The distinct behavior of the spindle pole bodies (SPBs) during spindle orientation in yeast metaphase does not result from them being differently mature, but astral microtubule organization correlates with the subcellular position rather than the age of the SPBs.
Abstract
In many asymmetrically dividing cells, the microtubule-organizing centers (MTOCs; mammalian centrosome and yeast spindle pole body [SPB]) nucleate more astral microtubules on one of the two spindle poles than the other. This differential activity generally correlates with the age of MTOCs and contributes to orienting the mitotic spindle within the cell. The asymmetry might result from the two MTOCs being in distinctive maturation states. We investigated this model in budding yeast. Using fluorophores with different maturation kinetics to label the outer plaque components of the SPB, we found that the Cnm67 protein is mobile, whereas Spc72 is not. However, these two proteins were rapidly as abundant on both SPBs, indicating that SPBs mature more rapidly than anticipated. Superresolution microscopy confirmed this finding for Spc72 and for the γ-tubulin complex. Moreover, astral microtubule number and length correlated with the subcellular localization of SPBs rather than their age. Kar9-dependent orientation of the spindle drove the differential activity of the SPBs in astral microtubule organization rather than intrinsic differences between the spindle poles. Together, our data establish that Kar9 and spatial cues, rather than the kinetics of SPB maturation, control the asymmetry of astral microtubule organization between the preexisting and new SPBs.
INTRODUCTION
During mitosis, the microtubule asters forming at each pole of the spindle of most eukaryotes, except plants, position the spindle within the cell. This process is of primary importance for the cell types that align their spindle with their polarity axis to divide asymmetrically (Siller and Doe, 2009). The formation of the microtubule asters is driven by the microtubule-organizing center (MTOC) present at each spindle pole. In many eukaryotes, these MTOCs are produced through conservative or semiconservative duplication (Bornens and Piel, 2002; Barral and Liakopoulos, 2009). In either case, this causes one aster to form around an MTOC inherited from the previous mitosis (older or preexisting MTOC), whereas the other aster contains the most recent MTOC (younger or new MTOC). The MTOC matures by accumulating MTOC-associated factors (e.g., pericentriolar material [PCM] in animal cells) and γ-tubulin complexes (γ-TuCs) to acquire microtubule nucleation capability (Meraldi and Nigg, 2002). Remarkably, in many asymmetrically dividing cells, like stem cells, the preexisting and young MTOCs segregate nonrandomly between daughter cells at mitosis (Lerit et al., 2013). How these cells specify the fate of MTOCs and segregate them in an age-dependent manner is only partially understood.
Based on the observation that generally the preexisting MTOC nucleates more astral microtubules than the new one, a recurring model proposes that MTOC specification is driven by one of them being mature and fully active, whereas the other one, the new one, is still immature (Lerit et al., 2013). For example, during the division of male germ stem cells of Drosophila, the older centrosome is surrounded by more abundant PCM and nucleates more microtubules than the younger one. This contributes to its anchorage to cortical cues near the stem cell niche and its maintenance in the renewing stem cell during division. In contrast, the younger centrosome originally organizes only few microtubules, preventing its retention. As a consequence, the newer MTOC is pushed to the opposite side of the cell and segregates into the differentiating daughter cell (Yamashita et al., 2007). Nevertheless, the amount of PCM around the centrioles is not necessarily a consequence of the centriole’s age. Indeed, Drosophila larval neural stem cells, called neuroblasts, start mitosis by carrying a similar amount of PCM and microtubules around both centriole pairs and subsequently strip the older centriole while maintaining the aster around the new one (Rebollo et al., 2007; Conduit and Raff, 2010; Januschke et al., 2011). These studies suggest that MTOC maturation is not an absolute determinant of the MTOC capacity to organize microtubules but that regulatory factors also play important roles.
In budding yeast, the MTOCs are formed by spindle pole bodies (SPBs), which are embedded in the nuclear envelope and consist of three plaques. The central plaque contacts the rim of the pore, which inserts it into the nuclear envelope and contains several proteins, including Spc42. It anchors the cytoplasmic outer plaque and the nuclear inner plaque of the SPB to form its final structure (Byers and Goetsch, 1974, 1975; Rout and Kilmartin, 1990; Jaspersen and Winey, 2004). Two of the main outer plaque components, Spc72 and Nud1, are conserved in metazoans (human TACC and Centriolin, respectively) (Keck et al., 2011), while no orthologue of the third main component, Cnm67, is known so far. The inner and outer plaques subsequently serve as platforms to carry the microtubule-nucleating machinery, including the γ-TuC (Lin et al., 2015). The γ-TuC is composed of γ-tubulin (Tub4 in yeast) and the scaffolding proteins Spc98 and Spc97 (respectively, TUBGCP3 and TUBGCP2 in humans) (Geissler et al., 1996; Knop and Schiebel, 1997; Murphy et al., 1998; Jaspersen and Winey, 2004). The mammalian GCP4-6 proteins are missing in budding yeast but are not required to form a ring-shaped template, called the γ-tubulin ring complex (γ-TuRC), to nucleate microtubules (Kollman et al., 2010, 2015). The γ-TuC is anchored to the inner and outer plaque via Spc110 and Spc72, respectively (Knop and Schiebel, 1997, 1998).
SPB duplication leads to the coexistence of a preexisting SPB and a newly assembled SPB at the opposite poles of the mitotic spindle (Pereira et al., 2001; Yoder et al., 2003; Menendez-Benito et al., 2013). Indeed, the duplication of the SPB is conservative for some of its core components: For example, preexisting Spc42 is kept within the preexisting SPB, whereas newly synthesized Spc42 is preferentially incorporated into the newly formed SPB (Pereira et al., 2001; Menendez-Benito et al., 2013). The maturation of the new SPB finishes once it possesses a full complement of the proteins that comprises the outer and inner plaques, enabling it to carry γ-TuCs and organize microtubules.
Strikingly, in the vast majority of budding yeast cells the two SPBs do not segregate randomly at mitosis. During metaphase, the preexisting SPB orients toward the bud, which inherits it on division (Pereira et al., 2001; Hotz et al., 2012). Since the bud is the stem cell equivalent in yeast (it renews its full division potential, while the mother ages), this segregation pattern is remarkably similar to what is observed in many animal stem cells (Yamashita et al., 2007; Barral and Liakopoulos, 2009; Wang et al., 2009). However, as for the centrosomes of stem cells, the mechanism underlying the nonrandom segregation of yeast SPBs is not fully understood.
Alignment of the yeast mitotic spindle along the mother–bud axis is driven by the asymmetric recruitment of Kar9 to astral microtubules emanating from the preexisting SPB. Subsequently, Kar9 drives microtubule plus ends into the bud along actin cables by binding to the type V myosin, Myo2 (Beach et al., 2000; Korinek et al., 2000; Miller et al., 2000; Yin et al., 2000; Hwang et al., 2003; Kusch et al., 2003; Liakopoulos et al., 2003). Thus, the recruitment of Kar9 to only a subset of astral microtubules is the main mechanism determining that the preexisting SPB specifically orients toward the bud. Accordingly, kar9∆ mutant cells segregate their SPBs nearly randomly (Pereira et al., 2001; Hotz et al., 2012). Recent studies have identified the SPB inheritance network (SPIN) (Lengefeld et al., 2017) and the yeast Hippo pathway (mitotic exit network; MEN) (Hotz et al., 2012) as key factors for directing Kar9 toward microtubules emanating from the old SPB. However, how these factors control Kar9 localization is not yet fully understood.
A possible clue to this process resides in the observation that the yeast metaphase spindle is also morphologically asymmetric: the astral microtubules emanating from the SPB oriented toward the bud are longer and more abundant than those emanating from the distal SPB (Shaw et al., 1997; Segal and Bloom, 2001; Grava et al., 2006). Furthermore, at least in some strain backgrounds the γ-TuC anchor on the outer plaque, Spc72, is more abundant on the preexisting than on the newly synthesized SPB (Juanes et al., 2013). This asymmetry in astral microtubule organization and abundance is thought to promote the recruitment of Kar9 preferentially toward the microtubules emanating from the preexisting SPB. How this observation and the role of the MEN and SPIN in Kar9 localization relate to each other to drive SPB inheritance is, however, unclear.
Indeed, the different behavior of the two SPBs might be caused by an incomplete maturation of the new SPB when the spindle orients itself in early metaphase; similarly to what is suggested for centrosome asymmetry (Yamashita et al., 2007; Morrison and Spradling, 2008; Yamashita and Fuller, 2008). This could cause the preexisting SPB to organize astral microtubules before the new one and drive the asymmetric recruitment of Kar9 to the preexisting SPB toward the bud (Juanes et al., 2013). In this case, SPIN might control the dynamics of SPB maturation. Alternatively, maturation might only marginally affect the differential behavior of the SPBs, which might rather be the consequence of SPIN and MEN regulating Kar9 distribution directly. To dissect these processes more precisely, we investigate here how maturation differences drive the distinct behaviors of the SPBs during spindle orientation.
RESULTS
The SPB inheritance network and mitotic exit network do not affect Spc72 localization
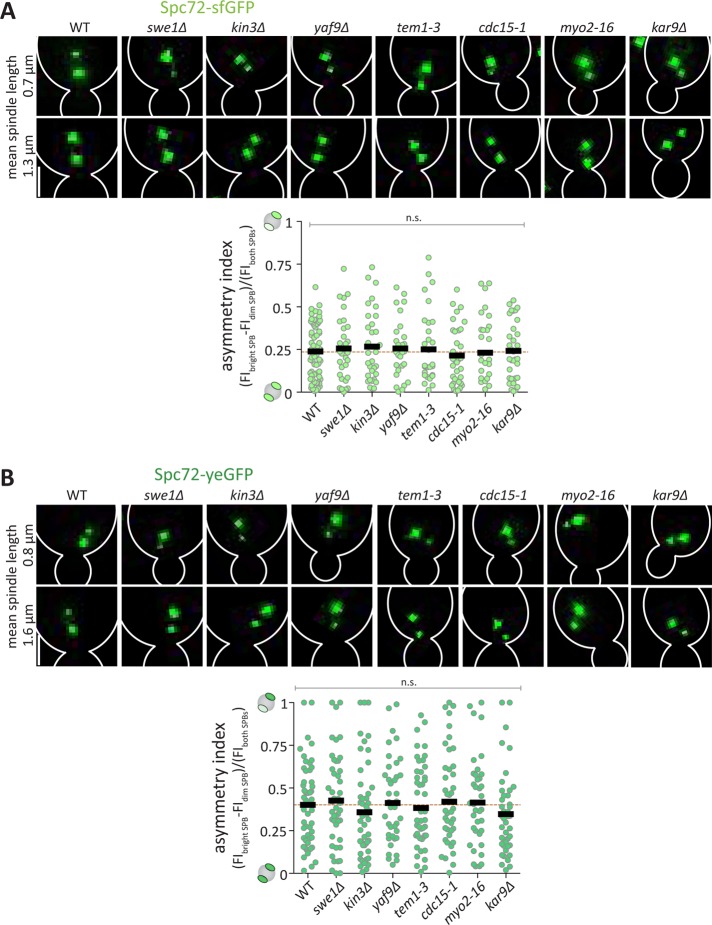
To clarify the relationships among SPIN (Lengefeld et al., 2017), MEN (Hotz et al., 2012), and the asymmetric distribution of Spc72 (Juanes et al., 2013), we investigated how SPIN and MEN affect Spc72 localization. Thus, we quantified the asymmetry index of Spc72-sfGFP and Spc72-yeGFP in SPIN and MEN mutant cells. Since we focused on spindle assembly, we first restricted our analysis to microscopy-visibly separated SPBs of spindles with an average length of 1.0 ± 0.2 μm (early metaphase) (Nazarova et al., 2013). At this stage, the spindle’s asymmetry governs its orientation, and Spc72 is expected to be less abundant at the new SPB. Spc72-sfGFP and Spc72-yeGFP were imaged in wild type, SPIN, and MEN mutant cells and the fluorescence intensity was measured at the SPBs. These values were used to compute the asymmetry index defined by dividing the difference between the fluorescence intensity of the bright and dim SPBs by the total fluorescence intensity of both SPBs (Figure 1, A and B). We quantified the asymmetry index using the intensity difference instead of the position of the SPBs, because many spindles misorient in SPIN and MEN mutant cells (compare difference from Figure 2). Theoretically, this asymmetry index reaches the value of 1 when the fluorescence is fully asymmetric toward one SPB and 0 when it is symmetric between the two SPBs. This analysis failed to reveal any significant difference in Spc72 asymmetry between the swe1∆, kin3∆, yaf9∆, tem1-3, and cdc15-1 mutant and the wild-type cells. Furthermore, Spc72 asymmetry was not affected in kar9∆ or myo2-16 control mutant cells. Thus, SPIN and MEN do not control Spc72 recruitment to SPBs, and hence, they act in spindle orientation after SPB maturation.
FIGURE 1:
The SPB inheritance network and mitotic exit network do not promote Spc72 localization. (A, B) Representative images of Spc72-sfGFP and Spc72-yeGFP at spindle-length (μm) of different genotypes and quantification of asymmetry index (dividing the difference between the fluorescence intensity of the bright and dim SPB by the total fluorescence intensity of both SPBs) of Spc72-sfGFP of metaphase cells of indicated genotype (n = 60 cells pooled from three independent experiments, mean ± SD). Statistical significance was calculated using one-way ANOVA; n.s. = nonsignificant. Scale bars, 2 μm.
FIGURE 2:
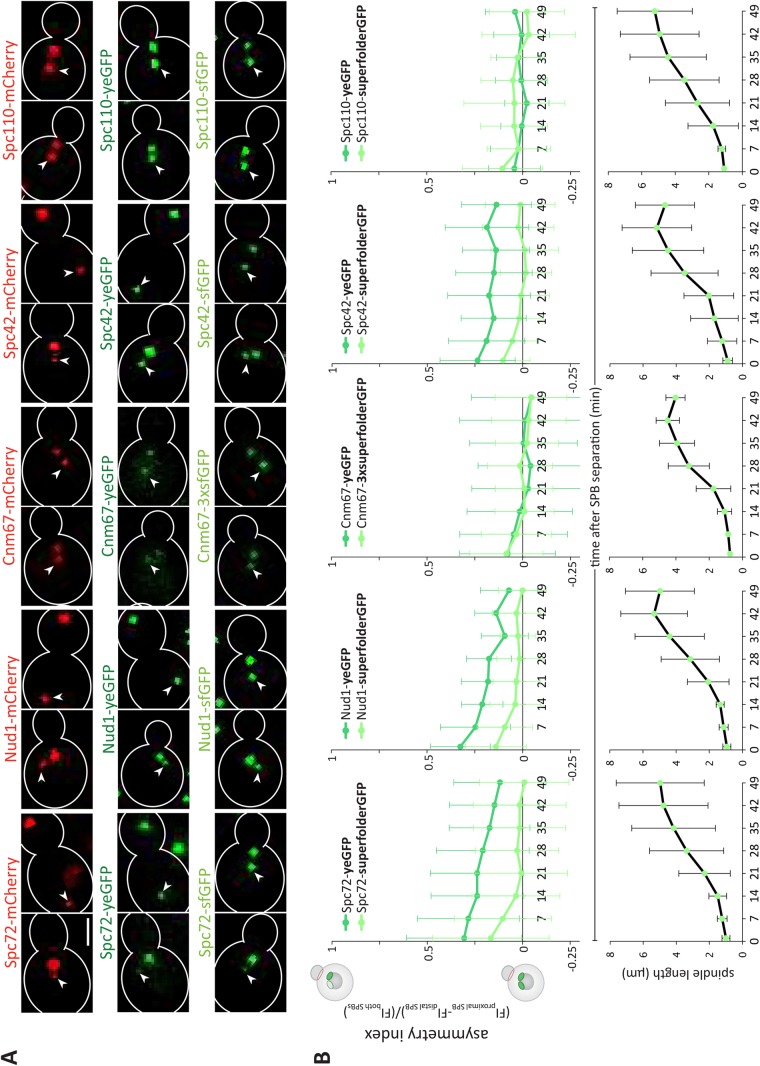
Analysis of outer plaque maturation kinetics. (A) Representative images and quantification of asymmetry index (dividing the difference between the fluorescence intensity of the proximal and distal SPB by the total fluorescence intensity of both SPBs) over time after SPB separation of Spc72, Spc42, Cnm67, and Spc110 tagged with sfGFP or yeGFP and quantification of corresponding spindle length (μm) (n = 120 cells analyzed at each time point from three independent experiments, mean ± SD). Scale bars, 2 μm. The distal SPB is pointed out with a white arrow.
Analysis of outer plaque maturation kinetics
Remarkably, in all these measurements the Spc72-sfGFP signal was systematically more symmetric between SPBs than that of Spc72-yeGFP (Figure 1A). Complete assembly of the outer plaque is a hallmark of SPB maturation. Thus, we investigated whether the abundance of Spc72 is influenced by its fluorescence tags or whether maturation differences of the fluorophore itself caused the observed difference. We analyzed the recruitment kinetics of outer plaque components on the new SPB, taking the preexisting SPB as a reference. To do this, we investigated the fluorescence signal of different fluorophores fused individually to Cnm67, Nud1 and Spc72 (outer plaque), Spc42 (central plaque), and Spc110 (central-inner plaque; Supplemental Figure S1A).
We reasoned that depending on the maturation time of each fluorophore, the fluorescence signal is indicative of protein abundance as well as the tendency of a protein to exchange at the SPB during SPB duplication. Indeed, proteins tagged with a fast-maturing fluorophore are assumed to most directly report about the protein abundance at SPBs. In contrast, proteins tagged with slow-maturing fluorophores are predicted to inform about protein age and not only about abundance. For example, preexisting Spc42-mCherry is maintained within the preexisting SPB while the newly synthesized Spc42-mCherry is mainly incorporated into the new SPB during SPB duplication. Owing to the relatively slow maturation properties of mCherry, the preexisting SPB appears significantly brighter than the new SPB (Pereira et al., 2001). This difference in fluorescence signal is not observed with a fast-maturing fluorophore (Figure 2A), indicating that Spc42 is present to similar levels on both SPBs. Hence, the asymmetric fluorescence of the slow-maturing fluorophore is not due to a difference in protein abundance but in protein age between the two SPBs (Supplemental Figure S1B). This further indicates that there is little or no exchange of Spc42 at the SPB. Thus, the difference in asymmetry observed between the fast- and slow-maturing fluorophores indicates that newly synthesized Spc42 is rapidly incorporated into the newly assembled SPB and that Spc42 is immobile at the SPBs. Put the other way around, when a slow-maturing fluorophore does not show more asymmetry than the fast-maturing one, one can assume that both SPBs contain as much new than old tagged protein, and hence that the protein exchanges between the SPBs (probably trough the cytoplasm) during or shortly after SPB assembly.
The estimated maturation half-times (t1/2, time required for fluorescence to reach half of its maximal value) of the fluorophores mCherry, yeast codon-usage optimized eGFP (yeGFP), and superfolder GFP (sfGFP) in vivo are 40, 25, and 6 min, respectively (Khmelinskii et al., 2012). In comparison, the formation of a 1- to 2-μm-long spindle takes ≈20 min (Segal et al., 2000). Thus, the proteins Spc72, Cnm67, Nud1, Spc42, and Spc110 were tagged individually with mCherry, yeGFP, and sfGFP. Overall, these C-terminal tags do not influence most epitope’s functions (Knop et al., 1999; Jaspersen et al., 2002; Yoder et al., 2003; Muller et al., 2005; Grava et al., 2006; Gryaznova et al., 2016). The central plaque protein Spc42 served as a reference as it assembles very early into the newly formed SPB and is present in similar amounts in both SPBs (Burns et al., 2015). The tagged strains were imaged, and the fluorescence intensity of the SPB originally localized proximally and distally from the bud neck was measured over time, starting as soon as the two SPBs were distinguishable from each other (see Materials and Methods for details). With these values, the asymmetry index was calculated, which was defined by dividing the difference between the fluorescence intensity of the proximal and distal SPB by the total fluorescence intensity of both SPBs. Unlike in Figure 1, this value ranges between 1 and -1, reaching 1 when the fluorescence is fully asymmetric toward the proximal SPB, 0 when it is symmetric, and -1 when it is asymmetric toward the distal SPB. In the case of the sfGFP- and yeGFP-tagged strains, these values were recorded every 7 min through the course of spindle assembly, and the asymmetry index was plotted as a function of time (Figure 2B). At each time point, the length of the spindle, that is, the distance between the two SPBs, was measured. Owing to rapid bleaching, this procedure was not possible for mCherry. Instead, the intensity was recorded in snapshot pictures and the computed asymmetry index was plotted for short and long spindles (mean length of 0.9 ± 0.3 and 4.8 ± 1.8 μm, respectively; Figure 3A).
FIGURE 3:
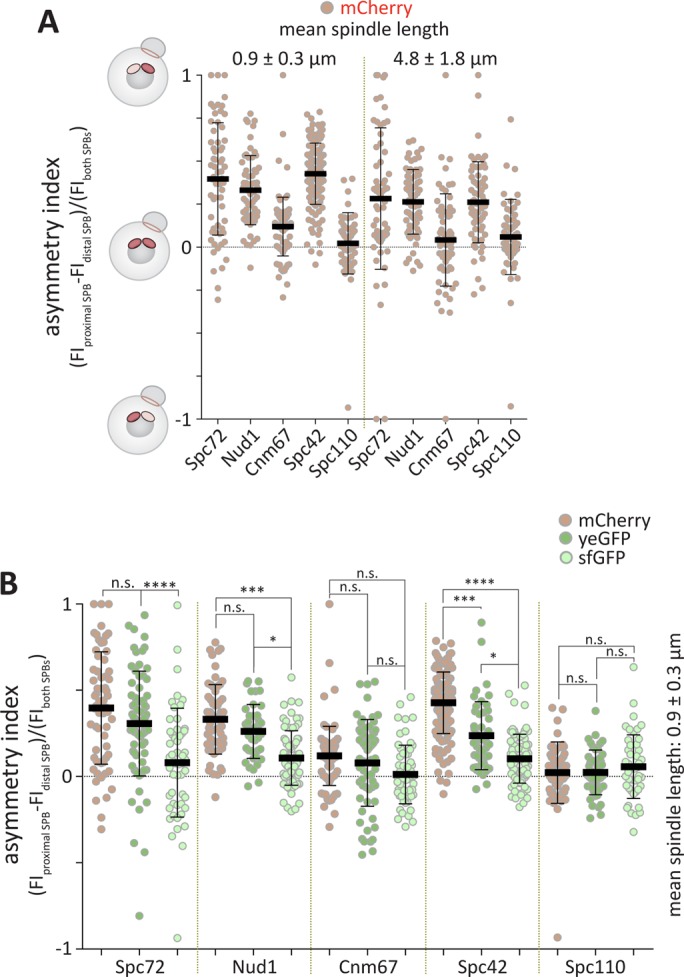
Analysis of outer plaque maturation kinetics. (A) Quantification of asymmetry index of mCherry tagged Spc72, Nud1, Cnm67, Spc42, and Spc110 for short spindles (mean spindle length 0.9 ± 0.3 μm) and long spindles (mean spindle length 4.8 ± 1.8 μm) (n = 60 cells pooled from three independent experiments, mean ± SD). (B) Comparison of Spc72, Nud1, Cnm67, Spc42, and Spc110 tagged with mCherry, yeGFP, or sfGFP on short spindles (mean spindle length 0.9 ± 0.3 μm) (n = 60 cells pooled from three independent experiments, mean ± SD). Data from A and Figure 2A. Statistical significance was calculated using Student’s t test. Scale bars, 2 μm. ****p < 0.0001, ***p < 0.001, *p < 0.05, n.s. = nonsignificant.
For Spc72, Nud1, and Spc42, the asymmetry index for all fluorophores tended to be highest in cells with the shortest spindles that is, taken immediately after SPB separation. Thus, either the new SPB takes more time to maturate or the fluorophore is not fully matured on the new SPB, and the tagged protein is immobile at the SPB. Supporting the latter model, the choice of the fluorophore influenced the asymmetry of the signal, with the slow-maturing ones displaying a higher asymmetry index than the fast ones on very short spindles (Spc72: mCherry = 0.37, yeGFP = 0.31, sfGFP = 0.16; Nud1: mCherry = 0.33, yeGFP = 0.32, sfGFP = 0.14; Spc42: mCherry = 0.42, yeGFP = 0.24, sfGFP = 0.10) (Figures 2 and 3, Supplemental Figure S1C). Remarkably, the asymmetry of Spc72 was not significantly different from that of Nud1 or Spc42, suggesting that right after SPB separation Spc72 is not more asymmetric than Nud1 or Spc42. Our data further suggest that the maturation of yeGFP or sfGFP was not influenced by the protein it was fused to since the asymmetry index similarly decreased over time when Spc72, Nud1, or Spc42 were tagged with either yeGFP or sfGFP (Supplemental Figure S1, D and E). We also tested whether photobleaching is constant for the different constructs fused to Spc72, Spc42, and Nud1 by comparing the intensity of the preexisting SPB over time (Supplemental Figure S2). The fluorescence intensities were indistinguishable over time, indicating that photobleaching is similar for the different fluorophores and is independent of the protein to which it is fused. In summary, the kinetics of fluorophore maturation considerably contribute to the observed asymmetry of Spc72.
These data suggest that, like Spc42 (Pereira et al., 2001) and Nud1 (Lengefeld et al., 2017), Spc72 is incorporated in a mainly conservative manner on SPBs, with the new SPB being substantially composed of newly synthesized protein and the preexisting one containing mainly older Spc72. Supporting this interpretation, in all cases the asymmetry of the signal decreased over time and the kinetics of this decrease correlated with the maturation speed of the fluorophore (Figures 2 and 3). Within ∼14 min, the fluorescent signals of Spc72-, Nud1-, and Spc42-sfGFP were as bright in the distal SPB (new) as in the proximal SPB (preexisting), whereas it took longer when the proteins were tagged with yeGFP. The same trend was observed when fluorescence was plotted against spindle length instead of time (Supplemental Figure S3). We concluded that shortly after SPB separation, Spc72 is present in similar amounts on both SPBs, and hence, the new SPB rapidly accumulates as much Spc72 as the preexisting one.
However, these assays led to a surprising observation for Cnm67. Independently of the fluorophore used and whether it was plotted against spindle length or the time elapsed after SPB separation, we always measured similar fluorescence intensities on both SPBs (Figures 2 and 3). The same observation was made when we fluorescently tagged Spc110. This suggests that, as described for Spc110 (Yoder et al., 2003), the outer plaque component Cnm67 tends to exchange between the SPBs during SPB maturation, leading to the equilibration of old and new proteins between both SPBs. Further, these results establish that, like the three other SPB components, Cnm67 and Spc110 are similarly abundant on both SPBs as soon as the preexisting and new SPBs have separated from each other.
Spc72 distribution is variable between strain backgrounds
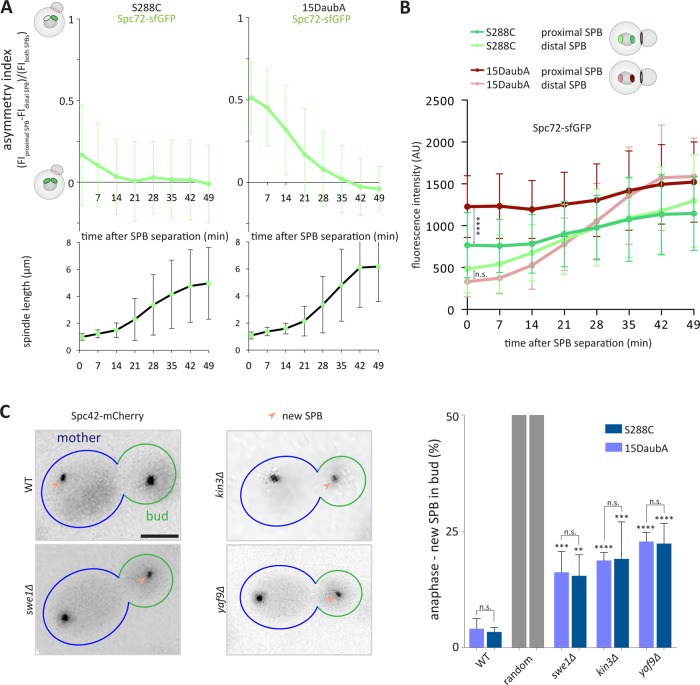
The observation that shortly after their separation both SPBs carry similar amounts of Spc72 is in contrast to a previous report indicating that Spc72 is distributed asymmetrically between SPBs at this cell-cycle stage (Juanes et al., 2013). These studies were carried out using yeGFP, which could explain the observed asymmetry but in cells of distinct genetic background (S288C here and 15DaubA in Juanes et al. [2013]). Thus, we reanalyzed Spc72-sfGFP fluorescence in 15DaubA cells, as described above. Interestingly, immediately after SPB separation, the Spc72-sfGFP asymmetry toward the proximal SPB was significantly stronger in the 15DaubA cells compared with S288C (Figure 4A). Interestingly, this asymmetry was caused by an increased intensity at the proximal SPB in 15DaubA cells compared with S288C cells, while the intensity of the distal SPB was indistinguishable between both strain backgrounds for very short spindles (Figure 4B). Therefore, we asked whether the 15DaubA cells used a different mechanism to bias SPB inheritance than the S288C cells. We tested whether the SPIN mutants randomize SPB inheritance in the 15DaubA background to a similar extent as in S288C. Spc42-mCherry served as a marker for distinguishing preexisting and newly synthesized SPBs as described (Pereira et al., 2001). Importantly, in the 15DaubA background the deletion of SWE1, KIN3, and YAF9 caused a significant increased fraction of mutant cells to segregate the new SPB into the bud in anaphase compared with wild-type cells. However, these effects were statistically indistinguishable from those observed in the S288C background (Figure 4C). Altogether, these data support the interpretation that the SPIN acts independently of Spc72 levels at SPBs and that at least in S288C cells differences in SPB maturation are not essential for driving SPB inheritance.
FIGURE 4:
Spc72 distribution is variable between strain backgrounds. (A) Quantification of asymmetry index (dividing the difference between the fluorescence intensity of the proximal and distal SPB by the total fluorescence intensity of both SPBs) over time after SPB separation of Spc72 tagged with sfGFP in S288C or 15DaubA strain background and quantification of corresponding spindle length (μm) (n = 120 cells analyzed at each time point from three independent experiments, mean ± SD). (B) Fluorescence intensity (AU) of Spc72 tagged with sfGFP in S288C or 15DaubA strain background at the preexisting or new SPB over time (min) (n = 120 cells analyzed at each time point from three independent experiments, mean ± SD). Raw data as in A. (C) Quantification of anaphase cells of indicated genotype segregating the new SPB into the bud (%) (n = 3 independent experiments with a total of >120 cells per genotype analyzed, mean ± SD). All statistical significances were calculated using Student’s t tests; ****p < 0.0001, ***p < 0.001, **p < 0.01, n.s. = nonsignificant. Scale bars, 2 μm.
The SPBs exchange Cnm67, but not Spc72, during duplication
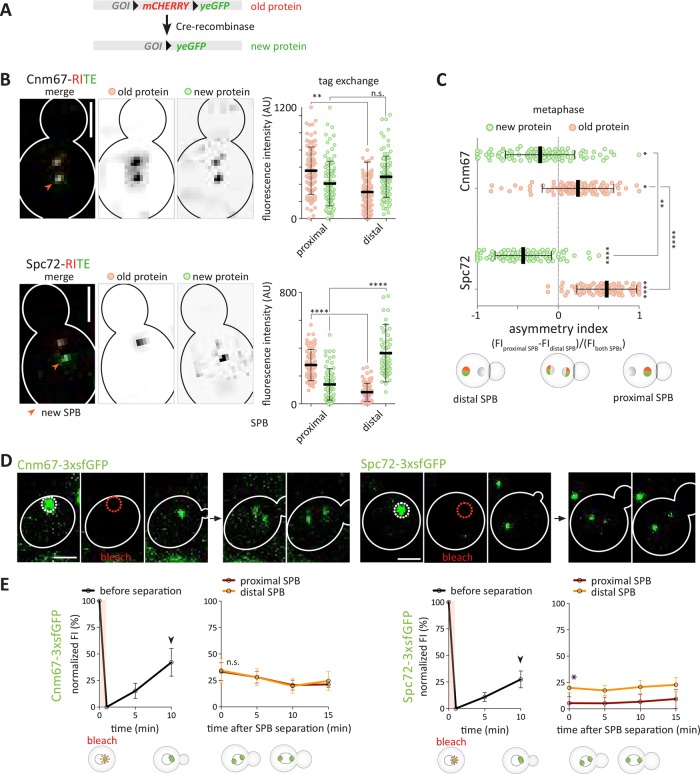
To further test the conclusions drawn from the studies above, we next investigated more directly whether Cnm67 indeed exchanges at the SPB while Spc72 does not. Thus, we assayed protein exchange using the recombinase-induced tag exchange (RITE) assay (Verzijlbergen et al., 2010; Hotz et al., 2012). In the RITE assay, the Cre recombinase excises the mCherry-coding sequence flanked with loxP-sites and fuses the yeGFP-coding sequence in-frame to the target open reading frame (Figure 5A). Thus, the preexisting gene-products are fused to mCherry and fluoresce in red. On recombination, newly synthesized protein is fused to yeGFP and fluoresces in green. The use of an estradiol-regulated form of Cre allows precise temporal control of the tag switching. Therefore, in case of a nonexchangeable protein, we expect that induction of recombination in cells in G1 phase would lead to the formation of a preexisting SPB mainly labeled with mCherry and a new SPB mainly labeled with yeGFP. Alternatively, if the new protein is incorporated into both SPBs, then we expect the preexisting SPB to contain both fluorophores and the new one to primarily contain yeGFP. Finally, if preexisting protein either from the preexisting SPB or the cytoplasm is incorporated into the newly built SPB, we expect both SPBs to carry mCherry and yeGFP. SPC72 and CNM67 genes were tagged with the RITE cassette and protein exchange was assayed. Strikingly, preexisting (red) and new (green) Cnm67 were rather equally abundant on both SPBs (Figure 5, B and C). These results are consistent with the hypothesis that both preexisting and newly synthesized Cnm67 protein are incorporated to fairly similar extents into both SPBs before they separate.
FIGURE 5:
Cnm67, but not Spc72, exchanges between the SPBs during its duplication. (A) Schema of the RITE: mCHERRY is flanked by loxP sites, which allows excision of mCherry-coding sequence by induction of Cre-recombinase and switch to yeGFP. Gene of interest (GOI). (B) Representative images and quantification of normalized fluorescence intensity of preexisting (mCherry) and new (yeGFP) Cnm67 and Spc72 in proximal and distal SPB before and during Cre-recombinase-induced tag exchange (n = 100 cells pooled from three independent experiments, mean ± SD). Statistical significance was calculated using one-way ANOVA. (C) Asymmetry indexes of preexisting (mCherry) and new (yeGFP) Cnm67 and Spc72 during tag exchange and SPB duplication (n = 100 cells pooled from three independent experiments, mean ± SD). Statistical significance was calculated using one-way ANOVA. (D) Representative image of Cnm67-3xsfGFP and Spc72-3xsfGFP during a FRAP experiment. Fluorescence signal (FI) was bleached in G1 phase and recovery was measured until metaphase. (E) Quantification of normalized FI (%) of Cnm67-3xsfGFP and Spc72-3xsfGFP of bleached SPBs from G1 phase for 15 min (SPB before separation) and after SPB separation for 15 min (distal or proximal SPB) (n = 12 cells pooled from two independent experiments, mean ± SEM). Statistical significance was calculated using Student’s t test. Arrow marks bud emergence. ****p < 0.0001, **p < 0.01, *p < 0.05, n.s. = nonsignificant. Scale bars, 2 μm.
The trend was different for the Spc72 protein. In this case, the preexisting fraction of Spc72 mainly remained within the proximal SPB (Figure 5, B and C). New Spc72 was preferentially incorporated into the distal SPB, although a limited fraction of it was visible on the proximal SPB, as observed for Spc42 (Hotz et al., 2012). The difference in behavior between Spc72 and Cnm67 was statistically significant. Of note, Cre-expression does not remove preexisting mCherry transcripts, potentially leading to some newly synthesized protein being tagged with mCherry after tag exchange. Together, although these data can be considered only as trends, they are consistent with the results obtained using fluorophores with different folding kinetics and support the notion that preexisting Cnm67 largely exchanges at SPBs, while preexisting Spc72 remains rather stable on the preexisting SPB.
To address whether the kinetics of fluorophore maturation cause the observed asymmetry of Spc72, we investigated at which cell-cycle stage newly synthesized Spc72 is incorporated into the new SPB, using fluorescence recovery after photobleaching (FRAP). The fluorescence signal of Cnm67-3xsfGFP and Spc72-3xsfGFP was photobleached specifically in G1 cells, that is, before the SPBs separated from each other (Figure 5D) (Liakopoulos et al., 2003; Lengefeld et al., 2017). The recovery traces indicate that in both cases newly synthesized material is incorporated in the SPB before and during bud emergence (Figure 5E). Subsequently, the same cells were followed as they entered the cell cycle, separated their SPBs, and started to assemble a spindle. Remarkably, in Cnm67-3xsfGFP-expressing cells, both SPBs showed similar fluorescence levels as soon as they began to separate from each other. The intensity of this fluorescence remained stable for the following 15 min, suggesting that little or no additional material was incorporated into the SPBs (Figure 5E, left panel). In contrast, for the Spc72-3xsfGFP-expressing cells, one SPB was significantly dimer. These dimer SPBs generally oriented toward the bud neck, consistent with being the old SPBs, and with containing mainly preexisting, immobile material that had been photobleached in G1 phase and to which little material had been added subsequently (Figure 5E, right panel). We also failed to see any further recovery on the newly synthesized SPB. Together, these data indicate that in relative terms Cnm67 and Spc72 behaved differently from each other. New and preexisting Spc72 was asymmetrically distributed with respect to its age and exchanged little at the SPBs. In contrast, preexisting and new Cnm67 exchange between SPBs at least during SPB duplication. Together with the results from the fluorophore-folding and RITE experiments, we conclude that newly synthesized Spc72 is incorporated mainly into the newly formed SPB and stably stays with that SPB thereafter. Furthermore, at least in S288C the outer plaque components of the SPB, including Spc72, are equally abundant on both SPBs on SPB separation.
The outer plaques of proximal and distal SPBs both have at least one γ-TuC regardless of metaphase spindle length
Given that our data suggest that the outer plaque component Spc72 is present similarly at both SPBs after their separation, we asked whether the γ-TuCs component, Spc97, is also present at the outer plaques of both SPBs in metaphase using super resolution microscopy.
We first used three-dimensional structured illumination microscopy (SIM), previously used to study the SPB duplication mechanism (Burns et al., 2015), to resolve the distribution of γ-TuCs components for metaphase spindles ranging from 0.5 to 2 μm in fixed cells. SIM captures both the interactions of high-frequency modulated illumination patterns and the high-frequency variations in the small structure’s fluorescence to provide superresolution. Imaging with a nonuniform wide-field illumination (a series of Moiré interference patterns in different orientations) and the subsequent computational reconstruction allows the extraction of subdiffraction sample information (Gustafsson, 2000; Schermelleh et al., 2008, 2010).
We used the γ-TuCs component Spc97 fused to the bright and fast-folding mNeonGreen fluorescent protein (Shaner et al., 2013) codon optimized for yeast and used the Spc97-mNG fusion protein to study the distribution of γ-TuCs on SPB inner and outer plaques. As expected, Spc97-mNG localizes to both SPBs of the metaphase spindle (Figure 6A). Using three-dimensional SIM, we could resolve distinct densities of Spc97-mNG at each SPB (Figure 6B). These densities, representing flat disks stacked within the 160-nm SPB structure (Bullitt et al., 1997; O’Toole et al., 1999), are separated by a distance of ∼165 nm (Bullitt et al., 1997; O’Toole et al., 1999) and were identified as inner and outer SPB plaques due to their relative intensities (inner plaque is brighter than outer) and orientation (dimmer plaque facing outward) (Figure 6C). We quantified the number of metaphase spindles exhibiting visually resolvable inner and outer Spc97-mNgr densities and found three classes of spindles; class 1 spindles (9 %; n = 6) exhibit resolvable inner and outer densities for both SPBs (Figure 6C), class 2 (47 %; n = 32) exhibit resolvable outer densities on one SPB (Figure 6D), and class 3 (44 %; n = 30) exhibit no resolvable outer density on either pole (Figure 6E). Owing to the limited axial resolution of three-dimensional SIM (>250 nm for wavelengths employed [ Schermelleh et al., 2008]), we expect to resolve inner and outer plaques when the SPB is aligned perpendicularly to the imaging plane. Thus, three-dimensional SIM limits visually resolvable densities of the inner and outer plaques to metaphase spindles with both SPBs equally aligned with imaging plane. A large number of spindles (30/68) do not present resolvable outer plaque densities on either SPB. We believe that this is primarily due to the orientation of the SPBs during SIM imaging as both astral and spindle microtubules in budding yeast are nucleated by a complete γ-TuC containing seven molecules of Spc97 (Kollman et al., 2015). In wild-type cells, 90% of metaphase spindles of 0.5–2 µm in length possess at least one astral microtubule, usually on the old SPB. Of these spindles, 80% will simultaneously possess an astral MT on the old and new SPBs for at least 60 s when observed over a 5-min time window (Shulist et al., 2017).
FIGURE 6:
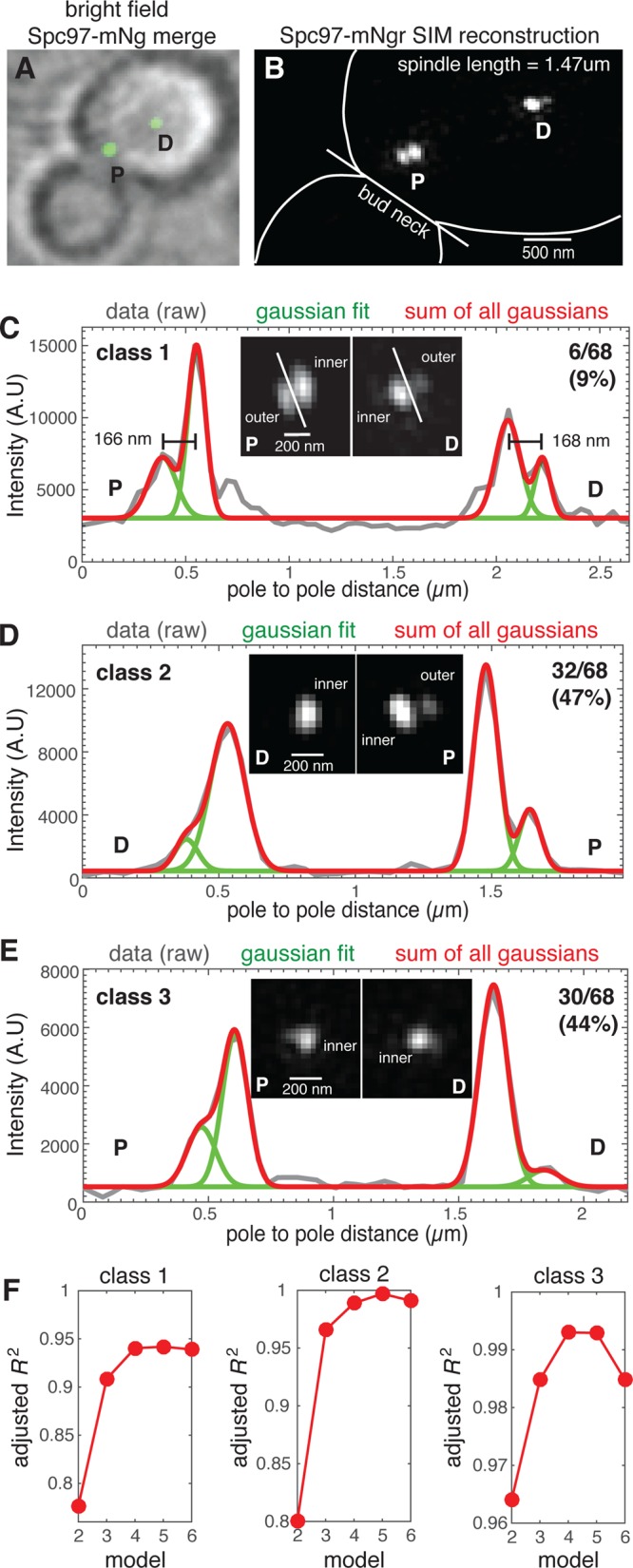
The γ-TuC protein Spc97 is present on the outer plaque of both SPBs of most metaphase spindles. (A) Spc97-mNG fusion protein localizes to both SPBs of metaphase spindles. (B) SIM reconstruction of Spc97-mNgr (spindle length = 1.47 µm) resolves densities representing the inner and outer plaques of SPBs that are proximal and distal to the bud neck. (C–E) The SIM reconstructions were used to classify 68 spindles with regard to visual detection of outer plaque densities; (C) both SPBs (class 1; 9%), (D) one SPB (class 2; 47%), or (E) neither SPB (class 3; 44%). For a representative spindle of each class, the fluorescence intensity (gray line) along the spindle axis of the SPBs shown, overlaid with the cumulative distribution (red) of the Gaussian mixture model for four individual peaks (green), with each peak representing distinct Spc97-mNgr densities. (F) The adjusted R2 for the Gaussian mixture model for peak numbers ranging from 2 to 6 for each class. The model fit improves with increasing peak number with goodness of fit increased at four peaks relative to two or three (densities for inner and outer plaques of both SPBs). This analysis reveals Spc97-mNgr outer plaque densities that are not visually resolved due to the axial resolution limit of SIM as well as a spindle axis that is frequently tilted relative to the focal plane.
When inner and outer plaques are well resolved, they have the characteristic shape of two side-by-side ellipses (Figure 6C, inset). In the cases where the outer plaques could not be resolved, the Spc97-mNgr formed a single density (Figure 6D, inset). This indicates that the SPB is not oriented perpendicularly to the imaging plane and the fluorescence emitted by Spc97-mNG on the inner and outer plaques overlaps. To determine whether Spc97 densities represent a single plaque or two unresolved plaques, we fit the SPB Spc97-mNG density profile to a Gaussian mixture model with a number of peaks ranging from two to six. For each SPB, if two Spc97 densities were unresolved, then the model with two Gaussian peaks would provide the best fit. We report the adjusted R2 as a measure of goodness of fit to the Gaussian mixture model (Figure 6F) and find that the nonelliptical densities of single SPBs are most appropriately described by a two-Gaussian model, even when accounting for the extra parameters in the fit. For both SPBs, the adjusted R2 values of the fit in all three classes of spindles increase significantly from 2 to 4 and then plateau or decrease. This analysis indicates that for spindles where one or both SPBs are a nonelliptical density there are two distinct densities of Spc97-mNG representing the SPB inner and outer plaques. Although these data support the model that Spc97 is present at the outer plaque of both SPBs in most if not all cells, we concluded that the axial resolution of three-dimensional SIM limits our capability of detecting and measuring the amount of Spc97-mNG located on the outer plaque.
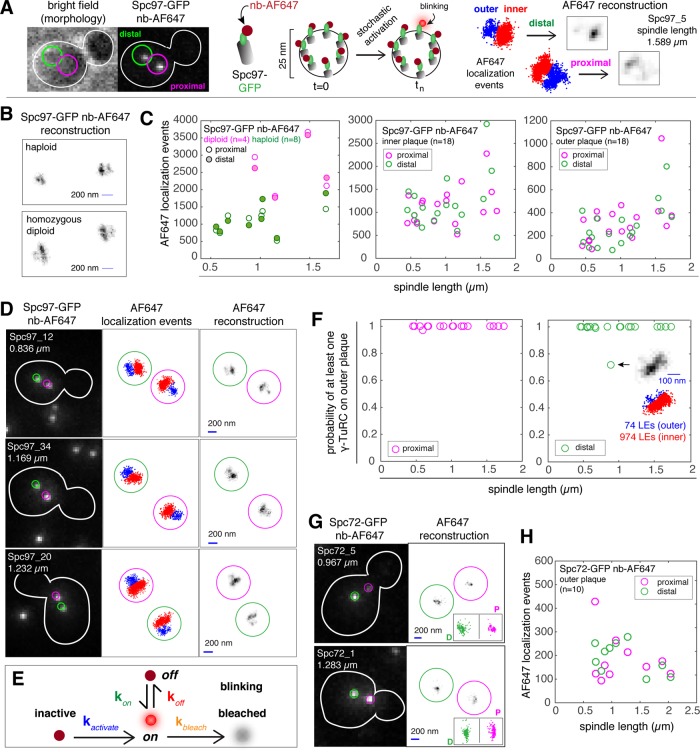
These data support the model that Spc97 is present at the outer plaque of both SPBs in most if not all cells, and this gave us confidence that we could further our analysis by asking whether sufficient γ-TuC components were present within a 25-nm-diameter spatial density to form a γ-TuC, which is required to nucleate an astral microtubule. SIM can be used to quantify intensity differences for spatially resolved objects (Demmerle et al., 2017; Kraus et al., 2017); however, the 25-nm-diameter structure of γ-TuC is below the 100-nm resolution limit of SIM. The γTuC can be resolved using direct stochastic optical reconstruction microscopy (dSTORM) (van de Linde et al., 2011; Ries et al., 2012), another quantitative superresolution technique. We therefore used dSTORM to analyze the distribution of Spc97, Tub4, and Spc72 bound to the inner and outer plaques of metaphase spindles ranging in length from 0.5 to 2 μm in fixed cells. Spc97-GFP, Tub4-GFP, and Spc72-GFP fusion proteins were detected using GFP-Trap nanobodies conjugated to the AlexaFluor 647 (AF647) fluorescent dye (nb-AF647) as previously described (Ries et al., 2012). While the Spc97-GFP and Tub4-GFP fusion proteins were functional based on genetic interaction tests, we found that the C-terminal fusion of GFP to Spc98 displayed functional defects (data not shown) and was not used for this study.
Our wide-field dSTORM approach is described in Figure 7A and in detail under Materials and Methods. Briefly, cell morphology is determined using bright field, and then the focal plane in which the SPBs reside and the distance of each SPB to the bud neck is found using confocal microscopy, using the AF647 channel. We refer to the SPB closest to the bud neck as the proximal SPB, with the other the distal SPB; however, the age of the SPB (new or old) cannot be determined using this method. Using wide-field dSTORM, stochastic activation of single AF647 molecules results in blinking that is computed as a single localization event (LE). Over time, many LEs are computed and these data are used for the reconstruction. The regime detects double blinks, which are rare, and these artifacts are removed from the prereconstruction data set used for AF647 reconstructions. Using dSTORM, we observed two densities for each SPB, representing the inner and the outer plaque when the SPBs were aligned perpendicularly to the imaging plane (Byers and Goetsch, 1975) and consistent with Figure 6, B and C, and previous studies using structured illumination microscopy (Burns et al., 2015) and electron microscopy (EM) (Byers and Goetsch, 1974, 1975; Rout and Kilmartin, 1990; Winey and Bloom, 2012). The two densities were observed irrespectively of which γ-TuC reporter protein (Spc97 or Tub4) was visualized. Of these two plaques, the lower density is almost always attributed to the outer plaque, based on the positions of the two SPBs of a bipolar spindle, and again using structural and spatial information for spindles gained from a multitude of tomography (Winey et al., 1995; O’Toole et al., 1997; Nazarova et al., 2013) and EM studies (Byers and Goetsch, 1975; Rout and Kilmartin, 1990).
FIGURE 7:
Outer plaques of proximal and distal SPBs contain at least one γ-TuC complex. (A) SPB inner and outer plaques are not resolvable by conventional light microscopy with Alexa-647-conjugated nanobodies (GFP-trap; Chromotek) used to label Spc97-GFP. We applied dSTORM, a blinking-based superresolution method that can resolve the inner and outer plaques, with at most one blink per frame. For each blink, a single molecule is localized and this localization event can be represented as a point in space. A two-dimensional distribution of localization events representing the inner (red) and outer (blue) plaques of both SPBs is the starting point to generate a dSTORM reconstructed image by discretizing the space into 24-nm pixels. (B) Representative Spc97-GFP nb-AF647 reconstructions of inner and outer plaques were used to compare the inner plaque dimensions of SPBs in haploid and diploid cells. Bar in reconstruction is 200 nm. (C) Number of localization events for Spc97-GFP-AF647 for the inner plaques of diploids and haploids (left panel) and on the inner and outer plaques of haploids (middle and right panel). (D) Representative images (confocal, distribution of localization events and reconstruction) of SPBs in fixed cells, with Spc97-GFP labeled with nb-AF647. Sample name and spindle length is provided in the confocal image. Bar in reconstruction is 200 nm. (E) A simplified kinetic model of one active (blinking) and six inactive Spc97-GFP-AF647 molecules within a single γ-TuC complex. Inactive and bleached states correspond to the long-lived dark state. We assume that the fluorophore exits and enters the long-lived dark state once and only once during the acquisition time. (F) Probability of having at least one complete γ-TuC on the outer plaques of proximal and distal SPBs. With one exception (shown in inset), all cells in the data set (n = 18) are predicted to have least one γ−TuC on the outer plaque of both the proximal and distal SPBs. (G) Confocal image and AF647 reconstruction for Spc72-GFP detected with nb-AF647. Localization events for proximal and distal SPBs are shown as insets in the reconstruction image. (H) Number of localization events for Spc72-GFP-AF647 on the outer plaque.
To validate the quantitative nature of the dSTORM method, we compared the reconstructions of haploid and homozygous diploid cells expressing Spc97-GFP. The inner plaque of a haploid SPB is ∼80 nm in diameter, and the inner plaque of a diploid SPB is ∼160 nm in diameter, based in EM measurements of longitudinal sections (Winey and Bloom, 2012). Representative reconstructions of a haploid and homozygous diploid cell expressing Spc97-GFP are shown in Figure 7B and Supplemental Figure S4, A and B, used the position of the SPBs in the confocal image as well as the LE counts for the inner plaque reconstructions of the proximal and distal SPBs (Figure 7C, left panel) to identify spindles where both SPBs are aligned with the focal plane. The length of the inner plaque reconstruction in haploid cells is 270 ± 23.9 nm (mean ± SD; n = 8), while the length of the inner plaque reconstruction in diploid cells is 430 ± 43.9 nm (mean ± SD; n = 4). The LE count for the inner plaque reconstructions of SPBs in haploid cells is also markedly lower than the corresponding LE counts in diploid cells (Figure 7C). LEs collected per feature (inner or outer plaque) for 18 haploid spindles are plotted in Figure 7C (middle and right panels). Confocal images, LE distribution plots, and reconstructions for representative Spc97-GFP spindles detected with the nb-AF647 probe are shown in Figure 7C and in Supplemental Figure S4B.
The number of intranuclear (spindle) microtubules per SPB ranges from 16 to 23 in wild-type cells and up to 40 in metaphase arrested cells (Winey et al., 1995; O’Toole et al., 1997). The number of spindle microtubules (20) is relatively stereotyped in comparison to cytoplasmic microtubules (none or one to three per SPB) and is on average ∼10:1. However, the biological variation for both cytoplasmic and spindle microtubule number in wild-type cells suggests a range from 1:5 to 0:20 (Winey et al., 1995; Winey, 1999; Nazarova et al., 2013; Shulist et al., 2017). If we assume the number of γ-TuCs is proportional to the number of microtubules, we estimate that the LE count for inner plaques will be ∼10-fold greater than for the outer plaque. On average, the inner plaque count was 1116 ± 407 (mean ± SD) LEs for the proximal poles and 1146 ± 574 LEs for the distal poles, while the average outer plaque count was 319 ± 204 LEs for the proximal poles and 275 ± 179.1 for the distal poles. The average ratio for inner:outer plaque LE count was 4.2 ± 1.86 for proximal poles and 5.7 ± 3.66 for distal poles; however, the distribution includes a distal SPB with an outer plaque with <100 LEs and an inner plaque with ∼1000 (1:10; spindle 97_21, 0.672 µm in length; Supplemental Figure S4B) as well as a distal SPB with an outer plaque with ∼500 LEs and an inner plaque with ∼400 LEs (spindle 97_8, 1.735 µm in length; Supplemental Figure S4B). For the latter spindle, a comparison of the inner plaque density of the distal SPB with the inner plaque of the proximal SPB suggests the spindle axis is not well aligned to the focal plane. However, for spindles that are well aligned to the focal plane, this analysis demonstrated that for all SPBs, we estimate that the LE count for the inner plaque is ∼5-fold greater than the LE count for the outer plaque.
In parallel, we performed the same analysis using Tub4-GFP labeled with the nb-AF647 probe (Supplemental Figure S5). Relative to Spc97, the overall number of localization events is lower than expected for both the inner and outer plaques. We attribute this to the combination of the high density of AF647 present on Tub4 and the ability of AF647 to quench itself (Berlier et al., 2003). The kinetic parameters determined from one haploid Tub4-GFP, one diploid Spc97-GFP, and three haploid Spc97-GFP data sets are provided in Table 1. Quenching for the Tub4-GFP AF647 is reflected by an increase in the measured kbleach for Tub4-GFP-AF647 (36.1) relative to Spc97-GFP AF647 (11.6). Lower-than-expected counts for Tub4 were obtained across all spindle lengths in our data set (0.5–2 µm). The kbleach of the Spc97 AF647 in homozygous diploid cells (36.5) was also increased relative to the kbleach of the Spc97-GFP AF647 in haploid cells. For this reason, we restricted our analysis to the three Spc97-GFP AF647 haploid data sets.
TABLE 1:
Kinetic model parameters for Spc97-GFP and Tub4-GFP dSTORM calculations.
| Data set | koff | kon | kbleached | kactivate |
|---|---|---|---|---|
| Tub4_haploid | 52.2730 | 9.3385 | 36.0948 | 0.0162 |
| 97_diploid1 | 58.8032 | 7.8385 | 36.4578 | 0.0059 |
| 97_haploid 1 | 45.8958 | 12.1792 | 5.4086 | 0.0056 |
| 97_haploid 2 | 46.2536 | 15.1589 | 9.1154 | 0.0025 |
| 97_haploid 3 | 47.9528 | 13.8517 | 20.4038 | 0.0116 |
| mean 97_haploid | 46.7 ± 1.09 | 13.7 ± 1.49 | 11.4 ± 7.81 | 0.007 ± 0.0046 |
Next, we asked whether the numbers of LEs between the distal and proximal SPBs are statistically different (Table 2). We applied a linear regression between the number of localizations and spindle length, with a categorical variable (inner or outer) for Spc97 and Tub4. The numbers of LEs between the distal and proximal outer plaques are not statistically significant (p = 0.94) based on a significance threshold ≤0.01. This was also the case for the inner plaques of the distal and proximal SPBs (p = 0.64). Altogether, our data suggest that the distribution of γ-TuCs associated with the outer plaques of the SPBs in metaphase cells cannot be distinguished based on their location relative to the bud neck (proximal or distal). Furthermore, our comparison of both SPBs for each spindle, one of which must be old and the other new, implies that the distribution of γ-TuC on outer plaques cannot be distinguished based on age.
TABLE 2:
The Tub4 and Spc97 content of the outer and inner plaques are indistinguishable between proximal and distal SPBs.
| Protein | Location | p value |
|---|---|---|
| Spc97 | Outer plaque | 0.94 |
| Spc97 | Inner plaque | 0.65 |
| Tub4 | Outer plaque | 0.76 |
| Tub4 | Inner plaque | 0.56 |
| Spc72 | Outer plaque | 0.49 |
The probability values for the linear regression between the number of localization events per feature, spindle length, and location (proximal or distal). A linear model was applied to the number of AlexaFluor 647 localization events per feature and spindle length with a categorical variable for either inner or outer plaques. The p values were obtained by comparing it to the null model without the categorical variable. p < 0.01 was used as significance threshold.
Next, we computed the probability that an outer plaque contains sufficient Spc97-GFP within a 25-nm radius to form one complete γ-TuC (Kollman et al., 2015). To estimate the probability of a single γ-TuC on the outer plaques, we generated a semiquantitative coarse-grained model of the expected number of localization events for the Spc97-GFP used to detect the γ-TuC. We assume that each γ-TuC forms a 25-nm ring structure (Kollman et al., 2010, 2015) and is complete (e.g., 7 Spc97-GFP and 14 Tub4), as partial assemblies are expected to be unstable relative to complete γ-TuCs (Marschall et al., 1996; Peng et al., 2015). We assume that labeling is 100% efficient, and, for this reason, our model will underestimate the number of γ-TuCs and thus provide a lower bound. The kinetic parameters for the three haploid Spc97-GFP data sets used for this analysis are provided in Table 1.
We first simulated the photophysical traces of 1000 blinking γ-TuCs, where blinking is the product of a single activated AF647 molecule attached to a single Spc97-GFP (Figure 7C; also see Materials and Methods and Supplemental Methods). On the basis of the dSTORM reconstruction of our simulated data, we expected 48 ± 18 (mean ± SD) Spc97-GFP AF647 localization events per complete γ-TuC. From this model, we calculated the probability that our observed LEs corresponded to at least 1 γ-TuC (Figure 7D). Both the proximal and distal SPBs have at least one γ-TuC, regardless of spindle length (Figure 7, D–F).
Spc72 acts as an outer plaque specific receptor for the γ-TuC (Knop and Schiebel, 1998). Our dSTORM analysis of Spc97 and Tub4 suggests that outer plaques of both SPBs contain at least one γ-TuC, and thus we hypothesized that this would also be the case for Spc72. Reconstructions of the SPBs of 10 cells showed a single density for each pole, as expected since Spc72 is specific to the outer plaque. On average, the LE count was 176.8 ± 97.69 (mean ± SD) for the proximal SPBs and 179.6 ± 70.52 for the distal SPB (Figure 7, G and H), with no significant difference found between the two (p = 0.49, significance threshold ≤0.01; Table 2). This result verified that the distal SPB accumulates as much Spc72 as the proximal SPB for spindles between 0.6 and 2 µm in length.
The SPB close to the bud neck carries functional astral microtubules independently of its age
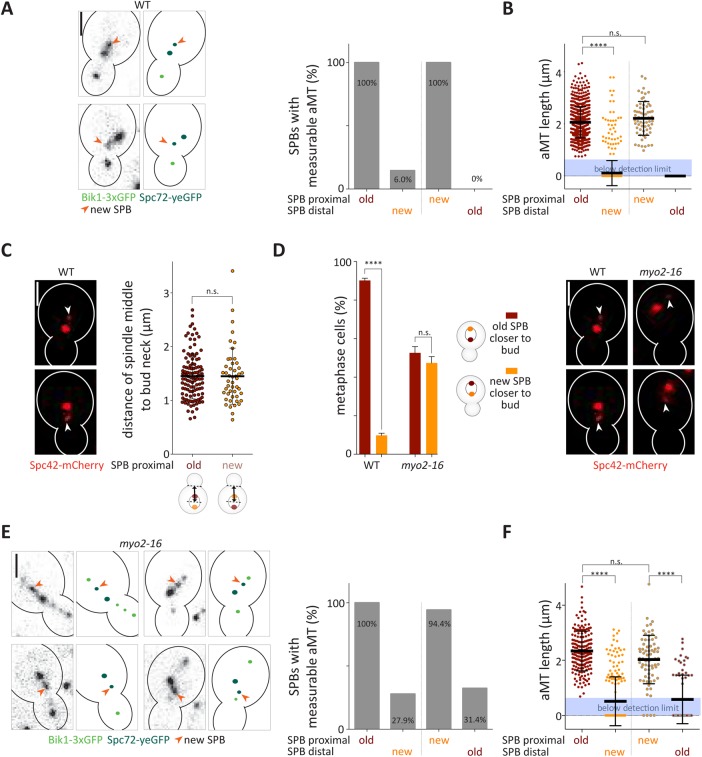
Our study shows that the outer plaques of both SPBs carry similar levels of γ-TuCs after their separation, enabling both to nucleate astral microtubules. Correctly positioned spindles organize long astral microtubules at the proximal SPB and fewer and shorter ones at the distal one (Shaw et al., 1997; Segal and Bloom, 2001). This suggests that the γ-TuCs on the distally located (new) SPB are either not yet functional or that their activity is down-regulated. To distinguish between these possibilities, we asked whether the differences observed in astral microtubule organization between the SPBs depend on the age or on the location of these SPBs. Therefore, we tested whether inverted spindles, that is, spindles that oriented the new SPB toward the bud instead of the preexisting one, still failed organizing astral microtubules from the new SPB. We measured the occupancy and length of astral microtubules on each side of the spindle by time-lapse microscopy, using Bik1-3xGFP (Lin et al., 2001) as microtubule plus-end marker (Carvalho et al., 2004) and Spc72-yeGFP to visualize SPBs. We assigned the SPB age based on their fluorescence intensities, the preexisting SPB being brighter than the new one, as shown above. We detected a measurable astral microtubule (length estimated >0.7 μm) on virtually all SPBs localized proximally to the bud neck, irrespective of whether it was the preexisting (92.1% of the cells) or the new (7.9% of cells) SPB (Figure 8A). In either case, very few spindles showed a measurable astral microtubule emanating from the distal SPB, irrespective of its age. Similarly, the average length of the astral microtubules emanating from the distal SPB was significantly shorter compared with the one emanating from the proximal SPB. Importantly, the astral microtubules emanating from the proximal SPB showed statistically undistinguishable length distributions whether this SPB was the newly assembled or the preexisting one (Figure 8B). To determine the functionality of the astral microtubule emerging from a new SPB, we asked whether cells orienting the new SPB toward the bud positioned their spindle less correctly. However, the orientation of the spindle had no impact on their position, measured as the distance between the middle of the spindle and the bud neck (Figure 8C). Thus, when oriented toward the bud, both new and preexisting SPBs are similarly competent to organize functional astral microtubules.
FIGURE 8:
The SPB close to the bud neck carries functional astral microtubules independently of its age. (A, E) Representative images and schematic of preexisting and new SPB localized proximally to the bud neck with Spc72-yeGFP as SPB age marker and Bik1-3xGFP as microtubule plus-end marker. Owing to the resolution limit of the microscope, an astral microtubule (aMT) was defined as measurable when its length was above 0.7 µm. Quantification of measurable astral microtubules from preexisting or new SPB of correctly or misoriented spindles (%) in wild-type (A) or myo2-16 mutant (E) cells (n = 500 measured cells from three independent experiments, mean). (B, F) Average length (μm) of astral microtubules (aMT) organized from preexisting or new SPB of correctly or misoriented spindles in wild-type (B) or myo2-16 mutant (F) cells. If the astral microtubule signal was below the detection limit (displayed as blue box), then the point was counted as 0 (n = 500 cells measured cells from three independent experiments, mean ± SD). Statistical significance was calculated using a two-tailed Student’s t test on a measurable data set. (C) Quantification of distance (μm) between the middle of the mitotic spindle and the bud neck in wild-type cells. Spc42-mCherry was used as SPB age marker (n = 200 measured cells from three independent experiments, mean ± SD). Statistical significance was calculated using Student’s t test. (D) Quantification of spindles with preexisting or new SPB closer to the bud neck in metaphase wild-type and myo2-16 mutant cells (%) (n = 3 independent experiments with a total of >120 cells per genotype analyzed, mean ± SD). Spc42-mCherry was used as SPB age marker. Statistical significance was calculated using Student’s t test. ****p < 0.0001, n.s. = nonsignificant. Scale bars, 2 μm. The new (dim) SPB is pointed out with an arrow. Wild-type or myo2-16 cells were shifted to 30°C for 2 h before imaging and were imaged at 30°C.
The ∼8% of spindles that misoriented in wild-type cells might carry an inappropriately mature new SPB. To investigate this possibility, we asked whether artificially misoriented spindles displayed the same behavior. We took advantage of the myo2-16 mutation, which prevents the interaction of Kar9 with actin and hence randomized spindle orientation (Schott et al., 1999; Yin et al., 2000). We observed that spindle orientation was already fully randomized at 30°C in myo2-16 mutant cells compared with wild-type (WT) cells (preexisting SPB closer to bud: WT 90.2 ± 1.2 %, myo2-16 52.6 ± 3.2%; Figure 8D). This indicated that interaction of the Myo2-16 protein with Kar9 is already functionally impaired at 30°C, while the cells are still viable (Supplemental Figure S6).
Strikingly, in myo2-16 mutant cells all correctly oriented and 94.4% of misoriented spindles showed measurable astral microtubules on the SPB closest to the bud neck and much fewer on the distal SPB (Figure 7E). In the myo2-16 mutant cells, the occupancy of astral microtubules at the distal SPB was higher compared with wild-type cells, but this effect was independent of whether the distal SPB was the preexisting or the new one. Furthermore, the average length of the proximal and distal astral microtubules, respectively, was not significantly different between myo2-16 mutant and wild-type cells, irrespective of whether the new or preexisting SPB was the proximal SPB (Figure 7F). Of note, Spc72 localization in metaphase was not affected in myo2-16 mutant cells (Figure 1). Altogether, these data indicate that in metaphase cells, the new SPB is as competent as the preexisting one in forming astral microtubules and the capacity of the SPBs to organize astral microtubules depends on their position relative to the bud neck rather than on their age.
Kar9 localization depends on the age of the SPB and not the orientation of the spindle
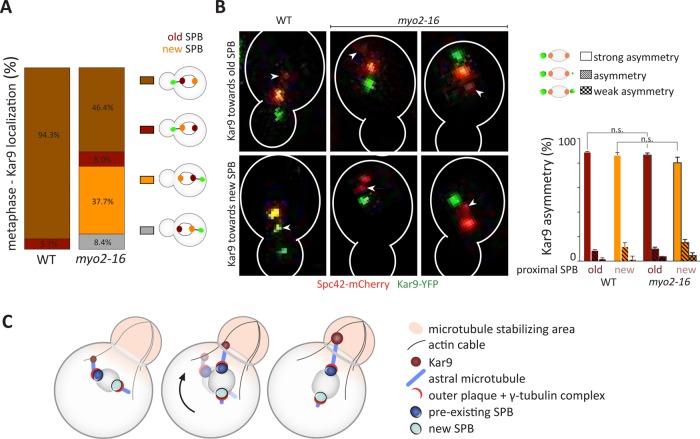
Our study establishes that the occupancy and length of astral microtubules on a given SPB correlates with its intracellular position rather than its age. Yet, in wild-type cells, the spindle is oriented specifically with the preexisting SPB toward the bud. This orientation depends on Kar9 function (Pereira et al., 2001; Hotz et al., 2012). Accordingly, in wild-type cells Kar9 localization is biased toward the preexisting SPB of metaphase spindles (Hotz et al., 2012; Lengefeld et al., 2017). Therefore, the question here is whether Kar9 accumulates on the microtubules emanating from the old SPB because they are long, and, hence, this spindle pole orients toward the bud, or whether it accumulates to these microtubules independently of their length and subsequently promotes their elongation as they become oriented toward the bud. To investigate this question, we asked whether in myo2-16 mutant cells grown at 30°C, Kar9 localizes preferentially toward the long astral microtubules emanating from the proximal SPB, independently of its age, or was preferentially associated with the preexisting SPB, independently of its localization, and, hence, of microtubule length. Therefore, we characterized the localization of Kar9-YFP relative to the preexisting and new SPB in cells with correctly or misoriented spindles in wild-type and in myo2-16 mutant cells. The brightness of Spc42-mCherry reported on the age of the SPBs. In all wild-type cells, Kar9 localized to the proximal aster, which emanated in 94.3% of the cells from the preexisting SPB (Figure 9A). Similarly, in the myo2-16 mutant cells, Kar9 localization correlated strongly with the age of the SPB (84.1% of the cells carried Kar9 on the aster of the preexisting SPB), irrespective of the position of the SPB relative to the bud neck. In 37.7% of cells, Kar9 localized toward a preexisting SPB that was distally positioned from the bud neck. Thus, Kar9 behavior is distinct from what we observed for astral microtubule occupancy and length. These data indicate that Kar9 localization is determined by the age of the SPBs and only to a lesser extent by the orientation of the spindle relative to the plane of the bud neck.
FIGURE 9:
Kar9 localization depends on the age of the SPB and not the orientation of the spindle. (A) Classification of wild-type and myo2-16 mutant metaphase cells with Kar9-YFP (class i) localized toward the proximal preexisting SPB, (class ii) localized toward the proximal new SPB, (class iii) localized toward the distal preexisting SPB, or (class iv) localized toward the distal new SPB (%). Spc42-mCherry was used as an SPB age marker (n = 3 independent experiments with a total of >100 cells per genotype analyzed, mean). (B) Representative images of wild-type and myo2-16 mutant cells and categorization of Kar9 asymmetry of spindles that carry Kar9 predominantly on the preexisting or new SPB: “strong asymmetry,” Kar9 only toward one SPB; “asymmetry,” Kar9 on both SPB but asymmetric; “weak asymmetry,” slightly more Kar9 on one SPB than the other (n = 3 independent experiments with a total of >100 cells per genotype analysed, mean ± SD). Statistical significance was calculated using a two-tailed Student’s t test. n.s. = nonsignificant. (C) Schematic drawing of Kar9-dependent spindle orientation and position-dependent asymmetry of the astral microtubules. Scale bars, 2 μm.
Next, we asked whether the orientation of the spindle influences the establishment of faithful Kar9 asymmetry. Our analysis revealed that in 87.9 ± 1.9 % of wild-type cells and in 83.4 ± 6.4 % of myo2-16 mutant cells Kar9 asymmetry was established faithfully regardless of whether the preexisting or new SPB was localized proximally to the bud neck (Figure 9B). This establishes that Kar9 asymmetry is established largely independently of the orientation of the spindle. Furthermore, these data indicate that Kar9 directs the morphological asymmetry of the asters toward the preexisting SPB by positioning this SPB close to the bud neck and not vice versa.
DISCUSSION
Our data establish that preexisting and new SPBs have a similar capacity to form functional astral microtubules early in mitosis. Furthermore, our findings indicate that SPB specification, that is, the process that distinguishes and orients the differently aged SPBs, is the result of regulation, consistent with recent studies (Hotz et al., 2012; Lengefeld et al., 2017), rather than SPB maturation. On the basis of our data, we propose the following model for how yeast cells associate a different number of microtubules, which are also of different length, to each SPB (Figure 9C).
First, our SIM, dSTORM and live-cell imaging data establish that in most cells, both SPBs have at least one γ-TuC. Cells with very short spindles (∼0.6 μm) already carry equivalent amounts of the outer plaque receptor Spc72 and one or more γ-TuCs on both SPBs. Either SPB is capable of forming a detectable astral microtubule. Regulation of microtubule dynamics and potentially also the γ-TuC, rather than the age of the SPB, intrinsically determines that the two SPBs will form astral microtubules that differ in both number and length. A recent study suggests that the number of astral microtubules associated with the preexisting and new SPBs is in part controlled by the phosphorylation of an invariant tyrosine residue of γ-tubulin (Y362) that is phosphorylated in vivo (Keck et al., 2011; Shulist et al., 2017).
Second, only one microtubule array becomes stably decorated with Kar9, generally that from the preexisting SPB. Kar9 localization is controlled by phosphorylation and sumoylation events, which affect Kar9 recruitment, maintenance, or removal from astral microtubules (Liakopoulos et al., 2003; Maekawa et al., 2003; Leisner et al., 2008; Hotz et al., 2012). Importantly, such studies suggested that Kar9 is capable of binding microtubules from either SPB in mutant cells (Liakopoulos et al., 2003; Cepeda-Garcia et al., 2010). Together, these data are consistent with Kar9 localization to only one microtubule array not reflecting incomplete maturation of the other SPB but rather differences in regulatory activities between the two SPBs. The MEN pathway, which is activated on SPBs (Valerio-Santiago and Monje-Casas, 2011; Rock et al., 2013) and the SPIN contribute to control Kar9 distribution (Hotz et al., 2012; Lengefeld et al., 2017).
Third, the association of Kar9 with the microtubules from one SPB ensures their interaction with the actin cytoskeleton and positioning of the connected SPB toward the bud (Theesfeld et al., 1999; Beach et al., 2000; Yin et al., 2000; Hwang et al., 2003; Liakopoulos et al., 2003). This orientation of the spindle subsequently subjects the microtubules of each pole to different spatial environments and regulation. Our data indicate that the SPB proximal to the bud neck is now positioned in a microtubule stabilizing area, making it more active as an MTOC, independently of its age. The nature of this regulation is unclear. It could involve interaction of the microtubules with cortical cues and regulatory proteins at these locations, such as kinases at the bud neck (Barral et al., 1999; Kusch et al., 2002; Grava et al., 2006), differences in cytoplasmic pH between mother and bud (Henderson et al., 2014), or a combination of such effects. This regulation ensures that astral microtubules projecting into the bud can establish stable contacts with the bud cortex during metaphase. This, in turn, promotes the dynein-dependent pulling of the SPB into the bud on anaphase onset (Adames and Cooper, 2000; Markus and Lee, 2011; Tang et al., 2012). Since Kar9 asymmetry is established prior to spindle positioning and formation of long microtubules on the bud side, this asymmetry does not originally depend on an asymmetry in microtubule length. Similarly, since the spindles of the myo2-16 mutant cells are still asymmetric, irrespective of the position of Kar9 and the age of the SPB, Kar9 asymmetry is not by itself what drives the asymmetry of microtubule length. However, the coordination of these two effects, the spatial environment on microtubule length and the function of Kar9 in pulling the preexisting SPB toward the bud, ensures that microtubule length asymmetry is coordinated with SPB age. Subsequently, the long microtubules oriented toward the bud are also predicted to recruit more Kar9 (Cepeda-Garcia et al., 2010). This subsequent process further enhances the asymmetry of the spindle (Cepeda-Garcia et al., 2010).
Regulation driving Kar9 toward the preexisting SPB
In the early stages of spindle assembly, a fraction of wild-type cells localize Kar9 toward both SPBs before polarizing its distribution toward the preexisting one (Cepeda-Garcia et al., 2010). This observation is in line with our finding that both SPBs are capable of organizing functional astral microtubules carrying Kar9 after their separation. The alignment of the mitotic spindle along the mother–bud axis is promoted by the asymmetric recruitment of Kar9 to the astral microtubules from only one SPB and its movement toward the bud neck (Liakopoulos et al., 2003; Maekawa et al., 2003). We show that in the myo2-16 mutant cells, both SPBs show an equal probability of being proximal to the bud and are equally competent to nucleate astral microtubules. Yet Kar9 still preferentially localizes toward the preexisting SPB. Thus, these data establish that SPBs control their own specification, that is, the process that distinguishes and orients the differently aged SPBs emanates from the SPBs themselves to regulate Kar9 localization. Indeed, previous studies suggest that Cdk1/Clb4 and MEN-dependent phosphorylation of Kar9 might be biased toward one SPB only (Liakopoulos et al., 2003; Hotz et al., 2012). In turn, this leads to the preferential orientation of the preexisting SPB toward the bud and the stabilization of the microtubules emanating from it.
Strain background differences might provide insights into regulatory mechanisms
We observed differences of Spc72 asymmetry between the yeast strain backgrounds S288C and 15DaubA. It will be important to understand in the 15DaubA background what prevents Spc72 of being symmetric at the SPBs during their duplication when its indirect adaptor, Cnm67, is symmetrically distributed (Juanes et al., 2013). Interestingly, SPC72 is not essential in the W303 strain background (Soues and Adams, 1998; Hoepfner et al., 2002), while it is essential in the S288C strain background (Chen et al., 1998; Knop and Schiebel, 1998). Thus, Spc72 might be differentially regulated in these strain backgrounds. Furthermore, Spc110 tagged with RFP (t1/2 = 42 min [ Bevis and Glick, 2002]) was observed to be asymmetric in the W303 background (Yoder et al., 2003). In contrast, in S288C derived strains, Spc110 tagged with mCherry (t1/2 = 40 min [ Khmelinskii et al., 2012]) appears symmetric on SPB separation. To what extent the differences in the strain backgrounds affect the regulation and functionality of the SPBs is yet to be determined. Nevertheless, these differences between strain backgrounds might be the source of important insights into how maturation, biased SPB inheritance, and spindle orientation are regulated. In any case, our data establish that at least S288C cells do not rely on Spc72 distribution being asymmetric to properly orient their spindle, suggesting that SPB maturation is not fundamentally relevant.
Do the dynamics of SPB proteins contribute to SPB specification?
The duplication of the SPB is conservative with the exception of Spc110 (central/inner plaque) (Yoder et al., 2003) and, as revealed here, Cnm67 (outer plaque). The maturation of the new SPB involves the incorporation of preexisting Cnm67 as well as newly synthesized one. The preexisting protein is recruited either from the preexisting SPB or a potential preexisting pool of Cnm67 in the cytoplasm. The nearly homogeneous distribution of Cnm67 during SPB duplication is surprising since Cnm67 is thought to be the anchor of Nud1, which in turn anchors Spc72 to the outer plaque of the SPB (Pereira et al., 1999; Gruneberg et al., 2000). In contrast, Nud1 (Lengefeld et al., 2017) and Spc72 itself exchange little between SPBs and, hence, with the cytoplasm. Preexisting Spc72 may be anchored to the half-bridge (Pereira et al., 1999) and specifically relocalizes toward the preexisting SPB once Cnm67 is built into both outer plaques. This model predicts that Spc72 relocalization is coordinated with the age of the SPB independently of the age of Cnm67. Alternatively, Spc72 interaction with the SPB might be only partly dependent on Cnm67. Interestingly, during SPB duplication, Cnm67 is detectable in only a fraction of cells that already carry Nud1 at the satellite structure (Burns et al., 2015), suggesting that Nud1 localization to the SPB is partially Cnm67 independent. Finally, Cnm67 might be in excess on the outer plaques relative to Spc72. On Spc72 binding to the outer plaque, the bound fraction of Cnm67 might stop exchanging, while the remainder of the protein still does.
The observation that some components are rather immobile at SPBs and others exchange during SPB duplication suggests that the stable components could play a role in carrying information for SPB specification. Indeed, the fusion of the γ-TuC-binding domain of Spc72 to Cnm67 (Spc72[γTuC]-Cnm67) causes abnormal astral microtubule organization and defects in Kar9 asymmetry (Juanes et al., 2013). Our data suggest that this fusion protein might randomize the distribution of preexisting and new Spc72 (γTuC) between the two SPBs. Therefore, we speculate that the spindle orientation defects caused by the Spc72 (γTuC)-Cnm67 fusion protein may have two possible causes. First, a redistribution of the preexisting and new Spc72 protein between the two SPBs might redistribute information (e.g., posttranslational modifications) carried by the protein. If this were the case, this would indicate that Spc72 is part of the regulatory network that controls SPB specification and Kar9 distribution. Nevertheless, whether Spc72 carries SPIN-dependent modifications in the γ-TuC-binding domain is unknown. Alternatively, the defect observed in the SPC72 (γTC)-CNM67 mutant cells may be caused by the loss of the rest of the Spc72 protein on fusing the γ-TuC-binding domain to Cnm67. Indeed, the lost part of the protein carries many phosphorylation sites (Keck et al., 2011), is a target of the SPIN acetyltransferase NuA4/Tip60, and contributes to the control of SPB specification (Lengefeld et al., 2017). Thus, the role of Spc72 regulation in SPB specification remains a task for future research.
MATERIALS AND METHODS
Strains and plasmids
Strains are isogenic to S288C or as indicated to 15DaubA. Yeast strains used in this study are listed in the Supplemental Material. Fluorescent proteins were tagged at endogenous loci (Knop et al., 1999) and verified by PCR. gdp:Cre-EBD78 was inserted at the LEU2 locus (Hotz et al., 2012). The loxP-mCherry-KanMX4-loxP-GFP cassette was integrated into a yeGFP-HisMX6 tagged GOI (Hotz et al., 2012). sfGFP:KanMX4 or 3sfGFP:KanMX4 was integrated in a yeGFP-HisMX6 tagged GOI.
Media, growth conditions, time course, and induction experiments
Cells were cultured in yeast extract peptone (YEPD: 2% glucose) or SC (synthetic medium: 2% glucose, lacking uracil, supplemented with extra 0.04 mg/ml adenine) at indicated temperature.
For microscopy, cells from a saturated culture were diluted in SC medium to O.D. 0.2 and cultivated at 25°C for 4 h. For 100-s time-lapse microscopy, a concentrated aliquot of the cells was used for acquiring microscopy fields of fluorescence images. For time-lapse microscopy, a concentrated aliquot of the cells was placed on an SC-medium agar patch for microscopy imaging at 30°C or the microfluidics system was used. Microfluidics experiments were carried out with the ONIX microfluidic perfusion platform with Y04C-02 microfluidic plates (CellAsic). SC medium was used as a medium with output pressure of 2 psi. Results of the same experiment obtained by agar-patch or microfluidics microscopy were not different.
For the exchange of the mCHERRY tag of RITE strains, cells from a saturated culture were diluted in SC medium to O.D. 0.2, estradiol was added in a final concentration of 1 µM (Sigma-Aldrich), and the cells were cultivated at 25°C for 4 h. For time-lapse microscopy imaging, a concentrated aliquot of the cells was placed on an SC-medium agar patch at 30°C. Only cells that displayed two mCherry-labeled SPB in the previous anaphase and showed GFP-signal after the entry in the cell cycle were analyzed.
Temperature-sensitive (ts) mutant strains (tem1-3, cdc15-1, myo2-16) were grown in SC medium at 25°C. For microscopy, ts-mutant strains were diluted in SC medium to O.D. 0.2, cultivated at 25°C for 3 h, and shifted for 50 min to 37°C (Figure 1) (Hotz et al., 2012) or 2 h to 30°C (Figures 8 and 9), and a concentrated aliquot was used for acquiring microscopy fields of fluorescence images. The MEN promotes Kar9 asymmetry directly in metaphase and not through its action in the previous anaphase (Hotz et al., 2012). As tem1-3 and cdc15-1 mutants arrest in anaphase at 37°C, we considered only cells that were in metaphase before they went into anaphase arrest.
Fluorescence microscopy and data analysis
Time-lapse microscopy (100 s; 10 Z stacks with step size 0.4 μm) was performed using an Olympus BX51 microscope equipped with an Andor’s iXon 885 EM-CCD camera and TILLVision software (TILLPhotonics). Time-lapse microscopy (2 min; time interval 20 s) and time-lapse microscopy (time interval 7–30 min) (10 Z stacks with step size 0.5 μm) was performed on a Personal Deltavision microscope (Applied Precision) equipped with a CCD HQ2 camera (Roper), 250-W Xenon lamps, with Softworx software (Applied Precision). For time-lapse microscopy, we analyzed cells that displayed a normal duration of cell-cycle progression during the microscopy imaging in SC media (137 ± 37 min, n > 50 [ Shaw et al., 1998]). For Figure 2, the SPB in the first measurement after their separation could be distinguished based on their fluorescence intensity (for Spc72, Spc42, and Nud1), and the same one was followed over time even if the spindle inverted. Images were analyzed with ImageJ. Maximum intensity projections of Z stacks were used. For fluorescence intensity, a region of interest (ROI) was drawn around the area of interest, and the integrated density was quantified. An identically sized ROI was put next to the area of interest to determine the background signal. Background intensity was subtracted from the area of interest intensity to yield the fluorescence intensity (AU), which was used to calculate the asymmetry index. Spindle length was measured by quantifying the distance between the two SPBs. Scale bars are 2 μm.
Photobleaching experiment
Photobleaching experiments were performed as in Boettcher et al. (2012): cells were grown to saturation at 30°C on agar plates of SD medium and thinly restreaked on an SC medium agar pad 4 h prior to a photobleaching experiment. A Zeiss LSM 510 confocal microscope with a 488-nm argon laser (maximum output: 25 mW) (Zeiss Microimaging) controlled by the ZEN 2010 software (Carl Zeiss) was used for imaging. Photobleaching was carried out at full intensity for 30 iterations on G1 phase cells. Afterward, the signal was detected by continuous acquisition (10 Z stacks with step size 0.4 μm) every 5 min for 10 min. The bleached signal (AU) during G1 was normalized to 0%. After SPB separation, the bleached traces were followed for 15 min every 5 min, as in this time span the spindle length ranges from 1 to 2 μm (metaphase). Bleaching was minimal as Spc72-3xsfGFP and Cnm67-3xsfGFP foci of unbleached cells were still visible. Images were analyzed using ImageJ.
SIM imaging
Haploid Spc97-mNeongreen strains were used for SIM. Strains were grown in minimal media (SC) to mid-log phase at 25°C. Cells were then fixed in 4% paraformaldehyde in 100 mM sucrose and washed twice with phosphate-bufferend saline (PBS) (pH 7.4) as in Burns et al. (2015). Cells were mounted on glass slide with a 1.5 coverslip (22 × 22 and 0.16–0.18 mm thick; Ted Pella). SIM images were acquired with an Applied Precision Deltavison OMX Blaze V4 on a EM-CCD (Photometrics Evolve) and a 100 × 1.4 oil immersion objective (Olympus). A 541/22 band-pass emission filter was used with a 488-nm (150-mW) laser line. SIM reconstruction was performed with the Applied Precision software Softworx with a Wiener filter of 0.001. Ten to 25 slices of 0.125 nm were acquired.
Image analysis of SIM reconstructed images was performed in FIJI. Spindles with a pole–pole length (measured in three dimensions) <2 µm were used for analysis. Raw SIM images were sum projected and line scans (3-pixel width) along the spindle axis were used for computing the intensity of Spc97-mNG densities. SPBs with two resolvable foci were identified as corresponding to inner and outer plaques based on orientation and intensity.
dSTORM
Sample preparation.
Yeast strains were prepared for dSTORM (van de Linde et al., 2011; Ries et al., 2012) superresolution as described previously (Ries et al., 2012). Briefly, cells were grown in YPED at 25°C. Cells were washed and loaded ont a Concanavalin A (Sigma-Aldrich)-coated 35-mm glass-bottom petri dish. Cells were fixed with 4% paraformaldehyde, 2% sucrose, and then permeabilized with 0.25% Triton X-100, 5% bovine serum albumin (BSA) and 0.004% NaN3 in PBS (pH 7.4) and stained with GFP-Trap nanobodies (ChromoTek) conjugated to AlexaFluor 647. The GFP-Trap:Alexafluor 647 probes were used to detect Spc97-GFP and Tub4-GFP fusion proteins (Huh et al., 2003).
Imaging system and method.
Imaging was performed using a Diskovery spinning disk confocal with a secondary wide field dSTORM mode (Quorum Technologies), equipped with a 63×-1.47NA Plan Apochromat oil-immersion Leica objective, 140-mW (638-nm) and 200-mW (405-nm) lasers, and a Hamamatsu ImagEM X2 EM-CCD camera. Metamorph software (Molecular Devices) was used for data acquisition. dSTORM imaging buffer consisted of 10% glucose, 1% 2-mercaptoethanol, and 2% glucose oxidase buffer (7% [wt/vol; Sigma-Aldrich], 0.4% catalase) and was applied to the sample immediately prior to imaging. Ten thousand frames with 20-ms exposure were acquired per field of view data set (one data set per cell). Spindle poles were detected as diffraction-limited pointlike objects in confocal mode using AlexaFluor 647. Spindle lengths, based on pole-to-pole distance in three dimensions, were measured from a confocal image Z stack as previously described (Nazarova et al., 2013).
Reconstructions.
Reconstruction was performed in two phases. In the first phase, the initial spots were found using a Gaussian filter with a kernel of size 2 × 3σ s + 1 pixels and parameter σ, corresponding to the measured point spread function (PSF) of the microscope (see the Supplemental Methods). Pixels with a signal 2 SD above average were considered as an initial estimate for the centroid of the Gaussian. If multiple connected pixels were identified, then the brightest was used as the centroid. In the second phase, the pixels around the centroid are analyzed with a fast Nvidia CUDA (Lindholm et al., 2008; Nickolls et al., 2008) implementation that fits localization events to a maximum likelihood function, with expected photon counts spatially derived from a Gaussian PSF model as described in Smith et al. (2010). This algorithm makes the fewest assumptions about the kinetics of the data and requires only the measured PSF as a user input. We implemented this method as a custom plugin (available on request) for the Objects Research Systems Dragonfly platform (ORS). Localization events were rejected if the Cramer–Rao lower bound, which reports the fitting accuracy for a maximum likelihood, was greater than 25 nm (Thompson et al., 2002). We then applied a density-based clustering method to discriminate localization events belonging to remove single localization events that are likely the result of unbound nanobodies or the free pool of proteins not bound to the SPB. Final reconstructed images are generated by discretizing the space into 24-nm boxes and counting the localizations that occurred in that box.
γ-TuC simulations.
To determine the presence of a complete γ-TuC, we modeled the photophysical states (see the Supplemental Methods) for each fluorophore expected on a γ-TuC based on structures described in Kollman et al. (2015). CryoEM structures of SPBs purified from yeast cells contain γ-TuCs (Kollman et al., 2015) with a structure similar to that of reconstituted budding yeast γ-TuC assembled in and purified from insect cells (Kollman et al., 2010). We assume, therefore, that γ-TuCs present at SPBs in fixed yeast cells are in the form of a ring structure. We divided the traces into camera frames according to the exposure time and simulated the images of emitting molecules using a pixel-integrated two-dimensional Gaussian model. The presence of a full γ-TuC in our experimental data was estimated from the probability of having at least one full γ-TuC according to the number of localization events in our simulated γ-TuCs. Given the short imaging regime, we assume that once the fluorophore enters the long-lived dark state, it does not convert back to the off state during our imaging regime and is akin to being photobleached.
Microtubule measurements
For determination of microtubule length, time-lapse movies were acquired using a back-illuminated EM-CCD camera (Evolve 512; Photometrics) mounted on a spinning disk microscope with a motorized piezo stage (ASI MS-2000) and 100 × 1.46 NA alpha Plan Apochromat oil immersion objective, driven by ZEN software (Carl Zeiss). Seventeen Z-section images separated by 0.24-µm increments were captured every 1.07 s for 85.6 s. To determine the length of astral microtubules, three-dimensional coordinates of microtubule plus-ends and the corresponding SPB were extracted with the Low Light Tracking Tool (Krull et al., 2014), and the distance between plus-end and SPB represents the length of microtubules.
Statistics
Each experiment was repeated with three independent clones or as indicated. Standard deviation (SD) of mean of three independent clones is shown in graphs or as indicated; n.s. (not significant) or stars indicate p values obtained from Student’s t test or one-way analysis of variance (ANOVA) to test significance. No statistical method was used to predetermine sample size.
Supplementary Material
Acknowledgments
We thank the Vogel and Barral lab members for discussions and comments on the manuscript, Vincent Pelletier and John Arbuckle of Quorum Technologies (Guelph, Canada) for access to the prototype and the optimized wide-field dSTORM Diskovery imaging system used for this study, and M. Segal and G. Pereira for strains. E.Y. was supported by a fellowship from the Cellular Dynamics of Macromolecular Complexes CREATE training program (Natural Sciences and Engineering Research Council of Canada [NSERC]). We acknowledge funding support from ETHZ and grants of ERC, SystemsX.ch, and SNF to Y.B. and operating grants from NSERC (262246) and the Canadian Institutes of Health Research (MOP-123335) awarded to J.V.
Abbreviations used:
- AF647
AlexaFluor 647
- dSTORM
direct stochastic optical reconstruction microscopy
- EM
electron microscopy
- FI
fluorescence intensity
- FRAP
fluorescence recovery after photobleaching
- LE
localization event
- MEN
mitotic exit network
- mNG
mNeongreen
- MTOCs
microtubule-organizing centers
- SIM
structured illumination microscopy
- SPB
spindle pole body
- SPIN
SPB inheritance network
- γ-TuC
γ-tubulin complexes.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-10-0725) on November 15, 2017.
REFERENCES
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y, Liakopoulos D. Role of spindle asymmetry in cellular dynamics. Int Rev Cell Mol Biol. 2009;278:149–213. doi: 10.1016/S1937-6448(09)78004-9. [DOI] [PubMed] [Google Scholar]
- Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Thibodeaux J, Maddox P, Yeh E, Bloom K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr Biol. 2000;10:1497–1506. doi: 10.1016/s0960-9822(00)00837-x. [DOI] [PubMed] [Google Scholar]
- Berlier JE, Rothe A, Buller G, Bradford J, Gray DR, Filanoski BJ, Telford WG, Yue S, Liu J, Cheung CY, et al. Quantitative comparison of long-wavelength Alexa Fluor dyes to Cy dyes: fluorescence of the dyes and their bioconjugates. J Histochem Cytochem. 2003;51:1699–1712. doi: 10.1177/002215540305101214. [DOI] [PubMed] [Google Scholar]
- Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- Boettcher B, Marquez-Lago TT, Bayer M, Weiss EL, Barral Y. Nuclear envelope morphology constrains diffusion and promotes asymmetric protein segregation in closed mitosis. J Cell Biol. 2012;197:921–937. doi: 10.1083/jcb.201112117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M, Piel M. Centrosome inheritance: birthright or the privilege of maturity? Curr Biol. 2002;12:R71–R73. doi: 10.1016/s0960-9822(01)00678-9. [DOI] [PubMed] [Google Scholar]
- Bullitt E, Rout MP, Kilmartin JV, Akey CW. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- Burns S, Avena JS, Unruh JR, Yu Z, Smith SE, Slaughter BD, Winey M, Jaspersen SL. Structured illumination with particle averaging reveals novel roles for yeast centrosome components during duplication. eLife. 2015;4:e08586. doi: 10.7554/eLife.08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Gupta ML, Jr, Hoyt MA, Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Cepeda-Garcia C, Delgehyr N, Juanes Ortiz MA, ten Hoopen R, Zhiteneva A, Segal M. Actin-mediated delivery of astral microtubules instructs Kar9p asymmetric loading to the bud-ward spindle pole. Mol Biol Cell. 2010;21:2685–2695. doi: 10.1091/mbc.E10-03-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XP, Yin H, Huffaker TC. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J Cell Biol. 1998;141:1169–1179. doi: 10.1083/jcb.141.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Raff JW. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr Biol. 2010;20:2187–2192. doi: 10.1016/j.cub.2010.11.055. [DOI] [PubMed] [Google Scholar]
- Demmerle J, Innocent C, North AJ, Ball G, Muller M, Miron E, Matsuda A, Dobbie IM, Markaki Y, Schermelleh L. Strategic and practical guidelines for successful structured illumination microscopy. Nat Protoc. 2017;12:988–1010. doi: 10.1038/nprot.2017.019. [DOI] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Grava S, Schaerer F, Faty M, Philippsen P, Barral Y. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev Cell. 2006;10:425–439. doi: 10.1016/j.devcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryaznova Y, Koca Caydasi A, Malengo G, Sourjik V, Pereira G. A FRET-based study reveals site-specific regulation of spindle position checkpoint proteins at yeast centrosomes. eLife. 2016;5:e14029. doi: 10.7554/eLife.14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- Henderson KA, Hughes AL, Gottschling DE. Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast. eLife. 2014;3:e03504. doi: 10.7554/eLife.03504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D, Schaerer F, Brachat A, Wach A, Philippsen P. Reorientation of mispositioned spindles in short astral microtubule mutant spc72Delta is dependent on spindle pole body outer plaque and Kar3 motor protein. Mol Biol Cell. 2002;13:1366–1380. doi: 10.1091/mbc.01-07-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz M, Leisner C, Chen D, Manatschal C, Wegleiter T, Ouellet J, Lindstrom D, Gottschling DE, Vogel J, Barral Y. Spindle pole bodies exploit the mitotic exit network in metaphase to drive their age-dependent segregation. Cell. 2012;148:958–972. doi: 10.1016/j.cell.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hwang E, Kusch J, Barral Y, Huffaker TC. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Llamazares S, Reina J, Gonzalez C. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Giddings TH, Jr, Winey M. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol. 2002;159:945–956. doi: 10.1083/jcb.200208169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Juanes MA, Twyman H, Tunnacliffe E, Guo Z, ten Hoopen R, Segal M. Spindle pole body history intrinsically links pole identity with asymmetric fate in budding yeast. Curr Biol. 2013;23:1310–1319. doi: 10.1016/j.cub.2013.05.057. [DOI] [PubMed] [Google Scholar]
- Keck JM, Jones MH, Wong CC, Binkley J, Chen D, Jaspersen SL, Holinger EP, Xu T, Niepel M, Rout MP, et al. A cell cycle phosphoproteome of the yeast centrosome. Science. 2011;332:1557–1561. doi: 10.1126/science.1205193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Keller PJ, Bartosik A, Meurer M, Barry JD, Mardin BR, Kaufmann A, Trautmann S, Wachsmuth M, Pereira G, et al. Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat Biotechnol. 2012;30:708–714. doi: 10.1038/nbt.2281. [DOI] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast gamma-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kollman JM, Greenberg CH, Li S, Moritz M, Zelter A, Fong KK, Fernandez JJ, Sali A, Kilmartin J, Davis TN. Ring closure activates yeast gamma TuRC for species-specific microtubule nucleation. Nat Struct Mol Biol. 2015;22:132–137. doi: 10.1038/nsmb.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek WS, Copeland MJ, Chaudhuri A, Chant J. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 2000;287:2257–2259. doi: 10.1126/science.287.5461.2257. [DOI] [PubMed] [Google Scholar]
- Kraus F, Miron E, Demmerle J, Chitiashvili T, Budco A, Alle Q, Matsuda A, Leonhardt H, Schermelleh L, Markaki Y. Quantitative 3D structured illumination microscopy of nuclear structures. Nat Protoc. 2017;12:1011–1028. doi: 10.1038/nprot.2017.020. [DOI] [PubMed] [Google Scholar]
- Krull A, Steinborn A, Ananthanarayanan V, Ramunno-Johnson D, Petersohn U, Tolic-Norrelykke IM. A divide and conquer strategy for the maximum likelihood localization of low intensity objects. Opt Express. 2014;22:210–228. doi: 10.1364/OE.22.000210. [DOI] [PubMed] [Google Scholar]
- Kusch J, Liakopoulos D, Barral Y. Spindle asymmetry: a compass for the cell. Trends Cell Biol. 2003;13:562–569. doi: 10.1016/j.tcb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Kusch J, Meyer A, Snyder MP, Barral Y. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 2002;16:1627–1639. doi: 10.1101/gad.222602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner C, Kammerer D, Denoth A, Britschi M, Barral Y, Liakopoulos D. Regulation of mitotic spindle asymmetry by SUMO and the spindle-assembly checkpoint in yeast. Curr Biol. 2008;18:1249–1255. doi: 10.1016/j.cub.2008.07.091. [DOI] [PubMed] [Google Scholar]
- Lengefeld J, Hotz M, Rollins M, Baetz K, Barral Y. Budding yeast Wee1 distinguishes spindle pole bodies to encode their pattern of age-dependent segregation. Nat Cell Biol. 2017;19:941–951. doi: 10.1038/ncb3576. [DOI] [PubMed] [Google Scholar]
- Lerit DA, Smyth JT, Rusan NM. Organelle asymmetry for proper fitness, function, and fate. Chromosome Res. 2013;21:271–286. doi: 10.1007/s10577-013-9350-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Lin H, de Carvalho P, Kho D, Tai CY, Pierre P, Fink GR, Pellman D. Polyploids require Bik1 for kinetochore-microtubule attachment. J Cell Biol. 2001;155:1173–1184. doi: 10.1083/jcb.200108119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TC, Neuner A, Schiebel E. Targeting of gamma-tubulin complexes to microtubule organizing centers: conservation and divergence. Trends Cell Biol. 2015;25:296–307. doi: 10.1016/j.tcb.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Lindholm E, Nickolls J, Oberman S, Montrym J. NVIDIA Tesla: a unified graphics and computing architecture. IEEE Micro. 2008;28:39–55. [Google Scholar]
- Maekawa H, Usui T, Knop M, Schiebel E. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 2003;22:438–449. doi: 10.1093/emboj/cdg063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Lee WL. Microtubule-dependent path to the cell cortex for cytoplasmic dynein in mitotic spindle orientation. Bioarchitecture. 2011;1:209–215. doi: 10.4161/bioa.18103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez-Benito V, van Deventer SJ, Jimenez-Garcia V, Roy-Luzarraga M, van Leeuwen F, Neefjes J. Spatiotemporal analysis of organelle and macromolecular complex inheritance. Proc Natl Acad Sci USA. 2013;110:175–180. doi: 10.1073/pnas.1207424110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Nigg EA. The centrosome cycle. FEBS Lett. 2002;521:9–13. doi: 10.1016/s0014-5793(02)02865-x. [DOI] [PubMed] [Google Scholar]
- Miller RK, Cheng SC, Rose MD. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol Biol Cell. 2000;11:2949–2959. doi: 10.1091/mbc.11.9.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller EG, Snydsman BE, Novik I, Hailey DW, Gestaut DR, Niemann CA, O’Toole ET, Giddings TH, Jr, Sundin BA, Davis TN. The organization of the core proteins of the yeast spindle pole body. Mol Biol Cell. 2005;16:3341–3352. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarova E, O’Toole E, Kaitna S, Francois P, Winey M, Vogel J. Distinct roles for antiparallel microtubule pairing and overlap during early spindle assembly. Mol Biol Cell. 2013;24:3238–3250. doi: 10.1091/mbc.E13-05-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickolls J, Buck I, Garland M, Skadron K. Scalable parallel programming with CUDA. Queue. 2008;6:40–53. [Google Scholar]
- O’Toole ET, Mastronarde DN, Giddings TH, Jr, Winey M, Burke DJ, McIntosh JR. Three-dimensional analysis and ultrastructural design of mitotic spindles from the cdc20 mutant of Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1–11. doi: 10.1091/mbc.8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Moritz M, Han X, Giddings TH, Lyon A, Kollman J, Winey M, Yates J, 3rd, Agard DA, Drubin DG, et al. Interaction of CK1delta with gammaTuSC ensures proper microtubule assembly and spindle positioning. Mol Biol Cell. 2015;26:2505–2518. doi: 10.1091/mbc.E14-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Grueneberg U, Knop M, Schiebel E. Interaction of the yeast gamma-tubulin complex-binding protein Spc72p with Kar1p is essential for microtubule function during karyogamy. EMBO J. 1999;18:4180–4195. doi: 10.1093/emboj/18.15.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Tanaka TU, Nasmyth K, Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat Methods. 2012;9:582–584. doi: 10.1038/nmeth.1991. [DOI] [PubMed] [Google Scholar]
- Rock JM, Lim D, Stach L, Ogrodowicz RW, Keck JM, Jones MH, Wong CC, Yates JR, 3rd, Winey M, Smerdon SJ, et al. Activation of the yeast Hippo pathway by phosphorylation-dependent assembly of signaling complexes. Science. 2013;340:871–875. doi: 10.1126/science.1235822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott D, Ho J, Pruyne D, Bretscher A. The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J Cell Biol. 1999;147:791–808. doi: 10.1083/jcb.147.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M, Bloom K. Control of spindle polarity and orientation in Saccharomyces cerevisiae. Trends Cell Biol. 2001;11:160–166. doi: 10.1016/s0962-8924(01)01954-7. [DOI] [PubMed] [Google Scholar]
- Segal M, Clarke DJ, Maddox P, Salmon ED, Bloom K, Reed SI. Coordinated spindle assembly and orientation requires Clb5p-dependent kinase in budding yeast. J Cell Biol. 2000;148:441–452. doi: 10.1083/jcb.148.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Maddox P, Skibbens RV, Yeh E, Salmon ED, Bloom K. Nuclear and spindle dynamics in budding yeast. Mol Biol Cell. 1998;9:1627–1631. doi: 10.1091/mbc.9.7.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulist K, Yen E, Kaitna S, Leary A, Decterov A, Gupta D, Vogel J. Interrogation of gamma-tubulin alleles using high-resolution fitness measurements reveals a distinct cytoplasmic function in spindle alignment. Sci Rep. 2017;7:11398. doi: 10.1038/s41598-017-11789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Smith CS, Joseph N, Rieger B, Lidke KA. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat Methods. 2010;7:373–375. doi: 10.1038/nmeth.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soues S, Adams IR. SPC72: a spindle pole component required for spindle orientation in the yeast Saccharomyces cerevisiae. J Cell Sci. 1998;111(Pt 18):2809–2818. doi: 10.1242/jcs.111.18.2809. [DOI] [PubMed] [Google Scholar]
- Tang X, Germain BS, Lee WL. A novel patch assembly domain in Num1 mediates dynein anchoring at the cortex during spindle positioning. J Cell Biol. 2012;196:743–756. doi: 10.1083/jcb.201112017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theesfeld CL, Irazoqui JE, Bloom K, Lew DJ. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1999;146:1019–1032. doi: 10.1083/jcb.146.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio-Santiago M, Monje-Casas F. Tem1 localization to the spindle pole bodies is essential for mitotic exit and impairs spindle checkpoint function. J Cell Biol. 2011;192:599–614. doi: 10.1083/jcb.201007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Linde S, Loschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- Verzijlbergen KF, Menendez-Benito V, van Welsem T, van Deventer SJ, Lindstrom DL, Ovaa H, Neefjes J, Gottschling DE, van Leeuwen F. Recombination-induced tag exchange to track old and new proteins. Proc Natl Acad Sci USA. 2010;107:64–68. doi: 10.1073/pnas.0911164107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M. Cell cycle: driving the centrosome cycle. Curr Biol. 1999;9:R449–R452. doi: 10.1016/s0960-9822(99)80279-6. [DOI] [PubMed] [Google Scholar]
- Winey M, Bloom K. Mitotic spindle form and function. Genetics. 2012;190:1197–1224. doi: 10.1534/genetics.111.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Fuller MT. Asymmetric centrosome behavior and the mechanisms of stem cell division. J Cell Biol. 2008;180:261–266. doi: 10.1083/jcb.200707083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HW, Pruyne D, Huffaker TC, Bretscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- Yoder TJ, Pearson CG, Bloom K, Davis TN. The Saccharomyces cerevisiae spindle pole body is a dynamic structure. Mol Biol Cell. 2003;14:3494–3505. doi: 10.1091/mbc.E02-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.