Cells expend energy to migrate. Metastatic cancer cell energy levels were investigated as a function of collagen architecture. In more migration-permissive environments or when migration is pharmacologically inhibited, cells reduce ATP:ADP levels. Changes in intracellular ATP:ADP levels during migration were associated with changes in cell speed. The data suggest that cells tune their energy production and utilization relative to their migration.
Abstract
Cell migration in a three-dimensional matrix requires that cells either remodel the surrounding matrix fibers and/or squeeze between the fibers to move. Matrix degradation, matrix remodeling, and changes in cell shape each require cells to expend energy. While significant research has been performed to understand the cellular and molecular mechanisms guiding metastatic migration, less is known about cellular energy regulation and utilization during three-dimensional cancer cell migration. Here we introduce the use of the genetically encoded fluorescent biomarkers, PercevalHR and pHRed, to quantitatively assess ATP, ADP, and pH levels in MDA-MB-231 metastatic cancer cells as a function of the local collagen microenvironment. We find that the use of the probe is an effective tool for exploring the thermodynamics of cancer cell migration and invasion. Specifically, we find that the ATP:ADP ratio increases in cells in denser matrices, where migration is impaired, and it decreases in cells in aligned collagen matrices, where migration is facilitated. When migration is pharmacologically inhibited, the ATP:ADP ratio decreases. Together, our data indicate that matrix architecture alters cellular energetics and that intracellular ATP:ADP ratio is related to the ability of cancer cells to effectively migrate.
INTRODUCTION
Cancer cell invasion and migration during metastasis are hallmarks of cancer progression (Hanahan and Weinberg, 2011; Pickup et al., 2014). There is an interdependent relationship between cell adhesion, migration, and the cellular microenvironment, where cells interact with their surrounding extracellular matrix (ECM), and in turn, the ECM mechanically changes in response to the cells’ behaviors (Paszek et al., 2005; Stevens and George, 2005). Mechanical cues from the ECM are sensed by the cell and converted to intracellular signals that direct cell protrusions, elongation, polarity, and migration efficacy (Ridley et al., 2003; Petrie et al., 2010; Carey et al., 2012a, 2014). Cells can sense and respond to an array of ECM cues, including matrix alignment, stiffness, protein composition, and structural heterogeneities, by altering their migration behavior (Provenzano et al., 2008; Hadjipanayi et al., 2009; Lu et al., 2012; Bordeleau et al., 2013). Moreover, metastasis and metastatic cell migration are associated with an epithelial-to-mesenchymal transition, which is affected by both biochemical and mechanical cues within the cell and its local microenvironment (Ingber, 2008; Zavadil et al., 2008; Kalluri and Weinberg, 2009; Hanahan and Weinberg, 2011; Gao et al., 2012; Polacheck et al., 2013). To effectively migrate through the complex architecture of the stromal ECM and ultimately to new locations throughout the body, cancer cells need to expend energy, generally via the dephosphorylation of ATP into ADP (Van Horssen et al., 2009). However, the energy needs of cells during migration are not yet well understood.
Proper regulation of ATP:ADP levels is critical for cell health and essential cell functions, as the ATP:ADP ratio serves as the principal regulator of cellular metabolism that drives many cellular reactions (Chen et al., 2007; Hanahan and Weinberg, 2011; Caneba et al., 2012; Liu et al., 2013; Yuan et al., 2014). ATP is not kept in abundance or stored in cells as other biomolecules are, but rather its production is increased to match the demand of cellular activity (Bergman, 1999; Kennedy et al., 1999; Alberts et al., 2002; Yaniv et al., 2013). Previous literature has used molecular ATP sensors, such as PercevalHR, to observe and quantify energy levels inside cells during various cellular activities with a focus on metabolic processes including glycolysis and oxidative phosphorylation (Merrins et al., 2016; Uslu and Grossmann, 2016). Not surprisingly, hindering, bypassing, or generating new metabolic pathways affects ATP levels (Hyslop et al., 1988; Thomas and Fell, 1998; Wu et al., 2007; Imamura et al., 2009). Changes in hypoxia also affect ATP and ADP levels (Berg et al., 2009; Wheaton and Chandel, 2011; Tarasov and Rutter, 2014). While our knowledge of ATP:ADP regulation is growing, currently there are no studies investigating ATP:ADP levels as a function of matrix architecture during migration.
Here we investigated the effects of collagen architecture on ATP utilization in metastatic cells. We employ a genetically encoded fluorescent biomarker, PercevalHR (Tantama et al., 2013), to determine how collagen density and fiber orientation affect energy regulation in MDA-MB-231 cancer cells during migration. PercevalHR binds to both conformations of ATP and ADP and can be utilized to determine a concentration-independent ATP:ADP ratio as a metric of intracellular energy (Tantama and Yellen, 2014; Yuan et al., 2014). Interestingly, an increase in collagen density decreases cellular migration while simultaneously increasing the intracellular ATP:ADP ratio, glucose uptake, and ATP hydrolysis. Time-lapse studies demonstrated that changes in intracellular ATP:ADP ratio were directly correlated with changes in cell stepwise velocity during migration. Aligned collagen matrices, known to facilitate cell migration (Provenzano et al., 2008; Carey et al., 2016), increased cellular migration while decreasing intracellular ATP:ADP ratio, glucose uptake, and ATP hydrolysis. Pharmacological inhibition of motility by inhibiting contractility and actin polymerization resulted in a decrease in ATP:ADP ratio. These results indicate that ECM architecture mediates energy utilization in cells and that the PercevalHR ATP:ADP probe is a promising tool for studying the thermodynamics of cancer migration in live-cell studies.
RESULTS and discussion
Cellular ATP:ADP levels respond to glucose and serum concentrations in two- and three-dimensional culture
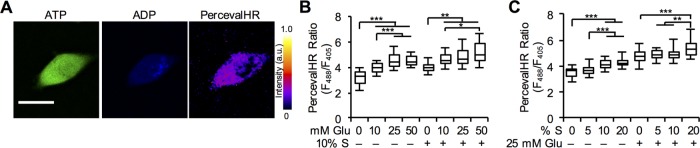
In this work, we employ the novel use of the PercevalHR sensor in metastatic MDA-MB-231 breast cancer cells to examine the relationship between the local cellular microenvironment and ATP utilization during migration. To validate that heightened metabolic activity correlates with increased ATP generation, MDA-MB-231 cells expressing PercevalHR were seeded on glass culture dishes and cultured in increased glucose and serum levels for 24 h. Glucose increases ATP production via increases to both the substrate supply and Ca2+-dependent activation of mitochondrial enzymes (Civelek et al., 1996; Kennedy et al., 1999). Here cells were cultured in media containing high excess glucose (25 mM), which is approximately five times higher than physiological glucose levels. The PercevalHR biomarker was imaged to quantify the relative ATP:ADP ratio inside migrating, live, single cells (Figure 1A). In high-glucose environments, which increase cellular metabolism (Vander Heiden et al., 2001), but in the absence of serum, ATP:ADP ratios increased as a function of glucose concentration (Figure 1B). Similarly, in standard (10%) serum levels, ATP:ADP levels increased with increasing glucose levels. Increased glucose levels with serum present resulted in overall higher ATP:ADP levels than without serum.
FIGURE 1:
Cellular ATP response to glucose and serum in two-dimensional culture. (A) Representative MDA-MB-231 cells expressing PercevalHR in two-dimensional culture demonstrating the sensor bound to ATP (green), ADP (blue), and PercevalHR ratiometric signal. (B) Quantification of PercevalHR ratio response to increasing glucose levels in the presence of 0 and 10% serum in two-dimensional culture (n = 30 cells from three independent experiments). (C) Quantification of PercevalHR ratio response to increasing percentage of serum in the presence of 0 and 25 mM glucose in two-dimensional culture (n = 45 cells from three independent experiments). Box-and-whisker plots show medians and 25th/75th and 5th/95th percentiles. *p < 0.05, **p < 0.01, ***p < 0.001 for one-way ANOVA with Tukey’s HSD post-hoc test. Scale bar = 20 μm.
A similar result was seen when cells were cultured without glucose and increasing serum levels, where increased serum concentration resulted in increased ATP:ADP ratio. In high glucose, the ATP:ADP ratio significantly increased with increased serum levels. Increasing serum levels overall resulted in higher ATP:ADP ratios when glucose was present compared with when glucose was absent (Figure 1C). Together, these data indicate that high levels of glucose and serum allow cells to generate more ATP.
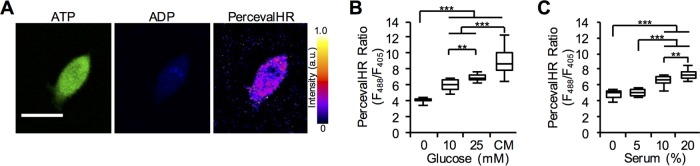
To investigate the effects of glucose and serum levels on the intracellular ATP:ADP ratio of cells seeded in three-dimensional environments, MDA-MB-231 cells expressing PercevalHR were cultured in various glucose and serum levels for 24 h in 1.5 mg/ml collagen matrices and imaged to quantify the ATP:ADP ratio (Figure 2A). Similarly to cells cultured on two-dimensional surfaces in the absence of serum, increased glucose levels resulted in increased intracellular ATP:ADP ratio (Figure 2B). In the absence of glucose, greater serum resulted in increased ATP:ADP ratio (Figure 2C). Together, these data indicate that stimulating cells embedded in three-dimensional matrices with glucose or serum, which are known to increase metabolic activity, results in an increase in cellular ATP:ADP.
FIGURE 2:
Cellular ATP response to glucose and serum in three-dimensional collagen matrices. (A) Representative MDA-MB-231 cells expressing PercevalHR in a 1.5 mg/ml three-dimensional collagen matrix demonstrating the sensor bound to ATP (green), ADP (blue), and PercevalHR ratiometric signal. (B) Quantification of PercevalHR ratio response to increasing glucose levels in the presence of 0% serum and complete media (CM; 25 mM glucose, 10% serum) in three-dimensional collagen gels (n ≥ 20 cells from three independent experiments). (C) Quantification of PercevalHR ratio in response to increasing serum levels in the presence of 0 mM glucose in three-dimensional collagen gels (n ≥ 13 cells from three independent experiments). Box-and-whisker plots show medians and 25th/75th and 5th/95th percentiles. **p < 0.01, ***p < 0.001 for one-way ANOVA with Tukey’s HSD post-hoc test. Scale bar = 20 μm.
Interestingly, we found higher intracellular ATP:ADP levels in cells cultured in three-dimensional matrices versus two-dimensional surfaces, when cultured with the same extracellular conditions. Cells differ greatly in two- and three-dimensional environments in characteristics such as morphology, migration, focal adhesions, or gene expression (Wozniak et al., 2004; Soares et al., 2012; Zschenker et al., 2012; Doyle et al., 2015). Furthermore, it has been shown that cells in three dimensions migrate faster and more often than cells in two dimensions (Hakkinen et al., 2011). As such, faster and higher-frequency migration events observed in three dimensions would require increased cytoskeletal and focal adhesion remodeling, which demand increased ATP hydrolysis (Bursac et al., 2005). Notably, greater motility and quicker cytoskeletal remodeling is observed in cancer cells with higher glycolytic activity (Shiraishi et al., 2015).
These results signify that the PercevalHR sensor is sufficiently sensitive and effective at detecting changes in MDA-MB-231 intracellular ATP:ADP ratio caused by changes in glucose and serum. Further, this effectiveness was independent from whether the cells were seeded in two- or three-dimensional matrices, making these sensors ideal for investigating the thermodynamic process of cancer cell migration in three-dimensional matrices.
Matrix density affects migration efficacy and intracellular ATP:ADP levels
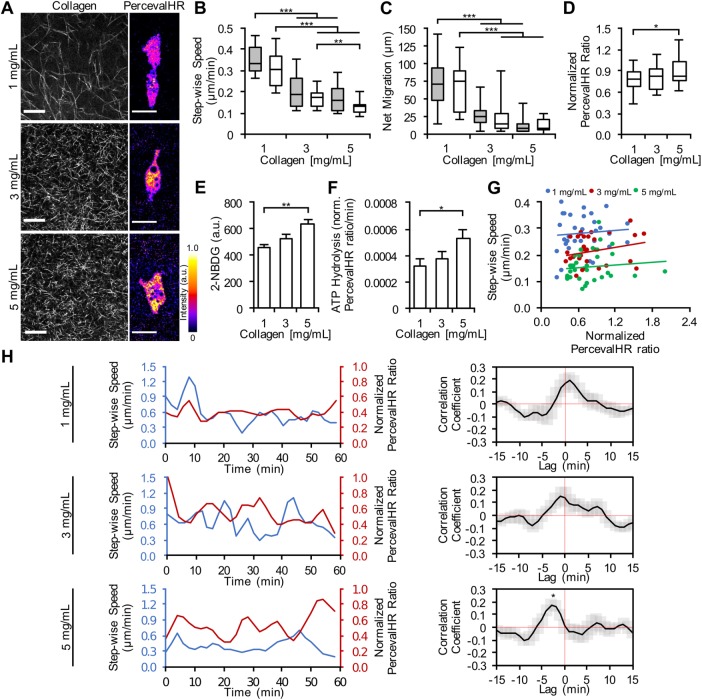
To determine how matrix density affects the energetic state of cells during migration, MDA-MB-231 cancer cells coexpressing PercevalHR and pHRed were seeded in three-dimensional collagen matrices of different densities (1, 3, and 5 mg/ml), and stepwise cell speed, net migration, and intracellular ATP:ADP ratio were measured. Many factors can alter the amount of ATP:ADP inside a cell, such as intracellular pH, stress, signaling activities, and metabolic and catabolic processes (Tantama et al., 2011, 2013; Carta et al., 2015; McKenna et al., 2016). Further, many fluorescent proteins and biosensors like PercevalHR are pH sensitive, and pH bias must be accounted for to avoid misinterpretation of sensor values. Here, pH bias was removed via use of the fluorescent pH biosensor, pHRed (Supplemental Figure S1). The pHRed sensor allows for the PercevalHR sensor to be normalized for pH variations due to various cell states (Supplemental Figure S2; Tantama et al., 2011). Confocal reflectance images were taken to visualize the collagen fibrillar structure, along with corresponding cells’ energy levels (Figure 3A). Cell speed and net migration decreased when matrix density increased for cells in both high and low glucose levels (Figure 3, B and C). Interestingly, cells exhibited significantly less intracellular ATP:ADP in three-dimensional collagen matrices of lower density (Figure 3D). These data suggest that cells require more energy for their migration and remodeling efforts in denser matrices.
FIGURE 3:
Effect of matrix density on ATP:ADP ratio and cell migration in three-dimensional collagen matrices. (A) Confocal reflectance images of collagen gels and representative pH-corrected PercevalHR ratios of MDA-MB-231 cells coexpressing PercevalHR and pHRed cultured in a 1, 3, and 5 mg/ml three-dimensional collagen matrix. Quantification of cell (B) stepwise cell migration speed and (C) net migration for increasing collagen density when cultured in DMEM with 10% serum and 25 mM glucose (gray) or DMEM with 10% serum and 0 mM glucose (white) (n = 30 cells per treatment from three independent experiments). Quantification of (D) pH-corrected PercevalHR ratiometric signal, (E) 2-NBDG uptake, and (F) ATP hydrolysis rate of cells cultured in three-dimensional collagen matrices of varying density (n = [D] 30, [E] 45, [F] 30 cells from three independent experiments). (G) Stepwise speed and accompanying pH-corrected PercevalHR ratiometric signal of individual cells cultured in three-dimensional collagen matrices of varying density averaged across 12–18 h of culture. Each data point represents an individual cell (n = 33 cells from three independent experiments). (H) Stepwise speed and pH-corrected PercevalHR ratiometric signal and of an individual, migrating cell cultured in 1, 3, and 5 mg/ml 3D collagen matrices over 1 h, and comparison of corresponding temporal cross-correlation analysis between stepwise speed and normalized PercevalHR ratiometric signal at each matrix density (n = 20 cells from three independent experiments). Box-and-whisker plots show medians and 25th/75th and 5th/95th percentiles. Bars denote mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 for one-way ANOVA with Tukey’s HSD post-hoc test, and one-sample t test, which was used to determine the significance of lag for temporal cross-correlation comparison. Scale bars = 20 µm.
The driving mechanism of cell metabolism under these conditions still remained unclear. For instance, if higher matrix density causes cells to reduce cellular activity that requires myosin turnover, then ATP hydrolysis would be expected to be reduced. In this case, relative to other conditions with higher ATP hydrolysis, the PercevalHR ratio may appear higher and create the illusion of the cell requiring more energy for migration and remodeling efforts in denser matrices. Similarly, if glucose uptake is increased, one would expect metabolic processes to increase the PercevalHR ratio as intracellular ATP stores are increased. To isolate these two variables, we first studied the relationship between glucose uptake and collagen density. Using the fluorescent glucose analogue 2-NBDG we observed that cells in higher-density collagen had a significant increase in their glucose uptake after 24 h relative to cells in lower-density matrices (Figure 3E). We next studied the rate of ATP hydrolysis as a function of matrix density by pharmacologically inhibiting oxidative phosphorylation and glycolysis and measuring the decrease in ATP:ADP levels over time. We found that cells cultured in higher-density matrices exhibited significantly increased ATP hydrolysis relative to those in lower-density matrices (Figure 3F). Together, these results support our original hypothesis that cells require more energy for migration and remodeling efforts in denser matrices; cells increase both their energy production and rate of energy consumption to transverse more demanding environments.
ATP usage and compartmentalization has been linked to cytoskeletal dynamics involved in migration (Bursac et al., 2005; Van Horssen et al., 2009). Recent work has also shown that localized energy production is required for cancer cell migration. Mitochondria traffic to pseudopodia at the leading edge of migrating cells to meet energy demands (Cunniff et al., 2016), and deficient mitochondrial trafficking to the cell cortex reduces ATP:ADP and disrupts actin-based membrane reorganization needed for migration (Schuler et al., 2017). To investigate further the relationship between cellular ATP:ADP and cytoskeletal usage, we captured PercevalHR measurements over time and correlated these with changes in stepwise cell speeds. The data revealed that, as expected, cell speed over 12–18 h of culture was reduced as matrix density is increased, and ATP:ADP levels increased as matrix density is increased (Figure 3G). Notably, cells in lower-density matrices achieved much higher stepwise speeds utilizing similar levels of intracellular ATP:ADP compared with cells in higher-density matrices. During migration over a 1 h interval, cell stepwise speed was found to be correlated with intracellular PercevalHR ratio across all matrix densities (Figure 3H). With increasing matrix density, lower velocity and higher ATP:ADP ratios were observed over 1 h, similarly to measurements averaged over 12–18 h. ATP:ADP ratio decreased during cell migration, indicating ATP consumption, and increased while cells were stationary, indicating ATP production (Figure 3H). The corresponding temporal cross-correlation comparison of stepwise speed and ATP:ADP ratio exhibited significant positive correlation independent of collagen density; however, only in a 5 mg/ml collagen matrix was significant temporal lag seen (Figure 3H). Together, these data suggest cells increase energy consumption for migration through more demanding environments, and they increase ATP production to compensate.
Our data have shown that increased levels of glucose in the extracellular space correlated with increased intracellular ATP:ADP levels. Curiously, migratory speeds and net migration in collagen matrices were not statistically significant with respect to glucose presence (Figure 3, B and C). The increased ATP:ADP level can be attributed to heightened metabolic activity and increased ATP production. Similar migration speeds in cells with glucose present or absent may indicate either that the amount of ATP needed for migration is met by cells in both conditions or that, in high-glucose environments, cells operate less efficiently such that the rate of ATP hydrolysis is higher relative to their lower glucose condition counterparts. The internal energy depot of the cell regulates the internal competition between developing an invasive or proliferative phenotype (Hecht et al., 2015). A cell needs to expend a certain amount of ATP to migrate; however, a cell in a high-glucose environment can generate higher ATP:ADP levels than a cell absent of extracellular glucose. Thus, the amount of ATP utilized by a cell can be independent of migration capability, as long as the cell meets the minimal ATP requirement to complete migration. In low-glucose environments, it is beneficial for cells to adapt an invasive phenotype due to the lower amount of energy necessary for migration compared with proliferation (Hecht et al., 2015). Interestingly, cells with an invasive phenotype demonstrated higher successful migration under low-energy conditions and are not as efficient at migration when energy resources are high (Hecht et al., 2015). Further, tumor cells have been shown to maintain energy production for migration under nutrient-deprived conditions through metabolic reprogramming of mitochondrial bioenergetics (Caino et al., 2013).
Energy is required for cell migration through the tumor stromal ECM in vivo and three-dimensional collagen gels in vitro. Our data indicated that more challenging ECM microenvironments (higher-density collagen) results in increased intracellular energy levels, where cells consume larger amounts of ATP during migration and overcompensate with increased ATP production. This result is in contrast to the inverse conclusion of Morris et al. (2016), where a decreased ATP level was observed within the cell population cultured in high-density matrices. One possible explanation is a selection bias between cells chosen for the studies. Here, live, single cells were selected such that it was clear they were migrating, while Morris et al. utilized a boiling water method to dissolve the entirety of the matrix, which selects for the entire cell population. The latter likely includes cells that are dying and/or dormant, possibly due to the high energy cost of migration through high-density collagen matrices and maintaining correspondingly low levels of intracellular ATP.
ECM alignment induces cell migration and reduces intracellular ATP:ADP levels
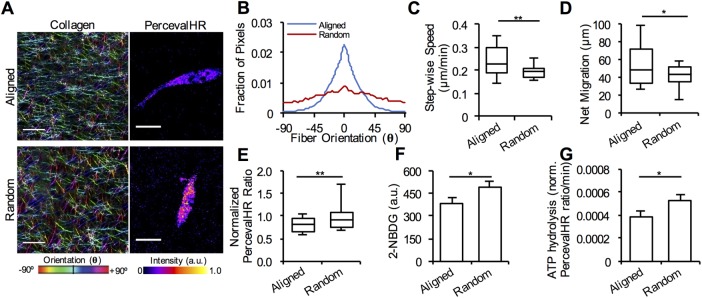
Given that increased collagen density results in decreased matrix pore size and an increased ATP:ADP ratio, we sought to determine whether preorganizing the matrix into a migration-permissive structure reduces the cellular energy demand and results in a lower ATP:ADP ratio in cells. To test this hypothesis, we aligned collagen fibers in a 1.5 mg/ml collagen matrix (Figure 4, A and B), which has been shown to facilitate migration both in vitro and in vivo (Stevens and George, 2005; Petrie et al., 2010; Pathak and Kumar, 2012; Polacheck et al., 2013; Carey et al., 2016). Cell migration velocity and net migration increased in aligned matrices compared with randomly oriented matrices of the same density (Figure 4, C and D). Accordingly, cells expressed significantly lower intracellular ATP:ADP ratios for aligned collagen matrices compared with those in random matrices (Figure 4D). Further, cells in aligned matrices had decreased 2-NBDG uptake and ATP hydrolysis, indicating lowered ATP production and ATP consumption, respectively. (Figure 4, F and G). These data indicate that easing the effort required by cells to migrate by aligning the collagen fibers lowered the ATP consumption required for migration. Overall, our results indicate that a cell’s local structural environment affects its regulation of ATP and ADP levels, as well as its ability to migrate in three-dimensional matrices. Taken together, these data suggest cells operate at demand-driven levels of energy to migrate through various matrix conditions.
FIGURE 4:
Reduction in ATP usage in cells cultured in three-dimensional collagen matrices with aligned fibers. (A) Confocal reflectance of an aligned and randomly oriented 1.5 mg/ml three-dimensional collagen matrix with representative pH-corrected PercevalHR ratios of MDA-MB-231 cells coexpressing PercevalHR and pHRed. Collagen is pseudo-colored corresponding to fiber angle. (B) Collagen fiber alignment in aligned and randomly oriented collagen matrices (n = 30 images from three independent experiments). Quantification of cell (C) stepwise cell migration speed and (D) net migration in aligned and random collagen matrices (n = 30 cells from three independent experiments). Quantification of (E) pH-corrected PercevalHR ratiometric signal, (F) 2-NBDG uptake, and (G) ATP hydrolysis of cells cultured in three-dimensional aligned and random collagen matrices (n = [E] 40, [F] 45, [G] 40 cells from three independent experiments). Box-and-whisker plots show medians and 25th/75th and 5th/95th percentiles. Bars denote mean ± SEM. *p < 0.05, **p < 0.01 for Wilcoxon rank test. Scale bars = 20 μm.
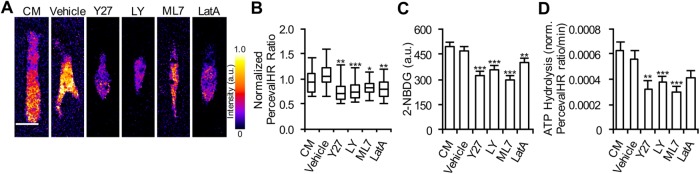
Contractility and migratory inhibitors decrease intracellular ATP:ADP levels
Since cells require energy to migrate and cells typically produce ATP to fulfill their energetic needs (Bergman, 1999; Yaniv et al., 2013), we hypothesized that if migration was pharmacologically inhibited, ATP:ADP levels would decrease. To inhibit migration, we employed pharmacological inhibitors against Rho-associated kinase (ROCK), phosphoinositide-3- (PI3) kinase, myosin light-chain (MLC) kinase, and actin polymerization. ROCK is a key regulator of cytoskeletal organization and cell contractility and therefore migration (Provenzano et al., 2008; Amano et al., 2010). By reducing cytoskeleton turnover, ROCK inhibition via Y27632 will impede cells from migrating. PI3-kinase is a major regulator of migration guidance in cells in relation to their extracellular environment (Jekely et al., 2005; Karam et al., 2010). Inhibiting PI3-kinase via LY294002 reduces a cell’s ability to direct and regulate its migration efforts. MLC kinase is a critical factor in regulating cell contractility (Tan and Leung, 2009; Shen et al., 2010). This contractility plays a pivotal role in cell protusion and retraction, and MLC kinase inhibition via ML7 subsequently affects cell migration. Latrunculin A (LatA) treatment prevents actin polymerization in the cytoskeleton by binding to free G-actin in the cytosol (Alberts et al., 2002; Bernstein and Bamburg, 2003; Shen and Turner, 2005). This inhibits the cell’s ability to remodel its cytoskeleton to facilitate migration. Each of these inhibitors was chosen as they reduce a cell’s ability to spread and migrate in three-dimensional matrices.
Although some of these inhibitors may indirectly block homeostatic ATP hydrolysis, which could cause an increase in intracellular ATP:ADP, it has been noted that cells will stop producing ATP as their ATP requirements decrease (Wu et al., 2013), and therefore an increase in ATP levels would not occur. All of these pharmacological drugs inhibit cell migration indirectly by hindering cytoskeleton dynamics. Drugs that stimulate actin cytoskeletal polymerization (e.g., jasplakinolide), stabilize the actin fibers (e.g., phalloidin), or stabilize microtubules (e.g., paclitaxel) were not tested here. However, these cytoskeletal stimulators may have interesting effects on ATP:ADP ratios, such as potentially increasing ATP as cells maximize utilization of their cytoskeleton for migration. Our results indicate that cells treated with contractility/migration inhibitors exhibited a statistically significant decrease in intracellular ATP:ADP in three-dimensional culture (Figure 5, A and B). Cells treated with inhibitors also had decreased 2-NBDG uptake and lowered ATP hydrolysis (Figure 5, C and D). These results demonstrate that ATP utilization is linked to cellular contractility and a cell’s ability to migrate. Together, these findings support the concept that energy generation is driven by demand based on cellular need, and inhibiting migration lowers the capacity for ATP utilization.
FIGURE 5:
Reduction of ATP usage in cells by migration inhibition. (A) Representative pH-corrected PercevalHR ratios of MDA-MB-231 cells coexpressing PercevalHR and pHRed cultured in a 1.5 mg/ml collagen matrix treated with complete media (CM), vehicle control (DMSO), Y27632 (Y27), LY294002 (LY), ML7, and Latrunculin A (LatA). Quantification of (B) pH-corrected PercevalHR ratiometric signal, (C) 2-NBDG uptake, and (D) ATP hydrolysis for cells cultured in 1.5 mg/ml collagen matrices across inhibitor treatments (n = [B] 40, [C] 45, [D] 30 cells from three independent experiments). Box-and-whisker plots show medians and 25th/75th and 5th/95th percentiles. Bars denote mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 for Wilcoxon rank test for each condition compared with control (CM, complete media). Scale bar = 20 μm.
Conclusion
There is significant research on how the tumor stromal ECM promotes tumor growth (Egeblad et al., 2010), metastasis from a primary tumor (Langley and Fidler, 2007; Polacheck et al., 2013), and tumor locoregional recurrence (Levental et al., 2010). However, little is known about how the ECM regulates metabolic processes. The PercevalHR ATP:ADP sensor is an effective and uniquely capable tool for studying the thermodynamics of live, single-cell cancer studies. Our data reveal that the local microenvironment affects cancer cells’ ability to efficiently migrate through the ECM and their energy generation and consumption. Denser collagen matrices impede a cell’s ability to migrate and therefore increase the amount of energy cells require to move. Additionally, aligned collagen promotes cell migration and reduces the energy needed for cells to migrate. Changes in intracellular ATP:ADP levels during migration were directly tied to changes in cell speed. Together, these results suggest that one mechanism by which the properties of the local ECM affect three-dimensional cancer cell metastatic invasion is through intracellular energy utilization. Future work should include understanding these differences more deeply as well as analyzing single-cell rates of ATP change as cells perform tasks associated with the various cancer hallmarks including proliferation, extravasation and intravasation, and collective invasion.
MATERIALS AND METHODS
Cell culture and reagents
Metastatic MDA-MB-231 breast adenocarcinoma cells (HTB-26; American Type Culture Collection, Rockville, MD) were maintained in DMEM (4.5 g/l [25 mM] glucose; Life Technologies, Grand Island, NY), supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA), and 100 U/ml penicillin and 100 µg/ml streptomycin (Life Technologies). HEK293T cells were cultured in MEM supplemented with 10% FBS and 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen). All cells were cultured at 37°C and 5% CO2, fed every other day, and passaged at 85% confluence. All cell culture, fluorescence imaging, and time-lapse imaging were performed at 37°C and 5% CO2. All cell lines used were tested for mycoplasma and deemed free of contamination.
DNA constructs and lentiviral transductions
FUGW-PercevalHR (Addgene plasmid #49083) and GW1-pHRed (Addgene plasmid #31473) were gifts from Gary Yellen (Harvard Medical School, Boston, MA), and both were coexpressed in the cell population. The pHRed sensor was inserted into the pFUW-CMV vector using BamH1 and EcoR1 restriction sites to generate pFUW-CMV-pHRed. PercevalHR and pHRed lentiviral particles were prepared by transient transfection of HEK293T cells with the lentiviral expression vectors and the second generation packing constructs psPAX2 and pMD2.G in the presence of TransIT-LT1 (Mirus). Lentiviral particles were harvested from the HEK293T supernatant at 48 and 72 h posttransfection and concentrated 100-fold with Lenti-X Concentrator (Clontech) followed by stable MDA-MB-231 cell transduction in the presence of 8 µg/ml polybrene overnight (Santa Cruz Biotechnology).
Two-dimensional cell seeding and three-dimensional cell encapsulation
In two-dimensional studies, MDA-MB-231 cells were seeded in 96-well glass-bottom dishes (MatTek Corporation, Ashland, MA) at 16,000 cells/cm2. Cells were overlaid with culture medium and allowed to adhere and spread overnight in a temperature-, humidity-, and CO2-controlled incubator. In three-dimensional studies, MDA-MB-231 cells were seeded at 125,000 cells/ml in three-dimensional collagen matrices prepared from acid-solubilized type I rat tail tendon collagen as previously described (Carey et al., 2012b, 2014). Collagen solutions were made from a 10 mg/ml collagen stock solution diluted with cooled DMEM complete and neutralized to pH 7.0 with 1 N NaOH. Neutralized collagen solution (300 µl) was pipetted into an 8-mm-diameter circular polydimethylsiloxane (PDMS) mold placed on a 50-mm glass-bottom dish (MatTek Corporation, Ashland, MA), and gels were then allowed to polymerize at 37°C for 30 min before being overlaid with culture medium. This created three-dimensional collagen matrices ∼10 mm thick. For glucose, serum, alignment, and inhibitor studies, a 1.5 mg/ml collagen gel was used. For density studies, collagen density was altered among 1, 3, and 5 mg/ml. The fiber structure of three-dimensional collagen matrices was visualized using a Zeiss LSM700 confocal microscope equipped with a 405-nm laser and 40×/1.1 N.A. water-immersion objective. For all experiments, cells were allowed to adhere and spread overnight in a temperature-, humidity-, and CO2-controlled incubator.
Matrix alignment
Collagen matrices were aligned by using a magnetic field to pull paramagnetic polystyrene beads (PM-20-10; Spherotech, Lake Forest, IL) through the collagen during matrix polymerization in a custom cell culture device (Guo and Kaufman, 2007). The custom cell culture device comprises a 20 × 20 mm PDMS square with two 8 × 8 mm wells removed from each side. The PDMS walls were then bonded to a No. 1 coverglass on the bottom and ends. Paramagnetic polystyrene beads were combined with the cell/collagen solution at 1% (vol/vol). This solution was added to one well of the custom cell culture device, while the other well was filled with the same solution without metal beads as a comparable random matrix control. The custom device was placed next to a neodymium magnet (BZX0Y0X0-N52; K&J Magnetics, Pipersville, PA) with surface field strength >4 kG to induce the magnetic field on the metal beads. Matrices were allowed to polymerize for 30 min at room temperature before being overlaid with culture medium.
The fiber structure of collagen matrices was visualized using a Zeiss LSM700 confocal microscope equipped with a 405-nm laser and 40×/1.1 N.A. water-immersion objective. Fiber orientation was then analyzed in ImageJ using OrientationJ. Pixel orientation relative to horizontal was used to pseudocolor collagen matrix images.
Live-cell fluorescence microscopy
For PercevalHR measurements, cells were imaged on a Zeiss LSM700 confocal microscope using a 40×/1.1 N.A. long working distance water immersion objective. PercevalHR was excited using a 488- and 405-nm laser, for the MgATP-bound conformation and ADP-bound conformation, respectively. Emission was collected through a 420–550-nm bandpass filter. pHRed was excited using a 488- and 405-nm laser, and emission was collected through a 560-nm longpass filter. Four channel image sets were taken with no delay between individual channel acquisitions. For time-lapse studies involving PercevalHR, cells were imaged on a Zeiss LSM700 confocal microscope using a 20×/0.8 N.A. objective. To account for color aberration between objectives, a correction factor was applied to PercevalHR and pHRed signals collected using the 20×/0.8 N.A. objective. For all other migration studies, time-lapse phase contrast imaging was performed on a Zeiss Axio Observer Z1 inverted microscope equipped with a Hamamatsu ORCA-ER camera and 10×/0.3 N.A. objective. All images were collected >200 μm above the bottom surface and edges of three-dimensional matrices to avoid edge effects.
Quantification of PercevalHR ratiometric signal
All images were analyzed using ImageJ. Background pixel intensity was measured for each channel from each image and subtracted from the entire image to minimize interference by background noise. To calculate ATP, ADP, pH1, and pH2 intensities, a region of interest was drawn around each cell, and mean pixel intensity of the whole cell was acquired. For this study, cells were randomly selected for analysis with the only requirement that they were elongated to ensure they were migrating and sufficiently isolated from other cells and surfaces to prevent any overlap of signal when traced or edge effects, respectively. PercevalHR ratios were calculated by dividing the ATP by ADP fluorescence intensities for a given cell. Owing to pH sensitives of the PercevalHR sensor, approximate removal of pH bias was performed using the RFP-based sensor, pHRed. pHRed ratios were calculated by dividing the pH1 by pH2 fluorescence intensities for a given cell. To remove pH bias, a pH calibration was performed as previously described (Tantama et al., 2011). Briefly, a treatment of 15 mM NH4Cl was applied to induce a transient alkalinization of the cytosol and maintain an approximately constant ATP:ADP while altering intracellular pH. The pH calibration was performed over a short period of time (3–4 min) to minimize metabolic stress on the cell. A linear correlation was then made between the uncorrected PercevalHR signal and pHRed signal to predict pH bias (Supplementary Figure S1). The linear relationship was used to transform pHRed signal to predict changes in PercevalHR due exclusively to change in pH, as previously reported (Tantama et al., 2011). The signal was then normalized by dividing the uncorrected PercevalHR by the transformed pH-corrected signal to approximately correct for predicted pH artifacts. pH correction was performed for density, alignment, and inhibitor studies. Unless otherwise specified, cells were imaged 24 h after seeding or encapsulation.
Glucose uptake analysis
After 24 h of culture, MDA-MB-231 cells were incubated in the presence of the fluorescent glucose-analogue 2-NBDG (Invitrogen). 2-NBDG was added to phenol-red free complete DMEM at a final concentration of 0.146 mM. Cells were incubated for 24 h and then fixed in 3.2% paraformaldehyde (Sigma-Aldrich) for 15 min at room temperature. Samples were washed overnight in 1× phosphate-buffered saline at 4°C and then imaged using a Zeiss LSM700 confocal microscope equipped with a 40×/1.1 N.A. water-immersion objective. 2-NBDG was excited using a 488-nm laser, and emission was collected through a longpass 490-nm filter. A region of interest was drawn around each cell, and mean pixel intensity of the whole cell was acquired to determine glucose uptake. Background pixel intensity was measured for each image and subtracted from the entire image. For this study, elongated and migrating cells were selected for analysis.
ATP hydrolysis analysis
MDA-MB-231 cell oxidative phosphorylation and glycolysis were inhibited using antimycin A (10 µM; Sigma-Aldrich) and iodoacetate (500 µM; Sigma-Aldrich), respectively, to inhibit ATP production. After 24 h of culture, culture media was replaced with complete media containing antimycin A and iodoacetate at the indicated concentrations. Treated cells were incubated for 1 h at 37°C, 5% CO2, and then PercevalHR and pHRed signals were collected every 10 min for 3 h using a Zeiss LSM700 confocal microscope equipped with a 20×/0.8 NA objective. The rate of ATP hydrolysis was calculated as the decrease in normalized PercevalHR ratiometric signal over time.
Cell migration analysis from time-lapse imaging
For migration studies, cells were analyzed between 12 and 18 h after seeding to allow the majority of the cell population enough time to adhere and migrate. Cells were imaged every 20 min during this 6 h period. Stepwise velocities were calculated by determining the displacement of the center of the cell body over time. Net migration was calculated as the maximum displacement of the center of cell body from its point of origin. To correspond the relative cellular ATP levels to cell stepwise velocity across conditions, PercevalHR and pHRed signals were also collected with cell body centroid measurements every 20 min and averaged for each cell. To determine how fluctuations in stepwise velocity correlate to changes in PercevalHR ratiometric signal, PercevalHR and pHRed signals in cells as well as cell body displacement was measured every 1 min for 1 h. To reduce noise, velocity and PercevalHR ratio were then smoothed in MATLAB (R2015b) using lowess regression. Temporal cross-correlation of cell velocity and PercevalHR ratio was calculated in MATLAB (R2015b).
Inhibition of cell migration
For inhibitor studies, the following reagents were used: ROCK inhibitor Y27632 (10 µM; VWR, Radnor, PA), PI3-kinase inhibitor LY294002 (20 µM; EMD Millipore, Billerica, MA), MLC kinase inhibitor ML7 (20 µM; EMD Millipore), and actin cytoskeleton inhibitor Latrunculin A (2 µM; Krackeler Scientific, Albany, NY). The culture media in each well was replaced with fresh complete media containing each inhibitor at the indicated concentration. Cells were also treated with 0.1% (vol/vol) dimethyl sulfoxide (DMSO) in culture media to serve as a vehicle control. Treated cells were incubated for 1 h at 37°C, 5% CO2 before being imaged.
Statistical analysis
All statistical analysis was performed using GraphPad Prism 7.0b (GraphPad Software, San Diego, CA). Data are presented as box-and-whisker plots showing the median, 25th/75th percentiles as the box edges, and 5th/95th percentiles as the bars, or bar graphs expressed as mean ± SEM. Data were compared by one-way analysis of variance (ANOVA) followed by a Tukeys honestly significant difference (HSD) post-hoc test or a nonparametric Wilcoxon rank test. A one-sample t test was used to determine significance of lag for temporal cross-correlation comparison of stepwise speed and PercevalHR ratio.
Supplementary Material
Acknowledgments
We gratefully acknowledge helpful discussions with Matthew Paszek. This work was supported by funding from the National Science Foundation (Award Numbers 1233827 and 1055502) to C.A.R., National Science Foundation Graduate Fellowships to M.R.Z., J.P.M., J.A.V., and M.C.L., and a scholarship for the Next Generation of Scientist from the Cancer Research Society to F.B.
Abbreviations used:
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- ECM
extracellular matrix
- 2-NBDG
2-[N-(7-nitrobenz-2-oxz-1,3-diazol-4-yl)amino]-2-deoxyglucose.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-01-0041) on November 8, 2017.
REFERENCES
- Alberts B, Johnson A, Lewish J. Molecular Biology of the Cell. 4th ed. New York: Garland Science; 2002. [Google Scholar]
- Amano M, Nakayama M, Kaibuchi K. Rho-kinase/ROCK: a key regulator of the cytoskeleton and cell polarity. Cytoskeleton. 2010;67:545–554. doi: 10.1002/cm.20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J, Pun Hung Y, Yellen G. A genetically encoded fluorescent reporter of ATP/ADP ratio. Nat Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman J. ATP: the perfect energy currency for the cell. Creat Res Soc Q. 1999;36 [Google Scholar]
- Bernstein BW, Bamburg JR. Actin-ATP hydrolysis is a major energy drain for neurons. J Neurosci. 2003;23:1–6. doi: 10.1523/JNEUROSCI.23-01-00002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau F, Tang LN, Reinhart-King CA. Togographical guidance of 3D tumor cell migration at an interface of collagen densities. Phys Biol. 2013;10:65004. doi: 10.1088/1478-3975/10/6/065004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursac P, Lenormand G, Fabry B, Oliver M, Weitz DA, Viasnoff V, Butler JP, Fredberg JJ. Cytoskeletal remodelling and slow dynamics in the living cell. Nat Mater. 2005;4:557–561. doi: 10.1038/nmat1404. [DOI] [PubMed] [Google Scholar]
- Caino MC, Chae YC, Vaira V, Ferrero S, Nosotti M, Martin NM, Weeraratna A, O’Connell M, Jernigan D, Fatatis A, et al. Metabolic stress control of cytoskeletal dynamics and metastasis. J Clin Invest. 2013;123:2907–2920. doi: 10.1172/JCI67841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caneba CA, Bellance N, Yang L, Pabst L, Nagrath D. Pyruvate uptake is increased in highly invasive ovarian cancer cells under anoikis conditions for anaplerosis, mitochondrial function, and migration. AJP Endocrinol Metab. 2012;303:E1036–E1052. doi: 10.1152/ajpendo.00151.2012. [DOI] [PubMed] [Google Scholar]
- Carey SP, D’Alfonso TM, Shin SJ, Reinhart-King CA. Mechanobiology of tumor invasion: engineering meets oncology. Crit Rev Oncol Hematol. 2012a;83:170–183. doi: 10.1016/j.critrevonc.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey SP, Goldblatt ZE, Martin KE, Romero B, Williams RM, Reinhart-King CA. Local extracellular matrix alignment directs cellular protrusion dynamics and migration through Rac1 and FAK. Integr Biol. 2016;8:821–835. doi: 10.1039/c6ib00030d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey SP, Kraning-Rush CM, Williams RM, Reinhart-King CA. Biophysical control of invasive tumor cell behavior by extracellular matrix microarchitecture. Biomaterials. 2012b;33:4157–4165. doi: 10.1016/j.biomaterials.2012.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey SP, Rahman A, Kraning-Rush CM, Romero B, Somasegar S, Torre OM, Williams RM, Reinhart-King CA. Comparative mechanisms of cancer cell migration through 3D matrix and physiological microtracks. AJP Cell Physiol. 2014;308:C436–C447. doi: 10.1152/ajpcell.00225.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta S, Penco F, Lavieri R, Martini A, Dinarello CA, Gattorno M, Rubartelli A. Cell stress increases ATP release in NLRP3 inflammasome-mediated autoinflammatory diseases, resulting in cytokine imbalance. Proc Natl Acad Sci USA. 2015;112:2835–2840. doi: 10.1073/pnas.1424741112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, Becker K, Yates JR, Felding-Habermann B. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 2007;67:1472–1486. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- Civelek VN, Deeney JT, Kubik K, Schultz V, Tornheim K, Corkey BE. Temporal sequence of metabolic and ionic events in glucose-stimulated clonal pancreatic β-cells (HIT) Biochem J. 1996;315:1015–1019. doi: 10.1042/bj3151015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunniff B, McKenzie AJ, Heintz NH, Howe AK. AMPK activity regulates trafficking of mitochondria to the leading edge during cell migration and matrix invasion. Mol Biol Cell. 2016;27:2662–2674. doi: 10.1091/mbc.E16-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AD, Carvajal N, Jin A, Matsumoto K, Yamada KM. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nat Commun. 2015;6:8720. doi: 10.1038/ncomms9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22:697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Vahdat LT, Wong S, Chang JC, Mittal V. Microenvironmental regulation of epithelial-mesenchymal transitionsin cancer. Cancer Res. 2012;72:4883–4889. doi: 10.1158/0008-5472.CAN-12-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Kaufman LJ. Flow and magnetic field induced collagen alignment. Biomaterials. 2007;28:1105–1114. doi: 10.1016/j.biomaterials.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Hadjipanayi E, Mudera V, Brown RA. Guiding cell migration in 3D: a collagen matrix with graded directional stiffness. Cell Motil Cytoskeleton. 2009;66:121–128. doi: 10.1002/cm.20331. [DOI] [PubMed] [Google Scholar]
- Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng. 2011;17:713–724. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hecht I, Natan S, Zaritsky A, Levine H, Tsarfaty I, Ben-Jacob E. The motility-proliferation-metabolism interplay during metastatic invasion. Sci Rep. 2015;5:13538. doi: 10.1038/srep13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyslop PA, Hinshaw DB, Halsey WA, Schraufstatter IU, Sauerheber RD, Spragg RG, Jackson JH, Cochrane CG. Mechanisms of oxidant-mediated cell injury: the glycolytic and mitochondrial pathways of ADP phosphorylation are major intracellular targets inactivated by hydrogen peroxide. J Biol Chem. 1988;263:1665–1675. [PubMed] [Google Scholar]
- Imamura H, Nhat KPH, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci USA. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber DE. Can cancer be reversed by engineering the tumor microenvironment. SeminCancer Biol. 2008;18:356–364. doi: 10.1016/j.semcancer.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jekely G, Sung HH, Luque CM, Rorth P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev Cell. 2005;9:197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam AK, Santiskulvong C, Fekete M, Zabih S, Eng C, Dorigo O. Cisplatin and PI3kinase inhibition decrease invasion and migration of human ovarian carcinoma cells and regulate matrix-metalloproteinase expression. Cytoskeleton. 2010;67:535–544. doi: 10.1002/cm.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy HJ, Pouli AE, Ainscow EK, Jouaville LS, Rizzuto R, Rutter GA. Glucose generates sub-plasma membrane ATP microdomains in single islet ß-cells. J Biol Chem. 1999;274:13281–13291. doi: 10.1074/jbc.274.19.13281. [DOI] [PubMed] [Google Scholar]
- Langley RR, Fidler IJ. Tumor cell-organ microenvironment interactions in the pathogenesis of cancer metastasis. Endocr Rev. 2007;28:297–321. doi: 10.1210/er.2006-0027. [DOI] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. 2010;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Duclos G, Sun B, Lee J, Wu A, Kam Y, Sontag ED, Stone HA, Sturm JC, Gatenby RA, et al. Minimization of thermodynamic costs in cancer cell invasion. Proc Natl Acad Sci USA. 2013;110:1686–1691. doi: 10.1073/pnas.1221147110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna JP, Ha J, Merrins MJ, Satin LS, Sherman A, Bertram R. Ca2+ effects on ATP production and consumption have key regulatory roles on oscillatory islet activity. Biophys J. 2016;110:733–742. doi: 10.1016/j.bpj.2015.11.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrins MJ, Poudel C, McKenna JP, Ha J, Sherman A, Bertram R, Satin LS. Phase analysis of metabolic oscillations and membrane potential in pancreatic islet β-cells. Biophys J. 2016;110:691–699. doi: 10.1016/j.bpj.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris BA, Burkel B, Ponik SM, Fan J, Condeelis JS, Aguire-Ghiso JA, Castracane J, Denu JM, Keely PJ. Collagen matrix density drives the metabolic shift in breast cancer cells. EBioMedicine. 2016;13:146–156. doi: 10.1016/j.ebiom.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Pathak A, Kumar S. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc Natl Acad Sci USA. 2012;109:10334–10339. doi: 10.1073/pnas.1118073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Mol Cell. 2010;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15:1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck WJ, Zervantonakis IK, Kamm RD. Tumor cell migration in complex microenvironments. Cell Mol Life Sci. 2013;70:1335–1356. doi: 10.1007/s00018-012-1115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano PP, Inman DR, Eliceiri KW, Trier SM, Keely PJ. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys J. 2008;95:5374–5384. doi: 10.1529/biophysj.108.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Schuler M-H, Lewandowska A, Di Caprio G, Skillern W, Upadhyayula S, Kirchhausen T, Shaw JM, Cunniff B. Miro1-mediated mitochondrial positioning shapes intracellular energy gradients required for cell migration. Mol Biol Cell. 2017;28:2159–2169. doi: 10.1091/mbc.E16-10-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Turner JR. Actin depolymerization disrupts tight junctions via caveolae-mediated endocytosis. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Rigor RR, Pivetti CD, Wu MH, Yuan SY. Myosin light chain kinase in microvascular endothelial barrier function. Cardiovasc Res. 2010;87:272–280. doi: 10.1093/cvr/cvq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi T, Verdone JE, Huang J, Kahlert UD, Hernandez JR, Torga G, Zarif JC, Epstein T, Gatenby R, McCartney A, et al. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget. 2015;6:130–143. doi: 10.18632/oncotarget.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares CP, Midlej V, de Oliveira MEW, Benchimol M, Costa ML, Mermelstein C. 2D and 3D-organized cardiac cells shows differences in cellular morphology, adhesion junctions, presence of myofibrils and protein expression. PLoS One. 2012;7:e38147. doi: 10.1371/journal.pone.0038147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MM, George JH. Exploring and engineering the cell surface interface. Science. 2005;310:1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- Tan I, Leung T. Myosin light chain kinases: division of work in cell migration. Cell Adhes Migr. 2009;3:256–258. doi: 10.4161/cam.3.3.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantama M, Hung YP, Yellen G. Imaging intracellular pH in live cells with a genetically-encoded red fluorescent protein sensor. J Am Chem Soc. 2011;133:10034–10037. doi: 10.1021/ja202902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantama M, Martínez-françois JR, Mongeon R, Yellen G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat Commun. 2013;4:1–25. doi: 10.1038/ncomms3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantama M, Yellen G. Imaging changes in the cytosolic ATP-to-ADP ratio. Methods Enzymol. 2014;547:355–371. doi: 10.1016/B978-0-12-801415-8.00017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov AI, Rutter GA. Use of genetically encoded sensors to monitor cytosolic ATP/ADP ratio in living cells. Methods Enzymol. 2014;542:289–311. doi: 10.1016/B978-0-12-416618-9.00015-7. [DOI] [PubMed] [Google Scholar]
- Thomas S, Fell DA. A control analysis exploration of the role of ATP utilisation in glycolytic-flux control and glycolytic-metabolite-concentration regulation. Eur J Biochem. 1998;258:956–967. doi: 10.1046/j.1432-1327.1998.2580956.x. [DOI] [PubMed] [Google Scholar]
- Uslu VV, Grossmann G. The biosensor toolbox for plant developmental biology. Curr Opin Plant Biol. 2016;29:138–147. doi: 10.1016/j.pbi.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Van Horssen R, Janssen E, Peters W, Van de Pasch L, Te Lindert MM, Van Dommelen MMT, Linssen PC, Ten Hagen TLM, Fransen JAM, Wieringa B. Modulation of cell motility by spatial repositioning of enzymatic ATP/ADP Exchange capacity. J Biol Chem. 2009;284:1620–1627. doi: 10.1074/jbc.M806974200. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Plas DR, Rathmell JC, Fox CJ, Harris MH, Thompson CB. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol Cell Biol. 2001;21:5899–5912. doi: 10.1128/MCB.21.17.5899-5912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton WW, Chandel NS. Hypoxia. 2. Hypoxia regulates cellular metabolism. Am J Physiol Cell Physiol. 2011;300:C385–C393. doi: 10.1152/ajpcell.00485.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochim Biophys Acta. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- Wu N, Zheng B, Shaywitz A, Dagon Y, Tower C, Bellinger G, Shen CH, Wen J, Asara J, McGraw TE, et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell. 2013;49:1167–1175. doi: 10.1016/j.molcel.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv Y, Spurgeon HA, Ziman BD, Lyashkov AE, Lakatta EG. Mechanisms that match ATP supply to demand in cardiac pacemaker cells during high ATP demand. Am J Physiol Heart Circ Physiol. 2013;304:H1428–H1438. doi: 10.1152/ajpheart.00969.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H-X, Xiong Y, Guan K-L. Nutrient sensing, metabolim, and cell growth control. Mol Biol Rep. 2014;49:379–387. doi: 10.1016/j.molcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J, Haley J, Kalluri R, Muthuswamy SK, Thompson E. Epithelial-mesenchymal transition. Cancer Res. 2008;68:9574–9577. doi: 10.1158/0008-5472.CAN-08-2316. [DOI] [PubMed] [Google Scholar]
- Zschenker O, Streichert T, Hehlgans S, Cordes N. Genome-wide gene expression analysis in cancer cells reveals 3D growth to affect ECM and processes associated with cell adhesion but not DNA repair. PLoS One. 2012;7:e34279. doi: 10.1371/journal.pone.0034279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.