Abstract
Antigen-presenting cells (APCs) occupy diverse anatomical tissues, but their tissue-restricted homeostasis remains poorly understood. Here, working in mouse models of inflammation, we found that mTOR-dependent metabolic adaptation was required at discrete locations. mTOR was dispensable for DC homeostasis in secondary lymphoid tissues but necessary to regulate cellular metabolism and accumulation of CD103+ DCs and alveolar macrophages in lung. Moreover, while numbers of mTOR-deficient lung CD11b+ DCs were not changed, they were metabolically reprogrammed to skew allergic inflammation from eosinophilic Th2 to neutrophilic Th17 polarity. The mechanism for this change was independent of translational control but dependent on inflammatory DC, which produced IL-23 and increased fatty acid oxidation. mTOR therefore mediates metabolic adaptation of APCs in distinct tissues, influencing the immunological character of allergic inflammation.
A fundamental property of the immune system is its ability to sense pathogens, through pathogen recognition receptors. Emerging evidence demonstrates that immune cells also sense environmental perturbations that induce cellular stress and metabolic changes (1, 2). However, the mechanisms underlying such environmental sensing are poorly understood. The amino acid sensor GCN2 plays a role in programming DCs to respond to viral vaccination (3, 4) and controls intestinal inflammation by suppressing inflammasome activation in the gut (5). In addition to GCN2, the mechanistic target of rapamycin (mTOR), a central orchestrator of cellular metabolism, regulates DC function (6).
mTOR functions in two supramolecular complexes termed mTORC1 and mTORC2, engaged to differing degrees by diverse stimuli including TLR signaling, growth factors, and nutrient availability. mTOR signaling impacts diverse downstream cellular processes including cell cycle progression, cell growth, differentiation, survival and metabolism (7). While mTOR is required for effector T-cell expansion (8) and germinal center B cell responses (9), it restrains T-memory generation (10). Within DCs, mTOR promotes type-I interferon production by plasmacytoid DCs (11, 12), but limits proinflammatory cytokine production by classical DCs (13). Moreover, mTOR promotes steady-state DC accumulation downstream of homeostatic cytokine signaling (14–16). The precise functions of mTOR have been proposed to be DC subset dependent (17), but the consequence of mTOR signaling in DCs and impact on immune responses in vivo is poorly understood.
Results
mTOR promotes lung APC homeostasis in vivo
We bred mice bearing floxed Mtor with a CD11c-Cre deleter strain (mTORΔAPC hereon), resulting in specific genetic ablation, reduced protein abundance and loss of mTOR pathway activity in CD11c+ cells (fig. S1, A to D). mTOR inhibition augments DC proinflammatory cytokine production (13, 17), which we observed in mTORΔAPC bone marrow (BM)–derived DCs and primary splenic DCs (fig. S1E), confirming functional deletion of mTOR.
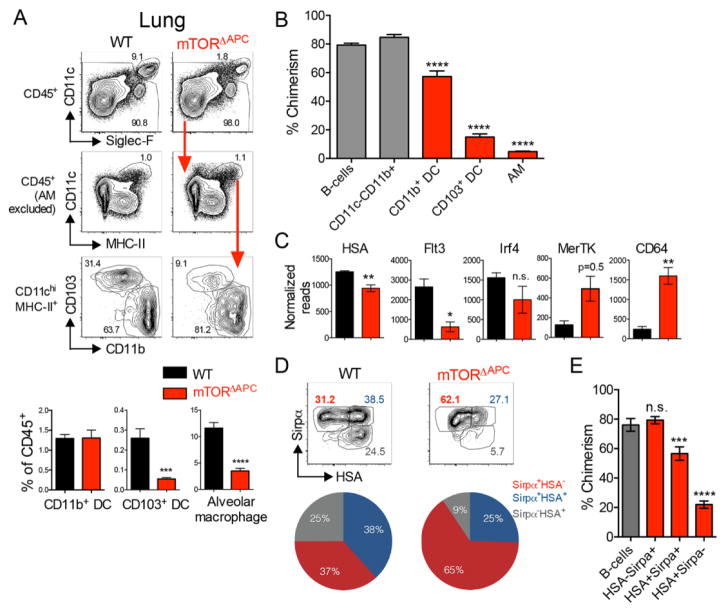
DCs can be subdivided into CD8α+/CD103+ and CD11b+ subsets. Globally, DCs require Flt3L and GM-CSF signals for homeostatic maintenance (18, 19), both of which activate mTOR (14). In addition, APCs occupy distinct microenvironments where mTOR may exhibit site-specific functions (15, 20). mTORΔAPC ablation did not exert global effects on APCs, but instead revealed defects in APC accumulation at specific anatomical sites. There was negligible requirement for cell-intrinsic mTOR in most tissues examined, however splenic CD11b+ DCs were modestly reduced and there was a skewed ratio of CD103+ to CD11b+ DCs in thymus (fig. S2). We also confirmed a substantial reduction in Langerhans cell frequency (15) (fig. S3). However, the most striking impact of mTORΔAPC deletion was observed in the lung. Here, CD103+ DC and CD11c+ alveolar macrophage (AM) populations were significantly reduced (Fig. 1A). Mixed BM chimeras showed homeostatic defects were cell intrinsic and additionally revealed a minor impairment in CD11b+ lung DC accumulation (Fig. 1B).
Fig. 1. Critical requirement for mTOR signaling to maintain lung innate immune homeostasis.
Lungs from naïve WT (black), mTORΔAPC (red) were analyzed by flow cytometry. (A) Bar charts show mean frequencies ± SEM of indicated lung APC populations, contour plots show representative flow cytometric data. Red arrows indicate gating hierarchies. (B) Mixed chimeras were established by reconstituting B6 CD45.1+/CD45.2+ hosts with donor mTORΔAPC (CD45.2) and B6 CD45.1 BM at a ~3:1 ratio. Bar chart shows the % contribution to chimerism of mTORΔAPC-derived donor cells after exclusion of residual host cells. Gray bars indicate control (CD11c− populations), red bars indicate CD11c+ APCs. (C) Bar charts show normalized read count for indicated genes in WT (black) and mTORΔAPC (red). (D) Contour plot shows expression of Sirpα and HSA. Pie chart shows average frequencies of indicated populations from one representative experiment (n = 4). (E) Bar chart shows the chimeric contribution of mTORΔAPC-derived donor cells. Gray bars denote control CD11c− populations, red bars indicate CD11c+ APCs. Data represents ≥3 independent experiments.
mTOR may simply regulate numerical accumulation of APCs, or alternatively may be central to control lung APC programming. Transcriptomes derived from mTORΔAPC lung APCs versus WT controls clustered distinctly by principal component analysis (PCA), whereas mTORΔAPC splenic populations clustered closely with their WT counterparts (fig. S4). Previous studies have identified core sets of lineage-specifying genes, enriched in either CD11b+ lung APCs or AMs, relative to other APC subsets (21, 22). mTORΔAPC AMs exhibited a broad loss of AM molecular identity (fig. S5A). Rather than disrupting the CD11b+ APC cellular identity, mTOR ablation skewed the balance of CD11c+CD11b+ lung APCs toward a macrophage/monocytic composition (fig. S5B) (22), typified by reduced HSA, Flt3 and a trend of lower Irf4 expression, with elevated MerTK and CD64 transcription (Fig. 1C). Utilizing an alternative gating scheme (22), we found that CD11c+Mhc-II+Sirpα+HSA+ DCs were reduced in mTORΔAPC mice (Fig. 1D), due to cell-intrinsic defects in accumulation (Fig. 1E). Thus, mTOR plays a central, cell-intrinsic role in APC homeostatic accumulation and subset composition in the lung.
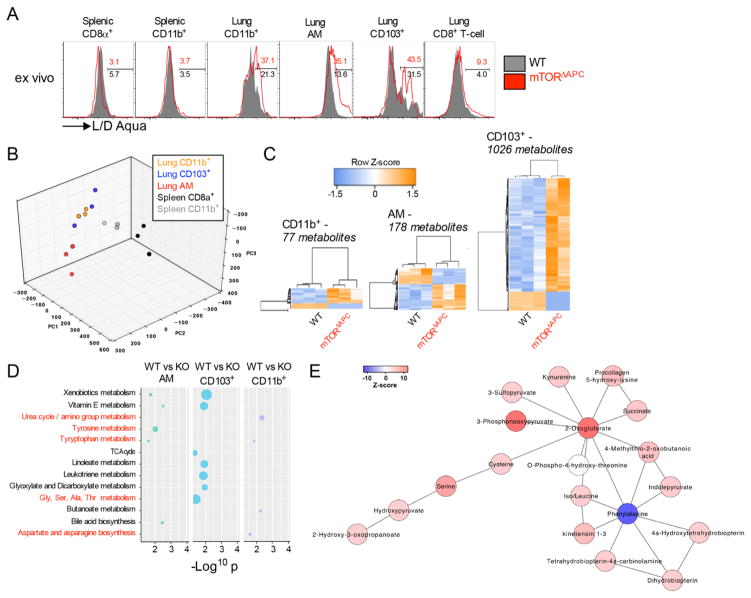
Steady-state DC homeostasis depends on cell development, proliferation, migration, and death rates. Ablation or inhibition of mTOR increased death-rates of lung APCs (Fig. 2A and fig. S6, A and B), whereas precursor frequency, proliferation rates and migration were unaffected (fig. S6, C to G). There was no evidence that GM-CSF, M-CSF or Flt3L expression was perturbed in mTORΔAPC mice, excluding a role for other cell extrinsic homeostatic factors (fig. S6H). mTOR associates with two molecular complexes (mTORC1/2). While mice lacking the critical mTORC1 protein Raptor phenocopied homeostatic defects seen in mTORΔAPC lung, APCs were unaffected in mTORC2-deficient RictorΔAPC mice (fig. S7A). mTORC1 signaling targets the translational regulator S6K1/2, and deactivates the translational repressor 4E-BP1/2 (fig. S7, B and C) (7). Curiously, S6k1−/−S6k2−/− mice exhibited normal lung APC homeostasis and genetic deletion of 4ebp1/2 did not rescue the survival of mTORΔAPC DCs (fig. S7, D and E). Thus, mTORC1 controls APC cell survival through a translation-independent mechanism.
Fig. 2. mTOR regulates lung APC survival via a metabolic mechanism Lung from WT (Black) or mTORΔAPC (red) mice was examined by flow cytometry and by cellular metabolomic profiling.
(A) Histograms show representative staining of L/D Aqua® on indicated ex vivo populations from WT (gray) and mTORΔAPC (red) lung. (B) PCA plot shows distinct clustering of lung and splenic APCs metabolomes from WT mice. (C) Heat maps show differentially abundant metabolite features between WT and mTORΔAPC subsets. The metabolite numbers include redundant mass spectral peaks. (D) Dot graph shows metabolic pathways distinctly perturbed by mTOR deficiency, with amino acid metabolism pathways highlighted in red. Dot size relates to the number of differentially expressed metabolites. (E) Network analysis reveals a higher representation of tentative amino acid metabolites within the mTORΔAPC CD103+ DC subset. Each link is from a known metabolic reaction. The network and tentative metabolite annotation was produced by the mummichog software. Data represents ≥3 independent experiments or 2–3 biological replicates.
APCs in the lung display a distinctive mTOR-dependent metabolic signature
The pronounced phenotype APCs in mTORΔAPC lung relative to other tissues was striking, particularly considering that development of CD103+ DCs in both lung and other tissues are regulated by the common transcription factor Batf3 (23). We hypothesized that the lung microenvironment may imprint a distinctive metabolic phenotype with heightened sensitivity to mTOR ablation. To investigate this, comprehensive metabolic profiling was performed on lung and splenic APCs from WT mice. Lung and spleen APCs were metabolically distinct (Fig. 2B), exhibiting differences in amino acid (AA) and nucleic acid metabolic intermediates (fig. S8, A and B), which were also reflected within observed transcriptional signatures (fig. S8, C and D). We next examined mTORΔAPC metabolomes, and the degree of metabolic perturbation in mTOR-deficient APC subsets correlated with the extent of their homeostatic disruption (Fig. 2C). Despite the skewing in the phenotypic composition of the lung CD11b+ DCs (Fig. 1D), there was a minor perturbation in the metabolome (Fig. 2C). Pathway analysis of differentially abundant metabolites revealed several AA metabolic pathways were disrupted in mTORΔAPC APCs (Fig. 2, D and E). mTORΔAPC AMs also exhibited alterations in lipid metabolite expression and corresponding changes in expression of the sterol regulatory binding protein (Srebp1/2) target genes (24) (fig. S9). Srebp1 orchestrates lipid anabolism and is a downstream target of mTOR, which directly links mTOR deficiency and metabolic perturbations in lung APCs (25, 26). To experimentally test this, we administered the Srebp1/2 inhibitor fatostatin, which substantially decreased AM frequency, a phenotype that was recapitulated by rapamycin treatment (fig. S10A). However, CD103+ DCs remained unaltered by either inhibitor, suggesting that inhibitor administration was insufficient to entirely pheno-copy mTORΔAPC mice, or that the Srebp pathways are not critical for CD103 DC survival. Finally, we investigated the role of another mTOR target, the glycolytic regulator Hif1α, observing that lung APC homeostasis was independent of Hif1α (fig. S10B). Thus, lung APCs have distinct mTOR-dependent metabolic signatures, and there appears to be a direct link between mTOR and Srebp-dependent lipid metabolism in AMs.
Ablation of mTOR in APCs reprograms allergic inflammation in the lung
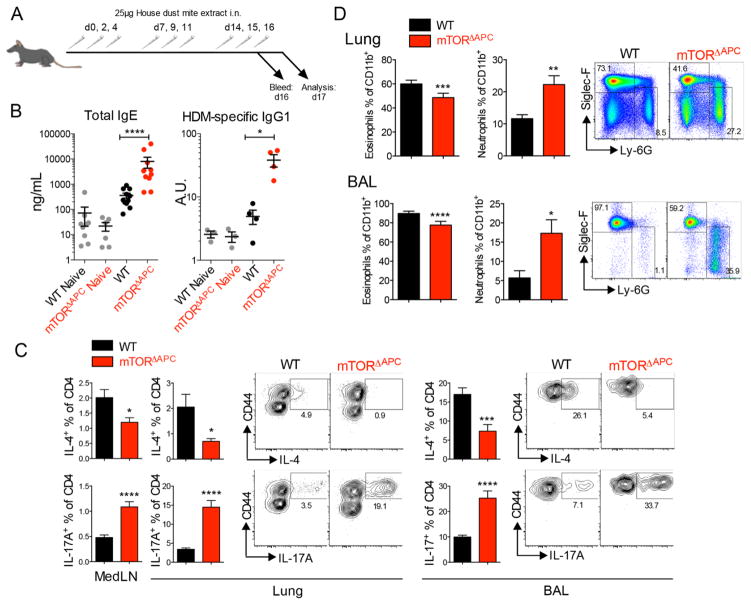
We next investigated whether lung immunity was functionally impacted in mTORΔAPC mice. Mice were intranasally (i.n.) challenged with house-dust mite (HDM) allergen, which is an established murine model for asthma (27, 28) (Fig. 3A). Despite reduced frequencies of select lung APC subsets, humoral responses in mTORΔAPC mice were unexpectedly increased (Fig. 3B and fig. S11A). Surprisingly, we observed a striking reduction in Th2 cells producing IL-4+ and IL5+/IL13+ and enhanced frequencies of Th17 cells in the lung and BAL (Fig. 3C and fig. S11B).
Fig. 3. mTOR ablation in APCs results in neutrophilic lung inflammation House dust mite (HDM) extract was administered to WT (black) or mTORΔAPC mice (red).
(A) Diagram shows the vaccination and analysis schedule. (B) Scatter bar charts show serum antibody titers in indicated mouse strains. (C) Bar charts show frequency of CD4+IL-4+ Th2 cells or CD4+IL-17A+ Th17 cells in indicated organs. Contour plots show representative flow cytometric data. (D) Bar charts show frequency of CD11b+Siglec-F+ eosinophils or CD11b+Ly-6G+ neutrophils. Pseudo-color plots show representative flow cytometric data. Data represents ≥3 independent experiments.
Th2 polarized allergy is typified by robust recruitment of eosinophils, whereas IL-17 can enhance neutrophils. Neutrophilia occurs in ~50% of human asthma cases, correlating with steroid-resistance and worsening prognosis (29, 30). Remarkably, we observed pronounced neutrophilia in mTORΔAPC lung and BAL (Fig. 3D). Skewed Th2 to Th17 modality was further observed in an alternative papain-induced model (fig. S12), whereas an anatomically discrete Th17-dependent experimental autoimmune encephalomyelitis (EAE) model was not significantly impacted by mTOR-deficiency in APCs (fig. S13). Th17 allergic phenotypes were driven by mTORC1 ablation (fig. S14A), but were independent of Hif1α (fig. S14B), or Srebp1/2 (fig. S14C). mTORC1 thus regulates innate programming of T-helper responses in lung, controlling the balance between eosinophilic and neutrophilic mediators of allergic inflammation.
mTOR-deficient CD103+ DCs and AMs are dispensable for neutrophilic allergy
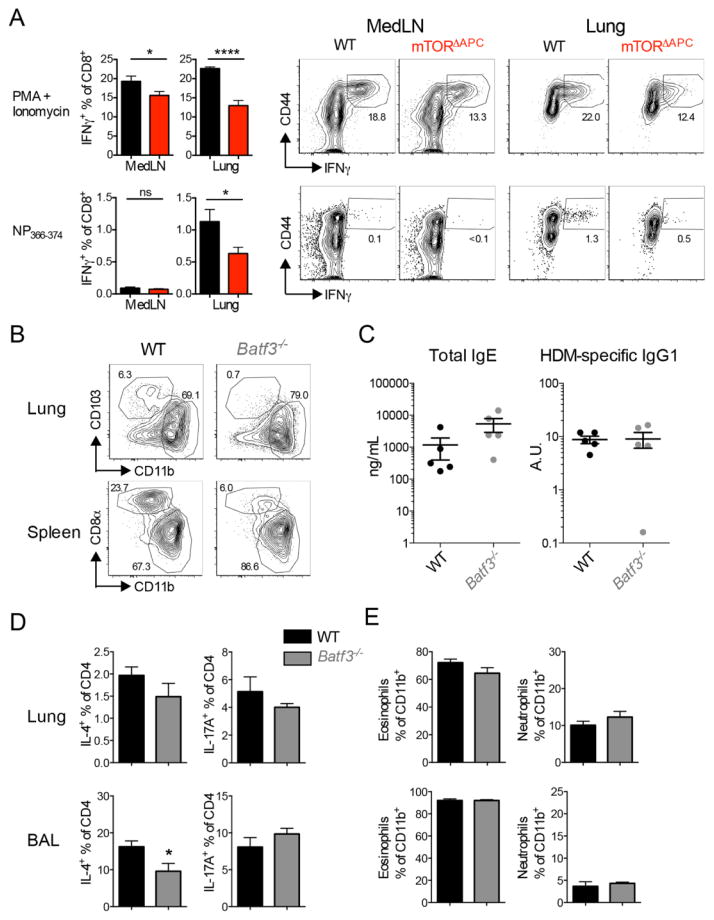
We next determined the APC subset responsible for promoting Th17/neutrophilic allergy in mTORΔAPC mice. CD103+ DCs have important antiviral functions, due to a superior ability to cross-present antigens (31). Consistent with this, mTORΔAPC mice exhibited impaired CD8+ T-cell responses to live-attenuated influenza vaccine challenge (Fig. 4A). If CD103+ DCs restrain Th17/neutrophilic allergy then we should observe a similar phenotype in Batf3−/− mice, which lack a transcription factor required for CD103+ DC development (Fig. 4B). However, Batf3−/− mice did not exhibit differential severity or modality of inflammation (Fig. 4, C to E), suggesting that impaired development CD103+ DC was not necessary to promote neutrophilic inflammation.
Fig. 4. mTOR-dependent CD103+ DCs are dispensable for neutrophilic lung inflammation but required for antiviral immunity.
(A) Flumist Quadrivalent vaccine (25 μL) was administered intranasally to WT (black) or mTORΔAPC mice (red). Vaccine response was examined 7 days post-infection. Bar charts show frequency of CD8+IFNγ+ cells in indicated organs. Contour plots show representative flow cytometric data. (B to E) HDM was chronically administered to WT (black) and Batf3−/− (gray) mice. (B) Contour plots show the absence of CD103+ and CD8α+ DCs in lung and spleen of Batf3−/− mice. (C) Serum ELISAs showing total IgE or representative HDM-specific IgG1 antibody titers. (D) Bar charts show Th2 (IL-4+) and Th17 (IL-17A+) frequencies in lung and BAL. (E) Bar charts show frequencies of eosinophils and neutrophils in lung and BAL. Pseudo-color plots show representative flow cytometric data. Data represents (A) ≥ 3 independent experiments, [(B) to (E)] n = 5 mice per group.
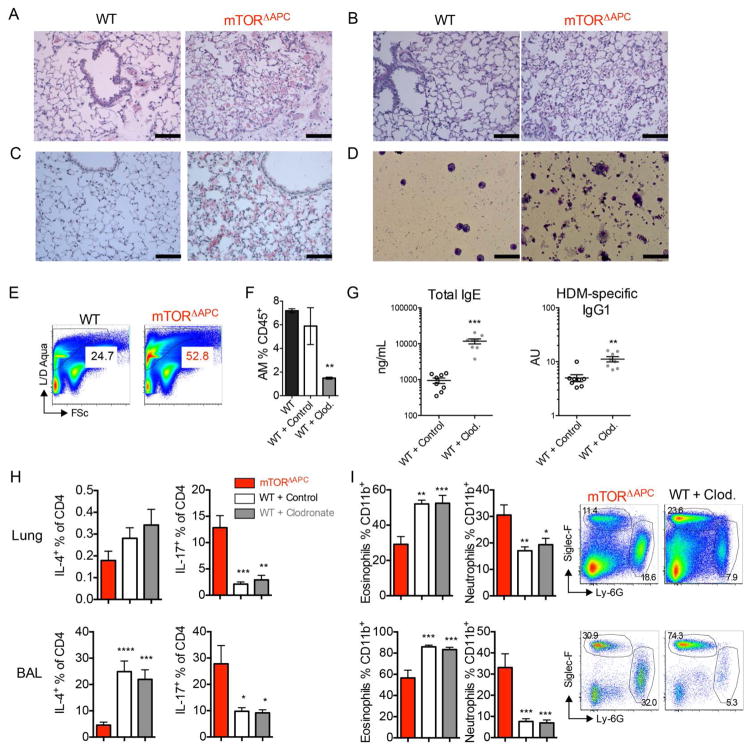
AMs reside in the lung alveolar lumen, and play a primary role in phagocytizing particulates and turning over lung surfactant. Defects in AM result in accumulated lung surfactant protein, termed pulmonary alveolar proteinosis (PAP) (32). Histological analysis of naïve mTORΔAPC lung showed airspace filling by eosinophilic material (Fig. 5A), elevated mucus (Fig. 5B), accumulation of lung surfactant protein (Fig. 5C), and increased debris and “foamy” macrophages in BAL (Fig. 5D). Increased proteinaceous matter was also evident in lung preparations by flow cytometry (Fig. 5E). These data represent hallmark features of PAP disease (33), representing a first link between mTOR signaling and PAP. We next determined whether functional loss of AM also contribute to neutrophilic lung inflammation. Using a previously described protocol (34), we observed ~75% depletion of AMs 3d after a single intratracheal (i.t.) dose of clodronate liposomes but not control liposomes (Fig. 5F). Following HDM challenge, humoral immunity was enhanced when AMs were depleted, in concordance with the phenotype observed in mTORΔAPC mice (Fig. 5G). However, Th17 polarization and neutrophilia were not present under these conditions (Fig. 5, H and I), consistent with observations that AMs cannot present antigen (28). Thus, absence of AM likely contributes to an enhanced magnitude of humoral immunity in mTORΔAPC mice, but cannot account for Th17/neutrophilic allergy.
Fig. 5. mTOR ablation in AMs results in PAP and enhances the antibody magnitude but not modality of allergy.
(A to C) Images show histopathological sections of WT or mTORΔAPC lung stained with (A) H&E, (B) PAS, (C) Immunohistochemical antibodies against surfactant protein-D. (D) BAL cellular fractions were prepared by cytospin, and analyzed after diff-quick staining. Slides are shown at [(A) to (C)] x200 magnification, where scale bars represent 100 μm, (D) x400 magnification, where scale bars represent 50 μm. (E) Pseudocolor plots show frequency of Live/Dead Aqua+ FSclo (dead cell/debris) events as determined by flow cytometry. (F to I) HDM was administered to indicated groups for 17d. Mice were treated with indicated clodronate or control liposome on day −3, 3 and 10. (F) AM frequency on day 0 prior to HDM administration. (G) Serum ELISAs showing total IgE or HDM-specific IgG1 antibody titers. (H) Bar charts show frequency of IL-4+ and IL-17+ T-cells in indicated groups and organs. (I) Bar charts show frequency of eosinophils and neutrophils in indicated groups and organs, pseudo color plots show representative data. Data represents [(A) to (D)] ≥3 slides from ≥2 biological replicates, [(E) to (I)] ≥2 representative experiments.
mTOR-deficiency reprograms inflammatory CD11b+ DCs to promote neutrophilic allergy
CD11b+ DCs directly promote Th2 allergy after HDM insult (28, 35), but can also prime Th17 immunity when activated by alternative stimuli (22, 36). To address whether mTORΔAPC CD11b+ DCs promoted neutrophilic allergy in a dominant fashion, we first generated mixed chimeras where donor WT and mTORΔAPC bone marrow was injected at a 1:10 ratio to allow repopulation of CD103+ DC and AM populations by WT cells, whereas ~80% of CD11c+CD11b+ DCs remained mTOR-deficient (fig. S15, A and B). HDM promoted elevated humoral responses and pronounced neutrophilia in chimeras (fig. S15, C and D). Moreover, primed CD11b+ DCs from mTORΔAPC mice directly promoted Th17/neutrophilic allergy in WT recipients (fig. S16). Thus, neutrophilic inflammation was dominantly conferred by mTORΔAPC CD11b+ DCs.
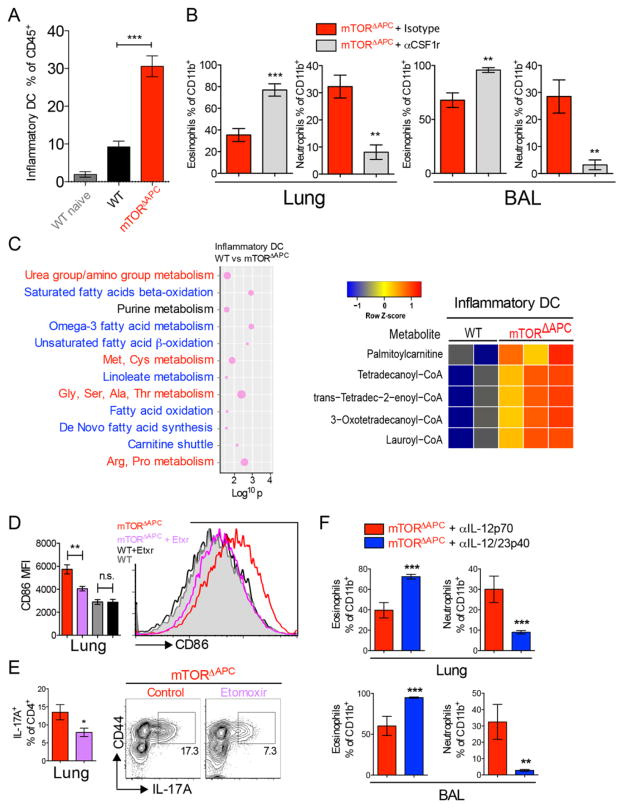
It was recently shown that mice deficient in Kruppel-like factor 4 (Klf4) have reduced frequencies of the Sirpα+HSA+ subset in lung CD11b+ DCs, and display impaired Th2 responses to HDM (37). We thus investigated how mTORΔAPC CD11b+ lung DCs differentially responded to allergy. HDM was instilled i.t. and mice examined after 3d. mTORΔAPC CD11b+ migratory DCs accumulated with enhanced frequency in MedLN (fig. S17A), and cellular inflammation was enhanced in the lung (fig. S17B). During allergic airway inflammation, a distinct CD11b+ Csf-1-dependent inflammatory DC population develops from monocytes (28, 38). Inflammatory DCs could be identified in allergic lung based on their surface CD11c+MHC-II+CD11b+ phenotype and Csf1-dependence (fig. S18, A and B) (28), and were elevated in mTORΔAPC mice (Fig. 6A). Lung inflammatory DCs were confirmed to be distinct from lung resident interstitial macrophages, AMs and classical DCs. They exhibited lower expression of F4/80, higher expression of MHC-II and CD11c compared to interstitial macrophages (fig. S18C), lower Siglec-F expression than AMs (fig. S18D), and high expression of CD64 and Ly6C relative to resident CD11b+ DCs (fig. S18E). Compared to WT, mTORΔAPC inflammatory DCs had higher expression of lineage-associated markers CD64 and Ly6C and elevated CD80 and CD86 costimulatory molecule expression (fig. S18, E and F), consistent with a more progressive differentiation/activation status. To directly test the requirement for inflammatory DCs in Th17/neutrophilic inflammation, we ablated this population by administering αCsf1r neutralizing antibodies, which reverted mTORΔAPC allergy to Th2/eosinophilic modality (Fig. 6B and fig. S19).
Fig. 6. CD11b+-intrinsic mTOR signaling restrains Th17/neutrophilic polarized HDM allergic responses 75 μg HDM was administered i.t. to WT and mTORΔAPC mice, on d0 and day 2. Lungs were analyzed by on d3.
(A) Bar chart shows total inflammatory DC cells in lung (B) HDM was administered to mTORΔAPC mice for three weeks along with αCsf1r neutralizing mAb or control. Bar charts show frequency of eosinophils or neutrophils in lung or BAL. (C) Dot graph shows significantly altered metabolic pathways between WT and mTORΔAPC inflammatory DCs with amino acid pathways highlighted in red, and fatty acid metabolism pathways highlighted blue. Dot size corresponds to the number of differentially expressed metabolites for a given pathway. The x-axis shows the log10 p-value. Heat map shows the relative abundance of metabolites involved in fatty acid oxidation pathways, based on tentative annotations produced by the mummichog software. (D) mTORΔAPC mice were treated with etomoxir (etxr) and HDM for 3d. Bar chart shows CD86 MFI, histogram shows representative flow cytometry data. (E) mTORΔAPC mice were treated with the Cpt1 inhibitor etomoxir and HDM for 17d. Bar chart shows expression of IL-17A+ T-cells in lung, contour plots show representative flow cytometric data. (F) HDM was chronically administered to mTORΔAPC mice for three weeks along with IL-12p70 control (gray) or IL12/23p40 (blue) neutralizing antibodies. Bar charts show frequency of eosinophils or neutrophils in lung or BAL. Data represents ≥2 independent experiments.
Given the observed mTOR-dependent metabolic phenotypes in steady-state lung APCs, we asked whether there was a metabolic basis for the Th17 allergic phenotype. Despite minimal differences between resting classical CD11b+ DCs in lungs of WT and mTORΔAPC mice, there was a striking perturbation in the metabolome of inflammatory DCs (fig. S20). In addition to differences in amino acid metabolism found in other DC subsets, we detected elevated fatty acid (FA) metabolites (Fig. 6C). mTOR inhibition in BMDCs can enhance fatty acid oxidation (FAO), and thereby regulate DC lifespan/cytokine expression profiles (39, 40). However, a physiological role for this mechanism in primary DCs has not been described. To directly test if elevated FAO underlies enhanced inflammatory DC activity, we treated mTORΔAPC mice with etomoxir, an inhibitor of the rate-limiting FAO enzyme Carnitine palmitoyltransferase 1 (Cpt1) (41). Etomoxir significantly reduced the activation phenotype of mTORΔAPC inflammatory DCs, without affecting WT (Fig. 6D), and also reduced Th17 polarization in mTORΔAPC lung (Fig. 6E). These observations are consistent with a model where mTOR deletion in inflammatory lung DCs increases FAO to promote Th17 polarizing activity.
Finally, we investigated whether dysregulated cytokine production by inflammatory DCs influences allergic polarity. The proinflammatory cytokines IL-6 and IL-1β function upstream of IL-23 to promote a Th17 polarizing cascade. Analysis of CD11b+ DCs in lung and MedLN after 3d HDM administration revealed evidence of elevated IL-12/23p40, IL-6 and IL-1β, all of which are associated with Th17 polarization and neutrophilia (fig. S21, A and B) (29). IL-23 neutralization reverted lung neutrophilia toward an eosinophilic phenotype (Fig. 6F), concomitant with restoration in T-cells of Th2 polarity (fig. S21C), identifying this cytokine as a key immunological player in mTORΔAPC-dependent neutrophilic inflammation.
Discussion
Here, we demonstrate a striking heterogeneity in the metabolic profiles of APCs in distinct tissues, and reveal a key role for mTOR in programming the homeostasis and function of APC subsets in the lung. In the steady state, mTORΔAPC mice exhibited tissue-dependent phenotypes with reduced numbers of AMs, and CD103 DCs in the lung and skin, but not elsewhere, unlike more global phenotypes observed when GM-CSF or Flt3l signaling is impaired (14, 18, 38). These effects were mediated by Raptor-dependent mTORC1 signaling but surprisingly independent of translational regulation of mRNA, the canonical signaling pathway downstream of mTOR. Instead, our data reveal a role for Srebp1/2 signaling downstream of mTOR in homeostasis of lung APCs in the steady state (fig. S10). The selective effects of mTOR deficiency in CD103 DCs in the lung and skin was surprising, since the development of this DC subset in diverse tissues is controlled by the transcription factor Batf3. Thus, these results suggest the homeostasis of DCs in tissues is orchestrated by the superimposition of a metabolic program on the transcriptional network that regulates development.
In addition to these effects in the steady state, mTOR deficiency also altered the character of allergic inflammation induced by lung CD11b+ DCs, skewing it toward the Th17/neutrophilic phenotype. This phenotype closely parallels aspects of human disease, where neutrophilic associates with higher clinical severity and worse prognosis (29). Interestingly, the allergic phenotype observed in mTORΔAPC mice was associated with elevated IgE. Such elevation in serum IgE is also observed in neutrophilic asthma patients (42) and can be enhanced by IL-23 (43), which is consistent with the IL-23-dependence of the mTORΔAPC neutrophilic inflammation phenotype. In normal circumstances, mTOR signaling in activated BMDCs suppresses oxidative phosphorylation (OXPHOS, in resting BMDCs) and is in part responsible for a switch to glycolytic metabolism, which rapidly depletes intracellular energy reserves and associates with a shortened cellular lifespan (1). In contrast, mTOR inhibition allows BMDCs to supply their energetic demands through a combination of glycolysis and fatty acid oxidation (FAO), increasing cell survival and proinflammatory phenotypes (39). We observed elevated FAO and increased production of several proinflammatory cytokines in mTORΔAPC lung. Indeed one of these cytokines, IL-23, plays an obligate role in Th17/neutrophilic inflammation. Together, our study reveals a key role for mTOR in programming the homeostasis and function of APCs in the lung. In the steady-state, mTOR supports accumulation of lung subsets by promoting lipid synthesis (anabolism) (fig. S22A). In the context of allergic inflammation, mTOR restrains catabolic processes such as FAO, preventing excessive inflammation (fig. S22B). The results herein identify tissue restricted metabolic adaptations of APCs as a critical determinant of immune regulation, raising the prospect of metabolic reprogramming of such tissue resident APCs for therapeutic benefit in various inflammatory disorders.
Supplementary Material
Acknowledgments
We thank V. Bliss and P. Sharma for histological support, G. Tharp and N. Patel for genomics analysis, V. Tran for metabolomics, B. Cervasi and K. Gill for FACS sorting, and H. Hartweger and E. Schweighoffer (Francis Crick Institute) for advice on B cell FACS staining. Finally, we thank Pulendran laboratory members for discussion and M. Sinclair for proofreading the manuscript. This work was supported by funding from Action Cycling Atlanta and from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007–2013) (C.S.) and from the U.S. National Institutes of Health (grants R37 DK057665, R37 AI048638, U19 AI090023, and U19 AI057266) (B.P.). We are not aware of any conflicts of interest. Additional data presented in this manuscript is available in the supplementary materials.
Footnotes
www.sciencemag.org/cgi/content/full/science.aaj2155/DC1
Materials and Methods
REFERENCES AND NOTES
- 1.Pearce EJ, Everts B. Dendritic cell metabolism. Nat Rev Immunol. 2015;15:18–29. doi: 10.1038/nri3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pulendran B. The varieties of immunological experience: Of pathogens, stress, and dendritic cells. Annu Rev Immunol. 2015;33:563–606. doi: 10.1146/annurev-immunol-020711-075049. [DOI] [PubMed] [Google Scholar]
- 3.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, Kennedy K, Wu H, Bennouna S, Oluoch H, Miller J, Vencio RZ, Mulligan M, Aderem A, Ahmed R, Pulendran B. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravindran R, Khan N, Nakaya HI, Li S, Loebbermann J, Maddur MS, Park Y, Jones DP, Chappert P, Davoust J, Weiss DS, Virgin HW, Ron D, Pulendran B. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343:313–317. doi: 10.1126/science.1246829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravindran R, Loebbermann J, Nakaya HI, Khan N, Ma H, Gama L, Machiah DK, Lawson B, Hakimpour P, Wang YC, Li S, Sharma P, Kaufman RJ, Martinez J, Pulendran B. The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature. 2016;531:523–527. doi: 10.1038/nature17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sukhbaatar N, Hengstschläger M, Weichhart T. mTOR-mediated regulation of dendritic cell differentiation and function. Trends Immunol. 2016;37:778–789. doi: 10.1016/j.it.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumont FJ, Staruch MJ, Koprak SL, Melino MR, Sigal NH. Distinct mechanisms of suppression of murine T cell activation by the related macrolides FK-506 and rapamycin. J Immunol. 1990;144:251–258. [PubMed] [Google Scholar]
- 9.Keating R, Hertz T, Wehenkel M, Harris TL, Edwards BA, McClaren JL, Brown SA, Surman S, Wilson ZS, Bradley P, Hurwitz J, Chi H, Doherty PC, Thomas PG, McGargill MA. The kinase mTOR modulates the antibody response to provide cross-protective immunity to lethal infection with influenza virus. Nat Immunol. 2013;14:1266–1276. doi: 10.1038/ni.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colina R, Costa-Mattioli M, Dowling RJO, Jaramillo M, Tai L-H, Breitbach CJ, Martineau Y, Larsson O, Rong L, Svitkin YV, Makrigiannis AP, Bell JC, Sonenberg N. Translational control of the innate immune response through IRF-7. Nature. 2008;452:323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- 12.Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, Murthy N, Pulendran B. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol. 2008;9:1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weichhart T, Costantino G, Poglitsch M, Rosner M, Zeyda M, Stuhlmeier KM, Kolbe T, Stulnig TM, Hörl WH, Hengstschläger M, Müller M, Säemann MD. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29:565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Sathaliyawala T, O’Gorman WE, Greter M, Bogunovic M, Konjufca V, Hou ZE, Nolan GP, Miller MJ, Merad M, Reizis B. Mammalian target of rapamycin controls dendritic cell development downstream of Flt3 ligand signaling. Immunity. 2010;33:597–606. doi: 10.1016/j.immuni.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellersch B, Brocker T. Langerhans cell homeostasis in mice is dependent on mTORC1 but not mTORC2 function. Blood. 2013;121:298–307. doi: 10.1182/blood-2012-06-439786. [DOI] [PubMed] [Google Scholar]
- 16.Nobs SP, Schneider C, Dietrich MG, Brocker T, Rolink A, Hirsch E, Kopf M. PI3-Kinase-γ has a distinct and essential role in lung-specific dendritic cell development. Immunity. 2015;43:674–689. doi: 10.1016/j.immuni.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Haidinger M, Poglitsch M, Geyeregger R, Kasturi S, Zeyda M, Zlabinger GJ, Pulendran B, Hörl WH, Säemann MD, Weichhart T. A versatile role of mammalian target of rapamycin in human dendritic cell function and differentiation. J Immunol. 2010;185:3919–3931. doi: 10.4049/jimmunol.1000296. [DOI] [PubMed] [Google Scholar]
- 18.McKenna HJ, Stocking KL, Miller RE, Brasel K, De Smedt T, Maraskovsky E, Maliszewski CR, Lynch DH, Smith J, Pulendran B, Roux ER, Teepe M, Lyman SD, Peschon JJ. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- 19.Vremec D, Lieschke GJ, Dunn AR, Robb L, Metcalf D, Shortman K. The influence of granulocyte/macrophage colony-stimulating factor on dendritic cell levels in mouse lymphoid organs. Eur J Immunol. 1997;27:40–44. doi: 10.1002/eji.1830270107. [DOI] [PubMed] [Google Scholar]
- 20.Nobs SP, Schneider C, Dietrich MG, Brocker T, Rolink A, Hirsch E, Kopf M. PI3-Kinase-γ has a distinct and essential role in lung-specific dendritic cell development. Immunity. 2015;43:674–689. doi: 10.1016/j.immuni.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma’ayan A, Chua W-J, Hansen TH, Turley SJ, Merad M, Randolph GJ, Gautier EL, Jakubzick C, Randolph GJ, Best AJ, Knell J, Goldrath A, Miller J, Brown B, Merad M, Jojic V, Koller D, Cohen N, Brennan P, Brenner M, Shay T, Regev A, Fletcher A, Elpek K, Bellemare-Pelletier A, Malhotra D, Turley S, Jianu R, Laidlaw D, Collins J, Narayan K, Sylvia K, Kang J, Gazit R, Garrison BS, Rossi DJ, Kim F, Rao TN, Wagers A, Shinton SA, Hardy RR, Monach P, Bezman NA, Sun JC, Kim CC, Lanier LL, Heng T, Kreslavsky T, Painter M, Ericson J, Davis S, Mathis D, Benoist C Immunological Genome Consortium. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, Ho AWS, See P, Shin A, Wasan PS, Hoeffel G, Malleret B, Heiseke A, Chew S, Jardine L, Purvis HA, Hilkens CMU, Tam J, Poidinger M, Stanley ER, Krug AB, Renia L, Sivasankar B, Ng LG, Collin M, Ricciardi-Castagnoli P, Honda K, Haniffa M, Ginhoux F. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edelson BT, Kc W, Juang R, Kohyama M, Benoit LA, Klekotka PA, Moon C, Albring JC, Ise W, Michael DG, Bhattacharya D, Stappenbeck TS, Holtzman MJ, Sung S-SJ, Murphy TL, Hildner K, Murphy KM. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F. SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung Y-L, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;19:R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saglani S, Mathie SA, Gregory LG, Bell MJ, Bush A, Lloyd CM. Pathophysiological features of asthma develop in parallel in house dust mite-exposed neonatal mice. Am J Respir Cell Mol Biol. 2009;41:281–289. doi: 10.1165/rcmb.2008-0396OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plantinga M, Guilliams M, Vanheerswynghels M, Deswarte K, Branco-Madeira F, Toussaint W, Vanhoutte L, Neyt K, Killeen N, Malissen B, Hammad H, Lambrecht BN. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38:322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Alcorn JF, Crowe CR, Kolls JK. TH17 cells in asthma and COPD. Annu Rev Physiol. 2010;72:495–516. doi: 10.1146/annurev-physiol-021909-135926. [DOI] [PubMed] [Google Scholar]
- 30.Douwes J, Gibson P, Pekkanen J, Pearce N. Non-eosinophilic asthma: Importance and possible mechanisms. Thorax. 2002;57:643–648. doi: 10.1136/thorax.57.7.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helft J, Manicassamy B, Guermonprez P, Hashimoto D, Silvin A, Agudo J, Brown BD, Schmolke M, Miller JC, Leboeuf M, Murphy KM, García-Sastre A, Merad M. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Invest. 2012;122:4037–4047. doi: 10.1172/JCI60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seymour JF, Presneill JJ. Pulmonary alveolar proteinosis: Progress in the first 44 years. Am J Respir Crit Care Med. 2002;166:215–235. doi: 10.1164/rccm.2109105. [DOI] [PubMed] [Google Scholar]
- 33.Shah PL, Hansell D, Lawson PR, Reid KB, Morgan C. Pulmonary alveolar proteinosis: Clinical aspects and current concepts on pathogenesis. Thorax. 2000;55:67–77. doi: 10.1136/thorax.55.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathie SA, Dixon KL, Walker SA, Tyrrell V, Mondhe M, O’Donnell VB, Gregory LG, Lloyd CM. Alveolar macrophages are sentinels of murine pulmonary homeostasis following inhaled antigen challenge. Allergy. 2015;70:80–89. doi: 10.1111/all.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y, Nish SA, Jiang R, Hou L, Licona-Limón P, Weinstein JS, Zhao H, Medzhitov R. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity. 2013;39:722–732. doi: 10.1016/j.immuni.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hägerbrand K, Marsal J, Gudjonsson S, Håkansson U, Reizis B, Kotarsky K, Agace WW. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Tussiwand R, Everts B, Grajales-Reyes GE, Kretzer NM, Iwata A, Bagaitkar J, Wu X, Wong R, Anderson DA, Murphy TL, Pearce EJ, Murphy KM. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 2015;42:916–928. doi: 10.1016/j.immuni.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greter M, Helft J, Chow A, Hashimoto D, Mortha A, Agudo-Cantero J, Bogunovic M, Gautier EL, Miller J, Leboeuf M, Lu G, Aloman C, Brown BD, Pollard JW, Xiong H, Randolph GJ, Chipuk JE, Frenette PS, Merad M. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity. 2012;36:1031–1046. doi: 10.1016/j.immuni.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amiel E, Everts B, Fritz D, Beauchamp S, Ge B, Pearce EL, Pearce EJ. Mechanistic target of rapamycin inhibition extends cellular lifespan in dendritic cells by preserving mitochondrial function. J Immunol. 2014;193:2821–2830. doi: 10.4049/jimmunol.1302498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amiel E, Everts B, Freitas TC, King IL, Curtis JD, Pearce EL, Pearce EJ. Inhibition of mechanistic target of rapamycin promotes dendritic cell activation and enhances therapeutic autologous vaccination in mice. J Immunol. 2012;189:2151–2158. doi: 10.4049/jimmunol.1103741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houten SM, Wanders RJ. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J Inherit Metab Dis. 2010;33:469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milovanovic M, Drozdenko G, Weise C, Babina M, Worm M. Interleukin-17A promotes IgE production in human B cells. J Invest Dermatol. 2010;130:2621–2628. doi: 10.1038/jid.2010.175. [DOI] [PubMed] [Google Scholar]
- 43.Peng J, Yang XO, Chang SH, Yang J, Dong C. IL-23 signaling enhances Th2 polarization and regulates allergic airway inflammation. Cell Res. 2010;20:62–71. doi: 10.1038/cr.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8-dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kellersch B, Brocker T. Langerhans cell homeostasis in mice is dependent on mTORC1 but not mTORC2 function. Blood. 2013;121:298–307. doi: 10.1182/blood-2012-06-439786. [DOI] [PubMed] [Google Scholar]
- 47.Brown J, Wang H, Suttles J, Graves DT, Martin M. Mammalian target of rapamycin complex 2 (mTORC2) negatively regulates Toll-like receptor 4-mediated inflammatory response via FoxO1. J Biol Chem. 2011;286:44295–44305. doi: 10.1074/jbc.M111.258053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, Schreiber RD, Murphy TL, Murphy KM. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1−/−/S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Bacquer O, Petroulakis E, Paglialunga S, Poulin F, Richard D, Cianflone K, Sonenberg N. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest. 2007;117:387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- 52.Kamisuki S, Mao Q, Abu-Elheiga L, Gu Z, Kugimiya A, Kwon Y, Shinohara T, Kawazoe Y, Sato S, Asakura K, Choo H-YP, Sakai J, Wakil SJ, Uesugi M. A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem Biol. 2009;16:882–892. doi: 10.1016/j.chembiol.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Sinclair C, Bains I, Yates AJ, Seddon B. Asymmetric thymocyte death underlies the CD4:CD8 T-cell ratio in the adaptive immune system. Proc Natl Acad Sci USA. 2013;110:E2905–E2914. doi: 10.1073/pnas.1304859110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock REW, Brinkman FSL, Lynn DJ. InnateDB: Systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 2013;41:D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(D1):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, Pulendran B. Predicting network activity from high throughput metabolomics. PLOS Comput Biol. 2013;9:e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CRH, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. A comprehensive classification system for lipids. J Lipid Res. 2005;46:839–862. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 58.Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, Chu FF, Randolph GJ, Rudensky AY, Nussenzweig M. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.