed that the average FEN1 mRNA level was significantly higher in tumor sections than that in the matched adjacent normal tissues (P < 0.05; n = 83) (Figure 1A). Consistently, FEN1 expression in lung cancer was also mined using mRNA expression data for both lung adenocarcinoma and lung squamous carcinoma from The Cancer Genome Atlas (https://cancergenome.nih.gov, last accessed January 8, 2016). Analysis revealed that FEN1 mRNA was significantly increased in both lung adenocarcinoma and lung squamous carcinoma samples by 2.8- and 4.7-fold compared with normal lung tissues (P = 3.1 × 10−59, P = 2.23 × 10−46, respectively) (Figure 1B).
Figure 1.
Overexpression of FEN1 in NSCLC tissues. A: Box-and-whisker plots for relative FEN1 mRNA expression in about 82 paired normal and NSCLC tissues measured by quantitative RT-PCR analysis. FEN1 mRNA in tumor samples normalized to that of adjacent normal sections. B: FEN1 mRNA expression in NSCLC based on The Cancer Genome Atlas data, the left panel for lung adenocarcinoma (LUAD), the right panel for lung squamous carcinoma (LUSC). The upper and lower limits of the boxes and the lines inside the boxes indicate the 75th and 25th percentiles and the median, respectively. C: Representative photomicrographs of FEN1 immunohistochemical staining in four NSCLC sections scored as −, +, ++, or +++ for <1%, 1% to <5%, 5% to 25%, or >25% positively stained tumor cells, respectively. ∗∗P < 0.01 versus normal. Original magnification, ×100 (C).
FEN1 protein levels were measured in about 136 NSCLC by IHC analysis. Representative FEN1 immunostaining of NSCLC tissues is shown in Figure 1C as a dominantly nuclear staining pattern in both tumor and normal tissues. Quantitative IHC analysis of FEN1 protein showed that the percentages of NSCLC tissues in which FEN1 staining were scored as −, +, ++, or +++ were 64.0% (87 of 136), 10.3% (14 of 136), 12.5% (17 of 136), and 13.2% (18 of 136), respectively, demonstrating FEN1 protein was substantially up-regulated in more than 36% of NSCLC tissues, whereas FEN1 was almost undetectable in the corresponding normal tissues.
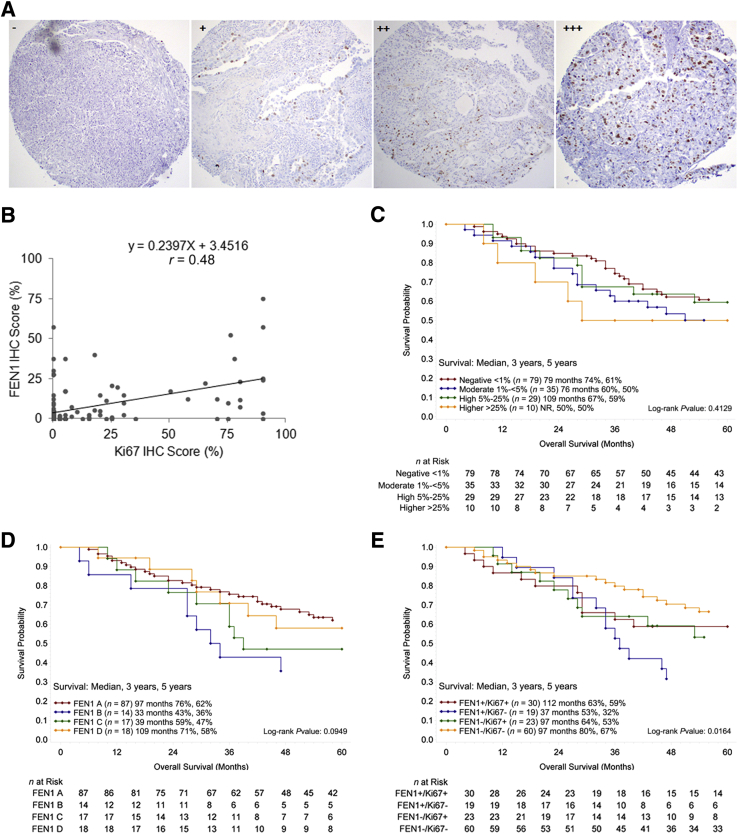
FEN1 Correlates with Enhanced Cell Proliferation and Poor Prognosis of NSCLC
The protein level of FEN1 significantly correlated to poor differentiation of NSCLC tissues (P = 0.012) (Table 1). The percentages of FEN1-positive cases in the well, moderately, and poorly differentiated groups were 8.3%, 40.7%, and 48.2%, respectively. The expression of Ki-67, a well-accepted factor of proliferation and poor prognosis for survival in NSCLC,30 was also measured by IHC staining (Figure 2A). Interestingly, a strong positive correlation between the protein levels of FEN1 and Ki-67 was identified in these NSCLC tissues (r = 0.485) (Figure 2B), suggesting overexpressed FEN1 conferred a proliferative advantage to NSCLC. Although log-rank test analysis showed that the overall survival rates among patients with differentially expressed Ki-67 (P = 0.4129) (Figure 2C) and FEN1 (P = 0.0929) (Figure 2D) were not significantly different, patients with FEN1-negative cancers tended to survive longer than patients with FEN1-positive cancers. All of NSCLC patients were further divided into four subgroups based on Ki-67 and FEN1 levels: double negative, single positive for either protein, or double positive by log-rank analysis, and the results indicated up-regulation of both Ki-67 and FEN1 was significantly related to poorer overall survival in NSCLC patients (P = 0.016) (Figure 2E). Therefore, the above findings indicated that overexpression of FEN1 was a potentially unfavorable factor to prognosis of NSCLC patients, and a combination of Ki-67 and FEN1 might better predict the prognosis in NSCLC patients.
Figure 2.
Overexpression of FEN1 is positively correlated with Ki-67 expression and poor prognosis of NSCLC. A: Representative photomicrographs of Ki-67 IHC staining in four NSCLC sections scored as −, +, ++, or +++ for <1%, 1% to <5%, 5% to 25%, or >25% positively stained tumor cells, respectively. B: The Pearson correlation coefficient analysis plot of the expression of FEN1 and Ki-67 in NSCL tissues demonstrate a strong positive correlation (r = 0.48; P < 0.01). C–E: Log-rank tests for overall survival of NSCLC patients based on IHC scores of Ki-67 (C), FEN1 (IHC scores: FEN1 A, −; FEN1 B, +; FEN1 C, ++; FEN1 D, +++) (D), and Ki-67 + FEN1 (E). The data show those patients with Ki-67–positive and FEN1-positive NSCLC exhibit significantly shorter survival. n = 136 (B). Original magnification, ×100 (A). IHC, immunohistochemistry; NR, not reached.
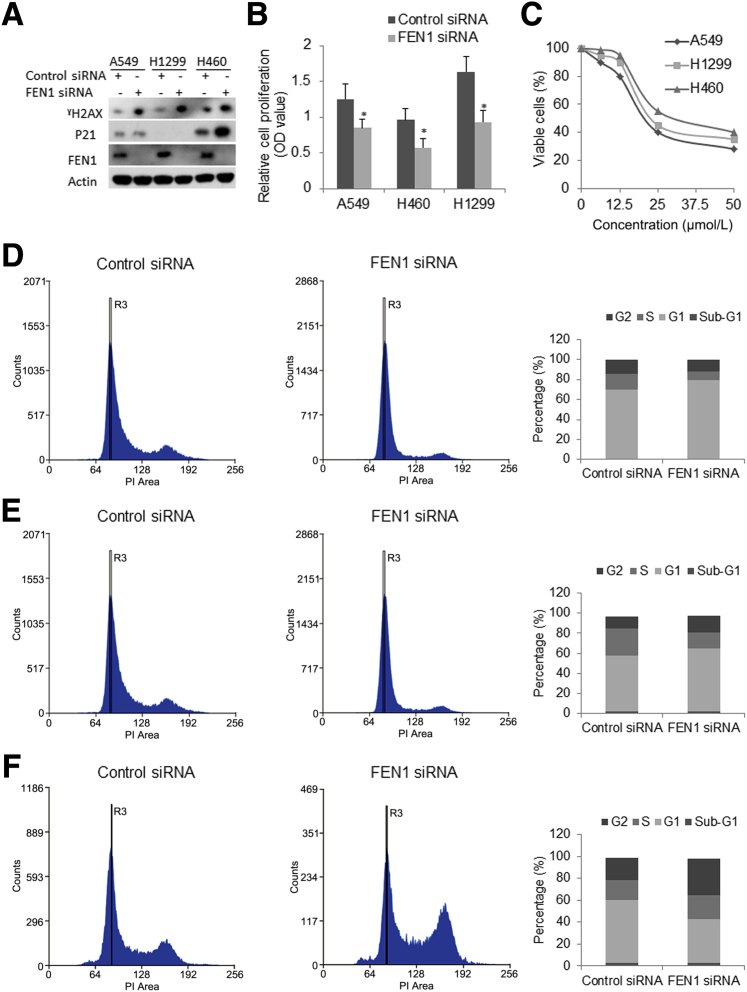
Knockdown of FEN1 Suppresses Cell Proliferation and Causes Cell Cycle Arrest in Lung Cancer Cells
The impact of siRNA-mediated FEN1 knockdown on in vitro cellular proliferation in NSCLC cell lines was examined. As demonstrated by Western blot analysis, FEN1 protein was completely depleted and γH2AX was increased in three human lung cell lines transfected with FEN1 siRNA, whereas P21 (CIP1/WAF1) was dramatically increased in A549 and H460 compared with cells transfected with control siRNA (Figure 3A). MTT proliferation assay showed that attenuation of FEN1 resulted in a significant decrease of in vitro cellular proliferation in both FEN1 siRNA–transfected cancer cells compared with controls transfected with scrambled siRNA over 72 hours (Figure 3B). The cytotoxic effect of a newly developed selective FEN1 inhibitor, SC13,26 in NSCLC cell lines was also examined. SC13 effectively suppressed cellular proliferation of the NSCLC cells in a dose-dependent manner (Figure 3C). The half maximal inhibitory concentration of the FEN1 inhibitor for the cells was around 20 to 30 μmol/L. Whether FEN1 knockdown would promote cell death in these lung cancer cells was further tested. Flow cytometry cell cycle analysis showed that knockdown of FEN1 slightly increased the sub-G1 population of all three cell lines transfected with FEN1-specific siRNA over 72 hours post-transfection. This was also supported by the elevated γH2AX. By using flow cytometry analysis, cell cycle change upon FEN1 knockdown in these three cells at 96 hours post-transfection of control siRNA or FEN1 siRNA was examined. Quantitative results specifically showed a dramatic increase in the G1 phase arrest in two p53–wild-type cancer cells, A549 (Figure 3D) and H460 (Figure 3E), and a dramatic increase in G2 phase arrest in p53-null H1299 cells (Figure 3F) transfected with FEN1 siRNA compared to control siRNA, suggesting FEN1 knockdown caused both G1/S and G2/M phase cell cycle arrests in these NSCLC cells.
Figure 3.
Knockdown of FEN1 suppresses in vitro cellular proliferation and causes cell cycle arrest in lung cancer cells. A: Western blot analysis for FEN1, P21, γH2AX protein in A549, H1299, and H460 at 72 hours post-transfection of control or FEN1 siRNA. B: Cell proliferation assay of A549, H1299, and H460 cells transfected with equal amounts of either control siRNA or FEN1 siRNA. Shown are the cell proliferation index for each measured by MTT assay at 72 hours after transfection. C: Cell viability assays of A549, H1299, and H838 treated with a selective FEN1 inhibitor for 72 hours. D–F: Representative cell cycle flow charts upon knockdown of FEN1 for A549 (D), H460 (E), and H1299 (F) cells. The lower histogram of cell cycle analysis shows the cell percentage in each phase of these cells transfected with control siRNA or FEN1 siRNA. ∗P < 0.05 compared with control siRNA. OD, optical density; PI, propidium iodide.
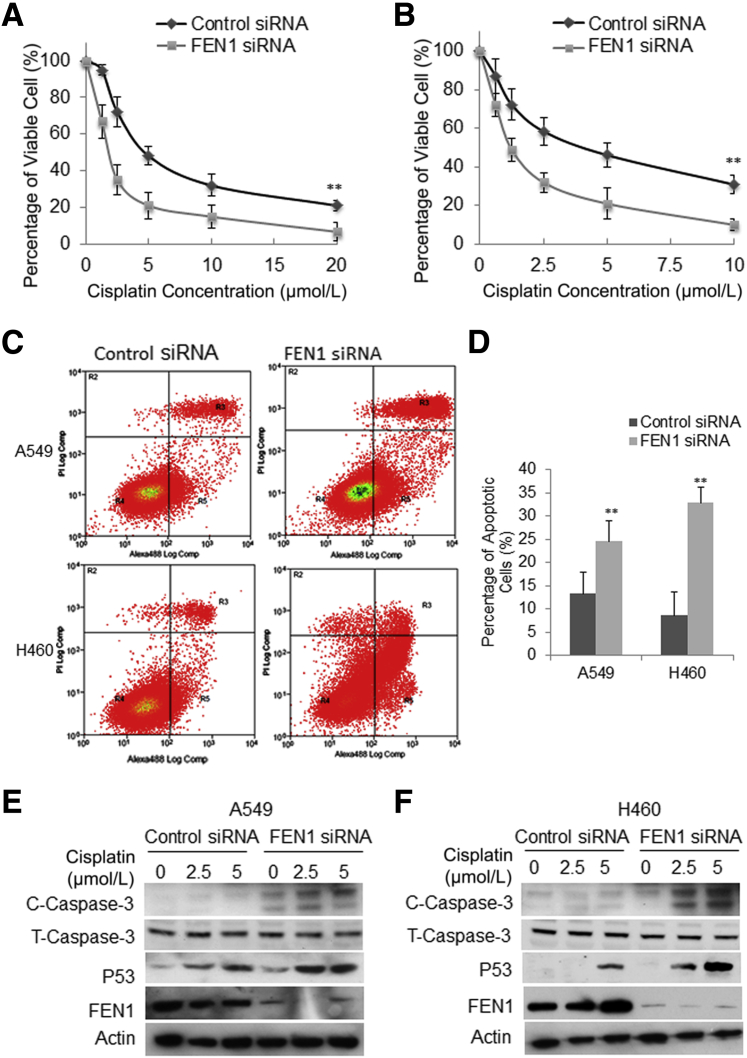
Considering the key function of FEN1 in DNA repair, it is pertinent to speculate that the high level of FEN1 expression contributes to intrinsic or acquired drug resistance in cancers. To gain insight into the effect of FEN1 on cell death by anticancer drugs and the therapeutic significance of FEN1, whether FEN1 knockdown would enhance the cell death caused by in vitro exposure to cisplatin was examined. The combined effect was tested on A549 and H460 cells that were treated with FEN1 siRNA or control siRNA for 24 hours, followed by exposure to various doses of cisplatin for a further 72 hours on A549 (0, 1.25, 2.5, 5, 10, 20 μmol/L) and H460 (0, 0.625, 1.25, 2.5, 5, 10 μmol/L) cells. Addition of FEN1 siRNAs to cisplatin-treated cells significantly reduced survival of A549 cells (Figure 4A) and H460 cells (Figure 4B) compared to control siRNA-treated cells. Annexin V flow cytometry analysis showed the percentage of annexin (+) cells increased in a linear manner with increasing cisplatin dose up to 10 to 20 μmol/L. Consistently, quantitative analysis showed that exposure of A549 transfected with FEN1 siRNA at 5 μmol/L cisplatin led to about 30% apoptotic cells, whereas the same concentration of cisplatin led to only about 12% of apoptotic cells in A549 cells transfected with the control siRNA (Figure 4, C and D). Similarly, exposure of 2.5 μmol/L cisplatin killed 8% of H460 cells transfected with control siRNA, whereas it killed about 36% of H460 cells transfected with FEN1 siRNA (Figure 4, C and D). In addition, Western blot analysis further demonstrated that knockdown of FEN1 induced higher levels of cleaved caspase-3 and P53 in both A549 cells (Figure 4E) and H460 cells (Figure 4F). The above data clearly showed knockdown of FEN1 significantly enhanced cisplatin-induced apoptosis in lung cancer cells, indicating alterations in the FEN1 level may have an impact on the cancer cell response to anticancer drugs, and FEN1 suppression may have the potential to be used in lung cancer treatment.
Figure 4.
Knockdown of FEN1 significantly enhances cisplatin-induced apoptosis in lung cancer cells. A and B: The viability of A549 (A) and H460 (B) cells was assessed after 72 hours of exposure to serially diluted cisplatin (for A549, from 20 to 1.25 μmol/L; for H460, from 10 to 0.625 μmol/L) and 24 hours pre-transfection with control or FEN1 siRNA. Cell survival was measured by MTT and expressed as a percentage of that in untreated cells. Error bars are representative of triplicated experiments. C: Representative flow cytometry charts for A549 and H460 24 hours pre-transfection with control or FEN1 siRNA, and then further treated with 5 μmol/L cisplatin for 72 hours. D: Quantitative analysis of apoptotic cells upon FEN1 knockdown. Error bars are representative of three individual treated samples of two duplicates. E and F: Western blot analysis of cisplatin-induced cleaved caspase 3 and P53 in A549 (E) and H460 (F) cells transfected with control or FEN1 siRNA. ∗∗P < 0.01 FEN1 siRNA versus control siRNA. C-Caspase-3, cleaved caspase-3; Log Comp, log compensation; PI, propidium iodide; T-Caspase-3, total caspase-3.
Effect of FEN1 Knockdown on DNA Double-Strand Break Repair
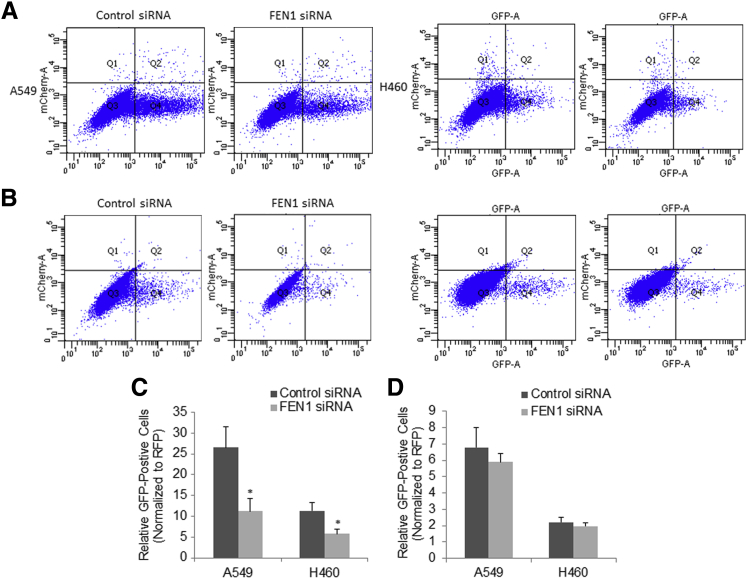
Chemoradiation induces DNA double-strand breaks (DSBs) in the context of chemotherapy-associated DNA aberrations/replication stress, and inhibits the growth and triggers the apoptotic death of cancer cells. DSBs can be repaired either by the HR or NHEJ pathways. A previous study showed that FEN1 eliminates heterologous sequences at DNA damage sites and facilitates DSB DNA repair by HR.31 Therefore, the effects of FEN1 depletion on both HR and NHEJ in lung cancer cells were further examined. Previous studies have shown that the DR-GFP and EJ5-GFP reporter assays could be used to measure the ability of HR and NHEJ repair, respectively.28 We then sought to elucidate the role of FEN1 in DSB DNA repair, using integrated DR-GFP (Figure 5A) and EJ5-GFP (Figure 5B) reporters in A549 and H460 cells. In this system, transient expression of I-Scel induces DSBs, which if repaired, results in the generation of GFP-positive cells. GFP signal indicated successful DSB repair, whereas red fluorescent protein signal indicated successful transfection. Consistent with the role of FEN1 as an essential component of HR repair, knockdown of FEN1 significantly suppressed the efficiency of HR (P < 0.05) (Figure 5C), but only slightly decreased NHEJ repair (Figure 5D) in A549 and H460 cells as shown by the reduction in the percentage of GFP+ cells in FEN1 siRNA-transfected DR-GFP assays. These data suggest that FEN1 depletion impaired HR repair pathways that may contribute to the accumulation of DSBs and enhanced genotoxic drug-induced apoptosis in lung cancers.
Figure 5.
Effect of knockdown of FEN1 on HR and NHEJ repair in lung cancer cells. A and B: Representative flow cytometry charts of A549-DRGFP/H460-DRGFP cells for HR reporter assay (A), and A549-EJ5GFP/H460-EJ5GFP cells for NHEJ repair reporter assay (B). C and D: Quantitative analysis of HR reporter (C) and NHEJ reporter (D) assays shows significant decrease of the relative HR reporter activity in both A549 and H460 upon knockdown of FEN1. Error bars are representative of three individual treated samples of two experimental duplicates. ∗P < 0.05 FEN1 siRNA versus control siRNA. RFP, red fluorescent protein.
Discussion
Previous studies showed that the expression of FEN1 was shut down in terminally differentiated cells.32 It has been indicated that elevated FEN1 in different types of cancers, including breast cancer17 and prostate cancer,33 may reflect a greater proliferation rate of cancer cells compared to normal cells. An early study on FEN1 knockout mice showed that FEN1 protein was required for S phase entry in trophoblast giant cells, and the FEN1−/− cells were unable to enter S phase and were arrested in the endocycle during embryonic development.34 Another early study showed that FEN1 was highly expressed by cycling cells and that it colocalizes with PCNA and polymerase alpha during the S phase. Consistent with the essential role of FEN1 in cellular proliferation, in the current study, we found up-regulation of FEN1 was positively associated with cell proliferation, and knockdown of FEN1 significantly inhibited cellular proliferation and induced significant cell cycle arrest at G1/S or G2/M transitions. In agreement with the observation in other cancers,20, 21 we showed for the first time that FEN1 overexpression was associated with poor differentiation and a trend to poor survival in NSCLC.
FEN1 plays a critical role in DNA repair; it has been proven that an up-regulated FEN1 contributes to intrinsic or acquired drug resistance, whereas depletion of FEN1 may potentially cause DNA damage to be unrepaired. For example, an early study demonstrated that FEN1 was induced by genotoxic stress in mouse fibroblasts and that overexpression of FEN1 attenuated UV light–induced DNA replication inhibition.35 Additionally, FEN1 and the nuclear excision repair (NER) protein XPG show homology in the DNA-binding domain, suggesting overexpression of FEN1 may confer resistance by enhanced NER.36 Because FEN1 is frequently overexpressed in various human cancers, it is clinically significant and interesting to examine whether targeting FEN1 has therapeutic benefits. Cisplatin is one of the most potent and useful antitumor agents for lung cancer that interacts with cellular DNA to form cross links that ultimately inhibit DNA replication and transcription, and if not repaired, lead to DNA breaks and cell death via the apoptotic pathway.37 In the current study, transient FEN1 knockdown did not result in dramatic apoptosis in these lung cancer cells, however, it significantly sensitized lung cancer cells to cisplatin. Repair of cisplatin–DNA intrastrand lesions is largely catalyzed by NER and long-patch base excision repair pathways.38 FEN1 was also shown to play a role in the final step of NER in a complex with ligase I.39 Therefore, FEN1 depletion enhanced cytotoxic effect of cisplatin in lung cancer cells.
Depletion FEN1 may also enhance cytotoxic effects of other chemoradiotherapy approaches. For example, Nikolova et al18 showed that knockdown of FEN1 by siRNA increased sensitivity to the methylating agent temozolomide and to cisplatin in LN308 glioma cells. Similarly, exposure of FEN1−/− blastocysts to gamma radiation caused extensive apoptosis, implying an essential role for FEN1 in the repair of radiation-induced DNA damage in vivo.34 A study also revealed that insect cells lacking FEN1 endonuclease were viable, but hypersensitive to DNA-damaging agents.40 Interestingly, another study showed that breast cancer cells with low FEN1 were much more sensitive to induction of apoptosis via polyamine depletion compared to breast cancer cells with a higher expressed FEN1.41 DNA double-strand breaks can arise spontaneously during the processing of DNA adducts, or single-strand breaks can arise through DNA repair or replication processes.42 A previous study showed FEN1 eliminates heterologous sequences at DNA damage site and facilitates DNA repair by HR.31 In our study, using HR and NHEJ report assays, we showed that FEN1 depletion hampers HR repair pathways in lung cancer cells. Interestingly, a recent study showed that small-molecule inhibitors of FEN1 selectively killed cancer cells with different genotypes, indicating FEN1 as a broad-spectrum target for anticancer therapeutic developments.24 Therefore, targeting FEN1 might have an impact on a broad range of DNA damage repair pathways and enhance the cytotoxic effects of multiple anticancer drugs. In conclusion, we found frequent overexpression of FEN1 was associated with poor prognosis of NSCLC patients. We further demonstrated that knockdown of FEN1 suppressed cancer proliferation and sensitized lung cancer cells to cisplatin chemotherapy. Therefore, FEN1 poses a marker of tumor progression and potential therapeutic target for NSCLC.
Acknowledgments
We thank the Baum Family Foundation for this research, Michael Lewallen and Lucy Brown (City of Hope National Medical Center) for immunohistochemistry staining, Lucy Brown for flow cytometry analysis, Dr. Zhigang Guo (Nanjing Normal University) for the generous gift of FEN1 inhibitor, and Prof. Binghui Shen (City of Hope National Medical Center) for critically reviewing the manuscript.
Footnotes
Supported in part by the V Foundation (D.J.R.) and the NIH National Cancer Institute grants 5K12CA001727-20 (D.J.R.) and P30CA33572 (Bioinformatics, Pathology, and Analytical Cytometry Cores).
Disclosures: None declared.
Contributor Information
Keqiang Zhang, Email: kzhang@coh.org.
Jae Y. Kim, Email: jaekim@coh.org.
References
- 1.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R., Ma J., Zou Z., Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Hakem R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008;27:589–605. doi: 10.1038/emboj.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fojo T. Cancer, DNA repair mechanisms, and resistance to chemotherapy. J Natl Cancer Inst. 2001;93:1434–1436. doi: 10.1093/jnci/93.19.1434. [DOI] [PubMed] [Google Scholar]
- 5.Parker R.J., Eastman A., Bostick-Bruton F., Reed E. Acquired cisplatin resistance in human ovarian cancer cells is associated with enhanced repair of cisplatin-DNA lesions and reduced drug accumulation. J Clin Invest. 1991;87:772–777. doi: 10.1172/JCI115080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christmann M., Tomicic M.T., Roos W.P., Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 7.Klungland A., Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waga S., Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 9.Zheng L., Jia J., Finger L.D., Guo Z., Zer C., Shen B. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 2011;39:781–794. doi: 10.1093/nar/gkq884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen B., Singh P., Liu R., Qiu J., Zheng L., Finger L.D., Alas S. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 2005;27:717–729. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- 11.Zheng L., Dai H., Hegde M.L., Zhou M., Guo Z., Wu X., Wu J., Su L., Zhong X., Mitra S., Huang Q., Kernstine K.H., Pfeifer G.P., Shen B. Fen1 mutations that specifically disrupt its interaction with PCNA cause aneuploidy-associated cancer. Cell Res. 2011;21:1052–1067. doi: 10.1038/cr.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng L., Dai H., Zhou M., Li M., Singh P., Qiu J., Tsark W., Huang Q., Kernstine K., Zhang X., Lin D., Shen B. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- 13.Wu Z., Lin Y., Xu H., Dai H., Zhou M., Tsao S., Zheng L., Shen B. High risk of benzo[alpha]pyrene-induced lung cancer in E160D FEN1 mutant mice. Mutat Res. 2012;731:85–91. doi: 10.1016/j.mrfmmm.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu H., Zheng L., Dai H., Zhou M., Hua Y., Shen B. Chemical-induced cancer incidence and underlying mechanisms in Fen1 mutant mice. Oncogene. 2011;30:1072–1081. doi: 10.1038/onc.2010.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L., Zhou C., Zhou L., Peng L., Li D., Zhang X., Zhou M., Kuang P., Yuan Q., Song X., Yang M. Functional FEN1 genetic variants contribute to risk of hepatocellular carcinoma, esophageal cancer, gastric cancer and colorectal cancer. Carcinogenesis. 2012;33:119–123. doi: 10.1093/carcin/bgr250. [DOI] [PubMed] [Google Scholar]
- 16.Yang M., Guo H., Wu C., He Y., Yu D., Zhou L., Wang F., Xu J., Tan W., Wang G., Shen B., Yuan J., Wu T., Lin D. Functional FEN1 polymorphisms are associated with DNA damage levels and lung cancer risk. Hum Mutat. 2009;30:1320–1328. doi: 10.1002/humu.21060. [DOI] [PubMed] [Google Scholar]
- 17.Singh P., Yang M., Dai H., Yu D., Huang Q., Tan W., Kernstine K.H., Lin D., Shen B. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol Cancer Res. 2008;6:1710–1717. doi: 10.1158/1541-7786.MCR-08-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikolova T., Christmann M., Kaina B. FEN1 is overexpressed in testis, lung and brain tumors. Anticancer Res. 2009;29:2453–2459. [PubMed] [Google Scholar]
- 19.LaTulippe E., Satagopan J., Smith A., Scher H., Scardino P., Reuter V., Gerald W.L. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 20.Wang K., Xie C., Chen D. Flap endonuclease 1 is a promising candidate biomarker in gastric cancer and is involved in cell proliferation and apoptosis. Int J Mol Med. 2014;33:1268–1274. doi: 10.3892/ijmm.2014.1682. [DOI] [PubMed] [Google Scholar]
- 21.Abdel-Fatah T.M., Russell R., Albarakati N., Maloney D.J., Dorjsuren D., Rueda O.M., Moseley P., Mohan V., Sun H., Abbotts R., Mukherjee A., Agarwal D., Illuzzi J.L., Jadhav A., Simeonov A., Ball G., Chan S., Caldas C., Ellis I.O., Wilson D.M., 3rd, Madhusudan S. Genomic and protein expression analysis reveals flap endonuclease 1 (FEN1) as a key biomarker in breast and ovarian cancer. Mol Oncol. 2014;8:1326–1338. doi: 10.1016/j.molonc.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J., Zhou L., Li Z., Zhang T., Liu W., Liu Z., Yuan Y.C., Su F., Xu L., Wang Y., Zhou X., Xu H., Hua Y., Wang Y.J., Zheng L., Teng Y.E., Shen B. YY1 suppresses FEN1 over-expression and drug resistance in breast cancer. BMC Cancer. 2015;15:50. doi: 10.1186/s12885-015-1043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearl L.H., Schierz A.C., Ward S.E., Al-Lazikani B., Pearl F.M. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer. 2015;15:166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]
- 24.van Pel D.M., Barrett I.J., Shimizu Y., Sajesh B.V., Guppy B.J., Pfeifer T., McManus K.J., Hieter P. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 2013;9:e1003254. doi: 10.1371/journal.pgen.1003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ba S., Zhang H., Lee J.Y., Wu H., Ye R., Huang D., Li T. Investigation of human flap structure-specific endonuclease 1 (FEN1) activity on primer-template models and exploration of a substrate-based FEN1 inhibitor. Bioorg Med Chem. 2016;24:1988–1992. doi: 10.1016/j.bmc.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 26.He L., Zhang Y., Sun H., Jiang F., Yang H., Wu H., Zhou T., Hu S., Kathera C.S., Wang X., Chen H., Li H., Shen B., Zhu Y., Guo Z. Targeting DNA flap endonuclease 1 to impede breast cancer progression. EBioMedicine. 2016;14:32–43. doi: 10.1016/j.ebiom.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K., Chu K., Wu X., Gao H., Wang J., Yuan Y.C., Loera S., Ho K., Wang Y., Chow W., Un F., Chu P., Yen Y. Amplification of FRS2 and activation of FGFR/FRS2 signaling pathway in high-grade liposarcoma. Cancer Res. 2013;73:1298–1307. doi: 10.1158/0008-5472.CAN-12-2086. [DOI] [PubMed] [Google Scholar]
- 28.Bennardo N., Cheng A., Huang N., Stark J.M. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierce A.J., Hu P., Han M., Ellis N., Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin B., Paesmans M., Mascaux C., Berghmans T., Lothaire P., Meert A.P., Lafitte J.J., Sculier J.P. Ki-67 expression and patients survival in lung cancer: systematic review of the literature with meta-analysis. Br J Cancer. 2004;91:2018–2025. doi: 10.1038/sj.bjc.6602233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kikuchi K., Taniguchi Y., Hatanaka A., Sonoda E., Hochegger H., Adachi N., Matsuzaki Y., Koyama H., van Gent D.C., Jasin M., Takeda S. Fen-1 facilitates homologous recombination by removing divergent sequences at DNA break ends. Mol Cell Biol. 2005;25:6948–6955. doi: 10.1128/MCB.25.16.6948-6955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim I.S., Lee M.Y., Lee I.H., Shin S.L., Lee S.Y. Gene expression of flap endonuclease-1 during cell proliferation and differentiation. Biochim Biophys Acta. 2000;1496:333–340. doi: 10.1016/s0167-4889(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 33.Lam J.S., Seligson D.B., Yu H., Li A., Eeva M., Pantuck A.J., Zeng G., Horvath S., Belldegrun A.S. Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score. BJU Int. 2006;98:445–451. doi: 10.1111/j.1464-410X.2006.06224.x. [DOI] [PubMed] [Google Scholar]
- 34.Larsen E., Gran C., Saether B.E., Seeberg E., Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol Cell Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christmann M., Tomicic M.T., Origer J., Kaina B. Fen1 is induced p53 dependently and involved in the recovery from UV-light-induced replication inhibition. Oncogene. 2005;24:8304–8313. doi: 10.1038/sj.onc.1208994. [DOI] [PubMed] [Google Scholar]
- 36.Herrero A.B., Martin-Castellanos C., Marco E., Gago F., Moreno S. Cross-talk between nucleotide excision and homologous recombination DNA repair pathways in the mechanism of action of antitumor trabectedin. Cancer Res. 2006;66:8155–8162. doi: 10.1158/0008-5472.CAN-06-0179. [DOI] [PubMed] [Google Scholar]
- 37.Henkels K.M., Turchi J.J. Induction of apoptosis in cisplatin-sensitive and -resistant human ovarian cancer cell lines. Cancer Res. 1997;57:4488–4492. [PubMed] [Google Scholar]
- 38.Koberle B., Grimaldi K.A., Sunters A., Hartley J.A., Kelland L.R., Masters J.R. DNA repair capacity and cisplatin sensitivity of human testis tumour cells. Int J Cancer. 1997;70:551–555. doi: 10.1002/(sici)1097-0215(19970304)70:5<551::aid-ijc10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 39.Mocquet V., Laine J.P., Riedl T., Yajin Z., Lee M.Y., Egly J.M. Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. EMBO J. 2008;27:155–167. doi: 10.1038/sj.emboj.7601948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuzaki Y., Adachi N., Koyama H. Vertebrate cells lacking FEN-1 endonuclease are viable but hypersensitive to methylating agents and H2O2. Nucleic Acids Res. 2002;30:3273–3277. doi: 10.1093/nar/gkf440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johansson V.M., Miniotis M.F., Hegardt C., Jonsson G., Staaf J., Berntsson P.S., Oredsson S.M., Alm K. Effect of polyamine deficiency on proteins involved in Okazaki fragment maturation. Cell Biol Int. 2008;32:1467–1477. doi: 10.1016/j.cellbi.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 42.Akyuz N., Boehden G.S., Susse S., Rimek A., Preuss U., Scheidtmann K.H., Wiesmuller L. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol Cell Biol. 2002;22:6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]