Abstract
There is increasing experimental evidence that cells can utilize biochemical noise to switch probabilistically between distinct gene-expression states. In this paper, we demonstrate that such noise-driven switching is dominated by tails of probability distributions and is therefore exponentially sensitive to changes in physiological parameters such as transcription and translation rates. Exponential sensitivity limits the robustness of noise-driven switching, suggesting cells may use other mechanisms in order to switch reliably. We discuss our results in the context of competence in the bacterium Bacillus subtilis.
1. Introduction
Recent experiments indicate that cells can use noise in gene expression to switch probabilistically between distinct gene-expression states [1]. For example, in the phenomenon of competence, the soil-dwelling bacterium Bacillus subtilis utilizes noise in gene expression to switch probabilistically between a vegetatively growing state and a competent state where a bacterium can take up exogenous DNA [2–6]. Noise is also likely responsible for the phenotypic heterogeneity found in mycobacteria [7] and the bistable behavior of B. subtilis in nutrient-rich media where genetically identical cells are found in two distinct phenotypes: motile swimmers and immobile chains [8]. Recent experiments also suggest that a noise-driven switch is at least in part responsible for the ability of both yeast [9] and the human fungal pathogen Candida albicans [10, 11] to exist in two distinct phenotypes. The role of stochastic fluctuations in many other biological phenomenon such as the lytic-lysogeny decision during bactriophage infection is still the subject of current debate [12–14].
In this paper, we demonstrate that in standard models of noise-driven switching [15–26] many biologically relevant quantities, such as the switching rate, exhibit an exponential sensitivity to physiological parameters. The root cause of this exponential sensitivity is that switching properties are dominated by rare concatenations of biochemical events [23, 24]. Consequently, great care must be taken when calculating switching properties using coarse-grained models [26]. In particular, we show that sources of noise often ignored in theoretical treatments, such as protein bursting [28–31], are extremely important for noise-driven switches. However, provided mRNA lifetimes are short, we show that switching can still be accurately simulated using protein-only models of gene expression.
The exponential sensitivity of switching may also have important biological consequences. For example, under many physiological conditions, B. subtilis cells can become competent only during a window of opportunity at the end of exponential growth [2, 3]. Thus, in standard models of switching, small changes in physiological parameter are likely to lead to large variations in the percentage of cells that become competent. It is then natural to ask if and how cells can achieve robust switching. We address this issue by discussing several alternative switching models that are more robust to changes in kinetics parameters.
2. Basic model of bistability
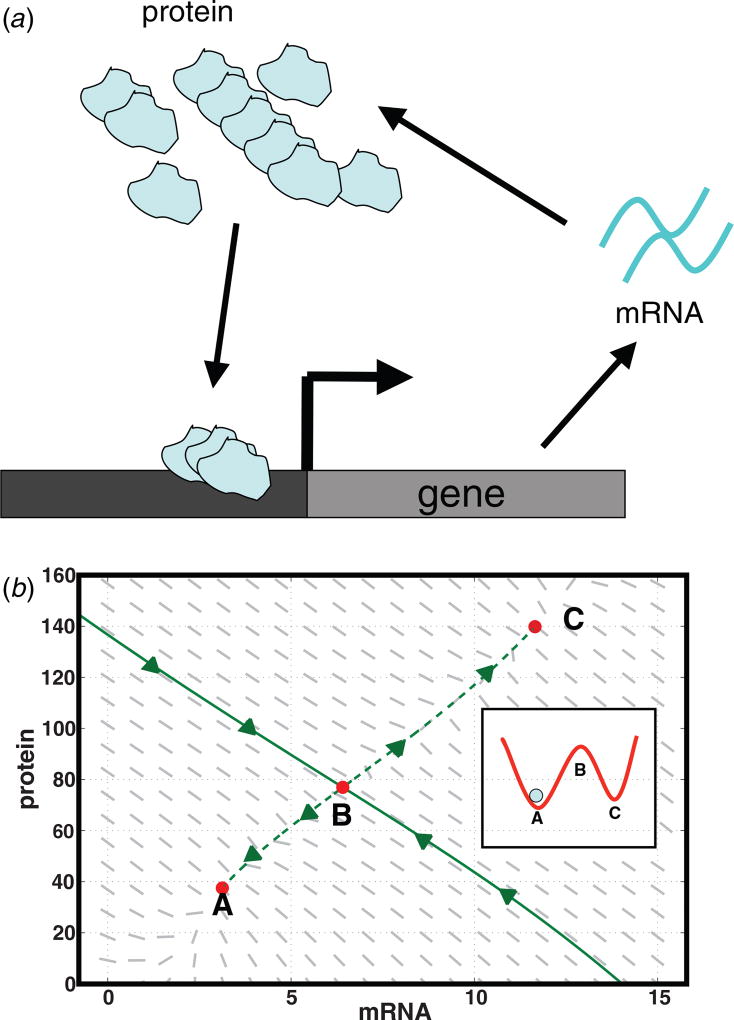
Perhaps the simplest example of a genetic switch is a genetic network in which a protein induces its own expression (see figure 1). If this feedback is nonlinear, e.g. if multiple copies of the protein are required to induce expression, such a network may be bistable with two distinct steady states, a low-expression state where the protein is present in small numbers and a high-expression state where the protein is present in large numbers. Noise from the inherent stochasticity in biochemical reactions can probabilistically switch a cell between these two gene-expression states [32, 33]. Such a simple bistable network is at the core of many noise-driven systems in biology [8, 10, 11]. For example, in competence, the master-regulator protein ComK promotes its own expression through a positive-feedback loop with noise in gene-expression switching bacteria from a low-ComK vegetative state to a high-ComK competent state in response to stress. It is worth noting that under certain physiological conditions, the high-ComK competent state is actually transient. However, since we are concerned only with entrance into competence this will not be important to our arguments below.
Figure 1.
(a) Schematic of a simple genetic network in which a protein activates its own transcription and (b) the phase portrait of the network dynamics. In mean-field theory, this network can exhibit bistable behavior with two stable fixed points (A and C), an intermediate unstable fixed point (B) and a separatrix (solid green curve) dividing the basins of attraction of the stable fixed points. Noise in transcription (gene → mRNA) and translation (mRNA → protein) can drive transitions between A and C. Inset: as discussed in the main text, switching is analogous to thermally-driven barrier crossing in a double well potential.
The average number of mRNAs, m̄, proteins, p̄, in such a genetic network can be described by the deterministic differential equations
| (1) |
| (2) |
where fm(p) is of the Hill form and is given by
| (3) |
with α0m being the basal rate of mRNA transcription and the second term representing positive feedback. Equation (1) describes the change in the mean number of mRNAs due to mRNA transcription and degradation. Equation (2) describes the change in the mean number of proteins due to protein translation and degradation. In the bistable regime, these equations have two stable fixed points divided by a separatrix containing an unstable intermediate fixed point (see figure 1).
The deterministic differential equations (1) and (2) ignore fluctuations in mRNA and protein numbers. To capture the effects of these fluctuations, we employ the exact Master Equation for the probability, P(m, p), of finding m mRNA molecules and p protein molecules [34]. Define shift operators, , which act on an arbitrary function g(m, p) according to and . In terms of these operators, the Master Equation describing our simple bistable switch is
| (4) |
The terms in equation (4) have the same biological meaning as the corresponding terms in equations (1) and (2). However, the Master Equation describes the full probability distributions P(m, p) and not just the mean numbers of mRNAs and proteins. In writing this equation, we have assumed that the noise in the system is due to intrinsic noise in transcription and translation [28] and we have ignored other sources of noise. In particular, we have modeled both protein and mRNA production with a simple Poisson model. This approximation may not hold in many biological systems where, in particular, transcription of mRNA may occur in bursts [35–37].
3. Exponential sensitivity of switching
An important property of biological switches is the mean first-passage time (MFPT)—in our case, the average time it takes to switch gene-expression states. The MFPT is also inversely related to the mean switching rate for a population. As discussed in the introduction, in competence the MFPT is directly related to the number of cells in a bacterial population that become competent during a window of opportunity at the end of exponential growth [2, 3]. We have calculated the MFPT for the simple positive-feedback network described by equation (4) by performing 5000 independent Gillespie simulations for each set of parameters and averaging the first-passage times (FPTs). (The Gillespie algorithm is a dynamic Monte Carlo method that numerically generates trajectories of a stochastic Master Equation [38].) The FPT for each simulation was calculated by initializing the system at the low-protein-number fixed point and calculating the time the trajectory took to reach the deterministic separatrix. Figure 2 shows the MFPT for two different ratios of the mRNA and protein lifetimes, holding the mean protein number fixed. When τm/τp = 1/16 ≪ 1, mRNAs are short-lived and typically many proteins are produced in a rapid burst from each mRNA molecule. In contrast, when τm/τp = 1, proteins are produced nearly continuously. In the two simulations, the mean number of proteins at the fixed points is identical. This is achieved by varying αp with τm so that b̄ = αpτm, the average number of proteins made per mRNA, is fixed.
Figure 2.
Mean first-passage time (MFPT) from the low-protein-number fixed point (A) to the separatrix (cf figure 1) as a function of the transcription-rate coefficient αm. The MFPT is shown for two distinct regimes: continuous protein synthesis (dashed purple curve) where mRNA lifetimes and protein lifetimes are equal (τm/τp = 1) and bursty protein synthesis (solid red curve) where mRNA lifetimes are much shorter than protein lifetimes (τm/τp = 1/16) so that proteins are made in bursts. All times are in minutes and rates in inverse minutes. Each MFPT was obtained by averaging the first-passage time from 5000 simulations, with parameters τp = 8, α0m = 0.5, Kd = 100, q = 3, b̄ = αpτm = 10. Inset: typical stochastic trajectory for protein number from a Gillespie simulation with bursty protein synthesis with αm = 2.6.
Note that the MFPT is exponentially sensitive to the transcription-rate coefficient αm. Similar sensitivity is observed with respect to other parameters such as the protein-degradation rate (data not shown). The MFPT in figure 2 also shows a strong dependence on the ratio of mRNA and protein lifetimes though the mean protein number is held fixed. In particular, the MFPT is significantly shorter when proteins are produced in bursts (τm/τp ≪ 1). Bursty protein production increases protein fluctuations, and as a result lowers the MFPT for switching. This dramatic reduction in MFPT due to bursting has also been observed in the context of a dimerizing genetic switch [27]. The increase in protein fluctuations due to bursts of production can be quite large. For example, the variance of the number of proteins for a fixed rate of mRNA transcription is given in the bursting limit by [28–31]
| (5) |
where b̄ = αpτm ≫ 1 is the mean burst size, i.e. the mean number of proteins made from each mRNA. This should be contrasted with the case where proteins are produced continuously and 〈δp2〉/p̄2 = 1/p̄ ≪ (1 + b̄)/p̄.
In the bursting limit (τm ≪ τp), gene regulation is often approximated using protein-only models. In these approximations, time derivatives of the mRNA species are set equal to zero, and the resulting mean mRNA concentrations are substituted into the equations describing protein dynamics. For our network, such a procedure yields the reduced equation
| (6) |
It is worth noting that since these approximations fail when τm ≥ τp, switching properties cannot be accurately described by protein-only models in the continuous protein synthesis regime.
4. Incorporating translational bursting in protein-only models
Even in the bursting regime, commonly used protein-only models often fail to accurately reproduce switching kinetics since these models neglect or oversimplify noise sources such as protein bursting [27]. Stochastic fluctuations are often included in protein-only models by assuming that proteins are produced probablistically one at a time. This assumption leads to the master equation
| (7) |
We have calculated the MFPT for switching between the low-expression and high-expression states for this Master Equation using the Gillespie algorithm and the result is shown in figure 3. Note that the MFPTs calculated in this way can be nearly four orders of magnitude longer than those calculated using the full mRNA-protein model, demonstrating that protein bursting significantly increases the rate of switching.
Figure 3.
Comparison of mean first-passage times (MFPTs) between the full mRNA-protein model and various protein-only models. The mean protein level at the fixed point is the same for all models and the MFPT is the average of 5000 independent simulations: (solid red curve) MFPT for bursty mRNA-protein model from figure 2; (dotted green curve) MFPT for a model where proteins are produced in bursts of size b, where b is drawn from a geometric distribution with mean b̄ = 10; (dashed blue curve) MFPT for a protein-only model where proteins are produced in bursts of size b = 10; (dot-dashed curve) MFPT for a model where proteins are produced without bursting (b = 1).
In fact, it is straightforward to include protein bursting in protein-only models in the bursting regime [39]. Since mRNA degradation and protein translation from an mRNA molecule are independent Poisson processes, it can be easily shown that the protein-burst size b (defined as the number of proteins produced from a single mRNA molecule) is geometrically distributed with mean burst size b̄ = αpτm and distribution
| (8) |
We consider two different approximations for incorporating bursting in a protein-only model. In the first, once an mRNA is produced, translation proceeds deterministically and b̄ proteins are produced. In the second, b proteins are produced from a single mRNA molecule with the geometric distribution, G(b). Both these approximations assume τm ≪ τp since all the proteins from a single mRNA molecule are considered to be produced instantaneously. The MFPTs computed within these two approximations are shown in figure 3. As for the full mRNA-protein model, the protein-only MFPTs are exponentially sensitive to changes in the transcription-rate coefficient αm. The MFPT computed using the geometric distribution of protein-burst sizes agrees well with the MFPT from the full mRNA-protein model. However, the protein-only model where proteins are produced with a fixed burst size b̄ is inadequate for calculating the MFPT even though this approximation correctly yields equation (5) for the variance in protein number at a fixed rate of mRNA transcription. Note that the MFPT computed within the geometric-burst-size approximation is smaller than that computed using the fixed-burst-size approximation. This is expected since geometrically distributed burst sizes increase the fluctuations in protein production.
5. Discussion
A qualitative understanding of our simulation results can be obtained from the Fokker–Planck approximation to the Master Equation (7). Gaussian approximations such as the Fokker–Plank and Langevin equations generally fail to accurately capture switching properties since they underestimate rare fluctuations [23], but nonetheless they allow us to identify the regime of exponential sensitivity. The Fokker–Plank equation that approximates our genetic network is [34]
| (9) |
where and . In deriving (9), we assumed that the state of the system is described by a single reaction coordinate, protein number, and have ignored fluctuations such as transcription-factor binding and unbinding. Additionally, we have made the simplifying approximation that proteins are produced in bursts of size b̄. We can calculate the MFPT using equation (9) and obtain [40]
| (10) |
where we have defined an effective potential U(p) using the equation and where p̄A(B) is the mean number of proteins at the low-expression stable fixed point (intermediate-expression, unstable fixed point). The result is derived assuming the constant-diffusion approximation where we replace a2(p) by its value at the low-expression fixed point a2(p̄A) and performing a saddle-point approximation. As expected, this approximate Fokker–Plank equation correctly determines the regime of exponential sensitivity but does not quantitatively reproduce the MFPT. Note that the MFPT resembles the FPT for barrier crossing by a particle moving in a potential U(p) with the effective diffusion constant D = a2(p̄A) and the barrier height UBA [41]. The analogy between barrier crossings and biochemical switches was discussed earlier in [19, 20]. We have extended this analogy to include protein bursting and shown that the burst size changes both the barrier height and the effective diffusion constant. When UBA/D ≫ 1, i.e. when the barrier height is large, the MFPT is exponentially sensitive to changes in the kinetic parameters that determine U(p) and D. Thus, exponential sensitivity indicates that switching is dominated by the tails of a probability distribution and the central limit theorem no longer applies. Thus, aspects of gene regulation, such as protein bursting which may not be important when considering small fluctuations around mean protein levels, may still play an important role in governing switching rates.
6. Conclusion and outlook
There is increasing evidence that the dynamic properties of bistable genetic switches are extremely sensitive to the details of gene regulation. This work, as well as others [25–27], shows that small changes in parameters can lead to large changes in the dynamics of switching. We have argued that this sensitivity occurs because the dynamics of switching depends exponentially on the effective barrier height and the effective diffusion coefficient for switching. Such exponential sensitivity has important theoretical implications for coarse-grained descriptions of biochemical networks [25]. For example, many sources of noise, such as mRNA bursting [35–37] and protein dimerization [16, 25, 26], likely play important roles in determining switching rates in some biological systems. Thus, great care must be taken theoretically when integrating out sources of noise [25]. This work complements and extends earlier theoretical studies by showing that when mRNA lifetimes are short, protein fluctuations can be correctly incorporated using protein-only models. In addition, it provides a simple understanding of exponential sensitivity using intuition from the statistical mechanics of barrier crossings.
The exponential sensitivity of genetic switches also has important physiological consequences. In recent experiments, Maamar et al [2, 3] showed that protein bursting is a major source of the noise driving the induction of competence. Furthermore, they were able to change the noise-driven switching rate (the inverse of the MFPT) in the B. subtilis competence system, while keeping mean protein numbers fixed, by manipulating transcription and translation rates. This suggests that evolution can tune switching rates while keeping mean protein levels fixed. (Note, however, that switching rates are also likely to be influenced by additional factors such as temperature, pH, fluctuations in the number of cellular components important for transcription and translation such as polymerases and ribosomes, and extra-cellular factors such as chemicals present in the environment.)
By our analysis, switching—because it is like barrier crossing in statistical mechanics—is likely to be exponentially sensitive to all these factors. Indeed, this may be one of the causes for the large variations observed in experiments on the number of swimming cells and chains in a cultures of B. subtilis [8]. However, it is then natural to ask if, and how, cells can achieve robust switching. For example, robust switching may be achieved using alternative switching mechanisms such as all-or-none events that switch the cell between the low- and high-expression states [42]. Such an all-or-none event could correspond to the disassembly of an entire repressor complex bound to the promoter of a gene, allowing multiple mRNA molecules to be made rapidly. If switching is dominated by these all-or-none events and not the tails of probability distributions, then switching is no longer necessarily exponentially sensitive to changes in parameters. This scenario may be relevant to competence since a repressor complex is known to be important in regulating the master competence regulator ComK [43]. Alternatively, cells could switch robustly by actively tuning noise to control switching rates. Recent experiments suggest that this may indeed be the case in mycobacteria [7]. A final possibility is that large variations in the switching rate may be acceptable or even desirable for cells living in certain environments [44, 45]. Thus, cells may exploit the exponential sensitivity of noise-driven switching to increase their fitness. It will be interesting to better understand the ecological and evolutionary implications of noise-driven switching.
Acknowledgments
We would like to acknowledge D Dubnau, H Maamar, A Raj, M Elowitz, G Suel, S Goyal and A Sengupta for useful discussions. This work was supported in part by US National Institutes of Health (NIH) grants R01 GM073186 and PSO GM071508, and by the Defense Advanced Research Project Agency (DARPA) under grant HR0011-05-0057.
Glossary
- Competence
A physiological state that allows a bacterial cell to take up exogenous DNA.
- Master equation
A mathematical equation that describes the time evolution of a probability distribution.
- Mean first-passage time (MFPT)
In our context, the average time it takes for a bistable genetic switch to transition from one gene-expression state to another due to fluctuations. The MFPT is inversely related to the switching rate.
References
- 1.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–8. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:463. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smits WK, Eschevins CC, Susanna KA, Bron S, Kuipers OP, Hamoen LW. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol. 2005;56:604–14. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- 4.Sel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 5.Sel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–9. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 6.Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003;149:9. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- 7.Sureka K, Ghosh B, Dasgupta A, Basu J, Kundu M, Bose I. Positive feedback and noise activate the stringent response regulator Rel in mycobacteria. 2007 doi: 10.1371/journal.pone.0001771. (Preprint 0802.1580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubnau D, Losick R. Bistability in bacteria. Mol. Microbiol. 2006;61:564. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 9.Kaufmann B, Yang Q, Mettetal J, van Oudernaarden A. Heritable stochastic switch revealed by single-cell genealogy. PLoS Biol. 2007;5:e239. doi: 10.1371/journal.pbio.0050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc. Natl Acad. Sci. USA. 2007;103:12659–60. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kourilsky P. Lysogenization by bacteriophage lambda: I. Multiple infection and the lysogenic response. Mol. Gen. Genet. 1973;122:183–95. doi: 10.1007/BF00435190. [DOI] [PubMed] [Google Scholar]
- 13.Kourilsky P, Knapp A. Lysogenization by bacteriophage lambda: III. Multiplicity dependent phenomena occurring upon infection by lambda. Biochimie. 1974;56:1517–23. [PubMed] [Google Scholar]
- 14.St-Pierre F, Endy D. Pre-existing variation controls the lysis-lysogeny decision in phage lambda infection; The First q-bio Conference on Cellular Information Processing.2007. [Google Scholar]
- 15.Karmakar R, Bose I. Positive feedback, stochasticity and genetic competence. Phys. Biol. 2007;4:29. doi: 10.1088/1478-3975/4/1/004. [DOI] [PubMed] [Google Scholar]
- 16.Walczak AM, Onuchic JN, Wolynes PG. Absolute rate theories of epigenetic stability. Proc. Natl Acad. Sci. USA. 2005;102:18926–31. doi: 10.1073/pnas.0509547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornos EM, Schultz D, Innocentini GCP, Wang J, Walczak AM, Onuchic JN, Wolynes PG. Phys. Rev. E. 2005;72:051907. doi: 10.1103/PhysRevE.72.051907. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q, Jia Y. Fluctuations-induced switch in the gene transcriptional regulatory system. Phys. Rev. E. 2004;70:41970. doi: 10.1103/PhysRevE.70.041907. [DOI] [PubMed] [Google Scholar]
- 19.Aurell E, Sneppen K. Epigenetics as a first exit problem. Phys. Rev. Lett. 2002;88:048101. doi: 10.1103/PhysRevLett.88.048101. [DOI] [PubMed] [Google Scholar]
- 20.Bialek W. Advances in Neural Information Processing. Vol. 13. Cambridge, MA: MIT Press; Stability and noise in biochemical switches. [Google Scholar]
- 21.Metzler R. The future is noisy: the role of spatial fluctuations in genetic switching. Phys. Rev. Lett. 2001;87:68103. doi: 10.1103/PhysRevLett.87.068103. [DOI] [PubMed] [Google Scholar]
- 22.Schultz D, Ben Jacob E, Onuchic JN, Wolynes PG. Molecular level stochastic model for competence cycles in. Bacillus subtilis Proc. Natl Acad. Sci. USA. 2007;104:17582–7. doi: 10.1073/pnas.0707965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roma DM, O’Flanagan RA, Ruckenstein AE, Sengupta AM, Mukhopadhyay R. Optimal path to epigenetic switching. Phys. Rev. E. 2005;71:11902. doi: 10.1103/PhysRevE.71.011902. [DOI] [PubMed] [Google Scholar]
- 24.Allen J, Warren P, ten Wolde PR. Sampling rare switching events in biochemical networks. Phys. Rev. Lett. 2005;94:018104. doi: 10.1103/PhysRevLett.94.018104. [DOI] [PubMed] [Google Scholar]
- 25.Morelli MJ, Tanase-Nicola S, Allen RJ, Wolde PR. Eliminating fast reactions in stochastic simulations of biochemical networks: a bistable genetic switch. J. Chem. Phys. 2008;128:45105. doi: 10.1063/1.2821957. [DOI] [PubMed] [Google Scholar]
- 26.Morelli MJ, Tanase-Nicola S, Allen RJ, ten Wolde PR. Reaction coordinates for the flipping of genetic switches. Biophys. J. 2008;94:3413–23. doi: 10.1529/biophysj.107.116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warren P, ten Wolde PR. Chemical models of genetic toggle switches. J. Chem. Phys. B. 2005;109:6812–23. doi: 10.1021/jp045523y. [DOI] [PubMed] [Google Scholar]
- 28.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1193. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 29.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nature Genet. 2002;34:69. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 30.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–3. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 32.Becskei A, Seraphin B, Serrano L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 2001;20:2528. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blake WJ, KAErn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–7. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 34.Van Kampen NG. Stochastic Processes in Chemistry and Physics. Amsterdam: North Holland: 2001. [Google Scholar]
- 35.Golding I, Paulsson J, Zawilski JM, Cox ED. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 36.Chubb JR, Trek T, Shenoy S, Singer R. Transcriptional pulsing of a developmental gene. Curr. Biol. 2006;16:1018–25. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gillespie DT. Stochastic simulations of coupled chemical reactions. J. Phys. Chem. 1977;81:2340. [Google Scholar]
- 39.Friedman N, Cai L, Xie X. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys. Rev. Lett. 2006;97:168302. doi: 10.1103/PhysRevLett.97.168302. [DOI] [PubMed] [Google Scholar]
- 40.Procaccia I, Ross J. Stability and relative stability in reactive systems far from equilibrium: II. Kinetic analysis of relative stability of multiple stationary states. J. Chem. Phys. 1977;67:5565. [Google Scholar]
- 41.Hanggi P, Talkner P, Borkovec M. Reaction rate theory: fifty years after Kramers. Rev. Mod. Phys. 1990;62:251. [Google Scholar]
- 42.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proc. Natl Acad. Sci. USA. 1957;43:553. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smits WK, Hoa TT, Hamoen LW, Kuipers OP, Dubnau D. Antirepression as a second mechanism of transcriptional activation by a minor groove binding protein. Mol. Microbiol. 2007;64:368–81. doi: 10.1111/j.1365-2958.2007.05662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thattai M, Oudenaarden A van. Stochastic gene expression in fluctuating environments. Genetics. 2004;167:523. doi: 10.1534/genetics.167.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]