Phosphorylation of eIF2 and the integrated stress response in response to UVB are cytoprotective by a mechanism featuring preferential translation of a specific splice variant of CDKN1A that facilitates G1 arrest and subsequent DNA repair.
Abstract
In response to sublethal ultraviolet B (UVB) irradiation, human keratinocytes transiently block progression of the cell cycle to allow ample time for DNA repair and cell fate determination. These cellular activities are important for avoiding the initiation of carcinogenesis in skin. Central to these processes is the repression of initiation of mRNA translation through GCN2 phosphorylation of eIF2α (eIF2α-P). Concurrent with reduced global protein synthesis, eIF2α-P and the accompanying integrated stress response (ISR) selectively enhance translation of mRNAs involved in stress adaptation. In this study, we elucidated a mechanism for eIF2α-P cytoprotection in response to UVB in human keratinocytes. Loss of eIF2α-P induced by UVB diminished G1 arrest, DNA repair, and cellular senescence coincident with enhanced cell death in human keratinocytes. Genome-wide analysis of translation revealed that the mechanism for these critical adaptive responses by eIF2α-P involved induced expression of CDKN1A encoding the p21 (CIP1/WAF1) protein. We further show that human CDKN1A mRNA splice variant 4 is preferentially translated following stress-induced eIF2α-P by a mechanism mediated in part by upstream ORFs situated in the 5′-leader of CDKN1A mRNA. We conclude that eIF2α-P is cytoprotective in response to UVB by a mechanism featuring translation of a specific splice variant of CDKN1A that facilitates G1 arrest and subsequent DNA repair.
INTRODUCTION
Ultraviolet B (UVB) irradiation from the sun is a major risk factor in the development of skin cancers such as squamous and basal cell carcinomas, which arise from epidermal keratinocytes (Melnikova and Ananthaswamy, 2005). To respond to environmental stressors such as UVB, eukaryotic cells have evolved a variety of mechanisms to regulate gene expression for adaptation and repair. Many of these adaptive mechanisms feature genome-wide changes in mRNA translation (Wek et al., 2006; Jackson et al., 2010; Baird and Wek, 2012). The process of protein synthesis involves distinct initiation, elongation, and termination stages, of which initiation is the rate-limiting and most highly regulated step (Jackson et al., 2010). Phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α-P) is induced following a variety of cellular stresses (including UVB) and is a central mechanism by which cells can control translation initiation. eIF2α-P decreases general protein synthesis by reducing the ability of eIF2 to bind GTP and deliver initiator Met-tRNAiMet to ribosomes at the onset of translation (Wek et al., 2006; Baird and Wek, 2012). Lowered global translation following cellular stress serves to conserve resources that are critical for the recovery of stressed cells. Because eIF2α-P and translational control can be induced by multiple protein kinases that are each activated by distinct stress conditions, this pathway is referred to as the integrated stress response (ISR) (Harding et al., 2003). There are four different eIF2α kinases expressed in mammals, with GCN2 (EIF2AK4) being the predominant version activated in response to UVB and UVC (Deng et al., 2002; Jiang and Wek, 2005; Powley et al., 2009; Dey et al., 2010).
Coincident with general repression of protein synthesis, eIF2α-P heightens translation of a cohort of prosurvival transcripts, including those encoding the transcription factor ATF4 and GADD34 (PPP1R15A) (Harding et al., 2000a; Vattem and Wek, 2004; Lee et al., 2009; Baird and Wek, 2012; Young et al., 2015), which facilitates feedback dephosphorylation of eIF2α-P (Connor et al., 2001; Novoa et al., 2001, 2003; Brush et al., 2003). Preferential translation directed by eIF2α-P utilizes upstream open reading frames (uORFs) in the 5′-leaders of these transcripts (Hinnebusch et al., 2016; Young and Wek, 2016). Multiple mechanisms involving combinations of inhibitory and positive acting uORFs have been described for genes that are translationally induced by eIF2α-P (Vattem and Wek, 2004; Palam et al., 2011; Young et al., 2015, 2016a, b; Young and Wek, 2016). While translational control through eIF2α-P has been shown to be protective against UVB-induced cell death (Powley et al., 2009; Collier et al., 2015), the underlying adaptive mechanisms are not fully understood.
UVB can directly create cross-linked DNA adducts such as cyclobutane dimers and 6,4 photoproducts in genomic DNA, which are resolved by nucleotide excision repair (NER) (Shuck et al., 2008; Cadet et al., 2012). Damaged DNA can lead to mutagenesis, which can cause cancer if mutations persist in tumor suppressors or oncogenes. As a consequence of UVB-induced DNA damage, keratinocytes transiently arrest progression in the cell cycle during G1 to prevent replication of damaged DNA (de Laat et al., 1996; Ortolan and Menck, 2013). If DNA damage is successfully repaired, cells can resume progression into S phase and complete the cell cycle. Prolonged arrest of the cell cycle can lead to cellular senescence, which in keratinocytes functions to preserve the barrier function of the epidermis while ensuring that injured keratinocytes do not replicate damaged DNA (Lewis et al., 2008). Finally, if genomic DNA in keratinocytes suffers irreparable damage, these cells will be lost in the epidermis by apoptosis. Each of these processes functions to eliminate potentially mutagenic cells that can develop into nonmelanoma skin cancers (NMSCs) (D’Errico et al., 2003; Campisi, 2005).
Mammalian cells have multiple checkpoint mechanisms that are important for determining cell fate. Expression of the cyclin-dependent kinase (CDK) inhibitor CDKN1A that encodes the p21 (CIP1/WAF1) protein is enhanced in response to UVB. The resulting elevated levels of p21 protein function to inhibit the activity of cyclin/CDK complexes, which ultimately blocks cell progression from the G1 to the S phase (Brugarolas et al., 1995; Deng et al., 1995; Mandal et al., 1998). Transcriptional expression of CDKN1A has been well characterized, with an emphasis on direct regulation by p53. Additional CDKN1A regulatory mechanisms, including epigenetic modifications, mRNA stability, and protein degradation, have also been described (Gartel and Tyner, 1999; Jascur et al., 2005; Scholpa et al., 2014). However, little is known about regulation of CDKN1A expression via translation. Furthermore, translation regulation involving CDKN1A-dependent processes, such as cell cycle regulation and senescence, has not been extensively studied.
In this report, we address the mechanisms by which eIF2α-P provides for cell protection against UVB exposure. Our results suggest that translational control through eIF2α-P is required for proper cell cycle control and cell fate determination. Loss of eIF2α-P diminished the length of UVB-induced G1 arrest, the rate of DNA repair, and the induction of cellular senescence, as well as increased apoptosis. Additionally, we utilized genome-wide studies to show that the human CDKN1A splice variant 4 mRNA, which contains a distinct 5′-leader, was preferentially translated during stress and eIF2α-P. These results define a mechanism by which eIF2α-P and translational control provide protection from stressors that require p21 protein expression, such as carcinogenic UVB irradiation.
RESULTS
Loss of eIF2α-P decreases G1 arrest and DNA repair in response to UVB
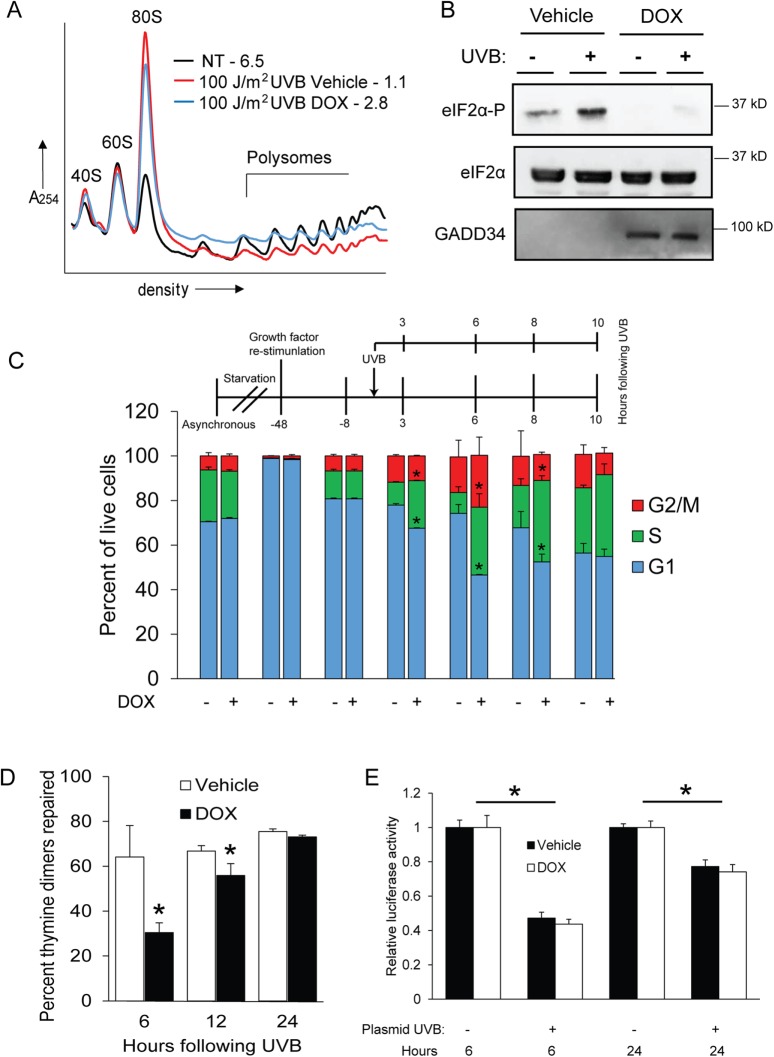
To investigate the effects of eIF2α-P deficiency on low-dose UVB–irradiated keratinocytes, we utilized a human keratinocyte cell line immortalized with hTERT (designated N-TERT) (Dickson et al., 2000) that was engineered to overexpress GADD34 via a doxycycline (DOX) inducible promoter. N-TERT cells are proliferative human epidermal keratinocytes that are useful and reliable surrogates for primary keratinocytes (Collier et al., 2015, 2017; Loesch et al., 2016) and therefore represent skin cells that can propagate UVB-induced mutations that initiate cancer. Elevated levels of GADD34 sharply reduce eIF2α-P in response to UVB, thwarting translational control and the ISR. Using these cells, we measured global levels of translation initiation via sucrose gradient centrifugation followed by polysome profiling. We chose to examine a low (100 J/m2) dose of UVB that can trigger cell cycle arrest and recovery as opposed to higher doses of UVB that instead induce significant levels of apoptosis (Lewis et al., 2008; Collier et al., 2015). Exposure of control N-TERT cells to a dose of 100 J/m2 UVB caused a sharp reduction in translation initiation as measured by an increase in mRNAs associated with 80s monosomes coincident with decreased levels of transcripts bound to heavy polysomes (Figure 1A). The polysome to monosome (p/m) ratio decreased from 6.5 to 1.1 following N-TERT exposure to UVB. This dose of UVB also elicited a robust increase in eIF2α-P levels (Figure 1B). GADD34 overexpression triggered by addition of DOX partially restored heavy polysomes (Figure 1A, blue line) as well as diminishing the amount of eIF2α-P induced by UVB (Figure 1B). Further kinetic analyses of GADD34 ovexpression in cells will be discussed later in Figure 5A. These findings support previous data showing that enhanced eIF2α-P following UVB exposure triggers a sharp reduction in the initiation of protein synthesis.
FIGURE 1:
Loss of eIF2α-P diminishes G1 arrest and DNA repair following UVB. Human N-TERT keratinocytes were treated with 100 J/m2 UVB in the absence or presence of DOX, which enhances GADD34 overexpression and diminishes eIF2α-P and the ISR. Alternatively, the cells were not subjected to treatment. (A) Lysates were prepared from the cells at 8 h post-UVB and subjected to polysome profiling. P/m values are indicated for each profile. (B) NTERT cells were subjected to vehicle or DOX to overexpress GADD34, followed by treatment with UVB (+) or no-stress treatment (–). Lysates were then prepared from the N-TERT cells and the indicated proteins were measured by immunoblot analysis. Protein size markers are indicated for each panel. (C) Cells were synchronized in G1 by a strategy highlighted in this panel, followed by irradiation with 100 J/m2 UVB. At the indicated times post-UVB treatment, cells were costained with EdU and PI to measure cell cycle phases via flow cytometry. Percentages of the indicated cell cycles are represented in the bar graphs. (D) Genomic DNA isolated from the synchronized cells was also subjected to immunoblot analysis to measure thymine dimer content. Cells were treated with UVB and then cultured for between 6 and 24 h in the presence of DOX to overexpress GADD34 or vehicle control. The DNA repair rate is represented as a percent decrease in thymine dimers compared with those in cells 15 min after UVB treatment. (E) A host-cell reactivation assay was performed at the indicated times posttransfection by measuring luciferase activity expressed from plasmid DNA that was treated with UVB (+) or no irradiation (-) prior to transfection into N-TERT cells. The N-TERT cells were cultured and treated with DOX to overexpress GADD34 or vehicle control for 6 or 24 h. Cells were lysed and assayed for luciferase activity. * indicates p < 0.05. Error bars represent mean ± SD of three separate experiments.
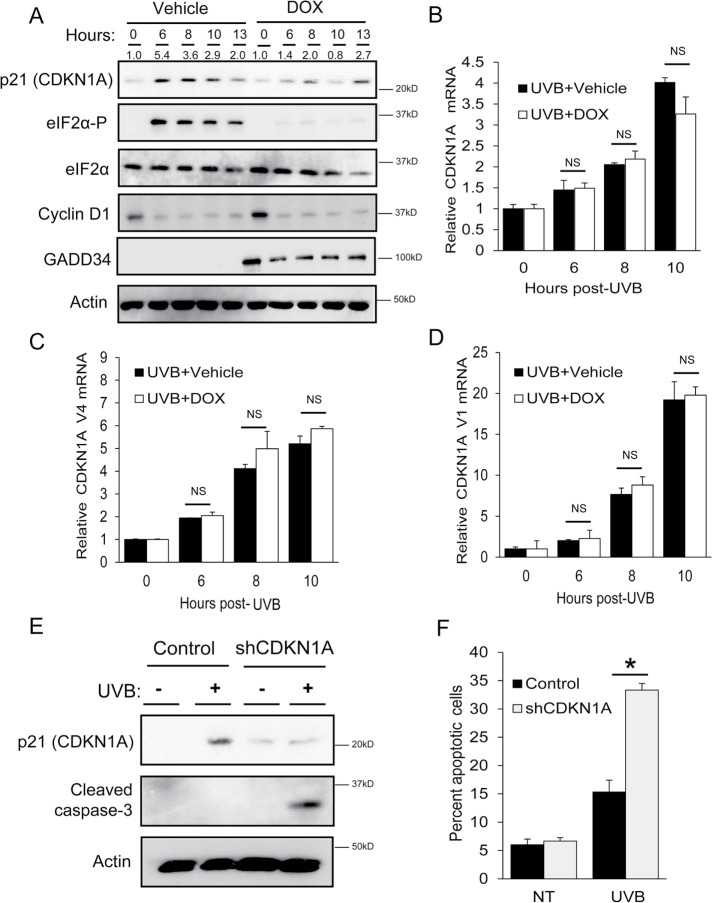
FIGURE 5:
Phosphorylation of eIF2α is required for induced expression of p21 protein following UVB. N-TERT keratinocytes were treated with either vehicle or DOX to induce overexpression of GADD34 and diminish eIF2α-P, followed by exposure to 0 or 100 J/m2 UVB. (A) Lysates were collected from cells at the indicated times following UVB treatment and then subjected to immunoblot analyses to measure the indicated proteins. Sizes of protein markers are indicated to the right of the panels. Additionally, qPCR was used to measure (B) total CDKN1A, (C) V4, or (D) V1 mRNAs in the N-TERT cells. To determine the effects of CDKN1A loss on cell death, control and CDKN1A knockdown N-TERTs were irradiated with 0 or 100 J/m2 UVB. Lysates were collected 8 h following the UVB treatment and subjected to (E) immunoblot analysis to measure cleavage of caspase 3 or (F) PI staining to measure DNA content and relative cell death. * indicates p < 0.05. Error bars represent mean ± SD of three separate experiments.
We next sought to address whether the ISR regulated the keratinocyte cell cycle in response to UVB. To determine the effect of eIF2α-P inhibition on G1 arrest triggered by UVB, N-TERT keratinocytes were synchronized in G1 as highlighted in Figure 1C and detailed in Materials and Methods. Synchronized cells were then irradiated with a dose of 100 J/m2 UVB, cultured for the indicated number of hours, and labeled with both EdU (a thymine analogue) and propidium iodide (PI). Six hours after UVB irradiation, cells containing a fully functional ISR displayed only 9% of live cells in S phase, while 74% remained in G1 (Figure 1C). In comparison, in DOX-treated cells following UVB, the number of cells in S phase was doubled (20%) and the G1 fraction was only 47%. These findings indicate that cells deficient for induced eIF2α-P display a lower percentage of cells in G1 and a corresponding higher percentage in S phase following UVB than for control cells that present with a fully functional ISR. This trend was also observed at 3 and 8 h following UVB treatment. However, by 10 h after UVB exposure, both control and DOX-treated cells were at steady-state proportions of the cell cycle. Therefore, in the absence of a functional ISR, UVB-irradiated keratinocytes entered S phase after little to no cell cycle arrest.
Because cell cycle arrest is important to allow time for repair of DNA damaged by UVB, we next measured the levels of DNA repair in control and DOX-treated cells following UVB. At both 6 and 12 h following exposure to 100 J/m2 UVB, DOX-treated cells showed significantly lower levels of genomic DNA repair as measured by loss of thymine dimers (Figure 1D). Lowered genomic repair was also observed when ISRIB, a pharmacological inhibitor of the ISR (Sidrauski et al., 2013), was added prior to treatment of wild-type N-TERT keratinocytes with UVB (Supplemental Figure 1A). To determine whether this effect was due to changes in the capacity for NER per se, we performed a host-cell reactivation assay. Briefly, luciferase plasmid DNA was irradiated outside of the cell and then transfected into vehicle or DOX-treated N-TERT cells overexpressing GADD34, followed by measurement of luciferase activity, which is indicative of NER activity. There was no significant difference in luciferase activity for the exogenously UVB-damaged luciferase plasmid DNA relative to the undamaged control in either vehicle or DOX-treated keratinocytes (Figure 1E). There was also no difference in repair of the damaged plasmid DNA when vehicle and DOX-treated cells were irradiated with UVB prior to transfection. These results suggest that loss of translational control through eIF2α-P diminishes UVB-induced G1 arrest in keratinocytes, resulting in decreased levels of DNA repair.
Inhibition of UVB-induced eIF2α-P drives keratinocyte fate toward apoptosis
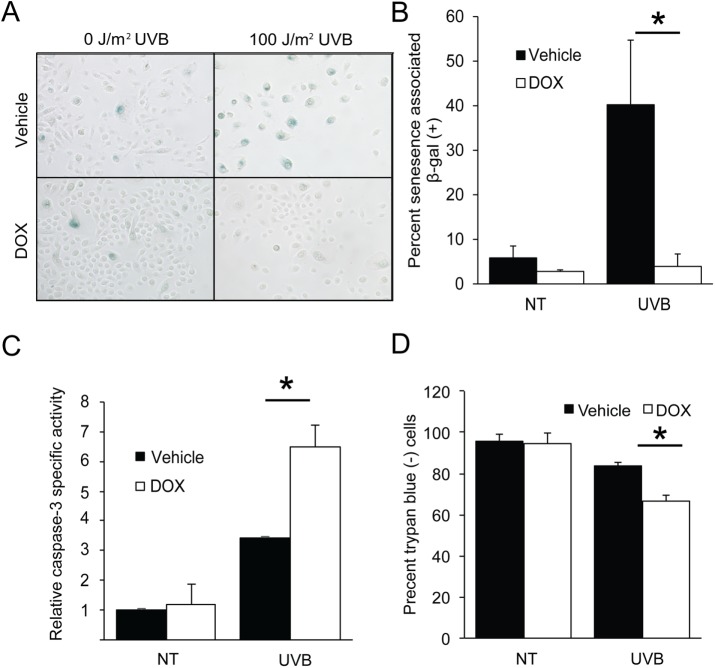
Given the importance of induced eIF2α-P in facilitating cell cycle arrest following a low dose of UVB, we next proposed that there would also be consequences of diminished levels of eIF2α-P for cell fate. Exposure to a dose of 100 J/m2 UVB is known to induce senescence in cultured human keratinocytes, which serves as a mechanism of protection from carcinogenesis (Lewis et al., 2008). Losing the ability to senesce in response to UVB can lead to UVB-induced DNA damage in proliferating keratinocytes (Lewis et al., 2010). To measure the effects of eIF2α-P deficiency on cellular senescence, we quantified the activity of a well-characterized marker, senescence-associated β-galactosidase (SA β-gal). Whereas UVB-treated control cells displayed a 40% increase in SA β-gal activity, there was minimal change in senescence following UVB in DOX-treated cells (Figure 2, A and B). This model was further supported by the finding that short hairpin RNA (shRNA) knockdown of the eIF2α kinase GCN2 in N-TERT cells also led to a sharp reduction in senescence following UVB (Supplemental Figure 1B). DOX-treated keratinocytes also had significantly higher levels of UVB-induced caspase-3 activity than irradiated cells with a fully functional ISR, indicating higher levels of apoptosis (Figure 2C). This increase in cell death equated to about a 20% reduction in the cell population, as measured by staining with trypan blue (Figure 2D). Furthermore, treatment of the TERT cells with the ISR inhibitor ISRIB led to increased caspase-3 activity following exposure to UVB (Supplemental Figure 1C). Knockdown of the eIF2α kinase GCN2 in these human keratinocytes also substantially increased capase-3 activity following UVB (Supplemental Figure 1D). In comparison, knockdown of PERK, an eIF2α kinase activated by endoplasmic reticulum stress (Harding et al., 1999, 2000b; Sood et al., 2000), did not significantly change caspase-3 activity in response to UVB (Supplemental Figure 1D). These results indicate that deficiency eIF2α-P and translational control shifts cells away from senescence and toward apoptosis in response to UVB.
FIGURE 2:
Loss of eIF2α-P switches cell fate away from senescence and toward apoptosis following UVB. (A) N-TERT keratinocytes were treated with 0 or 100 J/m2 UVB and maintained in culture for 72 h. Cells were then fixed and stained for senescence-associated β-galactosidase activity. Quantification of senescent cells (dark blue) is indicated in B. Alternatively, cells were collected 6 h post-UVB and assayed for (C) caspase-3 specific activity or (D) trypan blue to measure loss of cells. Error bars represent mean ± SD of three separate experiments, and * indicates p < 0.05.
Genome-wide analysis of mRNA translation following UVB irradiation
Because disruption of eIF2α-P in response to UVB impaired the arrest of the cell cycle, decreased induction of senescence, and led to increased cell death, we hypothesized that the ISR is responsible for translational control of individual mRNAs that regulate these processes. To determine which transcripts are translationally regulated following UVB exposure, we performed RNA-Seq on sucrose fractions collected during polysome profiling of N-TERT cells irradiated with 0 or 100 J/m2 UVB. RNA was pooled into two groups representing association with light (fractions 3–4) and heavy (fractions 5–7) polysomes. Translational efficiency was defined as the percentage of the total for each gene transcript that was present in the heavy polysome fraction. Changes in the percentage of a given mRNA in the polysome fraction between N-TERT cells exposed to 100 J/m2 UVB or no treatment were then determined. In parallel, we also performed RNA-Seq on total cell lysates from untreated or irradiated cells to determine changes in the mRNA levels that occur in response to UVB.
UVB irradiation caused changes in both the transcriptome and translatome (Figure 3A). In response to UVB, the majority of transcripts either displayed a significant decrease in association with heavy polysomes, indicative of a general repression of initiation of mRNA translation, or exhibited negligible changes in polysome association (Figure 3B). In comparison, ∼15% of genes showed enhanced translation efficiency following UVB, as delineated by at least a 10% shift toward heavy polysomes. This propensity for repression of gene expression was as observed for changes in mRNA in response to UVB, with 71% of those genes showing significant differences displaying lower transcript levels (Figure 3B). It is noteworthy that among those genes displaying lowered mRNAs following UVB treatment, 31% also showed lowered translation efficiencies (Figure 3C). In comparison, only 13% of the genes showing induced mRNA levels in response to UVB also showed enhanced translation (Figure 3C); this combination would be expected to further amplify the expression of these genes. It is also notable that many genes that are preferentially translated in response to UVB irradiation also displayed sharply lowered mRNA levels (Figure 3A). In this case, smaller amounts of mRNA would be available for preferential translation, leading to sharply reduced protein expression. We previously reported that this linkage between dynamic changes in mRNA levels and preferential translation would allow differential gene expression that is best suited for cell adaptation to a given stress condition (Dey et al., 2010; Baird and Wek, 2012).
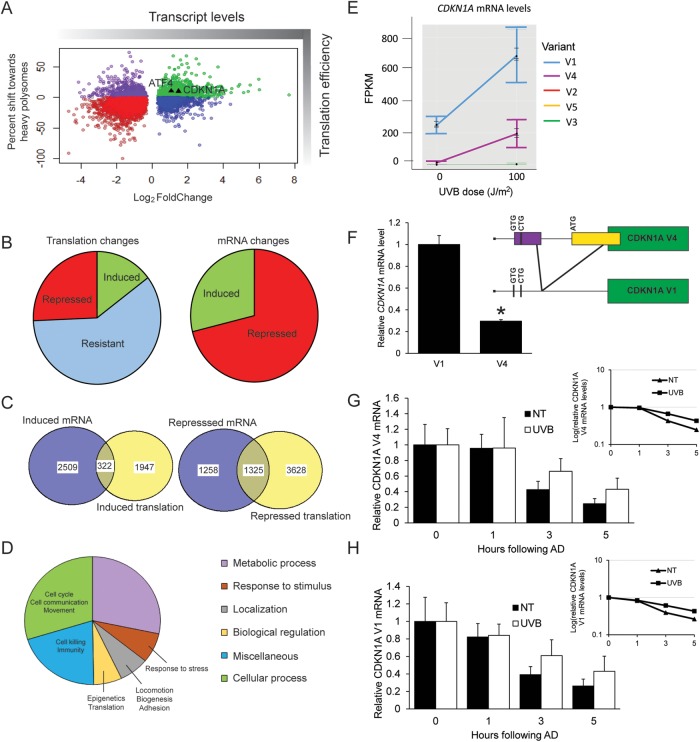
FIGURE 3:
Genome-wide analyses reveal preferential translation of CDKN1A following UVB. N-TERT keratinocytes were irradiated with 0 or 100 J/m2 UVB, followed by lysate preparation and polysome profiling. RNA was isolated from total cell lysate and light or heavy polysome fractions, and each was analyzed by RNA-Seq. (A) Scatterplot illustrating the shift toward heavy polysomes vs. relative mRNA fold changes following UVB irradiation. Only genes with significant mRNA fold changes (p < 0.05) are shown. (B) On the left side of the panel is a pie chart indicating the numbers of genes whose translation was induced (>10% shift), resistant (between -10% and 10% shift), or repressed (less than -10% shift) following UVB irradiation. On the right side of the panel is the number of genes whose transcripts were significantly induced or repressed after UVB treatment. (C) Venn diagram comparing the gene transcripts with significantly induced mRNA following UVB treatment and those genes that are subject to induced translation, as judged by a >10% shift of the encoded mRNAs toward large polysomes. Additionally, a Venn diagram is featured showing those gene transcripts that were significantly repressed following UVB irradiation, along with those genes displaying translation repression (>10% shift toward small polysomes). (D) Pie chart illustrating the different cellular functions of genes that are suggested to be preferentially translated in response to UVB. (E) FPKM values to indicate relative expression level for each CDKN1A transcript variant after the indicated UVB dose. (F) Pattern of alternative splicing of the CDKN1A, leading to V1 and V4 transcripts that display the uORFs and coding sequences (green boxes). Potential initiation codons are illustrated in the 5′-leader of the V1 transcript. Primers that recognize both V1 and V4 were used to perform PCR on N-TERT keratinocytes to determine relative transcript abundance. Bands were quantified by densitometry and normalized to PCR product size. N-TERTs were exposed to 0 or 100 J/m2 UVB, and after 8 h, RNA was prepared from the cells for measurement of V4 (G) and V1 (H) mRNAs by qPCR. Additionally, cells were treated with 10 μM actinomycin D (AD) and cultured for an additional 1, 3, or 5 h, and levels of the V1 and V4 mRNAs were determined to assess the rate of decay for these transcripts. Transcript levels were measured in no treatment (NT) or UVB exposed cells following actinomycin treatments were normalized to the respective cells measured in the absence of actinomycin D treatment (0). A semilog plot of each decay rate is shown in the inset plot for each transcript. Error bars represent mean ± SD of three biological replicates. * indicates p < 0.05.
To identify the cellular processes associated with preferentially translated genes (>10% shift toward heavy polysomes), we performed a gene ontology analysis that showed involvement in a range of biological processes, including metabolism, response to stress, cell localization, and regulatory processes (Figure 3D). There did not appear to be enrichment for genes involved in DNA repair, which aligns with results shown in Figure 1E, suggesting that DNA repair per se is not translationally regulated. It is noteworthy that among the preferentially translated genes there was enrichment for those involved in cell cycle regulation. One such gene was CDKN1A (WAF1 encoding the p21 protein), whose primary functions are to inhibit the progression from G1 to S phase of the cell cycle and to initiate cellular senescence (Brugarolas et al., 1995; Georgakilas et al., 2017). The 10% increase in the efficiency of CDKN1A translation following UVB was similar to that seen with ATF4, a gene previously shown to be preferentially translated in the ISR in response to a variety of stressors. Both genes are denoted as triangles in Figure 3A. There was also a sharp increase in CDKN1A mRNA levels, which is consistent with its previously reported transcriptional induction by UVB (Gartel and Tyner, 1999). Given our findings that induced eIF2α-P facilitates the arrest of the cell cycle, we decided to further address the regulatory significance of CDKN1A in the ISR following UVB irradiation.
Preferential translation of CDKN1A mRNA splice variant in response to UVB and eIF2α-P
Five different CDKN1A mRNA splice variants have been reported (RefSeq RNA: NM_000389.4, NM_078467.2, NM_001291549.1, NM_001220778.1, NM_001220777.1), each of which contributes to significant differences in the 5′-leader (Figure 3E). CDKN1A variants 1 and 4 (denoted V1 and V4, respectively) were readily detected in the RNA-Seq analysis and showed increased mRNA levels following UVB exposure (Figure 3E). The other three variants showed negligible expression and induction in N-TERT cells. V1 expression was ∼70% higher than V4, with almost undetectable V4 levels in untreated samples. We further addressed this difference in V1 and V4 mRNA levels in the untreated N-TERT cells by using semiquantitative PCR and primers that recognize both variants (Figure 3F). Following gel electrophoresis and staining, we were able to detect distinct DNA bands for both CDKN1A transcript variants, with V1 appearing to be ∼70% more abundant than V4 following normalization to PCR product size. This proportion of V1 and V4 transcripts as judged by PCR was unchanged following UVB treatment (unpublished data). As will be discussed more fully below, a critical difference between the CDKN1A transcript variants is that the V4 contains a functional uORF that overlaps out of frame with the CDS (coding sequence) (Figure 3F).
To determine the role of mRNA decay in the induction of V1 and V4 mRNAs upon UVB exposure, N-TERT cells were exposed to 0 or 100 J/m2 UVB, followed by treatment with an RNA polymerase II inhibitor, actinomycin D, for an additional 1, 3, or 5 h. Levels of V1 and V4 mRNAs were then measured by quantitative PCR (qPCR), revealing that the half-lives of V1 and V4 mRNAs were each 3 h in cells not subjected to UVB (Figure 3, G and H). In comparison, there was increased stabilization of both the V1 and V4 transcripts following exposure to UVB, with half-lives of 5 h. These results suggest that induction of CDKN1A mRNAs upon UVB irradiation involves enhanced stabilization of both V1 and V4 transcripts and well as increased transcription.
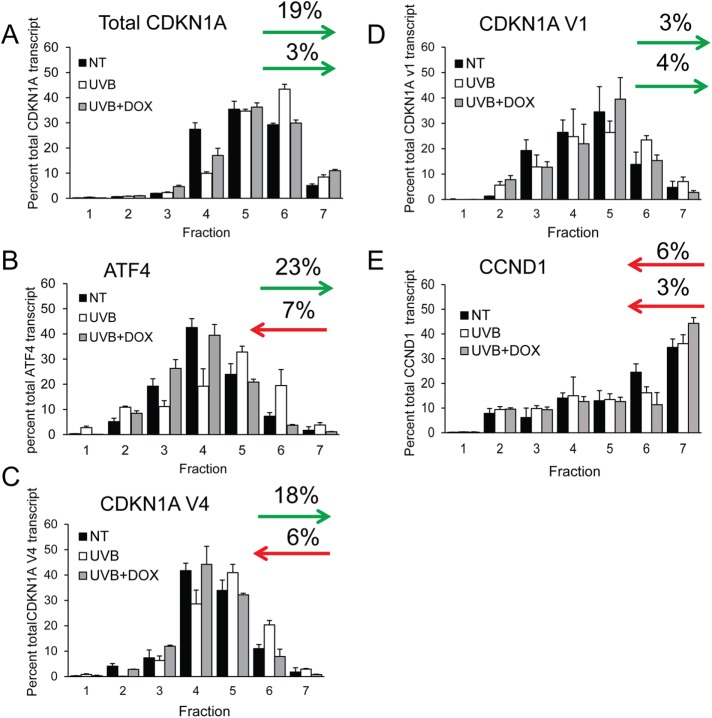
Because induced eIF2α-P can direct preferential translation through 5′-leaders of mRNAs, we next wished to determine whether there were differences in translational control between the two CDKN1A splice variants. As noted earlier, levels of CDKN1A V4 in the untreated cells were too low to be accurately measured in our RNA-Seq analysis. We therefore determined the changes in ribosome association of CDKN1A mRNAs using qPCR on sucrose fractions collected during polysome profiling. In response to UVB irradiation, total CDKN1A mRNA shifted 19% toward heavy polysome fractions (5–7) compared with control cells not subjected to irradiation (Figure 4A). Notably, the shift of CDKN1A mRNA toward heavy polysomes was diminished to only 3% when UVB irradiation was combined with DOX treatment, indicating that preferential translation of CDKN1A is regulated by induced eIF2α-P. These changes were comparable to shifts for the ISR regulator ATF4 mRNA following exposure to 100 J/m2 UVB exposure (Figure 4B). These findings indicate that CDKN1A is subject to preferential translation in response to UVB and induced eIF2α-P. CDKN1A V4 displayed an 18% shift toward heavy polysomes following UVB exposure (Figure 4C), whereas V1 displayed a negligible 3% shift (Figure 4D). Translation of V4 following UVB exposure is positively regulated by induced eIF2α-P, as DOX treatment reversed the preferential translation of V4, instead shifting 6% away from polysomes toward monosomes compared to no treatment. DOX treatment had no effect on V1 mRNA polysome association. Polysome association of CCND1 mRNA (encoding cyclin D1 protein) showed negligible changes in response to UVB with or without DOX treatment (Figure 4E), suggesting that this mechanism of translational control is not widespread among cell cycle genes. These results indicate that an individual CDKN1A transcript variant 4 is subject to preferential translation in response to induced eIF2α-P and UVB.
FIGURE 4:
Human CDKN1A splice variant 4 is preferentially translated following UVB. N-TERT cells were irradiated with 0 or 100 J/m2 UVB in the presence or absence of DOX to trigger depletion of eIF2α-P and the ISR. Cells were collected 8 h following UVB treatment and lysates were prepared and analyzed by polysome profiling. The amounts of the CDKN1A (A), ATF4 (B), CDKN1A V4 (C), CDKN1A V1 (D), and CCND1 (E) mRNAs in the indicated sucrose gradient fractions were measured by qPCR. Levels of the gene transcripts in each fraction are represented as a percentage of the total gene transcript to discount any changes in the specific mRNA levels. Arrows represent the percent of each transcript that shifts toward or away from heavy polysomes (fractions 5–7). For each panel, the top arrow indicates the shift toward heavy polysomes in response to UVB, whereas the bottom arrow is in response to UVB and DOX treatment. Error bars represent mean ± SD of three separate experiments.
To further test the idea that preferential translation of CDKN1A V4 elicits changes in p21 protein expression following UVB exposure, we performed immunoblot analyses in vehicle and DOX-treated keratinocytes. In the control cells, UVB enhanced the levels of p21 protein (Figure 5A). However, lowering of eIF2α-P levels by DOX diminished the amount of p21 protein by over 50% at 6, 8, and 10 h following UVB treatment. Furthermore, shRNA knockdown of GCN2 blocked induction of p21 in response to UVB, whereas there was induction of p21 protein in N-TERT cells depleted of PERK (Supplemental Figure 2). These results indicate that in response to UVB irradiation, induction of eIF2α-P is required for enhanced expression of p21 protein. In contrast, disruption of the ISR by DOX treatment had no effect on the levels of cyclin D1 protein (Figure 5A), a cell cycle regulator that was reported to be translationally repressed by eIF2α-P in response to endoplasmic reticulum (ER) stress (Hamanaka et al., 2005). These results indicate that the ISR facilitates induced p21 protein expression upon UVB irradiation.
To address the specificity of CDKN1A mRNA induction by UVB irradiation, we performed qPCR to measure mRNA levels of each individual CDKN1A variant in the keratinocytes at various times following UVB exposure. While total CDKN1A and both transcript variants 1 and 4 were induced in response to UVB over time, neither the total CDKN1A nor splice variants showed significant change in the induction of mRNA levels with GADD34 overexpression (Figure 5, B–D). These results indicate that diminished p21 protein in DOX-treated cells was not due to decreased induction of CDKN1A mRNA.
Our results indicate that the ISR facilitates cell cycle arrest and is required for CDKN1A expression following UVB. We next asked whether the loss of CDKN1A mimics the cell death phenotype associated with eIF2α-P deficiency. N-TERT keratinocytes were created to stably express shRNA against CDKN1A or a shRNA control and these cells were treated with 0 or 100 J/m2 of UVB. Keratinocytes depleted for CDKN1A showed increased UVB-induced cell death as measured by increased cleaved caspase-3 (Figure 5E), as well as increased apoptotic cell populations as measured by flow cytometry (Figure 5F). These results suggest that loss of the ISR mimics the consequences of loss of CDKN1A in the viability of keratinocytes exposed to UVB.
The 5′-leader of CDKN1A V4 mRNA confers preferential translation in response to induced eIF2α-P
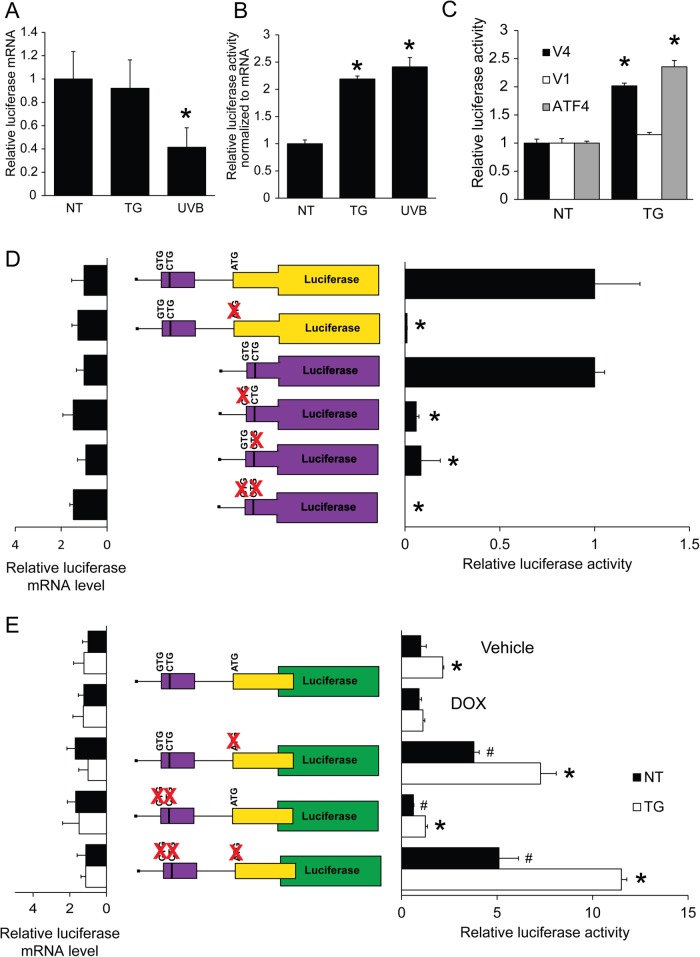
Because CDKN1A V4 was subject to preferential translation and differed from V1 only in the 5′-leader sequences, we hypothesized that translational control was mediated by uORFs. CDKN1A V4 mRNA contains two putative uORFs that feature one predicted canonical codon and two noncanonical start codons. Notably, the noncanonical initiation codons in the uORFs of CDKN1A V4 mRNA were identified in genome-wide ribosome profiling studies as functional sites of initiation (Lee et al., 2012). The 5′-leader of V1 contains three putative noncanonical CUG start codons that are in poor context and have not been identified as functional sites of translation. To determine whether the translational control of CDKN1A V4 is mediated through the 5′-leader and uORFs, we inserted the cDNA segments encoding the 5′-leader for either CDKN1A V1 or V4 between a minimal thymidine kinase (TK) promoter (PTk) and the firefly luciferase CDS. The arrangement of these reporter constructs was similar to that in other ISR transcripts tested for preferential translation (Vattem and Wek, 2004; Palam et al., 2011; Young et al., 2015, 2016b). We first measured mRNA levels of the CDKN1A V4 reporter following treatment with UVB or thapsigargin (TG), a known inducer of ER stress and eIF2α-P. Levels of luciferase reporter mRNA were reduced over 50% in response to UVB, indicating technical issues with UVB-specific repression of the TK promoter and/or reporter mRNA decay that were not apparent with the inducible endogenous CDKN1A gene and associated promoter (Figure 6A). In contrast, TG had no effect on reporter mRNA levels. We then measured luciferase activity of the CDKN1A V4 reporter and normalized these data to changes in mRNA levels in order to analyze changes independent of transcription or transcript decay. Luciferase activity of CDKN1A V4 increased twofold following treatment with either UVB or TG, indicating that preferential translation is mediated through the V4 5′-leader (Figure 6B).
FIGURE 6:
The 5′-leader of CDKN1A V4 mRNA directs preferential translation in response to eIF2α-P. N-TERTs were transfected with a CDKN1A V4 luciferase reporter followed by treatment with TG or 100 J/m2 UVB, as indicated. Measurements of (A) mRNA and (B) luciferase reporter activity are represented as histograms. (C) Translational control was determined for V4 and V1 variants of CDKN1A or an ATF4 luciferase in response to 6 h of treatment with TG. Firefly luciferase activity was determined using a dual luciferase assay. (D) N-TERTs were transfected with the depicted reporter plasmids encoding the indicated uORFs fused with the firefly luciferase CDS. Luciferase activity was measured after 24 h of transfection. (E) Wild type and the indicated mutant versions of the CDKN1A V4 reporter were transfected into N-TERT cells, followed by treatment with TG for 6 h. Firefly luciferase activity was measured using the dual luciferase assay. Relative amounts of luciferase reporter mRNA as measured by qPCR are shown to the left in D and E. The red “x” indicates mutations of the putative indication codons. * indicates p < 0.05 and # indicates p < 0.05 compared with the untreated reporter. Error bars represent mean ± SD of three separate experiments.
We next wished to determine the underlying mechanism for CDKN1A preferential translation in response to induced eIF2α-P. To circumvent the complication caused by UVB effects on the luciferase reporter mRNA in our analysis of preferential translation in response to eIF2α-P, we elected to carry out our experiments using TG, which did not affect levels of endogenous CDKN1A or reporter mRNA levels (Supplemental Figure 3A). Of interest, treatment of N-TERT cells with TG led to an increase in p21 protein levels (Supplemental Figure 3B), further supporting its usefulness as a tool for studying CDKN1A translational control. CDKN1A V4 and V1 and ATF4 luciferase reporters were treated with TG and collected after 6 h. While V4 luciferase activity increased with TG to a degree similar to that for ATF4, there was no change in reporter activity for V1 in response to stress (Figure 6C). These findings indicate that CDKN1A V4, but not V1, is subject to preferential translation by mechanisms involving its 5′-leader.
To determine whether uORFs in CDKN1A V4 mRNA are indeed translated, we created in-frame fusions between the 5′-proximal uORF1 and uORF2 and the firefly luciferase CDS. These constructs did not contain the luciferase translation start site, so any quantifiable luciferase activity was due to initiation at the in-frame uORF. The fusion of both uORF1 and uORF2 had significant luciferase activity, suggesting functional initiation codons for each uORF (Figure 6D). Substitution of AGG for ATG in uORF2 substantially ablated reporter expression. In comparison, mutations in uORF1 in either of the less frequent initiation codons, CTG and GTG, caused a substantial decrease in luciferase activity, indicating that each start codon is functional and required for translation of each uORF. The combination of mutations in both initiation codons resulted in minimal detectable reporter expression (Figure 6D). We conclude that both uORF1 and uORF2 can be effectively translated in the V4 transcript.
To determine the relative contribution of each uORF and initiation codon to preferential translation for CDKN1A V4, we performed a series of mutational analyses using variations of the luciferase reporter. Importantly, the increase in CDKN1A V4 reporter activity with TG was abolished during overexpression of GADD34 (Figure 6E), suggesting that the translation control mediated by the 5′-leader is dependent on induced eIF2α-P. Mutation of the ATG in uORF2 caused a significant increase in basal reporter activity, indicating that uORF2 serves to repress downstream translation. This mutation, however, had no effect on the ratio of induction of the reporter by TG treatment. Mutation of either the uORF1 GTG or CTG caused a decrease in basal reporter activity, but again had no effect on the proportion of induced luciferase expression upon stress. Mutation of both the uORF1 GTG and CTG showed similar results. There was no significant change in mRNA levels among the different reporters, suggesting that the changes in luciferase activity are a result of translational control. These results indicate that uORF1 can act as a moderate enhancer of downstream translation, whereas uORF2 represses CDKN1A translation. Because mutation of uORF initiation codons did not appreciably affect the induction ratio with stress, these results suggest that there are other features of the 5′-leader of the CDKN1A V4 mRNA that are also instrumental in preferential translation by induced eIF2α-P.
DISCUSSION
This study demonstrates that translational control through induced eIF2α-P is required for the keratinocyte response to UVB. Specifically, levels of eIF2α-P increased by UVB enhanced translation of a CDKN1A transcript variant 4, which is required for a G1 cell cycle arrest. Loss of eIF2α-P shifts cell fate away from senescence and toward apoptosis, due to decreased DNA repair as a result of cell cycle deregulation. We also show that translational control of human CDKN1A transcript variant 4 is mediated through its 5′-leader. These results highlight a previously unrecognized mechanism for induced eIF2α-P and translational control in response to UVB, and point toward its potential involvement in diseases initiated by UVB damage, such as NMSCs.
These results provide a comprehensive landscape for genome-wide changes in translation that occur following UVB irradiation. Following exposure to a 100 J/m2 dose of UVB, there is a general repression of both translation and cellular mRNA levels as measured by RNA-Seq, which correlates with previously published results (Deng et al., 2002; Williamson et al., 2017). There is also an increase in mRNA levels, as well as translation, for a collection of genes involved in apoptosis, cell cycle regulation, and stress response. These stress-responsive genes include the ISR effectors ATF4, IBTKα, and GADD34, which we have previously shown to be preferentially translated during stress by a mechanism dependent on induced eIF2α-P (Vattem and Wek, 2004; Baird et al., 2014; Young et al., 2015). Notably, ATF4 transcript levels were slightly increased following irradiation with 100 J/m2 UVB, which contrasts with our previously published report that focused on high, apoptotic UVB doses (600 J/m2) (Collier et al., 2015). Consistent with previous reports, CHOP (DDIT3) mRNA was not induced by UVB (Dey et al., 2010; Collier et al., 2015). Of the cell cycle genes found to be preferentially translated and transcribed following UVB, we focused on CDKN1A, as it is known to be involved in cell cycle and senescence regulation. Of interest, there was no change in most DNA repair genes in the UVB-exposed keratinocytes, including those previously suggested to be under translational regulation (DDB1, ERCC1, and ERCC5; Powley et al., 2009). An explanation for differences in expression of DNA repair enzymes between the earlier report and the present one could be different cell types (HeLa vs. human keratinocytes) and UVB doses (275 vs. 100 J/m2).
In response to UVB, p21 protein expression is prosurvival in that it halts proliferation to allow DNA repair. CDKN1A regulation has previously been characterized at the level of transcription, with p53 being the most prominent regulator (Macleod et al., 1995). A recent study described a GCN2-dependent mechanism for preferential translation of a mouse CDKN1A transcript variant in response to amino acid starvation (Lehman et al., 2015). The 5′-leader of the mouse CDKN1A transcript described in the previous study also featured a uORF, but had no appreciable sequence similarities to human CDKN1A V4. This finding suggests that while there might be major differences between the mouse and human CDKN1A and its alternative splicing, there are mechanisms shared between the mammalian CDKN1A genes and the ISR that provide for regulation of the cell cycle in response to diverse stress conditions. Alternative splicing of human CDKN1A also appears to ensure that the protein is induced even in cases where eIF2α-P does not occur, by allowing transcriptional upregulation of CDKN1A variants without significant 5′-leaders. These data provide a new role for splicing in providing new 5′-leader sequences for selective preferential translation of key proteins regulating the cell cycle and appropriate adaptation to stress.
One of the main drivers of cellular transformation is loss of cell cycle control (Nakanishi et al., 2006). Here we describe a mechanism by which the cell cycle is regulated in response to UVB through preferential translation in the ISR. Our data demonstrate that perturbation of translational control has severe consequences for the fate of the stressed cell, and provide support for the idea that induced eIF2α-P is important in the prevention of cancer initiation. Depending on the cellular context, CDKN1A has been suggested to act as a tumor suppressor or as an oncogene. Induced CDKN1A has also been shown to be required for cellular senescence, which is considered an anticancer response, as it permanently halts proliferation of DNA-damaged cells (Roninson, 2002). On the other hand, senescent cells can contribute to carcinogenesis through paracrine signaling to transformed cells (Krtolica et al., 2001). According to our results, induced eIF2α-P enhances CDKN1A translation in response to either UVB or ER stresses. These findings support the notion that ISR perturbations resulting in either increased or decreased levels of eIF2α-P could play a role in cancer initiation and/or progression.
Our study focuses specifically on the human keratinocyte response to UVB, which is a major risk factor in the formation of NMSCs. High expression of p21 protein in head/neck, esophageal, and oral squamous cell carcinomas has been correlated with poor prognosis, indicating that loss of CDKN1A regulation could promote cancer progression in epithelial cells (Rigberg et al., 1998; Ng et al., 1999; Choi et al., 2003). As highlighted above, although CDKN1A expression is often dysregulated in a variety of cancers, its expression has been shown to be either increased or decreased depending on the cancer and cell type (Abbas and Dutta, 2009). Therefore, therapies targeting CDKN1A have proven problematic (Eastman, 2004), and targeting key signaling factors regulating CDKN1A expression may help circumvent these complexities. Our study provides a rationale for targeting translational control through induced eIF2α-P to prevent the initiation or halt the progression of NMSCs by modulating aberrant CDKN1A expression. A GADD34 inhibitor, salubrinal, has been shown to be effective in mouse models of neurodegenerative disease and osteoporosis (Boyce et al., 2005; Fullwood et al., 2012). Alternatively, ISRIB, which blocks translational control in response to increased levels of eIF2α-P and can improve memory function in mice, could serve as a means to inhibit induced hyperlevels of eIF2α-P in NMSCs (Sidrauski et al., 2013). Taken together, our report demonstrates a mechanism of translational control in human keratinocytes that could be implicated in UVB-mediated carcinogenesis, providing new targets for skin disease therapy.
MATERIALS AND METHODS
Cell culture
N-TERT keratinocytes (Dickson et al., 2000) were cultured in EpiLife media (Invitrogen) supplemented with human keratinocyte growth supplement (HKGS; Invitrogen) and 1000 U penicillin–streptomycin (Roche). Cultured cells were treated with 1 µM thapsigargin, 1 µg/ml doxycycline, 200 nM ISRIB (Tocris Bioscience), or 20 µM actinomycin D (Sigma-Aldrich), as indicated. Stable overexpression of GADD34 in N-TERT cells was carried out as previously described (Collier et al., 2015), and doxycycline was added to the culture media for 24 h prior to experimental setups. CDKN1A, GCN2, and PERK stable knockdowns were generated by shRNA and lentivirus delivery as described previously (Teske et al., 2013) using validated Sigma Mission shRNA clones TRCN0000287091, TRCN0000300850, and TRCN0000262380, respectively. We previously reported knockdown and characterization of these eIF2α kinases using these shRNAs in N-TERT cells (Collier et al., 2017). UVB irradiation of N-TERT keratinocytes was carried out using Philips FS20T12 UVB broadband light sources as described previously (Lewis et al., 2010). An IL1700 radiometer and an SED240 UVB detector (International Light) were used to measure UVB intensity prior to each experiment, for a distance of 8 cm from the light source to the culture dish. Cells were always irradiated in EpiLife media, which eliminates any UVC wavelengths, followed by normal incubation settings (37°C and 5% CO2).
Polysome profiling by sucrose gradient ultracentrifugation
Polysome profiling was performed as described previously (Teske et al., 2011). Polysome analysis used 10–50% sucrose gradients containing 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, and 50 μg/ml cycloheximide. Cells were rinsed twice with an ice-cold phosphate-buffered saline (PBS) solution containing 50 μg/ml cycloheximide and then lysed with 500 μl of cold lysis buffer. Supernatants were applied to the top of the 10% and 50% sucrose gradients, which were then subjected to ultracentrifugation in a Beckman SW41Ti rotor at 40,000 rpm for 2 h at 4°C. Polysome profiles and fractions for each sample were collected with a piston gradient fractionator and a 254-nm ultraviolet monitor with Data Quest software.
Immunoblot analysis
Immunoblots were performed as described previously (Teske et al., 2013; Collier et al., 2015). Antibodies include eIF2α-P (Abcam, Cambridge, UK; #32157), eIF2α total (Scott Kimball, Pennsylvania State University College of Medicine), GADD34 (Proteintech; #10449), p21 and cleaved caspase-3 (Cell Signaling; #2947 and #9661, respectively), β-actin (Sigma-Aldrich; A5441), and cyclin D1 (Millipore; DCS-6).
Cell cycle analysis
S phase labeling was accomplished using a Click-iT EdU Alexa Fluor 488 Imaging Kit (Life Technologies) following manufacturer instructions. N-TERT keratinocytes were synchronized by removing growth factors for 48 h, followed by recovery with growth factors for 8 h. Following UVB treatments as described in the text, cells were labeled with 10 μM EdU for 1 h, followed by termination of EdU labeling by trypsinization. Cells were then fixed with 4% paraformaldehyde, permeabilized, and added to the Click-iT Plus reaction cocktail to detect EdU incorporation according to manufacturer instructions. Following incubation with the reaction cocktail, cells were spun out of solution and resuspended in 500 μl of FxCycle PI/RNase Staining Solution (Life Technologies) in the dark for 30 min. EdU staining and DNA content were measured on an Attune acoustic focusing flow cytometer (Life Technologies) using 530/30 BL1 or 574/26 BL2 filters, respectively. Live cells were gated based on forward (FSC) and side (SSC) scattering light area. For each sample, 100,000 live events were captured and characterized. Appropriate cell populations were calculated using Attune Cytometric software and presented as a percentage of total live cells.
DNA repair and luciferase assays
Thymine dimers were measured in genomic DNA prepared from UVB-treated N-TERT cells as described previously (Loesch et al., 2016). DNA was loaded per well for each sample and bound to a membrane using a vacuum, followed by incubation with thymine dimer antibody (KTM53; Kamaya Biomedical). Levels of DNA repair are represented as changes in thymine dimer densitometry between the time indicated and 15 min post-UVB (no repair). Host cell reactivation was performed by transfecting 1 μg of a PGL3 control plasmid (Promega) alongside 50 ng Renilla transfection control plasmid (Promega) into each well of a six-well dish containing 50,000 cells using Fugene-6. DNA damage was induced by irradiating the PGL3 plasmid with 600 J/m2 UVB. Luciferase assays were performed as described previously (Young et al., 2015).
Cell survival and senescence assays
Senescence-associated β-galactosidase assays were performed using N-TERT cells 72 h after UVB treatment as described previously (Lewis et al., 2008). Caspase-3 specific activity was measured 6 h post-UVB using a synthetic fluorogenic substrate (DEVD-AMC; Alexic Biochemicals) as described previously (Collier et al., 2015). Trypan blue assays were performed using a TC20 automated cell counter (BioRad). A 10-μl cell suspension was added to 10 μl of 0.4% trypan blue dye (BioRad). A sample of 10 μl of the mixture was added to a counting slide and live cells were quantified by the cell counter.
RNA-Seq analysis
Sucrose gradient fractions 3–4 or 5–7 were pooled following polysome profiling. RNA was isolated as described above. Library preparation and RNA sequencing were carried out in the Center for Medical Genomics at the Indiana University School of Medicine. RNA sequencing was performed using Illumina NextSeq 500 with paired-end 75 nucleotide reads. FASTQ files were mapped to the human genome using Tophat (Galaxy version 2.1.0), followed by calculation of FPKM (fragments per kilobase of transcript per million mapped reads) using cufflinks (Galaxy version 2.1.1.0). Differential expression analysis was performed using cuffdiff (Galaxy version 2.2.1.3) on transcript assemblies generated by cufflinks. Plots were generated using cummeRbund (Galaxy version 1.0.1) and edited using Rstudio (version 1.0.143). Scatterplots and pie charts involving changes in total transcript levels included only those genes with significant (p < 0.05) fold changes as determined by cuffdiff. RNA-Seq data are deposited in the Gene Expression Omnibus under the series record number GSE99745.
Measurement of mRNA levels by qPCR
Total mRNA and polysome mRNA levels were measured using the SYBR Green method as previously described (Collier et al., 2015). mRNA levels were normalized to spike-in luciferase RNA control. Primer sequences are as follows (5′- to 3′-):
ATF4 F-TCAAACCTCATGGGTTCTCC, R-GTGTCATCCAACGTGGTCAG;
CCND1 F-GAGGAAGAGGAGGAGGAGGA, R-GAGATGGAAGGGGGAAAGAG;
Firefly luciferase F: CCAGGGATTTCAGTCGATGT; R: AATCTCACGCAGGCAGTTCT;
CDKN1A total F: CAGCAGAGGAAGACCATGTG, R: GGCGTTTGGAGTGGTAGAAA;
CDKN1A V1 F: AGGCACTCAGAGGAGGCGCCA, R: GGTGACAAAGTCGAAGTTCCA;
CDKN1A V4 F: TGTTTCTGCGGCAGGCGCCAT, R: CCGCCATTAGCGCATCACAGT; and
CDKN1A V1 and V4 F: AGCAGCTGCCGAAGTCAGTTC, R: GGACATCCCCAGCCGGTTCTG.
Quantification of CDKN1A transcript variants was carried out using cDNA generated from untreated N-TERT keratinocytes. PCR was performed for 35 cycles with gene-specific primers (CDKN1A V1/V4) using Bullseye Taq Polymerase (MIDSCI). PCR samples were separated by electrophoresis using a 2% agarose gel, and DNA was visualized by staining with ethidium bromide. Representative bands were quantified with ImageJ software.
Luciferase translation reporter assays and plasmid constructs
PTk-CDKN1A-Luc reporters were constructed by inserting a cDNA fragment containing the wild-type or mutated 5′-leader of CDKN1A V1 or V4 using HindIII and NcoI sites between the TK promoter and luciferase CDS in a derivative of PGL3 plasmid (Vattem and Wek, 2004). Fusion constructs were made by inserting cDNA fragments between HindIII and NarI sites on the same PGL3 plasmid, which ensured removal of the luciferase start codon. A sample of 1 μg of each construct and 50 ng of a Renilla transfection control plasmid (Promega) were cotransfected into 50,000 N-TERT keratinocytes for 24 h using Fugene-6 as described previously (Young et al., 2015). Luciferase activity was measured as described previously following treatment with either 100 J/m2 UVB or 0.1 μM TG for 6 h. Changes made in mutated constructs are listed below:
CDKN1A V4 uORF1 ΔATG: AGGATGCGT to AGGAGGCGT;
CDKN1A V4 uORF2 ΔGTG: CTTGTGGAG to CTTGGGGAG;
CDKN1A V4 uORF2 ΔCTG: GAGCTGGGC to GAGCGGGGC.
Statistical analyses
Experimental data are depicted as ± SD, with biological replicates of three or more for each experiment. Differences between paired samples were determined by a Student’s t test. For all figures, p < 0.05 was considered statistically significant and is marked with an asterisk (*), while treatment groups with significant differences are marked with a number sign (#).
Supplementary Material
Acknowledgments
We acknowledge Sheree Wek and David Southern for their technical assistance and support and Jeanette McClintick for her advice with genome-wide analyses. Support for this work was supplied by the National Institute of Environmental Health Sciences (ES020866 to D.F.S. and F31ES026517 to A.E.C.), the National Institute on Aging (AG048946 to D.F.S.), and the National Institute of General Medical Sciences (GM049164 to R.C.W.).
Abbreviations used:
- ATF4
activating transcription factor 4
- CDKN1A
cyclin-dependent kinase inhibitor 1A
- CDS
coding sequence
- DOX
doxycycline
- Edu
5-ethynyl-2’-deoxyuridine
- eIF2α
eukaryotic initiation factor 2 subunit α
- eIF2α-P
phosphorylation of eukaryotic initiation factor 2 subunit α
- GADD34
growth arrest and DNA damage-inducible 34
- GCN2
general control nonderepressible 2
- ISR
integrated stress response
- ISRIB
integrated stress response inhibitor
- NER
nucleotide excision repair
- NMSCs
nonmelanoma skin cancers
- ORF
open reading frame
- PI
propidium iodide
- qPCR
quantitative PCR
- SA β-gal
senescence-associated β-galactosidase
- TG
thapsigargin
- TK
thymidine kinase
- uORF
upstream open reading frame
- UVB
ultraviolet B
- V1
CDKN1A mRNA splice variant 1
- V4
CDKN1A mRNA splice variant 4.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0362) on November 8, 2017.
REFERENCES
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird TD, Palam LR, Fusakio ME, Willy JA, Davis CM, McClintick JN, Anthony TG, Wek RC. Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKalpha. Mol Biol Cell. 2014;25:1686–1697. doi: 10.1091/mbc.E14-02-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J, Mouret S, Ravanat JL, Douki T. Photoinduced damage to cellular DNA: direct and photosensitized reactions. Photochem Photobiol. 2012;88:1048–1065. doi: 10.1111/j.1751-1097.2012.01200.x. [DOI] [PubMed] [Google Scholar]
- Campisi J. Suppressing cancer: the importance of being senescent. Science. 2005;309:886–887. doi: 10.1126/science.1116801. [DOI] [PubMed] [Google Scholar]
- Choi HR, Tucker SA, Huang Z, Gillenwater AM, Luna MA, Batsakis JG, El-Naggar AK. Differential expressions of cyclin-dependent kinase inhibitors (p27 and p21) and their relation to p53 and Ki-67 in oral squamous tumorigenesis. Int J Oncol. 2003;22:409–414. [PubMed] [Google Scholar]
- Collier AE, Wek RC, Spandau DF. Translational repression protects human keratinocytes from UVB-induced apoptosis through a discordant eIF2 kinase stress response. J Invest Dermatol. 2015;135:2502–2511. doi: 10.1038/jid.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier AE, Wek RC, Spandau DF. Human keratinocyte differentiation requires translational control by the eIF2alpha kinase GCN2. J Invest Dermatol. 2017;137:1924–1934. doi: 10.1016/j.jid.2017.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol Cell Biol. 2001;21:6841–6850. doi: 10.1128/MCB.21.20.6841-6850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat A, van Tilburg M, van der Leun JC, van Vloten WA, de Gruijl FR. Cell cycle kinetics following UVA irradiation in comparison to UVB and UVC irradiation. Photochem Photobiol. 1996;63:492–497. doi: 10.1111/j.1751-1097.1996.tb03075.x. [DOI] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Deng J, Harding HP, Raught B, Gingras AC, Berlanga JJ, Scheuner D, Kaufman RJ, Ron D, Sonenberg N. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol. 2002;12:1279–1286. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- D’Errico M, Teson M, Calcagnile A, Proietti De Santis L, Nikaido O, Botta E, Zambruno G, Stefanini M, Dogliotti E. Apoptosis and efficient repair of DNA damage protect human keratinocytes against UVB. Cell Death Differ. 2003;10:754–756. doi: 10.1038/sj.cdd.4401224. [DOI] [PubMed] [Google Scholar]
- Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J Biol Chem. 2010;285:33165–33174. doi: 10.1074/jbc.M110.167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman A. Cell cycle checkpoints and their impact on anticancer therapeutic strategies. J Cell Biochem. 2004;91:223–231. doi: 10.1002/jcb.10699. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Zhou W, Shenolikar S. Targeting phosphorylation of eukaryotic initiation factor-2alpha to treat human disease. Prog Mol Biol Transl Sci. 2012;106:75–106. doi: 10.1016/B978-0-12-396456-4.00005-5. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp Cell Res. 1999;246:280–289. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- Georgakilas AG, Martin OA, Bonner WM. p21: a two-faced genome guardian. Trends Mol Med. 2017;23:310–319. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA. PERK and GCN2 contribute to eIF2alpha phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol Biol Cell. 2005;16:5493–5501. doi: 10.1091/mbc.E05-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000a;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translation regulation and cell survival during the unfolded protein response. Mol Cell. 2000b;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jascur T, Brickner H, Salles-Passador I, Barbier V, El Khissiin A, Smith B, Fotedar R, Fotedar A. Regulation of p21(WAF1/CIP1) stability by WISp39, a Hsp90 binding TPR protein. Mol Cell. 2005;17:237–249. doi: 10.1016/j.molcel.2004.11.049. [DOI] [PubMed] [Google Scholar]
- Jiang HY, Wek RC. Gcn2 phosphorylation of eIF2α activates NF-kB in response to UV irradiation. Biochem J. 2005;385:371–380. doi: 10.1042/BJ20041164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci USA. 2012;109:E2424–E2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YY, Cevallos RC, Jan E. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation. J Biol Chem. 2009;284:6661–6673. doi: 10.1074/jbc.M806735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman SL, Cerniglia GJ, Johannes GJ, Ye J, Ryeom S, Koumenis C. Translational upregulation of an individual p21Cip1 transcript variant by GCN2 regulates cell proliferation and survival under nutrient stress. PLoS Genet. 2015;11:e1005212. doi: 10.1371/journal.pgen.1005212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Travers JB, Somani AK, Spandau DF. The IGF-1/IGF-1R signaling axis in the skin: a new role for the dermis in aging-associated skin cancer. Oncogene. 2010;29:1475–1485. doi: 10.1038/onc.2009.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Yi Q, Travers JB, Spandau DF. UVB-induced senescence in human keratinocytes requires a functional insulin-like growth factor-1 receptor and p53. Mol Biol Cell. 2008;19:1346–1353. doi: 10.1091/mbc.E07-10-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesch MM, Collier AE, Southern DH, Ward RE, Tholpady SS, Lewis DA, Travers JB, Spandau DF. Insulin-like growth factor-1 receptor regulates repair of ultraviolet B-induced DNA damage in human keratinocytes in vivo. Mol Oncol. 2016;10:1245–1254. doi: 10.1016/j.molonc.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- Mandal M, Bandyopadhyay D, Goepfert TM, Kumar R. Interferon-induces expression of cyclin-dependent kinase-inhibitors p21WAF1 and p27Kip1 that prevent activation of cyclin-dependent kinase by CDK-activating kinase (CAK) Oncogene. 1998;16:217–225. doi: 10.1038/sj.onc.1201529. [DOI] [PubMed] [Google Scholar]
- Melnikova VO, Ananthaswamy HN. Cellular and molecular events leading to the development of skin cancer. Mutat Res. 2005;571:91–106. doi: 10.1016/j.mrfmmm.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Nakanishi M, Shimada M, Niida H. Genetic instability in cancer cells by impaired cell cycle checkpoints. Cancer Sci. 2006;97:984–989. doi: 10.1111/j.1349-7006.2006.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng IO, Lam KY, Ng M, Regezi JA. Expression of p21/waf1 in oral squamous cell carcinomas–correlation with p53 and mdm2 and cellular proliferation index. Oral Oncol. 1999;35:63–69. doi: 10.1016/s1368-8375(98)00083-9. [DOI] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 2003;22:1180–1187. doi: 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortolan TG, Menck CF. UVB-induced cell death signaling is associated with G1-S progression and transcription inhibition in primary human fibroblasts. PLoS One. 2013;8:e76936. doi: 10.1371/journal.pone.0076936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palam LR, Baird TD, Wek RC. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem. 2011;286:10939–10949. doi: 10.1074/jbc.M110.216093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powley IR, Kondrashov A, Young LA, Dobbyn HC, Hill K, Cannell IG, Stoneley M, Kong YW, Cotes JA, Smith GC, et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 2009;23:1207–1220. doi: 10.1101/gad.516509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigberg DA, Kim FS, Blinman TA, Cole MA, Lane JS, So J, McFadden DW. p21 expression is increased by irradiation in esophageal squamous cell carcinoma. J Surg Res. 1998;76:137–142. doi: 10.1006/jsre.1998.5308. [DOI] [PubMed] [Google Scholar]
- Roninson IB. Oncogenic functions of tumour suppressor p21(Waf1/Cip1/Sdi1): association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Lett. 2002;179:1–14. doi: 10.1016/s0304-3835(01)00847-3. [DOI] [PubMed] [Google Scholar]
- Scholpa NE, Zhang X, Kolli RT, Cummings BS. Epigenetic changes in p21 expression in renal cells after exposure to bromate. Toxicol Sci. 2014;141:432–440. doi: 10.1093/toxsci/kfu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res. 2008;18:64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, Gamache K, Gallagher CM, Ang KK, Wilson C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife. 2013;2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Porter AC, Ma K, Quilliam LA, Wek RC. Pancreatic eukaryotic initiation factor-2alpha kinase (PEK) homologues in humans, Drosophila melanogaster and Caenorhabditis elegans that mediate translational control in response to ER stress. Biochem J. 2000;346:281–293. [PMC free article] [PubMed] [Google Scholar]
- Teske BF, Baird TD, Wek RC. Methods for analyzing eIF2 kinases and translational control in the unfolded protein response. Methods Enzymol. 2011;490:333–356. doi: 10.1016/B978-0-12-385114-7.00019-2. [DOI] [PubMed] [Google Scholar]
- Teske BF, Fusakio ME, Zhou D, Shan J, McClintick JN, Kilberg MS, Wek RC. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol Biol Cell. 2013;24:2477–2490. doi: 10.1091/mbc.E13-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Williamson L, Saponaro M, Boeing S, East P, Mitter R, Kantidakis T, Kelly GP, Lobley A, Walker J, Spencer-Dene B, et al. UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell. 2017;168:843–855. doi: 10.1016/j.cell.2017.01.019. e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Baird TD, Wek RC. Translation regulation of the glutamyl-prolyl-tRNA synthetase gene EPRS through bypass of upstream open reading frames with noncanonical initiation codons. J Biol Chem. 2016a;291:10824–10835. doi: 10.1074/jbc.M116.722256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Palam LR, Wu C, Sachs MS, Wek RC. Ribosome elongation stall directs gene-specific translation in the integrated stress response. J Biol Chem. 2016b;291:6546–6558. doi: 10.1074/jbc.M115.705640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Wek RC. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J Biol Chem. 2016;291:16927–16935. doi: 10.1074/jbc.R116.733899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Willy JA, Wu C, Sachs MS, Wek RC. Ribosome reinitiation directs gene-specific translation and regulates the integrated stress response. J Biol Chem. 2015;290:28257–28271. doi: 10.1074/jbc.M115.693184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.