Abstract
Protein phosphorylation is a dynamic post-translational modification. Mass spectrometry–based quantitation was performed to determine the phosphoproteome profile of epithelial cells in response to injury, nucleotide, or epidermal growth factor. Phosphotyrosine enrichment used immunoprecipitation and immobilized metal affinity chromatography. Nucleotides released after scratch wounding activate purinergic receptors, leading to a distinct phosphorylation profile on epidermal growth factor receptor (EGFR) compared with its natural ligand. ATP induced a 2- to 15-fold phosphorylation increase over control on EGFR Y974, Y1086, and Y1148, with minimal phosphorylation intensity on EGFR Y1173 compared with the level measured in response to epidermal growth factor. Differential phosphorylation induced by epidermal growth factor or ATP was site specific on Src, Shc, phospholipase Cγ, protein kinase C, focal adhesion kinase, paxillin, and mitogen-activated protein kinases 1, 12, and 13. After wounding, the P2Y2 receptor mRNA expression increased, and after knockdown, migration and Ca2+ mobilization were impaired. To examine phosphorylation mediated by P2Y2, cells were cultured in media containing stable isotope-labeled amino acids, the receptor was knocked down, and the cells were stimulated. Mass spectrometry–based comparison of the phosphorylation profiles of control versus transfected cells revealed a 50-fold decrease in phosphorylation of EGFR Y974 and 1086, with no decrease in Y1173 phosphorylation. A similarfold decrease in Src Y421 and Y446 and paxillin Y118 was detected, indicating the far-reaching importance of the P2Y2 receptor in mediating migration.
Physical injury, mechanical stimuli, and cellular stress induce the release of endogenous nucleotides to the extracellular milieu.1, 2, 3 These nucleotides mediate a wide range of signaling on activation of purinergic receptors, which have two distinct classes: the ligand-gated P2X receptors and the G-coupled P2Y receptors. To date, eight P2Y receptor subtypes (P2Y1, 2, 4, 6, and 11–14) have been cloned.4, 5 The receptor subtypes (P2Y1, 2, 4, 6, and 11) are expressed in corneal epithelial cells.3 Activation of the P2Y receptors results in the recruitment of the heterotrimeric G-proteins, and changes in distinct signaling pathways, by either inducing Ca2+ release from intracellular stores or directly affecting intracellular signaling proteins, resulting in changes in cell migration and wound repair.3, 6, 7, 8
The P2Y receptor subtypes are cell-type and ligand specific, enabling them to play several regulatory roles.9 P2Y2 and P2Y4 receptors mediate ATP-induced activation of the protein kinase C/mitogen-activated protein kinase (MAPK) and protein kinase C/SRC pathways in the human breast cancer epithelial cell line MCF-7.10 The P2Y2 receptor has been shown to mediate metalloproteinase-dependent phosphorylation of epidermal growth factor receptor (EGFR) in human salivary gland cells,11 whereas the P2Y2 and P2Y6 receptors are responsible for the Ca2+ response in neuronal and glial cells, respectively.12 In corneal epithelial cells, activation via nucleotides or injury induces a rapid and transient phosphorylation of extracellular signal–related kinase (ERK). Likewise, treatment of the wound media with apyrase inhibits the mobilization of Ca2+ and results in decreased p-ERK, p-EGFR, and cell migration.3, 8
The EGFR facilitates rapid wound healing in corneal epithelium, and studies showed that nucleotide-induced EGFR activation is mediated, in part, by the activation of membrane-anchored metalloproteinases, followed by ectodomain shedding of HB-EGF.3, 13, 14 However, HB-EGF is only one of the components of the wound media.14 Furthermore, stimulation of epithelial cells by injury or with exogenous nucleotides causes activation of purinergic receptors, followed by transient EGFR phosphorylation and internalization that is distinct from that induced by EGF.14 Because EGFR phosphorylation sites serve as docking sites for downstream signaling proteins,14, 15, 16, 17, 18 the phosphorylation of EGFR tyrosine residues may result in either an increase or a decrease in the phosphorylation of neighboring residues. Although activation of the EGFR with EGF and HB-EGF causes recruitment of Shc, growth factor receptor-bound protein 2, and Src, activation of purinergic receptors elicits a differential recruitment of Shc and growth factor receptor-bound protein 2; however, there is no apparent difference in Src.14 Cross communication between different signaling pathways serves to integrate responses to different external stimuli the cell receives.19, 20, 21 We hypothesize that activation of the P2Y2 receptor results in distinct post-translational modification of the EGFR, non–receptor-type kinases, and other structural proteins that serve as a key link between the stimulus and signaling pathways that lead to corneal wound repair.
The goal of the present study was to ask if differential phosphorylation occurs in response to nucleotides or EGF, and to distinguish the responses. Although activation of the EGFR by its natural ligand yielded prototypical phosphorylation of tyrosine residues, ATP stimulation caused a 2- to 15-fold increase over control in EGFR Y974, Y1086, and Y1148 residues and a distinct difference in phosphorylation pattern of downstream signaling proteins, with ATP inducing a greaterfold increase in phosphorylation than EGF on Src Y421 but less at other sites on Src and phospholipase C (PLC)-γ. In addition, ATP induced a greaterfold increase in phosphorylation than EGF on specific residues of the kinases common to the P2YR and EGFR signaling networks [protein kinase C (PKC), focal adhesion kinase (FAK), and MAPKs 1, 12, and 13]. Additional post-translational modification changes were detected in paxillin, in which four residues showed a higher phosphorylation. Analysis of cell migration after a scratch wound revealed an up-regulation in P2Y2 receptor mRNA. Stimulation with UTP (agonist for P2Y2) after siRNA knockdown resulted in a decrease in phosphorylation of major downstream signaling proteins, including Y1086 and Y974 residues of EGFR, Src, and paxillin. Our results indicate that the initial activation of receptors plays a major role in the differential phosphorylation of signaling molecules, permitting cells to mediate downstream events.
Materials and Methods
Materials
Keratinocyte serum-free medium (K-SFM), bovine pituitary extract, fetal bovine serum, and penicillin/streptomycin were purchased from Mediatech (Manassas, VA). Fungizone, Lipofectamine, antibodies directed against site-specific phosphotyrosine residues of EGFR (pY845, pY1068, pY1086, and pY1173), Dynabeads protein G, and PCR kits (oligo-dT primers, 10× reaction buffer, MgCl2, dNTPs, Moloney Murine Leukemia Virus Reverse Transcriptase, and random primers) were purchased from Invitrogen (Carlsbad, CA). Acetoxymethyl (AM) ester derivatives of fluorescent indicators (Fluo-3 AM) were purchased from Invitrogen. C-18-Stage tips and CaCl2 were from Thermo Fisher Scientific (Cambridge, MA). Phosphotyrosine antibody T66, ATP, ADP, UTP, dithiothreitol, ammonium acetate, trifluoroacetic acid, acetic acid, Nonidet P-40 (NP-40), glycine, 2-amino-2-hydroxymethyl-propane-1,3-diol⋅HCl (Tris-HCl), sodium chloride (NaCl), sodium deoxycholate, EDTA, bicinchoninic acid (BCA), and urea were purchased from Sigma Aldrich (St. Louis, MO). siRNA sequences targeted against P2YR2, P2YR4, or a scrambled sequence [nontargeting (NT)] were from Dharmacon (Lafayette, CO). DNase I was from New England Biolabs (Ipswich, MA). RNase inhibitor, complete protease inhibitor cocktail, and PhosSTOP Phosphatase Inhibitor Cocktail were purchased from Roche Applied Science (Indianapolis, IN). Polyvinylidene difluoride membrane was from Pierce (Rockford, IL). Anti-phosphotyrosine antibody 4G10 was purchased from Millipore (Billerica, MA). Anti-EGFR antibody and horseradish peroxidase–conjugated secondary antibody were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). Western Lightning was purchased from GE Healthcare Bio-Sciences Corp (Piscataway, NJ). Phosphotyrosine antibody (PY100) was from Cell Signaling Technology (Danvers, MA). Stable isotope-labeled lysine and arginine containing 13C and/or 15N were purchased from Cambridge Isotope Laboratories (Andover, MA). C-18 reversed-phase material (MagicC18AQ) of 3- and 5-μm particle size and 200-Å pore size was purchased from Michrom Biosciences (Auburn, CA). Microcapillary columns of 75/100 μm inner diameter and 360 μm outer diameter were obtained from Polymicro Technologies (Phoenix, AZ). Trypsin was purchased from Worthington (Lakewood, NJ). The RNA extraction kit was purchased from Qiagen (Valencia, CA). TaqMan probes were purchased from Applied Biosystems (Foster City, CA). Iodoacetamide and sample buffer were purchased from Bio-Rad Laboratories (Irvine, CA).
Cell Culture
Human corneal limbal epithelial cells (HCLEs) were cultured in K-SFM supplemented with 30 μg/mL bovine pituitary extract, 0.032 nmol/L EGF (Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (Mediatech).14 Twenty-four hours before experimentation, EGF and bovine pituitary extract were removed.
siRNA Transfection
Cells were cultured until 50% to 60% confluency and transfected as described.7 Optimization experiments with multiple siRNA sequences were used individually or as part of a four-sequence pool, and the sequences resulting in the largest down-regulation of transcripts, as assayed by real-time PCR, were used for further experimentation. Experiments were performed using the same four-sequence pool or the two individual sequences (D-003688-01 or D-003688-09 and D-005693-05 or D-005693-03), which showed responses equivalent to the pool described.7 Sequences used for P2Y2 include the following: (01) 5′-CAACAUGGCCUACAAGGUUUU-3′ (sense) and 5′-AACCUUGUAGGCCAUGUUGUU-3′ (antisense); (09)5′-GAACUGACAUGCAGAGGAUUU-3′ (sense) and 5′-AUCCUCUGCAUGUCAGUUCUU-3′ (antisense); (04) 5′-GCAGAGGCUCGUACGCUUUUU-3′ (sense) and 5′-AAAGCGUACGAGCCUCUGCUU-3′ (antisense); and (902) 5′-GGAAUGCGUCCACCACAUAUU-3′ (sense) and 5′-UAUGUGGUGGACGCACAUAUU-3′ (antisense). Control nontargeting sequences were transfected as control. Lipofectamine and siRNA sequences were incubated separately in K-SFM supplemented with 30 μg/mL bovine pituitary extract and 0.032 nmol/L EGF without antibiotics, at room temperature. Mixtures were combined, incubated, and added to cells at a final concentration of 2 μL/mL Lipofectamine and 20 nmol/L siRNA. After 6 hours, the transfection medium was replaced with growth medium.
Real-Time Quantitative PCR
Total RNA was extracted using a Qiagen RNeasy kit, according to the manufacturer’s protocol. Genomic DNA was removed by incubation with DNase I in the presence of 1 U/mL RNase inhibitor. First-strand cDNA was enzymatically synthesized from the DNase-treated RNA templates using the Moloney murine leukemia virus–reverse transcriptase, and the control RT reaction lacked the reverse transcriptase. The TaqMan Gene Expression Master Mix (Applied Biosystems, Grand Island, NY) was used, and real-time PCR was performed using an ABI 7300 (Applied Biosystems, Foster City, CA). The following TaqMan probes were used: Hs00704965_s1 for P2Y1, Hs00602525_m1 for P2Y2, Hs00267404_s1 for P2Y4, and the eukaryotic 18S ribosomal RNA endogenous control for the 18S ribosomal subunit (VIC/MGB Probe, Primer Limited; Applied Biosystems, Grand Island, NY). cDNA templates were incubated at 50°C for 2 minutes and 95°C for 10 minutes, and that was followed by 40 to 50 amplification cycles of 95°C for 15 seconds and 60°C for 1 minute. Results were presented as relative expression normalized to 18S ribosomal RNA and calculated using the ΔΔCT method.7
Western Blot Analysis
Cells were washed with cold PBS and lysed in radioimmunoprecipitation assay buffer (150 mmol/L NaCl, 50 mmol/L Tris-HCl, pH 7.4, 1% NP-40, 1% deoxycholate, and 5 mmol/L EDTA) supplemented with complete protease inhibitor cocktail and PhosSTOP Phosphatase Inhibitor Cocktail (Roche Applied Science). Lysates were incubated at −80°C for 20 minutes and centrifuged at 4°C at 13,500 × g for 20 minutes, and supernatants were transferred to a new tube. Total protein concentration was determined using a BCA-based method, samples were subjected to SDS-PAGE, and proteins were transferred to a polyvinylidene difluoride membrane. Immunoblots were blocked using 5% bovine serum albumin in Tris-buffered saline and Tween-20 (TBS-T) for 2 hours at room temperature or overnight at 4°C, as appropriate. Membranes were probed with primary antibodies (1:1000) directed to phosphotyrosine or site-specific phosphotyrosine residues of EGFR (pY845, pY1068, pY1086, and pY1173) overnight at 4°C, washed with TBS-T, and incubated with horseradish peroxidase–conjugated secondary antibody (1:2000) for 1 hour at room temperature. Membranes were washed with TBS-T and exposed using Western Lightning (Perkin Elmer, Waltham, MA), and signals were quantified using ImageJ software version 1.48 (NIH, Bethesda, MD).
Scratch Wound Assay
The scratch wound assay and imaging analysis have been described.14 Scratch wounds were made using a gel loader tip, and the medium was removed and replaced with unsupplemented medium (control) or supplemented medium containing one of the following ligands (ADP, ATP, UDP, UTP, or EGF). In additional experiments performed to determine the response to UTP, cells were lysed at time points to determine the expression of P2Y receptors. Cultures were transfected with NT or siRNA to P2Y2 receptor following the protocol previously described and then wounded and cultured in media in the presence or absence of UTP. Slides were placed on the stage of a laser-scanning confocal microscope (Zeiss Axiovert 200M LSM 510; Zeiss, Thornwood, NY), and wound locations were marked. Slides were placed in an environmental chamber, and cells were incubated at 37°C and 5% CO2. Cell migration was monitored using the multitime module of the LSM software version 4.0 (Zeiss). Images were taken at each location every 20 minutes for 16 hours, as described.14 Wound areas at various time points were measured using the proprietary LSM software version 4.0, and percentage wound closure was calculated.
Ca2+ Mobilization
Briefly, cells were transferred into HEPES-buffered saline solution and loaded with 5 μmol/L Ca2+ indicator dye fluo 3-AM solution containing 0.02% pluronic acid/dimethyl sulfoxide.3, 6, 7 Cells were washed to remove excess dye and imaged using a Zeiss Axiovert 100M LSM 510 confocal microscope equipped with argon and two HeNe lasers (Zeiss). Cells were perfused with HEPES before injury with a micromanipulator to establish a baseline fluorescence reading. All perturbations were conducted while cells were continuously scanned using a Harvard apparatus perfusion chamber.
Preparation of Cell Lysates for Mass Spectrometry
HCLEs were stimulated with ATP, EGF, or media change (control) for 5 minutes and lysed with 8 mol/L urea in 50 mmol/L Tris-HCl and 75 mmol/L NaCl, pH 8.2, supplemented with a complete protease inhibitor cocktail and PhosSTOP Phosphatase Inhibitor Cocktail. For absolute quantitative phosphorylation analysis, cells were cultured in the presence of stable isotope-labeled amino acids in cell culture (SILAC), using a custom-made K-SFM supplemented with isotope-labeled lysine (12C6,14N2 = light and 13C6,15N2 = heavy) and arginine (12C6,14N4 = light and 13C6,15N4 = heavy). Protein concentration was determined by BCA. An average of 10 mg total protein was reduced, with a final concentration of 10 mmol/L dithiothreitol in 100 mmol/L ammonium acetate, pH 8.9, for 1 hour at 56°C, and free cysteines were alkylated, with a final concentration of 55 mmol/L iodoacetamide in 100 mmol/L ammonium acetate, pH 8.9, for 1 hour in the dark. After the concentration of urea was reduced to 1.5 mol/L with 100 mmol/L ammonium acetate, pH 8.9, proteins were digested with trypsin at a 1:50 trypsin/protein ratio for 16 hours at room temperature. Digested lysates were acidified to pH <3 with trifluoroacetic acid, desalted with a C18 Sep-Pak Plus cartridge (Waters Corp, Milford, MA), eluted with 25% acetonitrile/0.1% acetic acid, and lyophilized to dryness.
Phosphopeptide Immunoprecipitation
Phosphopeptide immunoprecipitation was performed, as described, with the following modification.22 Briefly, a 60-μL volume of Dynabeads protein G (Life Technologies, Grand Island, NY) was washed with immunoprecipitation buffer (100 mmol/L Tris, 100 mmol/L NaCl, and 1% NP-40, pH 7.4) and incubated with a combination of three anti-phosphotyrosine antibodies (PY100, T66, and 4G10) at 4°C for 8 hours on a rotator. The antibody-bead conjugates were centrifuged, and the beads were removed using a magnetic bar and washed with immunoprecipitation buffer. Lyophilized peptides were resuspended and incubated with the antibody-conjugated beads at 4°C in a rotator overnight, and the bead-antibody-peptide complex was separated using a magnetic bar. Beads were washed with 100 mmol/L Tris-HCl, pH 7.4. Phosphopeptides were eluted twice with 100 mmol/L glycine, pH 2.5, for 30 minutes at room temperature, combined, and vacuum dried in a SpeedVac (ThermoScientific, Waltham, MA).
Phosphopeptide Enrichment and Liquid Chromatography–Tandem Mass Spectrometry
Dried peptides were phosphoenriched using immobilized metal affinity chromatography (IMAC) and desalted using a stage tip (C-18), as described.23 Phosphoenriched peptides were redissolved in 99% water, 1% CH3CN, and 0.1% formic acid and injected into a nanoAcuity Ultra Performance Liquid Chromatography (Waters Corp) coupled through a TriVersa NanoMate robot (Advion, Ithaca, NY) to an Linear Ion Trap-Orbitrap Discovery Electron Transfer Dissociation (Thermo Fisher Scientific, San Jose, CA) mass spectrometer. The metal components of the sample introduction pathway had been replaced with inert materials. Trapping was performed using a self-packed C-18 reversed-phase microcapillary column (MagicC18AQ; 5-μm particle size and 200-Å pore size; 10-cm bed length; 360-μm OD × 100-μm ID). A 10-cm microcapillary analytical column (360-μm OD × 75-μm ID; 10-cm bed length) was packed with C-18 reversed-phase material (MagicC18AQ) of 3-μm particle size and 100-Å pore size. The mobile buffer A, consisting of 99% H2O, 1% CH3CN, and 0.1% PBS, was run with the elution buffer B, of 99% CH3CN, 1% H2O, and 0.1% PBS over a 90-minute gradient with a flow rate of 0.5 μL/minute. The LTQ-Orbitrap was operated in a data-dependent mode in which each full MS scan (m/z 300 to 2000 with 60,000 resolution at m/z 400) was followed by ion-trap MS/MS scans of the five most abundant ions.
Mass Spectrometry Data Analysis
The data from triplicate runs for each sample were used for label-free quantitative phosphopeptide analysis using Progenesis liquid chromatography–mass spectrometry (LC-MS; Nonlinear Dynamics, Durham, NC). Modified peptides with identification power of ≥0.8 and P ≤ 0.05 were filtered and exported for a database search. The database search was performed against the SwissProt human database using the Mascot search engine, version 2.3.02 (Matrix Science, Boston, MA). Carbamidomethylation of cysteine was set as a fixed modification. Oxidation of methionine and phosphorylation of tyrosine, serine, and threonine were set as variable modifications. A maximum of two missed cleavages was set, with trypsin as the protease. Mascot results were filtered with 10-ppm precursor ion mass tolerance and MS/MS tolerance of 0.8 Da for peptide fragment ions. Data from SILAC-labeled samples were analyzed using Proteome Discoverer version 1.2 (Thermo Fisher Scientific, San Jose) using similar search parameters as previously described. For quantitative analysis, the ratio (heavy/light) of phosphosite occupancy was calculated.
Statistical Analysis
For each identified phosphorylation site, fold increase over control was calculated from the normalized average relative intensity of the phosphorylation profile generated using the mean peak area of the total ion chromatogram. The results obtained for a subset of the phosphorylated proteins and modifications are given in Table 1, and the significant differences in phosphorylation between treatments are calculated. The data were filtered based on P values (P < 0.05), and fold change significance was calculated using analysis of variance.
Table 1.
Label-Free Quantitative Tyrosine Phosphorylation Profile of a Subset of Average Normalized Abundances in the Tyrosine Phosphorylation Profile Determined from HCLEs Treated with Control (Media Change), ATP, or EGF
| Protein | Modifications | m/z | Running time (minutes) | Charge | ANOVA (P) (×10-2) | Fold | Average normalized abundances (×103) |
||
|---|---|---|---|---|---|---|---|---|---|
| Control | ATP | EGF | |||||||
| EGFR | p-Tyr-974 | 548.9004 | 30.4 | 3 | 0.0000104 | 46.7 | 13.8 | 69.5 | 325 |
| p-Tyr-1086 | 827.0708 | 24.7 | 3 | 0.000147 | 39.7 | 6.96 | 51.9 | 312 | |
| p-Tyr-1148 | 772.6703 | 33.9 | 3 | 0.000435 | 142.2 | 13.1 | 266 | 1870 | |
| p-Tyr-1173 | 759.0297 | 27.4 | 3 | 0.0000255 | 351.7 | 4.53 | 2.43 | 856 | |
| KPCD | p-Tyr-313 | 940.9012 | 29.7 | 2 | 0.00000962 | 13.67 | 6.65 | 91.2 | 23.3 |
| p-Ser-304 | 706.2921 | 29.2 | 3 | 0.000751 | 12.4 | 2.85 | 35.3 | 21.3 | |
| MAPK-13 | p-Tyr-182 | 765.3261 | 24.3 | 2 | 2 | 9.7 | 5.29 | 51.7 | 11.2 |
| p-Tyr-258 | 574.5929 | 25.7 | 3 | 0.0172 | 53.1 | 0.565 | 0.132 | 7.02 | |
| MAPK-12 | p-Tyr-185 | 816.8103 | 27.6 | 2 | 0.1 | 29.8 | 2.59 | 10.4 | 0.348 |
| MAPK-1 | p-Tyr-187 | 741.9948 | 29.7 | 3 | 1.7 | 23.7 | 130 | 1730 | 731 |
| p-Thr-190 | 1112.489 | 29.7 | 2 | 0.000184 | 88.7 | 8.74 | 70 | 0.789 | |
| p-Thr-185 | 768.6503 | 30.9 | 3 | 1 | 9.1 | 39.4 | 359 | 143 | |
| PLCG1 | p-Tyr-771 | 846.3689 | 29.1 | 2 | 0.000818 | 15.3 | 7.7 | 73.2 | 118 |
| p-Tyr-227 | 896.3942 | 36.5 | 2 | 1 | 8 | 1.66 | 3.65 | 13.3 | |
| RIN1 | p-Tyr-36 | 948.1138 | 27.9 | 3 | 0.2 | 11.9 | 26 | 310 | 133 |
| CDK2 | p-Tyr-15 | 541.2668 | 26.6 | 3 | 0.00000816 | 192.2 | 1.46 | 0.369 | 71 |
| CDC42-K | p-Tyr-827 | 739.3634 | 27.5 | 2 | 0.0676 | 29.1 | 1.6 | 11.3 | 0.389 |
| p-Tyr-859 | 605.3012 | 30.4 | 4 | 0.00207 | 49.5 | 2.3 | 14.6 | 114 | |
| p-Tyr-860 | 806.7332 | 30.5 | 3 | 0.0251 | 10.4 | 3.01 | 3.43 | 31.3 | |
| SHC1 | p-Tyr-427 | 1102.007 | 34.3 | 2 | 0.00303 | 196.7 | 2.45 | 1.62 | 319 |
| p-Tyr-315 | 457.5704 | 21.2 | 3 | 0.000967 | 401.5 | 0.546 | 0.122 | 48.8 | |
| SHB | p-Tyr-246 | 818.8424 | 25.7 | 2 | 0.000301 | 34.4 | 4.07 | 38.9 | 1.33 |
| p-Tyr-268 | 641.9358 | 24.1 | 3 | 0.7 | 8.2 | 4.38 | 3.27 | 26.8 | |
| p-Tyr-98 | 817.3299 | 28.5 | 2 | 0.0306 | 9.1 | 5.89 | 31.4 | 53.8 | |
| GAB1 | p-Tyr-659 | 636.7968 | 24.1 | 2 | 0.000000287 | 29.3 | 1.11 | 30.8 | 1.05 |
| p-Tyr-259 | 828.8849 | 31.5 | 2 | 0.01 | 14.8 | 4.86 | 72.2 | 19 | |
| p-Tyr-626 | 787.8527 | 33.8 | 2 | 0.00433 | 16.2 | 4.99 | 80.9 | 6.06 | |
| SRC8 | p-Tyr-446 | 750.98 | 27.9 | 3 | 0.008 | 11.5 | 1.84 | 20 | 21.1 |
| p-Tyr-421 | 530.9075 | 31.2 | 3 | 0.0411 | 27.3 | 8.55 | 23 | 6.36 | |
| PAXI | p-Tyr-88 | 676.6452 | 22.3 | 3 | 3.7 | 7.9 | 47.4 | 374 | 99 |
| p-Tyr-118 | 538.8987 | 24.8 | 3 | 4.5 | 12 | 357 | 4280 | 375 | |
| FAK | p-Tyr 576/577 | 783.3149 | 21.9 | 2 | 0.0575 | 64.6 | 1.75 | 0.193 | 12.5 |
| p-Tyr-861 | 880.432 | 28.2 | 4 | 0.000252 | 23.1 | 7.08 | 163 | 7.27 | |
| TYK2 | p-Tyr-259 | 627.8057 | 25.1 | 4 | 0.00481 | 35.8 | 8.11 | 79.3 | 290 |
| p-Tyr-292 | 800.3649 | 28.5 | 2 | 3.6 | 7.3 | 46.2 | 225 | 30.9 | |
| P85B | p-Tyr-464 | 794.8136 | 29.5 | 2 | 0.5 | 4.4 | 11.2 | 49.6 | 40.6 |
| p-Tyr-467 | 794.8136 | 29.5 | 2 | 0.5 | 4.4 | 11.2 | 49.6 | 40.6 | |
| FAK2 | p-Tyr-579/580 | 956.3759 | 24.4 | 2 | 0.00625 | 37.6 | 6.65 | 31.7 | 69.3 |
| PTK6 | p-Tyr-447 | 663.2683 | 29 | 2 | 0.2 | 30.8 | 18.9 | 147 | 583 |
| PTN11 | p-Tyr-62 | 908.3773 | 29.3 | 2 | 0.3 | 42.8 | 25.3 | 114 | 2.67 |
| p-Tyr-63 | 908.3773 | 29.3 | 2 | 0.3 | 42.8 | 25.3 | 114 | 2.67 | |
| PRP4B | p-Tyr-849 | 1259.0596 | 34.1 | 2 | 1.3 | 6.1 | 15.6 | 95.4 | 28 |
| p-Tyr-849 | 839.7086 | 34.2 | 3 | 1.4 | 10.3 | 105 | 1080 | 443 | |
| STAT3 | p-Tyr-705 | 646.2804 | 24.9 | 4 | 0.00455 | 15.4 | 15.3 | 122 | 18.7 |
| ITB4 | Y-1207 | 714.299 | 29 | 3 | 2.6 | 3.5 | 8.89 | 25.5 | 31.4 |
| CBL | p-Tyr-674 | 787.4292 | 33.3 | 3 | 0.0231 | 56 | 5.43 | 38.4 | 304 |
Statistical significance is calculated within the triplicate runs for each condition. Data were filtered based on P values (P < 0.05). Fold change significance was calculated using ANOVA.
ANOVA, analysis of variance.
Results
Dynamic protein phosphorylation is critical for the regulation of many cellular signaling mechanisms. Because there are multiple signaling pathways induced by injury that involve the activation of EGFR, knowledge of the specific changes will enhance our understanding in regulation of cell migration and wound repair.
Role of Nucleotides on Cell Migration
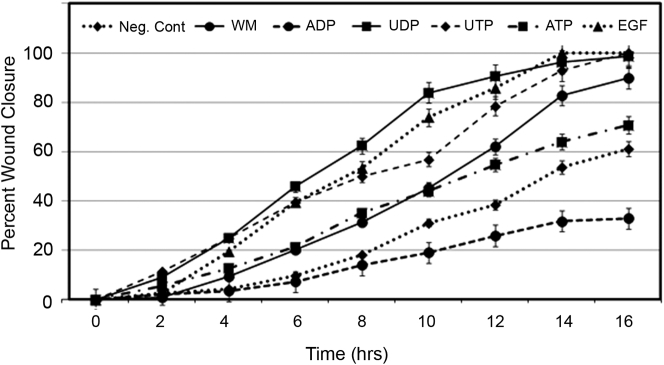
Previous studies have shown that nucleotides and growth factors enhance cell migration in Boyden chamber assays.3 In addition, we have shown that EGFR is necessary for nucleotide-induced cell migration by either using tyrosine kinase inhibitors or mutating specific residues in scratch wound assays.14 To determine the response to individual nucleotides, scratch wounds were made and migration was monitored every 20 minutes for 16 hours. At the final time point, the gap generated by the scratch wound had decreased by >60% in response to all agonists, except for ADP (Figure 1). The positive control, in which wounded cultures contained the wound media, demonstrated an 80% gap closure (Figure 1).
Figure 1.
Role of individual nucleotides on cell migration. HCLEs grown to confluence on 8-well glass-bottom chambers were serum starved for 16 hours. Scratch wounds were made using a gel loader tip, and medium was immediately aspirated and replaced with medium alone (Neg. Cont) or medium containing one of the following ligands (100 μmol/L ADP, 100 μmol/L ATP, 100 μmol/L UDP, 100 μmol/L UTP, or 0.5 nmol/L EGF). Positive controls were cultures that were wounded and in which the wound media were retained for the duration of the time course [wound media (WM)]. Live cell migration was monitored using a laser-scanning confocal microscope equipped with an environmental chamber that maintained the cell environment at 37°C and 5% CO2. Images were taken at each location every 20 minutes for 16 hours. Wound closure was determined using the region of interest function of the LSM software version 4.0, and percentage wound closure was calculated. Wound closure is indicated by the average closure at each time point ± SEM. Data are representative of three independent experiments.
MS-Based Quantitative Analysis of Phosphorylation Profile
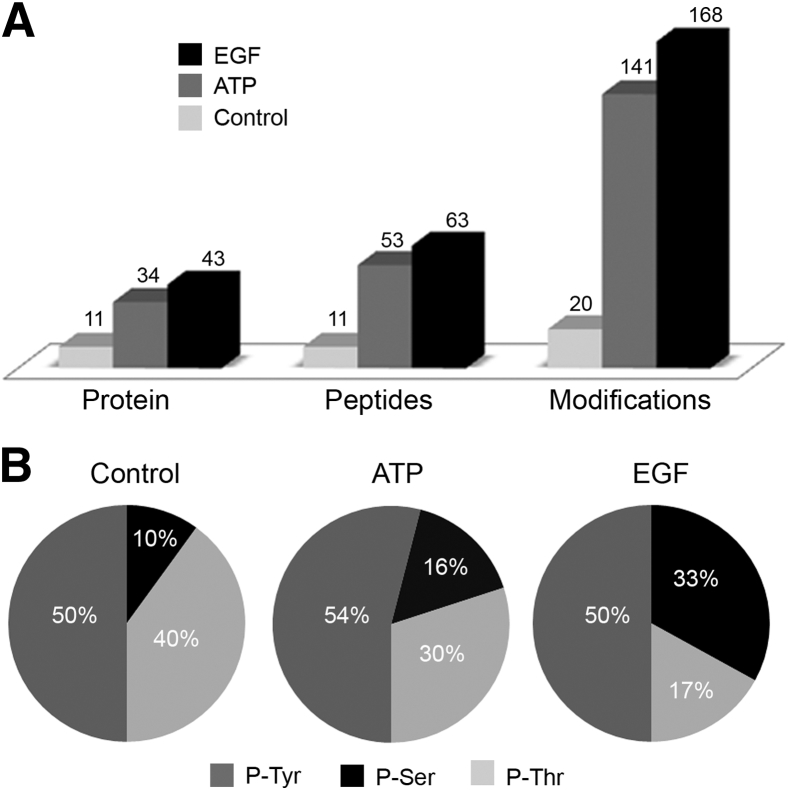
In global MS-based phosphoproteome studies,24, 25, 26 detected levels of phosphotyrosine modification in peptides were lower than the levels of phosphoserine and phosphothreonine modifications after treatment of cells with agonist for 5 minutes. Therefore, we used phosphotyrosine peptide immunoprecipitation using three anti-phosphotyrosine antibodies to enrich the detection level of phosphotyrosine residues in the samples subjected to LC-MS analysis. Initial analysis using database searches on each data set with Proteome Discoverer generated a total of 20, 141, and 168 phosphorylation modification sites in the control, ATP, and EGF treatment groups, respectively (Figure 2A). ATP was used to stimulate cells because it activates most purinergic receptors and, thus, gave a global response. The phosphotyrosine contribution observed under all treatment conditions was approximately 50% of the total phosphopeptides identified (Figure 2B). The immunoprecipitates were dried and further phosphoenriched using IMAC, yielding a percentage distribution of phosphotyrosine containing peptides >30% of the phosphopeptides identified by MS. Label-free quantitative analysis using Progenesis LC-MS (Nonlinear Dynamics) generated a list of >300 phosphorylation sites on 139 proteins. A total of 130 phosphopeptides and proteins and average normalized abundances are given (Supplemental Table S1). The analysis is described in Materials and Methods.
Figure 2.
Quantitative ligand-specific phosphotyrosine profile analysis. HCLEs were stimulated with a media change (control), ATP, or EGF. Cells were lysed and enzymatically digested, and phosphotyrosine peptides were immunoprecipitated using three anti-tyrosine antibodies in a 1:1 solution (PY-100, 4G10, and T66). A: The number of proteins and the corresponding peptides and modifications are presented. B: Percentage distribution of the phosphorylation sites (p-Tyr, p-Ser, and p-Thr). Data are representative of two experiments, each including triplicate UPLC-MS analysis.
Phosphorylation of EGFR and Downstream Signaling Proteins
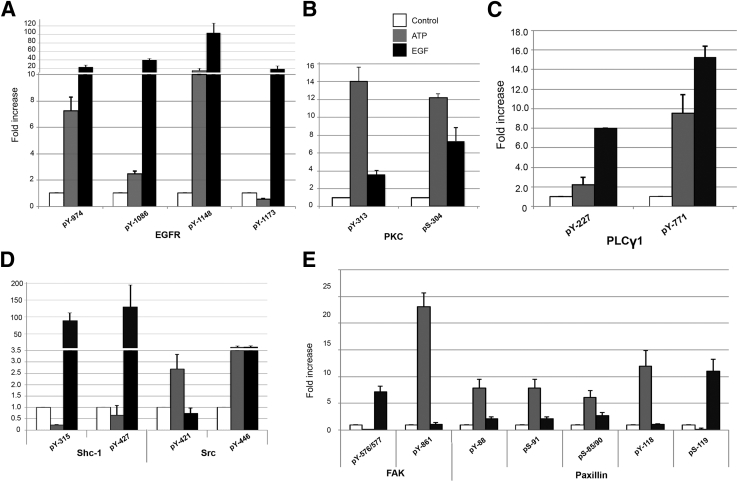
Previously, we showed that EGFR was phosphorylated indirectly by activation of purinergic receptors in a dose-dependent manner.14, 19 Five minutes after stimulation, the data from internalization studies of the EGFR were similar in response to EGF, nucleotides, or a scratch wound. In addition, when cells were immunoprecipitated with EGFR and probed for p-EGFR in response to EGF or nucleotide, there was a significant change in p-EGFR over untreated.14 In the current experiments, there was a differential phosphorylation at 5 minutes in response to ATP or EGF (Table 1). EGF caused a 10- to 100-fold increase in phosphorylation over control on residues EGFR Y974, Y1086, Y1148, and Y1173, whereas ATP caused a 2- to 15-fold increase over control on EGFR Y974, Y1086, and Y1148 residues (Figure 3A). In contrast, ATP induced less phosphorylation compared with media control on Y1173 (Figure 3A). Other residues may be phosphorylated, but the fold change over control did not fit the stringency requirement of the analysis of variance because the data were filtered based on P < 0.05.
Figure 3.
Label-free quantitative analysis of signaling proteins using Progenesis LC-MS. Normalized average relative abundances of the phosphorylation profile generated by calculating the mean peak area of the total ion chromatogram of each peptide are shown. Fold change over control of the following phosphotyrosine and serine residues: EGFR (A), PKCΔ (B), PLC-γ1 (C), Shc and Src (D), and FAK and paxillin (E). Phosphopeptide abundances are calculated from the normalized average relative abundances for each phosphopeptide. Data are representative of two experiments, each including triplicate UPLC-MS analysis (see Table 1).
Kinases common to purinergic and EGFR signaling networks showed site-specific distinct phosphorylation patterns in response to activation. We speculated that PKC plays a role in the phosphorylation of EGFR via changes in mobilization of Ca2+ after activation of purinergic receptors or after cleavage of phosphatidylinositol 4,5-bisphosphate 2 to diacylglycerol and inositol triphosphate. For PKCΔ, both sites showed higher fold increases in phosphorylation in response to ATP compared with EGF. For example, ATP caused phosphorylation at pY313 and pS304, with 14- and 12-fold increases in phosphorylation over control and 3- and 1.5-fold increases over EGF stimulation, respectively (Figure 3B).
Previously, we demonstrated that the phosphorylation of PLC-γ in response to EGF decreased in the presence of ATP.7, 14 When we evaluated its phosphorylation profile in response to ATP or EGF (as positive control), there was a differential response on two residues. Y227 showed an eightfold increase over control in response to EGF and a twofold increase over control in response to ATP. Likewise, Tyr-771 showed a 14-fold increase over control, and a ninefold change over control in response to ATP (Figure 3C). In addition, with ATP, we detected changes in the extent of oxidation of methionine.
When downstream signaling proteins shown to display a differential recruitment with Western blot analyses were examined, we detected a differential phosphorylation in Shc and Src. Previously, we had detected little or no change in association of Src14 with EGFR. At a higher resolution in these studies, we found a significant fold increase in phosphorylation of Y421 in response to ATP, whereas the response to EGF was less than control. For Y446, ATP and EGF stimulation resulted in almost equivalent, 11-fold increases over control. Previously, we had shown that there was a recruitment difference among the three Shc isoforms.14 Herein, we detected differences in phosphorylation intensity at Y315 and Y427 (Figure 3D). Tyr-315 showed a negligible increase in phosphorylation in response to ATP and was lower than control. In contrast, there was an 89-fold increase in phosphorylation over control in response to EGF. Tyr-427 showed a similar profile, with no increase in phosphorylation over control in response to ATP, in contrast to a 130-fold increase in phosphorylation over control after EGF stimulation. These results indicate that association with EGFR does not correlate with fold changes in phosphorylation.
The changes in the phosphorylation profile of signaling proteins, such as PLC-γ and Src, were speculated to influence the profile of FAK and paxillin, adaptor adhesion kinases that mediate cytoskeletal changes and cell migration. For FAK Y576/577, there was greater than fivefold increase in response to EGF but a smaller change than control in response to ATP. In contrast, Y861 showed a 20-fold increase in response to ATP and a minimal response to EGF. Y397 showed a negligible response to ATP (data not shown). Downstream of FAK, paxillin showed a greater fold response to ATP than EGF on Y88, S85/90, S9, and Y118, with a 10-fold greater response to EGF than ATP on S119 (Figure 3E).
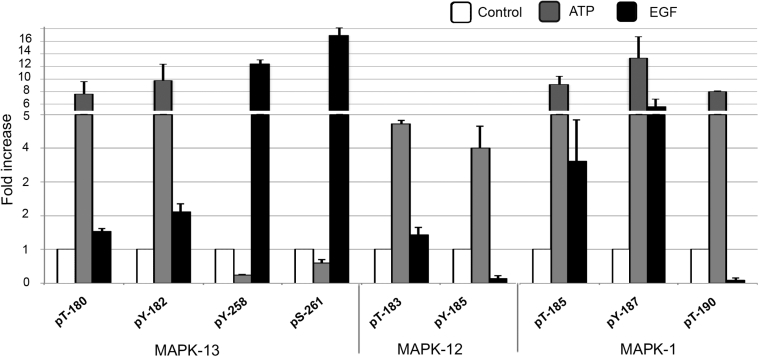
Previously, we demonstrated that nucleotides induced a rapid transient phosphorylation (2 to 10 minutes) of ERK in response to injury that contrasted with the long-lasting phosphorylation in response to EGF.8 Herein, we detected ligand-specific differences in nine phosphorylation sites in three isoforms of the MAPK family: MAPK-1, MAPK-12, and MAPK-13. Seven of the mapped residues (T185, Y187, and T190 of MAPK-1, Y185 and T183 of MAPK-12, and T180 and Y182 of MAPK-13) showed a minimum of a fourfold increase in phosphorylation in response to ATP over control and a twofold increase over EGF stimulation, whereas Y258 and S261 of MAPK-12 were highly phosphorylated only in response to EGF (Figure 4).
Figure 4.
Label-free quantitative analysis of MAPK using Progenesis LC-MS. Normalized average relative abundances of the phosphorylation profile are generated by calculating the mean peak area of the total ion chromatogram of each peptide are shown. Fold changes over control of MAPK phosphopeptide abundances are calculated from the normalized average relative abundances of each phosphopeptide. Data are representative of two experiments, each including triplicate UPLC-MS analysis (see Table 1).
Together, these data indicate that stimulation directly and through cross activation causes a distinct response, and when it occurs through the secondary route, recruitment of signaling molecules and activation are modified.
P2Y Receptor Expression Changes during Wound Repair
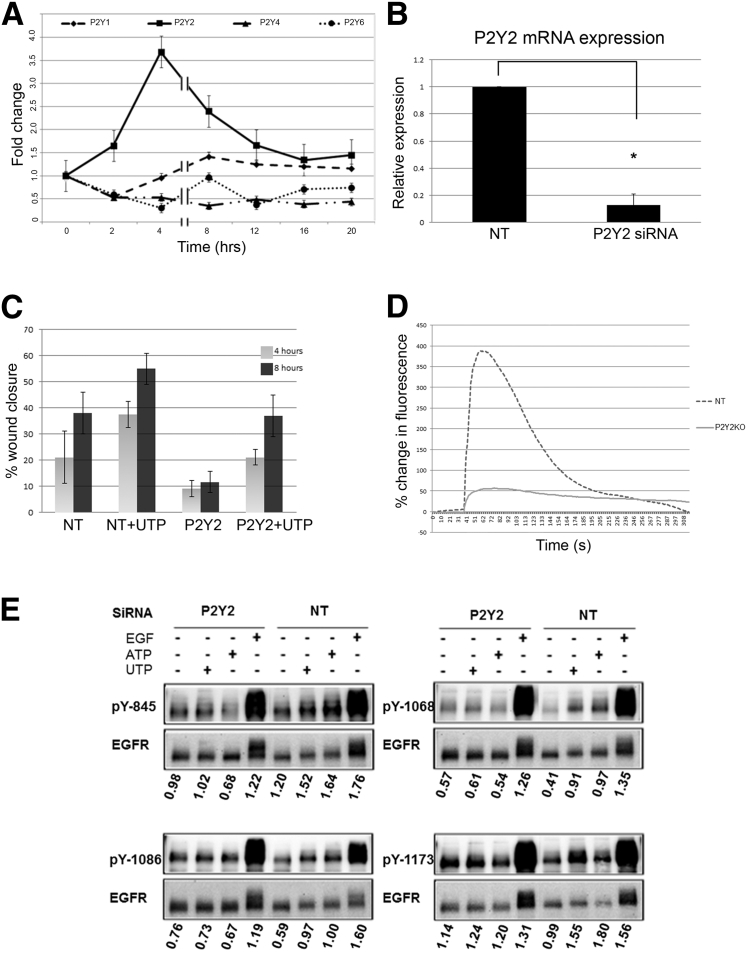
P2Y receptor mRNA expression levels were evaluated at 0, 2, 4, 8, 12, 16, and 20 hours after injury (Figure 5A). We found an increase in P2Y2 receptor mRNA expression 4 hours after injury and little change in P2Y1, P2Y4, and P2Y6 receptor mRNA expression over the time course of 20 hours (Figure 5A). Because the expression of P2Y2 receptor increased, we performed knockdown experiments to understand the role of P2Y2 on epithelial function after injury. Verification of transfection with siRNA to P2Y2 receptor was compared with nontargeting sequence (Figure 5B) and had been shown previously by our laboratory.7, 14 To examine the role of the receptor on migration, cells were transfected with siRNA to P2Y2 or a nontargeting sequence, and wounded and cultured in the presence of UTP, which was used because it has the greatest potency for the P2Y2 receptor.6 Cells were imaged over 20 hours using live cell microscopy under environmental conditions, and percentage closure was determined at 4 and 8 hours, time points near maximal expression and during active cell migration. Cultures transfected with an NT sequence and stimulated with UTP showed a 1.8-fold change in closure of the gap over non-stimulated at 4 hours (Figure 5C). When cells were transfected with the siRNA to the P2Y2 receptor, closure of the gap was minimal at either time point. We did detect an increase in closure in response to UTP under transfected conditions, and we attribute this to activation of P2Y4. Studies were performed to determine Ca2+ mobilization of injured transfected cultures. Cultures transfected with siRNA directed to the P2Y2 receptor exhibited minimal Ca2+ mobilization after scratch wound compared with cultures transfected with siRNA to an NT sequence (Figure 5D).
Figure 5.
Purinergic receptor activation mediates cell function. A: Expression of purinergic receptors (P2Y1, P2Y2, P2Y4, and P2Y6) after scratch wound. HCLE cultures were subjected to scratch wounds, and at 0, 2, 4, 8, 12, 16, and 20 hours, RNA was isolated. Real-time PCR was conducted to determine relative expression levels of the P2Y1, P2Y2, P2Y4, and P2Y6 receptor mRNA transcripts. Data are representative of three independent runs from three independent experiments. Values are given as the means ± SEM. B: Relative expression of cells transfected with siRNA to P2Y2 or a scrambled siRNA sequence (NT) using real-time PCR (t-test, *P < 0.05). C: Role of P2Y2 receptor on cell migration. HCLEs were transfected with siRNA to P2Y2 receptor or NT, and scratch wounds were made and stimulated with UTP. Cells were monitored and images were taken at 4 and 8 hours, wound areas were measured using the region of interest of the LSM software version 4.0, and percentage wound closure was calculated. Data are representative of three independent experiments. D: P2Y2 receptor knockdown attenuates Ca2+ mobilization after injury. HCLEs transfected with siRNA to P2Y2 receptor or NT were incubated in 5 μmol/L fluo 3-AM and imaged in a flow-through apparatus on a Zeiss LSM 510 confocal microscope. Cells were perfused with HEPES to establish baseline fluorescence. Data are representative of three independent runs, and a representative run is graphed as percentage change in average fluorescence over time. E: Role of P2Y2 receptor activation on EGFR phosphorylation. HCLEs were cultured, transfected with siRNA to P2Y2 or NT, and stimulated with medium alone, UTP, ATP, or EGF. Proteins were resolved by SDS-PAGE, and immunoblot analysis was performed using primary antibodies directed to site-specific phosphotyrosine residues of EGFR (pY845, pY1068, pY1086, and pY1173). The resultant ratio of phosphorylation over the total EGFR signals was calculated, and the normalized ratio was used to calculate fold changes over medium control. Data are representative of three independent experiments.
We then evaluated the role of the P2Y2 receptor on phosphorylation of EGFR tyrosine residues using Western blot analysis. Cells transfected with siRNA to the P2Y2 receptor or NT were immunoblotted using site-specific antibodies directed against EGFR tyrosine residues evaluated previously (Y845, Y1068, Y1086, and Y1173).14 When cells were transfected with the NT sequence, there was an increase in the phosphorylation of Y845, Y1068, Y1086, and Y1173 after stimulation with UTP or EGF. When cells were transfected with siRNA to the P2Y2 receptor, there was a detectable increase in phosphorylation in response to EGF; however, there was minimal or no increase in the phosphorylation in response to nucleotides (Figure 5E). These data implicated the P2Y2 receptor as a major player in epithelial cell migration and EGFR cross activation. In addition, they demonstrated that the phosphorylation is complex and these results led to stable-isotope labeling of epithelial cells to further delineate the role of the receptor.
Role of P2Y2 Receptor on EGFR Phosphorylation
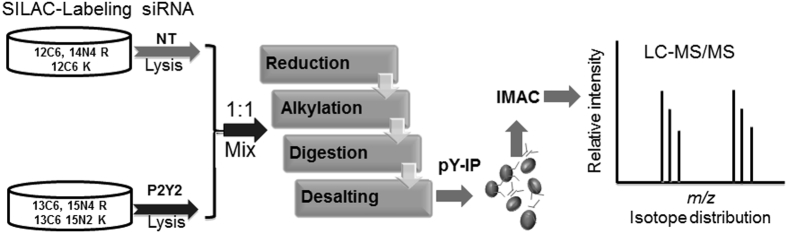
SILAC is an accurate and sensitive method for quantifying peptide and protein modifications.27, 28 To test the role of the P2Y2 receptor in regulating the nucleotide-induced phosphorylation profile, cells were SILAC labeled with isotopically labeled lysine (K) and arginine (R) and the groups were designated as light [12C6,14N2(K), 12C6,14N4(R)] or heavy [13C6,15N2(K), 13C6,15N4(R)]. Cells were transfected with siRNA directed against P2Y2, or an NT siRNA sequence, stimulated with 100 μmol/L UTP and lysed. Equal amounts of total protein from each condition were mixed and digested using trypsin. Desalted peptides were immunoprecipitated, and the double-label data reflected changes in phosphorylation because of the P2Y2 receptor (Figure 6 and Table 2). For example, in Figure 3, we detected a distinct increase in phosphorylation of three EGFR tyrosine residues (Y974, Y1086, and Y1148) in response to nucleotides, albeit lower than in response to EGF. After P2Y2 receptor knockdown, there was a >50-fold decrease in phosphorylation of EGFR tyrosine residues Y974 and Y1086, with a lesser decrease in phosphorylation of Y1148 after UTP stimulation (Table 2). In addition, there was no change in phosphorylation of Y1173 (Figure 6 and Table 2). Furthermore, there was a decrease in phosphorylation on Src, Shc, MAPK-1, and paxillin. Differences in the phosphorylation states of the residues indicate the high degree of complexity caused by the simultaneous transactivation and direct activation by ligands that occurs with injury.
Figure 6.
Schematic for SILAC-based quantitative analysis. The experimental scheme for SILAC-based quantitative phosphoproteome analysis includes culturing HCLEs with SILAC-labeled with heavy- or light-labeled lysine and arginine. The heavy [13C6,15N2 (K), 13C6,15N4 (R)] and light [12C6,14N2(K), 12C6,14N4(R)] isotope-labeled cells were transfected with siRNA directed to P2Y2 receptor and NT (scrambled sequence), respectively. Cells were stimulated with 100 μmol/L UTP for 5 minutes and then lysed, and equal amounts of total protein from each condition were mixed. Lysate was reduced, alkylated and trypsin digested, and subjected to immunoprecipitation using anti-phosphotyrosine antibody. Eluted peptides were further phosphoenriched using IMAC and analyzed using a nanoAcuity UPLC (Waters Corp) coupled through a TriVersa NanoMate (Advion) to an LTQ-Orbitrap mass spectrometer (ThermoFisher Scientific).
Table 2.
Relative Role of P2Y2 Receptor on Tyrosine Phosphorylation
| Sequence | Modifications | Ratio H/L |
|---|---|---|
| EGFR | p-Tyr-974 | 0.345 |
| p-Tyr-1086 | 0.230 | |
| p-Tyr-1148 | 0.779 | |
| p-Tyr-1173 | 1.215 | |
| Src | p-Tyr-421 | 0.339 |
| p-Tyr-446 | 0.128 | |
| SHC | p-Tyr-42 | 0.694 |
| MAPK-1 | p-Tyr-187 | 0.789 |
| Paxillin | p-Tyr-118 | 0.100 |
Subset of SILAC-based quantitative phosphorylation profile from cells transfected with siRNA to P2Y2 receptor and stimulated with UTP (H) compared with cells transfected with siRNA to nontargeting sequence (L) (see Figure 6). Phosphorylation is presented as the ratio of H/L.
Discussion
Re-epithelialization is important to the healing process, and proper closure of wounds requires neighboring cells to signal and communicate. The release of nucleotides on injury activates the purinergic receptors of the cornea, resulting in Ca2+ mobilization and cell-cell communication. This article demonstrates the novel contribution from activation of purinergic receptors on phosphorylation of specific residues of EGFR and downstream signaling proteins. Moreover, the results of this work highlight the critical role of the P2Y2 receptor in mediating epithelial migration. Knowledge of the specific changes in phosphorylation that occur with injury is essential to understanding how cell migration and wound repair can be compromised.
More than 300 phosphorylation sites on 139 proteins were identified and quantified. We demonstrated that the use of a combination of phosphopeptide fractionation and phosphoenrichment strategies is important to achieve a comprehensive analysis of complex biological samples. We speculate that the qualitative and quantitative differences in the levels of phosphorylation at each individual site may cause distinct effects on their respective downstream signaling pathways and the biological response(s) that they mediate. As shown, EGFR tyrosine residues exhibit spatiotemporal dynamics in phosphorylation,26, 29 and the modifications are dose dependent.30 An important factor is the concentration of agonist. Phosphorylation is often examined under conditions in which the levels of agonist are much higher than would be encountered in vivo. Although these types of studies facilitate the analysis, they can mask important changes. Therefore, we used concentrations of EGF at least fivefold lower than those used in most of the previously reported mass spectrometry–based phosphoproteome studies.22, 29, 30, 31 Although the threshold at which the phosphorylation level of proteins results in a biological response is not yet known, increasing evidence implicates a direct relationship between the degree of phosphorylation and the corresponding signaling strength. These changes in phosphorylation may occur in response to distinct ligands or under unique cellular environments. In addition, there are other modifications that we have not yet addressed, including changes in O-GlcNAcylation that will be examined in future studies. Changes in glycosylation may cause cryptic sites and modify signaling.
Although the release of nucleotides and activation of purinergic receptors have been known to cause transactivation of EGFR and recruitment of downstream signaling proteins that is distinct from EGF, the previous reliance on phosphorylated antibodies targeted to generic modifications had not permitted determination of site-specific modifications.14 Herein, we demonstrate that direct activation of EGFR with EGF and transactivation of EGFR via activation of purinergic receptors result in distinct phosphorylation profiles of EGFR residues. We have shown that the activation of purinergic receptor subtypes is ligand specific; the patterns of polar amino acid residues in the extracellular domains of the transmembrane loops 6 and 7 of the P2Y receptors allow one to account for ligand specificity.32 Gan et al24 used a glioblastoma tumor cell line to demonstrate that mutation of one or more EGFR tyrosine residues affected the degree of phosphorylation of sites. They showed that mutation of EGFR tyrosine residue Y1173F led to increased phosphorylation of other sites. Although the EGFR tyrosine residue Y1173 is probably the most reported site of phosphorylation on EGFR, in HCLEs, this site has a basal level of phosphorylation, and although direct activation of EGFR results in its phosphorylation, the process does not appear to be mediated by activation of the P2Y2 receptor. This was verified in our SILAC experiments, in which knockdown of the P2Y2 receptor and stimulation with UTP resulted in decreased phosphorylation of Y974, Y1086, and Y1148 but no change in Y1173 phosphorylation compared with control. Thus, this method of experimentation and analysis allowed us to compare protein expression and modification in cells and, despite the low stoichiometry of tyrosine phosphorylation and the use of low levels of agonist, the analytical sensitivity yielded highly confident results.
We further determined that the agonist-specific induced phosphorylation of EGFR selectively modified downstream signaling proteins (eg, PLC-γ1, Shc, PKC, Src, paxillin, FAK, and MAPK). For instance, among the PKC isoforms, only PKCΔ was more highly phosphorylated by ATP than EGF on Y313 and S304. In addition, we found that paxillin was more highly phosphorylated by ATP at four tyrosine and serine residues compared with EGF, whereas FAK has one site more highly phosphorylated by ATP and another by EGF. These data, along with the increase in the P2Y2 receptor mRNA with migration, indicated the importance of this receptor in wound repair. After culture of cells in stable isotope-labeled amino acids and knockdown of the P2Y2 receptor, we detected that the phosphorylation of specific residues on proteins described previously (eg, Y446 on Src, Y118 on paxillin, and Y1068 on EGFR) was reduced by >50%, indicating that their signaling correlated with the activation of this receptor. Previous studies have shown that mutation of Y1068 on EGFR reduces directional migration.14 Furthermore, this site is reduced under hypoxic conditions in which migration is attenuated in vitro and in organ culture models (data not shown). These indicate that the P2Y2 receptor does mediate a subset of downstream signaling proteins directly correlated to migration. The modifications of other proteins that are phosphorylated on activation by nucleotides may be mediated by other purinergic receptors and are a topic of future studies. The results from our current studies indicate residues that will be of interest to investigate. Moreover, the differential phosphorylation of specific residues on EGFR and downstream molecules indicates that it is imperative to examine specific sites on these and other dynamically modified phosphoproteins in pathological tissue, where wound repair is compromised.
Acknowledgments
We thank Dr. Amanda Del Rosario and the members of Dr. Forest White Laboratory (MIT, Cambridge, MA) for sharing their expertise and protocols in phosphotyrosine immunoprecipitation and enrichment.
Footnotes
Supported by NIH grants EY06000 and EY06000S and the New England Corneal Transplant Fund (V.T.-R.); NIH grants S10 RR017967, P41 RR10888/GM104603, S10 RR15942, and S10 RR020946 and NIH–National Heart, Lung, and Blood Institute contract HHSN268201000031C (C.E.C.); and a Massachusetts Lions Eye Research Fund Ophthalmology Departmental grant.
Current address of A.K., Buck Institute, Novato, CA.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2013.08.015.
Contributor Information
Catherine E. Costello, Email: cecmsms@bu.edu.
Vickery Trinkaus-Randall, Email: vickery@bu.edu.
Supplemental Data
References
- 1.Gomes P., Srinivas S.P., Van Driessche W., Vereecke J., Himpens B. ATP release through connexin hemichannels in corneal endothelial cells. Invest Ophthalmol Vis Sci. 2005;46:1208–1218. doi: 10.1167/iovs.04-1181. [DOI] [PubMed] [Google Scholar]
- 2.Burnstock G. Purinergic signaling and vascular cell proliferation and death. Arterioscler Thromb Vasc Biol. 2002;22:364–373. doi: 10.1161/hq0302.105360. [DOI] [PubMed] [Google Scholar]
- 3.Klepeis V.E., Weinger I., Kaczmarek E., Trinkaus-Randall V. P2Y receptors play a critical role in epithelial cell communication and migration. J Cell Biochem. 2004;93:1115–1133. doi: 10.1002/jcb.20258. [DOI] [PubMed] [Google Scholar]
- 4.Erb L., Liao Z., Seye C.I., Weisman G.A. P2 receptors: intracellular signaling. Pflugers Arch. 2006;452:552–562. doi: 10.1007/s00424-006-0069-2. [DOI] [PubMed] [Google Scholar]
- 5.Burnstock G. Purine and pyrimidine receptors. Cell Mol Life Sci. 2007;64:1471–1483. doi: 10.1007/s00018-007-6497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinger I., Klepeis V.E., Trinkaus-Randall V. Tri-nucleotide receptors play a critical role in epithelial cell wound repair. Purinergic Signal. 2005;1:281–292. doi: 10.1007/s11302-005-8132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boucher I., Rich C., Lee A., Marcincin M., Trinkaus-Randall V. The P2Y2 receptor mediates the epithelial injury response and cell migration. Am J Physiol Cell Physiol. 2010;299:C411–C421. doi: 10.1152/ajpcell.00100.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L., Cranson D., Trinkaus-Randall V. Cellular injury induces activation of MAPK via P2Y receptors. J Cell Biochem. 2004;91:938–950. doi: 10.1002/jcb.10774. [DOI] [PubMed] [Google Scholar]
- 9.Abbracchio M.P., Burnstock G., Boeynaems J.-M., Barnard E.A., Boyer J.L., Kennedy C., Knight G.E., Fumagalli M., Gachet C., Jacobson K.A., Weisman G.A. International Union of Pharmacology LVIII: update on the P2Y G protein-coupled nucleotide receptors: from molecular mechanisms and pathophysiology to therapy. Pharmacol Rev. 2006;58:281–341. doi: 10.1124/pr.58.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scodelaro Bilbao P., Boland R., Santillán G. ATP modulates transcription factors through P2Y2 and P2Y4 receptors via PKC/MAPKs and PKC/Src pathways in MCF-7 cells. Arch Biochem Biophys. 2010;494:7–14. doi: 10.1016/j.abb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Ratchford A.M., Baker O.J., Camden J.M., Rikka S., Petris M.J., Seye C.I., Erb L., Weisman G.A. P2Y2 nucleotide receptors mediate metalloprotease-dependent phosphorylation of epidermal growth factor receptor and ErbB3 in human salivary gland cells. J Biol Chem. 2010;285:7545–7555. doi: 10.1074/jbc.M109.078170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calvert J.A., Atterbury-Thomas A.E., Leon C., Forsythe I.D., Gachet C., Evans R.J. Evidence for P2Y1, P2Y2, P2Y6 and atypical UTP-sensitive receptors coupled to rises in intracellular calcium in mouse cultured superior cervical ganglion neurons and glia. Br J Pharmacol. 2004;143:525–532. doi: 10.1038/sj.bjp.0705959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin J., Yu F- S.X. ERK1/2 mediate wounding- and G-protein-coupled receptor ligands-induced EGFR activation via regulating ADAM17 and HB-EGF shedding. Invest Ophthalmol Vis Sci. 2009;50:132–139. doi: 10.1167/iovs.08-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boucher I., Kehasse A., Marcincin M., Rich C., Rahimi N., Trinkaus-Randall V. Distinct activation of epidermal growth factor receptor by UTP contributes to epithelial cell wound repair. Am J Pathol. 2011;178:1092–1105. doi: 10.1016/j.ajpath.2010.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yarden Y., Sliwkowski M.X. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 16.Huang F., Sorkin A. Growth factor receptor binding protein 2-mediated recruitment of the RING domain of Cbl to the epidermal growth factor receptor is essential and sufficient to support receptor endocytosis. Mol Biol Cell. 2005;16:1268–1281. doi: 10.1091/mbc.E04-09-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batzer A.G., Rotin D., Ureña J.M., Skolnik E.Y., Schlessinger J. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol Cell Biol. 1994;14:5192–5201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okutani T., Okabayashi Y., Kido Y., Sugimoto Y., Sakaguchi K., Matuoka K., Takenawa T., Kasuga M. Grb2/Ash binds directly to tyrosines 1068 and 1086 and indirectly to tyrosine 1148 of activated human epidermal growth factor receptors in intact cells. J Biol Chem. 1994;269:31310–31314. [PubMed] [Google Scholar]
- 19.Boucher I., Yang L., Mayo C., Klepeis V., Trinkaus-Randall V. Injury and nucleotides induce phosphorylation of epidermal growth factor receptor: MMP and HB-EGF dependent pathway. Exp Eye Res. 2007;85:130–141. doi: 10.1016/j.exer.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas S.M., Bhola N.E., Zhang Q., Contrucci S.C., Wentzel A.L., Freilino M.L., Gooding W.E., Siegfried J.M., Chan D.C., Grandis J.R. Cross-talk between G protein–coupled receptor and epidermal growth factor receptor signaling pathways contributes to growth and invasion of head and neck squamous cell carcinoma. Cancer Res. 2006;66:11831–11839. doi: 10.1158/0008-5472.CAN-06-2876. [DOI] [PubMed] [Google Scholar]
- 21.Schäfer B., Marg B., Gschwind A., Ullrich A. Distinct ADAM metalloproteinases regulate G protein-coupled receptor-induced cell proliferation and survival. J Biol Chem. 2004;279:47929–47938. doi: 10.1074/jbc.M400129200. [DOI] [PubMed] [Google Scholar]
- 22.Huang P.H., Miraldi E.R., Xu A.M., Kundukulam V.A., Del Rosario A.M., Flynn R.A., Cavenee W.K., Furnari F.B., White F.M. Phosphotyrosine signaling analysis of site-specific mutations on EGFRvIII identifies determinants governing glioblastoma cell growth. Mol Biosyst. 2010;6:1227–1237. doi: 10.1039/c001196g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villén J., Gygi S.P. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gan C.S., Guo T., Zhang H., Lim S.K., Sze S.K. A comparative study of electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) versus SCX-IMAC-based methods for phosphopeptide isolation/enrichment. J Proteome Res. 2008;7:4869–4877. doi: 10.1021/pr800473j. [DOI] [PubMed] [Google Scholar]
- 25.Chen X., Wu D., Zhao Y., Wong B.H.C., Guo L. Increasing phosphoproteome coverage and identification of phosphorylation motifs through combination of different HPLC fractionation methods. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:25–34. doi: 10.1016/j.jchromb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 27.Hilger M., Mann M. Triple SILAC to determine stimulus specific interactions in the Wnt pathway. J Proteome Res. 2012;11:982–994. doi: 10.1021/pr200740a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanucara F., Eyers C.E. Mass spectrometric-based quantitative proteomics using SILAC. Meth Enzymol. 2011;500:133–150. doi: 10.1016/B978-0-12-385118-5.00008-6. [DOI] [PubMed] [Google Scholar]
- 29.Schulze W.X., Deng L., Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100012. 2005.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo L., Kozlosky C.J., Ericsson L.H., Daniel T.O., Cerretti D.P., Johnson R.S. Studies of ligand-induced site-specific phosphorylation of epidermal growth factor receptor. J Am Soc Mass Spectrom. 2003;14:1022–1031. doi: 10.1016/S1044-0305(03)00206-X. [DOI] [PubMed] [Google Scholar]
- 31.Zoumaro-Djayoon A.D., Heck A.J.R., Muñoz J. Targeted analysis of tyrosine phosphorylation by immuno-affinity enrichment of tyrosine phosphorylated peptides prior to mass spectrometric analysis. Methods. 2012;2:268–274. doi: 10.1016/j.ymeth.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Von Kügelgen I., Wetter A. Molecular pharmacology of P2Y-receptors. Naunyn Schmiedebergs Arch Pharmacol. 2000;362:310–323. doi: 10.1007/s002100000310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.