Transcriptomic profiling of cells treated with Golgi-disrupting compounds reveals that some target genes including several spliceosome components are controlled by ELK1, GABPA, and ETS1, the activity of which is regulated by MEK/ERK signaling. Furthermore, brefeldin A and golgicide A cause increased splicing of the proapoptotic MCL1-S isoform.
Abstract
The secretory pathway is a major determinant of cellular homoeostasis. While research into secretory stress signaling has so far mostly focused on the endoplasmic reticulum (ER), emerging data suggest that the Golgi itself serves as an important signaling hub capable of initiating stress responses. To systematically identify novel Golgi stress mediators, we performed a transcriptomic analysis of cells exposed to three different pharmacological compounds known to elicit Golgi fragmentation: brefeldin A, golgicide A, and monensin. Subsequent gene-set enrichment analysis revealed a significant contribution of the ETS family transcription factors ELK1, GABPA/B, and ETS1 to the control of gene expression following compound treatment. Induction of Golgi stress leads to a late activation of the ETS upstream kinases MEK1/2 and ERK1/2, resulting in enhanced ETS factor activity and the transcription of ETS family target genes related to spliceosome function and cell death induction via alternate MCL1 splicing. Further genetic analyses using loss-of-function and gain-of-function experiments suggest that these transcription factors operate in parallel.
INTRODUCTION
Perturbations to the homeostatic maintenance of compartments following a specific stress stimulus must be integrated at the cellular level by coordinating responses of and between different organelles to adjust compartmental capacity and overall cellular fitness for appropriate cell fate decisions. Impairment of the function of organelles can lead to the activation of stress-regulated pathways, which, depending on the duration and severity of the stress, can be either adaptive or cell death–promoting. For instance, accumulation of misfolded proteins in the endoplasmic reticulum (ER) lumen stimulates the unfolded protein response (UPR), which initially protects cells against further insult through enhancement of adaptive processes, including increased protein chaperone activity, enhanced ER-associated degradation (ERAD) of terminally misfolded proteins, and reduced protein synthesis, but upon chronic ER stress transforms into a signaling network that favors the elimination of stressed cells (Urra et al., 2013). Observable changes in organelle morphology, size and number, or organelle-specific induction of cell death in response to various external and cell-intrinsic cues suggest that several subcellular structures harbor stress-responsive signaling pathways (Galluzzi et al., 2014; Mast et al., 2015). The ER, mitochondria, peroxisomes, and lysosomes/autophagosomes have evolved signaling cascades leading to transcriptomic changes in response to stimuli to regulate organelle capacity according to need. For instance, accumulation of unfolded proteins in the ER lumen or mitochondria triggers transcriptional responses mediated by the UPR and mtUPR. Similarly, lysosomal stress results in the dephosphorylation and nuclear translocation of transcription factor EB (TFEB), leading to expression changes in lysosomal biogenesis genes (Sardiello et al., 2009; Roczniak-Ferguson et al., 2012; Settembre et al., 2012; Martina et al., 2014). Transcriptional reprogramming also underlies metabolic changes manifesting in malignancy, including cancer development. An example is pancreatic ductal adenocarcinoma, a cancer type in which the family of MITF, TFE3, and TFEB3 transcription factors directs the transcriptional induction of autophagy and lysosome-related genes for maintenance of intracellular amino acid pools (Perera et al., 2015).
Surprisingly, much less is known about whether the Golgi complex may possess a stress-responsive pathway that is triggered by stress cues sensed at or transmitted to the Golgi (Machamer, 2015; Taniguchi and Yoshida, 2017). A number of physiological and pathological conditions have been shown to be associated with changes in the size, subcellular localization, and organization of the Golgi (Sengupta and Linstedt, 2011; Machamer, 2015). For instance, progression through the cell cycle requires the Golgi to dismantle into ministacks at the G2/M phase, a finding that was named “Golgi mitotic checkpoint” (Corda et al., 2012). Thus, a comprehensive description of Golgi stress–regulated gene expression changes and signaling mechanisms will help us to better understand basic processes such as growth or cell cycle progression and might offer great potential for novel therapeutic interventions.
Here, we have used gene expression profiling in response to three Golgi-dispersing compounds: brefeldin A (BFA), golgicide A (GCA), and monensin (MON). Through transcription factor–binding motif enrichment analysis of promoter regions of genes regulated by Golgi stress treatments, we uncovered significant enrichment for DNA-binding motifs of the three ETS transcription factor family members ELK1, ETS1, and GABPA/B, suggesting that they might control a subset of the transcriptional output in response to pharmacological Golgi disruption. Further, we demonstrate that Golgi stress activates MEK1/2 and ERK1/2, which promote cell death and regulate the stability and/or activity of aforementioned ETS factors. We have further investigated the signaling mechanisms underlying the activation changes of these transcription factors and their loss-of-function and gain-of-function effects in response to Golgi stress treatments and found that knockdown of any of the three ETS transcription factors provides resistance to these small molecules, a phenotype further enhanced in double-knockdown cells. Conversely, their overexpression confers sensitivity to Golgi stress. These effects might be related to differential regulation of some splicing factors and ELK1-mediated splicing of MCL1 isoforms in particular. Thus, we have uncovered a novel signaling module in response to Golgi stress involving MEK/ERK leading to increased ELK1, ETS1, and GABPA/B stability/activity and subsequent remodeling of the transcriptome.
RESULTS
Transcriptomics of A549 cells treated with BFA or GCA reveals up-regulation of Golgi apparatus and spliceosome components
We have previously described a signaling cascade switched on by several Golgi disruptors leading to CREB3/Luman-mediated transcriptional up-regulation of ARF4, encoding for a small GTP-binding protein involved in vesicular trafficking (Reiling et al., 2013). To identify global changes in the transcriptional landscape in response to BFA, GCA, or MON, we performed gene expression profiling using cDNA microarrays after 8 or 20 h of small-molecule treatment. The compounds were chosen based on their ability to induce Golgi morphological changes (i.e., dispersal), as illustrated in Figure 1A, and to up-regulate ARF4 (Fujiwara et al., 1988; Pan et al., 2008; Sáenz et al., 2009; van der Linden et al., 2010; Reiling et al., 2013; Ignashkova et al., 2017). All three chemicals can entail ER stress leading to UPR activation at concentrations frequently used in the literature for acute treatment (Moon et al., 2012; Reiling et al., 2013; Yoon et al., 2013; Sandow et al., 2014). For BFA, we used a considerably lower concentration than commonly applied to cells in studies using short-term treatments, because we were interested in determining whether a transcriptional stress response might be triggered due to effects predominantly occurring on the Golgi. Treatment of A549 cells with different concentrations of BFA, GCA, or MON for 8 or 20 h revealed negative dose-dependent effects on Gaussia luciferase reporter protein secretion. The extent of inhibition using the same concentration as for the microarray experiment (Figure 1A) at both time points was strongest with GCA, and comparable between BFA and MON (Supplemental Figure S1).
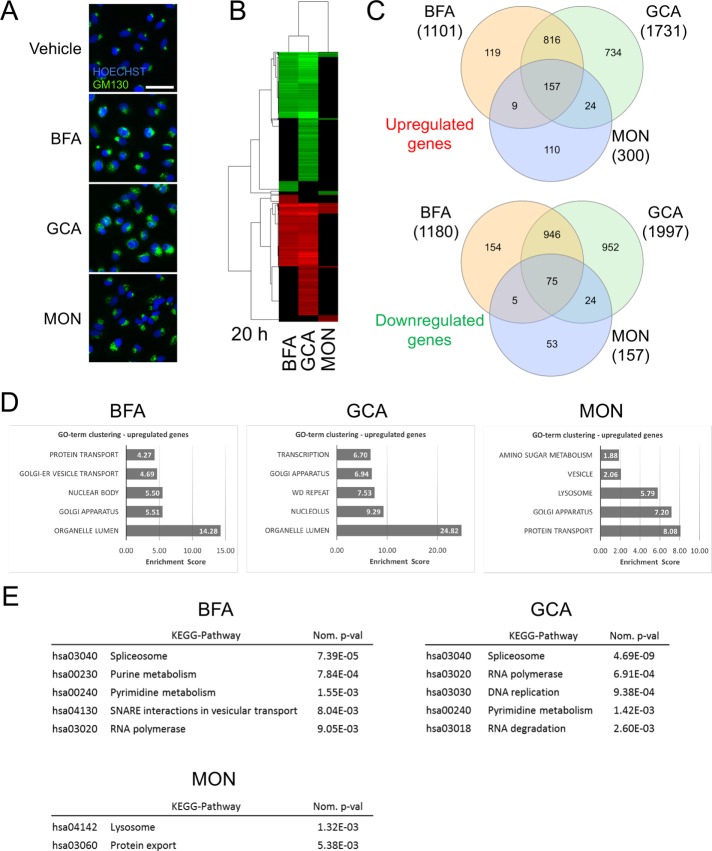
FIGURE 1:
Gene expression profiling of A549 cells treated for 20 h with Golgi stress–inducing compounds. (A) Immunofluorescence images displaying A549 cells treated for 24 h with vehicle, 71 nM (20 ng/ml) BFA, 5 μM GCA, or 10 μM MON (green: anti-GM130, a cis-Golgi marker; blue: Hoechst). Scale bar: 50 μm. (B) Heat-map representation of significantly regulated genes (FDR p value cutoff < 0.05, fold change ≥ |1.5|) by BFA, GCA, or MON used at the concentrations indicated in A. (C) Venn diagram indicating shared genes between the three treatments. Ten genes that are up-regulated by one compound and down-regulated by another are not captured by this representation. (D) GO-term enrichment analysis of significantly regulated genes by BFA, GCA, and MON. The graphs display the top five results of a GO-term clustering analysis using DAVID (see Materials and Methods). (E) KEGG-pathway enrichment analysis of significantly regulated genes by BFA, GCA, and MON (p < 0.05).
We surveyed the drug-induced transcriptional changes at an early (8 h; Supplemental Figure S2 and Supplemental Table S1) and a late (20 h; Figure 1 and Supplemental Table S2) time point. After 8 h of treatment, a substantial number of genes were regulated by GCA, while BFA and MON affected only a relatively small group of genes. For this reason, we focused our analysis on the 20-h time point. The transcriptional profiles of BFA- and GCA-treated cells after 20 h of treatment overlap considerably, which was not unexpected, given that both compounds inhibit the large ARF guanine nucleotide exchange factor GBF1, while MON, which acts as an ionophore that leads to stoichiometric H+/Na+ exchange followed by osmotic swelling and fragmentation of the Golgi (Dinter and Berger, 1998), appears to regulate only some of the genes also affected by the presence of BFA and GCA (Figure 1, B and C). All three treatments influence a significant number of genes related to the GO terms “protein and vesicle transport” and “Golgi apparatus” but lack a significant enrichment of ER stress or UPR-related GO terms (Figure 1D). BFA- and GCA-regulated genes are significantly enriched for members of the KEGG-pathway “spliceosome” and nucleotide metabolism-related processes, while MON shows significant enrichment for the “lysosome” and “protein export” pathways (Figure 1E).
BFA, GCA, and MON up-regulate transcriptional programs orchestrated by ELK1, ETS1, and GABPA/B
To evaluate whether there are hidden commonalities among the three compounds, we performed transcription factor–binding motif enrichment analysis of the promoter regions of significantly regulated genes 2 kb up- and downstream of the transcription start site. Figure 2A shows all enriched binding motifs with a family-wise error rate p value of less than 0.05. All three gene expression profiles show a highly significant enrichment for genes with the binding motif SCGGAAGY_V$ELK1_02, which is recognized by the ETS family transcription factor ELK1. Additionally, BFA and GCA share the significant enrichment of another ELK1-based motif (V$ELK1_02) as well as a significant number of genes regulated by the ETS family transcription factors GABPA/B (MGGAAGTG_V$GABP_B, V$NRF2_01, V$GABP_B) and ETS1 (V$CETS1P54) (Figure 2A). Many of the genes that contribute to this enrichment are regulated by more than one of these transcription factors, as illustrated by the thickness of the green edges in Figure 2B. Because the SCGGAAGY_V$ELK1_02 motif is significantly enriched in BFA-, GCA-, and MON-regulated genes, we further analyzed the entirety of this gene set via GO-term enrichment analysis (Figure 2C). Genes with this target sequence in their promoter region show significant enrichment for GO terms like “Golgi apparatus,” “protein transport,” “protein catabolic process,” and “organelle lumen,” strengthening the notion that this is a secretory stress-responsive gene set.
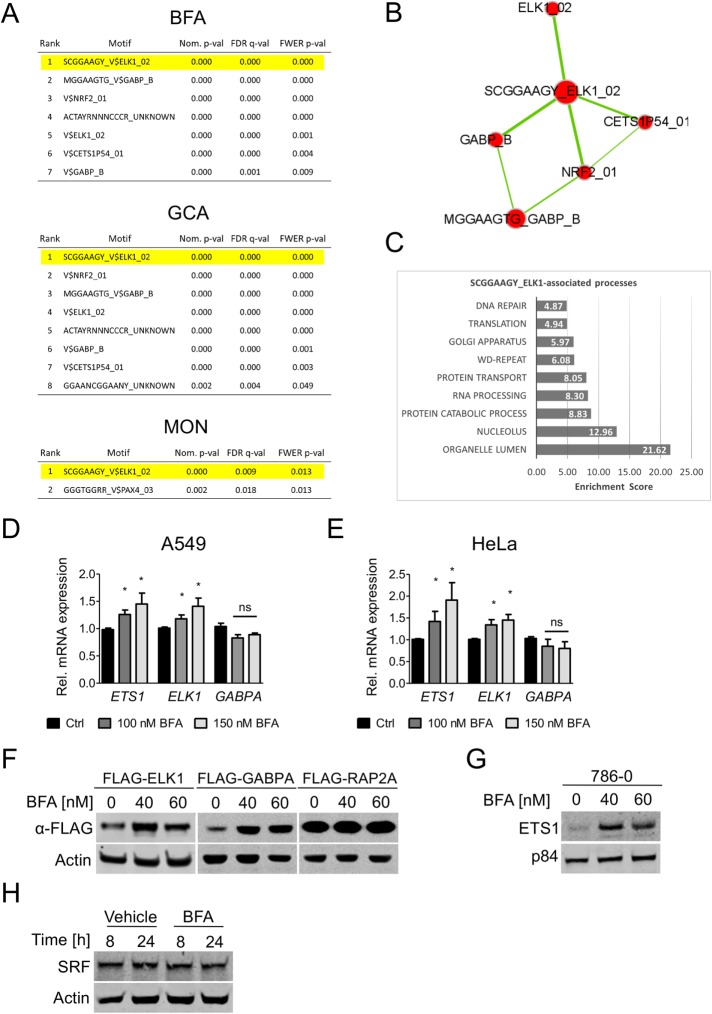
FIGURE 2:
Transcription factor–binding motif enrichment analysis suggests activation of ETS family transcription factors ELK1, ETS1, and GABPA by Golgi stress. (A) Transcription factor–binding motif enrichment analysis of significantly regulated genes by BFA, GCA, and MON. Some p values are 0, because the decimal value exceeds the number of digits in the 64-bit space. (B) Enrichment map representation of the top hits in A. Nodes represent binding motifs; edges represent gene member overlap between the groups. Node size and edge thickness are proportional to the number of genes in each group. (C) GO-term enrichment analysis using all genes in the human genome containing the SCGAAGY_ELK1 binding motif as input. (D, E) ELK1, ETS1, and GABPA mRNA expression was measured by qPCR in A549 (D) and HeLa (E) cells after 24 h BFA treatment. Data are shown as mean and SD of three independent biological replicates; *, p ≤ 0.05, Student’s t test. (F, G) Representative blots of three independent experiments are shown. (F) Western blot of A549 cells stably overexpressing the indicated transcription factors and treated with BFA. (G) Western blot analysis of 786-0 cells treated with BFA. (H) Western blot analysis of SRF expression in A549 cells treated with 60 nM BFA for the indicated duration.
Based on their target gene expression, ELK1, GABPA/B (GABPA encodes the DNA-binding subunit, and GABPB1 or GABPB2 encodes the transactivating subunit of the GABP heterodimer), and ETS1 might play an important role in governing gene expression in response to Golgi stress treatments; however, these transcription factors were not themselves part of the differentially expressed genes in our transcriptomic analysis. A follow-up analysis via quantitative PCR (qPCR) detected a small but significant up-regulation when higher levels of BFA were used (Figure 2, D and E). Nonetheless, these transcription factors seem to rather be regulated at the protein level. Figure 2F demonstrates that, unlike the FLAG-Rap2a control protein, FLAG-tagged constructs of ELK1 and GABPA display enhanced protein levels upon treatment with BFA for 20 h. Interestingly, we were not able to generate stable ETS1-overexpressing cells. Despite multiple attempts at lentiviral infection of A549, HeLa, and MCF7 cells, all cells that survived the selection process had lost ETS1 expression. Because 786-0 cells display higher ETS1 levels, they were used to assess the effects of BFA on the endogenous expression of this transcription factor. In line with the results presented in Figure 2F, endogenous ETS1 levels were up-regulated following BFA treatment (Figure 2G). ELK1 and ETS1 belong to the ETS transcription factor subfamily of ternary complex factors that are known to induce transcription from serum response elements when in complex with another transcription factor, serum response factor (SRF) (Sharrocks, 2001). However, SRF expression was not altered in response to Golgi stress (Figure 2H).
Loss of ELK1, ETS1, or GABPA independently protects cells from BFA, GCA, and MON, while overexpression induces sensitivity
Remarkably, loss of ELK1, ETS1, and GABPA via short hairpin RNA (shRNA)-mediated knockdown protected A549 and HeLa cells from BFA, GCA, or MON (Figure 3, A–F, and Supplemental Figure S3, A–F); however, ELK1 and ETS1 knockdown cells were not resistant to two frequently used ER stress–inducing agents, the N-glycosylation inhibitor tunicamycin or the sarco/endoplasmic reticulum Ca2+ ATPase inhibitor thapsigargin (Supplemental Figure S3, G–J). This suggests that these three ETS transcription factors are predominantly involved in a Golgi stress–induced response, but not necessarily in ER stress–associated pathways. ELK1, ETS1, and GABPA/B are able to initiate transcription either alone or in a coordinated manner with other members of the ETS family, for example, via ELK1/GABPA heterodimers (Odrowaz and Sharrocks, 2012a, b). To evaluate whether these transcription factors depend on one another in response to Golgi stress, we generated ELK1/GABPA and ELK1/ETS1 double-knockdown cell lines and challenged them with BFA and GCA. In both A549 and HeLa cells, an additional protective benefit could be detected when ELK1 knockdown was combined with loss of either GABPA or ETS1, suggesting that ELK1 is operating in parallel to the other two transcription factors (Figure 3, G and H, and Supplemental Figure S3, K–N). Next, we complemented the knockdown studies with gain-of-function experiments. Overexpression of FLAG-ELK1 in A549 cells induced sensitivity to BFA (Figure 4A), similar to the effects seen with FLAG-CREB3 gain of function, which was previously shown to induce BFA sensitivity (Reiling et al., 2013). Overexpression of GABPA or GABPB1 alone did not cause increased sensitivity to Golgi stress (Figure 4, A and B). Only when both components of the heterodimer were coexpressed did cells become sensitized to BFA or GCA (Figure 4B). Congruent with our results using double-knockdown of ELK1/GABPA, expressing ELK1 simultaneously with GABPA and GABPB1 further enhanced the sensitivity to BFA and GCA (Figure 4C). The triple-overexpressing cell line also displayed sensitivity toward thapsigargin but not to the DNA damage–inducer doxorubicin or the second-generation ATP-competitive mTOR inhibitor torin2 (Supplemental Figure S4, A–C), suggesting that ELK1- and GABPA/B-mediated sensitization to cell death may occur in the context of Golgi/secretory pathway stress but not necessarily in response to all stressors.
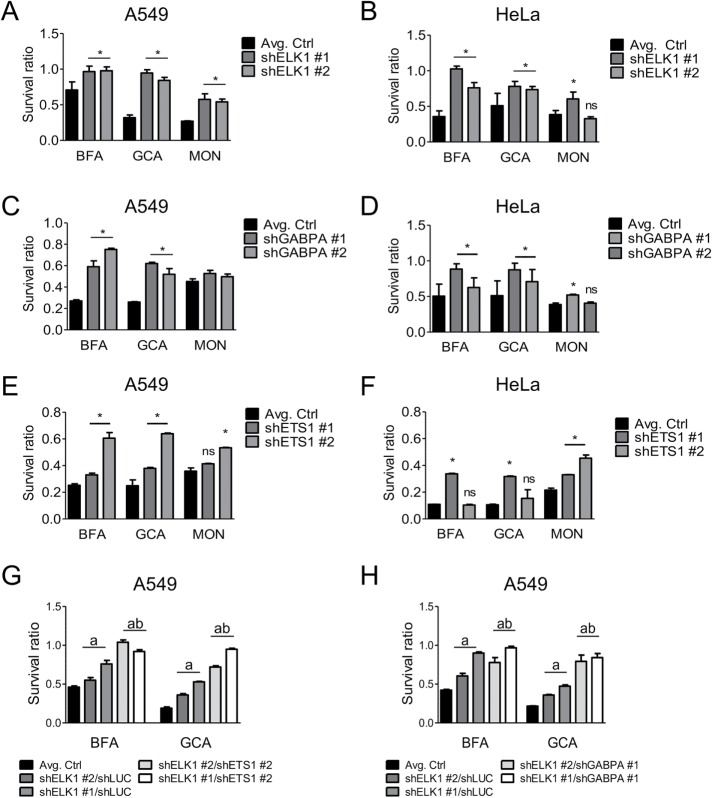
FIGURE 3:
Knockdown of ELK1, ETS1, and GABPA protects A549 and HeLa cells from Golgi stress–induced cell death. A549 (A, C, E) and HeLa (B, D, F) cells were transduced with lentiviral vectors to stably express shRNAs against ELK1, ETS1, or GABPA and subsequently treated with BFA, GCA, or MON. Avg. Ctrl represents the combined average survival ratio of cells infected with either of two separate control hairpins (shLUC and shRFP). (G) Survival ratios of A549 cells stably expressing shRNAs targeting ELK1 and ETS1. (H) Survival ratios of A549 cells stably expressing shRNAs against ELK1 and GABPA. (G, H) a, significant compared with Avg. Ctrl (Avg. Ctrl = shLUC/shLUC); b, significant compared with the corresponding single knockdown; (A, G, H) BFA = 40 nM, GCA = 1.8 µM, MON = 2 µM; (C, E) BFA = 60 nM, GCA = 1.8 µM, MON = 2 µM; (B, D, F) BFA = 25 nM, GCA = 1.8 µM, MON = 0.5 µM. (A–H) Data are shown as mean and SD of one representative experiment measuring six wells per genotype and condition of three independent experiments; *, a, b, p ≤ 0.05, two-way analysis of variance (ANOVA) with Bonferroni posttest.
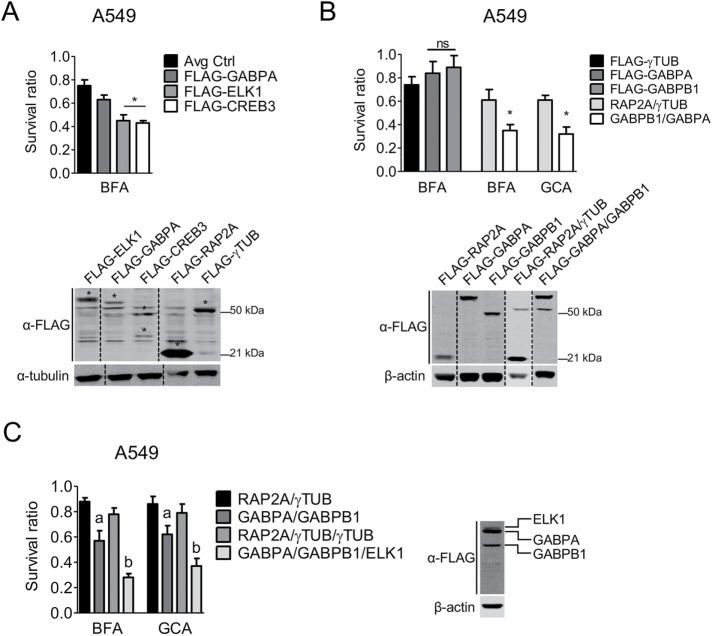
FIGURE 4:
Overexpression of ELK1 and GABPA/B1 leads to sensitivity to Golgi stress–induced cell death. (A) Survival ratios and expression validation of A549 cells stably expressing FLAG-ELK1, FLAG-GABPA, or FLAG-CREB3, treated with 40 nM BFA. Asterisks (*) denote specific overexpression bands. Avg Ctrl refers to the average survival ratio of control cells infected with either FLAG-RAP2A or FLAG-γTUB. (B) Survival ratios and expression validation of A549 cells stably expressing FLAG-GABPA, FLAG-GABPB1, or the combination of both, treated with 40 nM BFA. (C) Survival ratios of A549 cells stably expressing the combination of FLAG-GABPA/FLAG-GABPB1 or FLAG-GABPA/FLAG-GABPB1/FLAG-ELK1, treated with 40 nM BFA. Expression validation shown for FLAG-GABPA/FLAG-GABPB1/FLAG-ELK1, for all other lines refer to A and B; a, significant compared with Avg Ctrl; b, significant compared with the corresponding single/double overexpression. (A–C) Data are shown as mean and SD of one representative example of three independent experiments measuring six wells per genotype and condition; *, a, b, p ≤ 0.05, two-way ANOVA with Bonferroni posttest. Protein lysates were run on the same gel, and dashed lines in blots indicate where unrelated samples were cropped out.
Small molecule inhibition or knockdown of the ELK1 upstream kinases MEK1/2 and ERK1/2 protects cells from BFA and GCA
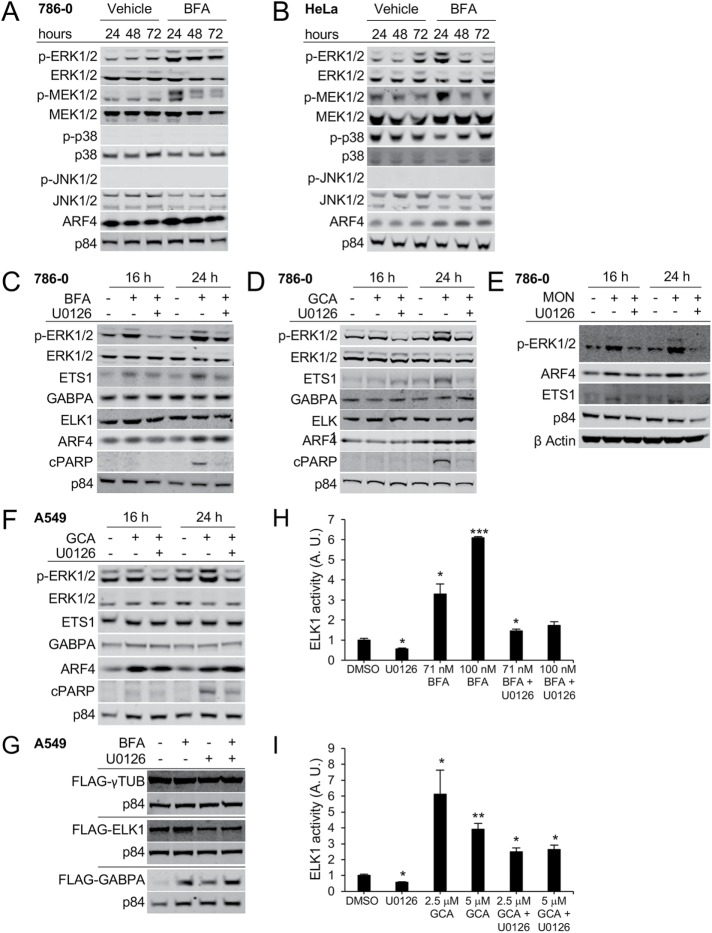
ELK1 is known to be activated by several MAP kinases (MAPKs) such as JNK1/2, p38, or ERK1/2 (Selvaraj et al., 2015). We therefore profiled HeLa and 786-0 cells via immunoblotting to detect these signaling events in response to Golgi stress. Both cell lines showed activation of MEK1/2 and ERK1/2, but not of JNK1/2 or p38, after 24 h of BFA treatment (Figure 5, A and B, and Supplemental Figure S5, A and B). ARF4 has been previously shown to be up-regulated by BFA, GCA, or MON and serves as a marker of Golgi stress induction (Reiling et al., 2013). Similar to BFA treatment, increased MEK/ERK signaling was also detected in both cell lines upon GCA or MON treatment. GCA treatment resembled the effects of BFA, as the highest phospho-ERK1/2 or phospho-MEK1/2 levels were detected after 24 h, which appeared to somewhat decline over the next 48 h. However, phospho-MEK1/2 and phospho-ERK1/2 levels remained steady in response to MON treatment (Supplemental Figure S5, C–F). Blocking MEK-ERK signaling using the MEK inhibitor U0126 prevented the activation of ERK1/2 by BFA, GCA, or MON; ameliorated proapoptotic PARP cleavage; and also reduced Golgi stress–mediated induction of ETS1, FLAG-ELK1, but not FLAG-GABPA (Figure 5, C–G, and Supplemental Figure S5, G–I). To test whether Golgi stress modulates ELK1 activity, and whether this is dependent on MEK/ERK signaling, we performed dual-luciferase reporter assays using a reporter construct containing multimerized ELK1-binding sites fused to luciferase (ELK1-luciferase) and a cotransfected control reporter for normalization of transfection efficiency. At 48 h after transfection of the ELK1-reporter, A549 cells were treated for 20 h with BFA or GCA in the presence or absence of U0126. Both Golgi stressors caused increased ELK1 transcriptional activity, which was substantially diminished upon addition of the MEK inhibitor, suggesting that ERK1/2 controls ELK1 activity in response to BFA or GCA treatment (Figure 5, H and I). An increase in ELK1 activity upon BFA treatment can also be inferred from the fact that BFA-induced up-regulation of the ELK1 target FOSB (ENCODE Project Consortium, 2012) was decreased in ELK1 A549 or HeLa knockdown cells (Supplemental Figure S6, A and B). Similar to pharmacological MEK inhibition, genetic knockdown of ERK1 or ERK2 by lentiviral hairpin transduction of HeLa or A549 cells provided protection against BFA-mediated cell death (Supplemental Figure S6, C and D). In contrast to ETS1, we were unable to detect obvious changes in total endogenous ELK1 and GABPA protein levels in the presence of BFA and/or U0126, which were observed using FLAG-tagged overexpression constructs (compare Figures 5, C, D, and G, and 2F). This could be due to only minor changes in endogenous expression levels that cannot be detected with these antibodies by immunoblotting or due high turnover rate and/or the inherent instability of ELK1 and GABPA.
FIGURE 5:
BFA treatment activates MEK1/2 and ERK1/2, but not p38 or JNK1/2 signaling. (A) 786-0 or (B) HeLa cells were treated with 40 nM BFA for the indicated duration. The indicated proteins were detected by Western blot. (C–E) 786-0 cells were treated for the indicated duration with 40 nM BFA (C), 1.75 µM GCA (D), or 500 nM MON in the presence or absence of the MEK inhibitor U0126, which was used at 10 μM. The specified proteins were detected by immunoblotting; cPARP: cleaved PARP. (A–E) Representative blots of one example out of three independent experiments are shown with the exception of E, where n = 1. (F) A549 cells were treated with 1.75 µM GCA in the presence or absence of the MEK inhibitor U0126 used at 10 μM for the indicated duration. (G) A549 cells stably expressing the indicated constructs were treated with 70 nM BFA in the presence or absence of 10 μM U0126 for 24 h. (H, I) ELK1 activity in response to 20-h treatment with the indicated concentrations of BFA (H) or GCA (I) in the absence or presence of 10 µM U0126 was measured using a dual-luciferase reporter assay. A representative example of two independent experiments (single compound treatment) is shown; the combinatorial treatment was done once. Three wells per condition were analyzed per experiment. A.U. indicates arbitrary units. Asterisks (*) represent significant differences between DMSO and treatment conditions: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Reiling et al. (2013) previously showed that overexpression of the small GTP-binding protein ARF1 protects cells from BFA-mediated Golgi dispersal and cell death. To test whether the observed MEK1/2 and ERK1/2 activation following Golgi stress is influenced by ARF1 expression and the extent of Golgi dispersal, we treated BFA-resistant ARF1-overexpressing cells and control cells with different doses of BFA and monitored ERK1/2 phosphorylation (Supplemental Figure S6E). Phospho-ERK1/2, which was readily induced in control cells treated with 40 or 60 nM BFA, was largely abolished in ARF1-myc overexpression cells. Likewise, ARF1 gain of function caused diminished GRP78 accumulation under the same treatment conditions, suggesting that increased MEK/ERK signaling activity in response to BFA is at least partially ARF1 dependent.
ELK1 regulates spliceosome components and together with ERK1/2 signaling regulates proapoptotic MCL-1 splicing
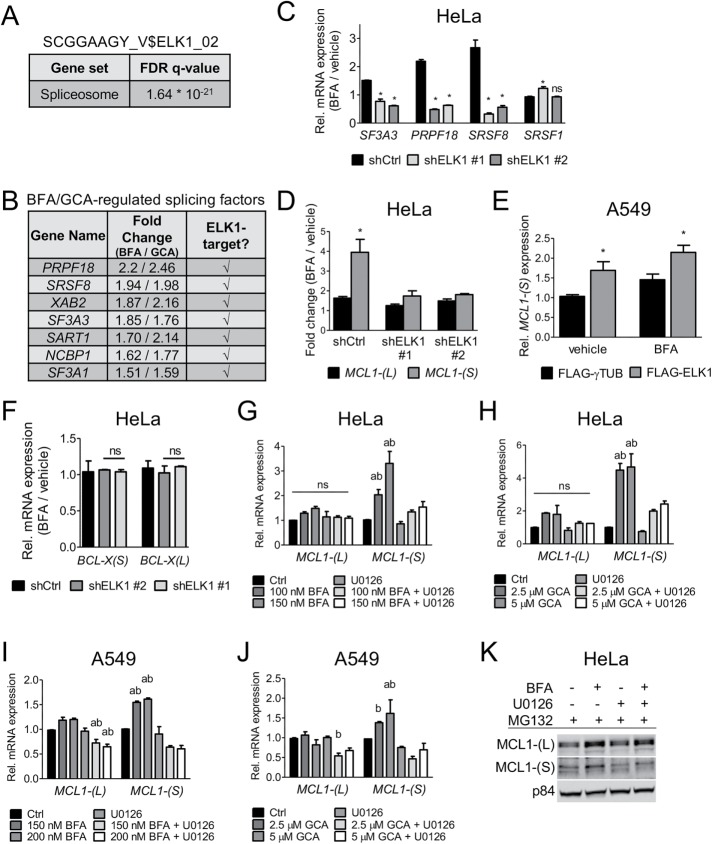
On the basis of the significant enrichment of spliceosome components observed in the microarray analysis (Figure 1E), we hypothesized that ELK1, as the most significant transcription factor predicted to bind to promoter regions of Golgi stress–regulated genes, may be involved in this process. Indeed, when we used the complete list of genes that contain the SCGGAAGY_V$ELK1_02-binding motif (1199 genes) to run a pathway-enrichment analysis, we found a highly significant false discovery rate (q value: 1.64 × 10−21) enrichment of spliceosome components (Figure 6A). Several BFA/GCA-induced spliceosome-associated genes are predicted to be ELK1 targets (Figure 6B). We therefore tested whether the BFA-mediated up-regulation of a chosen subset of these genes is affected when ELK1 is knocked down. Indeed, in the absence of ELK1 expression of SF3A3, PRPF18, and SRSF8, but not SRSF1, the latter of which is not a predicted ELK1 target, was severely abrogated (Figure 6C). Because knockdown of ELK1 protects cells from Golgi stress, we speculated that ELK1 is involved in the generation of a potentially proapoptotic splicing isoform of an apoptosis-regulating protein such as BCL-X or MCL1 (Boise et al., 1993; Bae et al., 2000; Bingle et al., 2000). We have previously demonstrated that BFA induces caspase activity (van Raam et al., 2017) and that BFA-associated cell death can be partially rescued by pan-caspase inhibition, suggesting involvement of apoptotic signaling in response to this compound (Ramírez-Peinado et al., 2017). Indeed, Golgi stress induction resulted in significant up-regulation of the proapoptotic short isoform MCL1-(S), and this up-regulation was prevented by knockdown of ELK1 (Figure 6D). Conversely, cells overexpressing FLAG-ELK1 showed significantly higher MCL1-(S) transcript levels under basal and BFA-treated conditions (Figure 6E). Interestingly, three of the significantly up-regulated spliceosome components in response to BFA/GCA treatment (SF3A1, SF3A3, and SART1) were previously shown to regulate alternative splicing of MCL1, suggesting that this BCL2 family member might be a critical target of the splice factors mentioned earlier that control apoptosis (Moore et al., 2010; Laetsch et al., 2014). We also assessed alternative splicing of BCL-X, which can generate either a long, antiapoptotic isoform [(BCL-X(L)] or a short form [(BCL-X(S)] with proapoptotic properties. BCL-X splicing, however, was unaffected by ELK1 knockdown or BFA treatment (Figure 6F). Our results thus far showed that genetic or chemical inhibition of MEK-ERK signaling reduces apoptotic PARP cleavage and makes cells more resistant to Golgi stress–induced cell death (Figure 5, C–F, and Supplemental Figure S6, C and D). In line with these data, combining U0126 with BFA or GCA prevented the Golgi stress–induced up-regulation of the proapoptotic MCL1-(S) isoform in HeLa and A549 cells, which was also reflected by endogenous MCL1-(S) protein expression (Figure 6, G–K). These data suggest that, upon Golgi disruption, ERK1/2 signaling and ELK1 transcriptional activity are up-regulated to switch MCL1 isoform expression toward the MCL1-(S) splicing variant to promote cell death.
FIGURE 6:
ELK1 expression levels and MEK1/2 activity alter MCL-1 alternative splicing. (A) KEGG-pathway enrichment analysis using all genes that are part of the SCGGAAGY_V$ELK1_02 gene set as input. (B) Excerpt from the microarray data described in Figure 1. List of genes that are targets of ELK1 (based on the gene lists SCGGAAGY_V$ELK1_02 and V$ELK1_02) and also part of the KEGG-pathway “Spliceosome.” Fold changes refer to 20-h 71 nM BFA and 5 μM GCA treatments, respectively. (C) qPCR expression analysis of candidate genes of the gene list shown in A of BFA-treated HeLa ELK1 knockdown cells (50 nM BFA). SRSF1 was chosen as non-ELK1 target-gene control. shCtrl corresponds to shLUC. (D) HeLa cells stably expressing ELK1 or nontargeting shRNAs were treated for 24 h with 50 nM BFA. MCL1 isoform expression was assessed by qPCR. shCtrl corresponds to shLUC. (E) A549 cells stably expressing FLAG-ELK1 or a control protein were treated with 100 nM BFA for 24 h, and MCL1-(S) expression was measured by qPCR. (F) Isoform expression and splicing of BCL-X(L)/BCL-X(S) was evaluated in BFA-treated HeLa ELK1 knockdown cells after 24 h of treatment (100 nM BFA). shCtrl corresponds to shLUC. HeLa (G, H) or A549 (I, J) cells were treated with BFA or GCA alone or in combination with 10 μM U0126. MCL1 isoform expression was measured by qPCR; a, significant compared with Ctrl; b, significant compared with the corresponding single treatment. Ctrl refers to vehicle treatment. (K) MCL1 protein expression analysis by immunoblotting of HeLa cells treated with 10 μM of the proteasome inhibitor MG132 in the presence of either 150 nM BFA, 10 μM U0126, or a combination thereof for 24 h. Representative blots of one example out of three independent experiments are shown. (C, D–J) Data are shown as mean and SD of three independent experiments; *, a, b, p ≤ 0.05, two-way ANOVA with Bonferroni posttest.
DISCUSSION
A surprisingly high number of host factors are implicated in the regulation of Golgi morphology as determined by genetic knockdown experiments (Chia et al., 2012). Nonetheless, the apical molecular mechanisms through which the Golgi senses homeostatic fluctuations to initiate reestablishment of organelle function or cell death are unknown. The Golgi is increasingly recognized as an organelle with important signaling functions (Farhan and Rabouille, 2011). Moreover, dynamic changes to its normal morphology occur in physiological conditions such as cell division or during the differentiation of B-cells into antibody-secreting plasma cells (Kirk et al., 2010) as well as in response to drug treatment and other pathological conditions. Several neurodegenerative diseases, including Alzheimer’s, amyotrophic lateral sclerosis, corticobasal degeneration, spinocerebellar ataxia type 2, and Creutzfeldt-Jacob disease, are associated with Golgi fragmentation (Gonatas et al., 2006; Fan et al., 2008). Likewise, bacterial pathogens, including Chlamydia, actively cause Golgi dispersal as a means of increased lipid acquisition (Heuer et al., 2009). Intriguingly, certain cancer cells with increased metastatic capabilities show an extended Golgi apparatus phenotype (Halberg et al., 2016). Other stressful conditions causing Golgi morphology rearrangements are metabolic stress, hypoxia, or cell senescence (Mukhopadhyay et al., 2006; Joshi et al., 2015; Machamer, 2015). Field and colleagues (Farber-Katz et al., 2014) reported that DNA damage triggers an adaptive nucleus-to-Golgi directed signaling module composed of DNA-activated protein kinase (DNA-PK) and the Golgi oncoprotein GOLPH3, thereby causing fragmentation of the Golgi complex. Stress-signaling pathways in general represent potential targets for therapeutic intervention in many disease processes. Morphological Golgi alterations, as mentioned earlier, are likely to be at least partially mediated through transcriptional changes leading to the remodeling of the organelle. Therefore, pharmacological disruption of the Golgi could be envisioned as a proxy to mimic disease-induced Golgi dispersal and could further be used to inform about transcriptome changes present in certain pathologies associated with Golgi dispersal or to obtain new insights for novel therapeutic intervention opportunities. For instance, we previously identified the small GTPase ARF4 not only as critical mediator of BFA-induced toxicity, but also as an important host factor for survival and growth of pathogenic bacteria that induce Golgi morphology alterations, suggesting that genetic modulators of chemically induced Golgi disorganization could potentially be explored in more clinically relevant settings (Reiling et al., 2013). Still, pharmacological Golgi-dispersing drugs such as those used in this study induce nonphysiological responses, and a future challenge will be to uncover those genes and processes shared between compound-induced and stress- or disease-associated Golgi morphology aberrations.
To begin to understand which genes are differentially regulated in response to Golgi stress and through which factors and signaling cascades these transcriptomic changes are governed, we used cDNA microarrays to study the gene expression responses of A549 cells at two different time points (8 and 20 h) after treatment with either of three classical Golgi-fragmenting agents, BFA, GCA, or MON. Interestingly, in our experimental system, the gene profiles induced by these small molecules did not reveal an obvious UPR signature nor did connectivity mapping reveal an overrepresentation of ER stress–inducing chemicals (Gendarme et al., 2017), suggesting that the gene expression changes we observed reflect effects only distantly related to ER stress signaling. However, we want to point out that some UPR components were identified in our study as well, which is not unexpected, given the close physical and functional relationship between the ER and Golgi apparatus. For instance, compromising Golgi homeostasis might lead to the accumulation of unfolded proteins in the endomembrane system, which as a consequence might trigger the UPR in the ER. The overlap of differentially regulated mRNAs was substantially bigger between BFA and GCA than between MON and BFA or GCA, which was not surprising, given their similar modes of action. Through promoter analysis of all significantly regulated genes (FDR p value < 0.05), we found an enrichment of ELK1-binding motifs after treatment with any of the three compounds and with GABPA/B as well as ETS1 in response to BFA and GCA. These motifs were predominantly found in up-regulated genes. In agreement with this notion, Golgi stress treatment causes the stabilization of ETS1, ELK1, and GABPA. At least for ETS1 and FLAG-ELK1, BFA-induced stabilization is MEK1/2 dependent (Figure 5, C, D, and G). Indeed, another important finding of this study is that BFA, GCA, and MON treatment causes increased MEK1/2- and ERK1/2-phosphorylation, which in turn leads to enhanced protein stability of the ETS transcription factors. Dual-luciferase assays using a genetic reporter for ELK1 activation furthermore corroborate that BFA and GCA induce increased ELK1 activity in a MEK/ERK-dependent manner. Interestingly, lentiviral-mediated knockdown of ERK1 or ERK2 was associated with cellular resistance to BFA similar to ELK1/GABPA/ETS1 depletion. Thus, it appears that, in the context of BFA/GCA-induced Golgi stress, MEK/ERK signaling mediates cell death–promoting signals (see also the model presented in Supplemental Figure S7). Therefore, it appears that increased ERK signaling, generally considered to be a prosurvival pathway, under certain stressful conditions, including Golgi stress, can also induce cell death, which could be related to the production of reactive oxygen species that promote sustained ERK activation (Cagnol and Chambard, 2010).
The importance of ELK1, GABPA/B, and ETS1 in the regulation of cell survival following Golgi stress is highlighted by their loss-of-function and gain-of-function phenotypes. Single knockdown or double-knockdown combinations of these three ETS family members were associated with increased resistance to BFA/GCA, whereas overexpression sensitized cells to the compound treatment. Because we observed additive effects on survival when ELK1 and GABPA were simultaneously depleted, it is likely that these two factors regulate an independent cohort of genes in addition to common targets, ultimately regulating a similar biological process, as has also previously been observed (Odrowaz and Sharrocks, 2012b). These findings suggest that ELK1, GABPA/B, and ETS1 direct a transcriptional program, which is proapoptotic in nature when combined with Golgi stress inducers. ELK1, GABPA/B, and ETS1 belong to the ETS (E26 transformation-specific) protein transcription factor family comprising 28 members in humans (Hollenhorst et al., 2011), all of which contain the conserved ETS DNA-binding domain. ETS transcription factors are involved in cell differentiation, proliferation, apoptosis, development, and tissue remodeling. Given the highly conserved DNA-binding domains, it is only poorly understood how individual ETS family members elicit specific and diverse biological effects. One possibility could be that specificity might depend on cooperative interactions with other cofactors and transcriptional regulators. Importantly, ETS transcription factors are downstream nuclear targets of RAS/MAPK signaling and play an important role in transformation of cells. Deregulation of ETS expression can be brought about by different means, including chromosomal rearrangements, as exemplified in Ewing’s sarcoma and leukemia resulting in neoplastic phenotypes (Kar and Gutierrez-Hartmann, 2013). Interestingly, the chimeric oncoprotein EWS/FLI, composed of the RNA-binding domain of EWS and the DNA-binding domain of the ETS family member FLI, which can be found in ∼85% of Ewing’s sarcomas, affects pre-mRNA splicing coinciding with its transformation ability (Knoop and Baker, 2000, 2001). Through gene set enrichment analysis (GSEA) of KEGG pathways, we found components of the “spliceosome,” of which seven factors (PRPF18, SRSF8, XAB2, SF3A3, SART1, NCBP1, and SF3A1) are predicted ELK1 targets, to be significantly overrepresented upon BFA/GCA treatment, suggesting that these Golgi disruptors might induce differential splicing profiles. The generation of the proapoptotic MCL-1(S) isoform in response to Golgi stress, an event that is largely suppressed in ELK1 knockdown cells or in cells treated with the specific MEK1/2 inhibitor U0126 (Supplemental Figure S7), appears to be of particular importance in this context with respect to cell survival. ELK1 was previously reported to regulate spliceosome components (Boros et al., 2009) and has been implicated in the regulation of splicing events in colon cancer (Hollander et al., 2016).
Considering that deregulation of some ETS transcription factors by gene fusion events, overexpression, or modulation of RAS/MAPK signaling can lead to malignancy, including proliferation, invasion, metastasis, and angiogenesis, our findings may suggest that treatment of such tumors with compounds that mimic BFA’s mode of action or that induce a similar gene expression profile could prove beneficial as an alternative treatment strategy to target cancer cells.
MATERIALS AND METHODS
Cell culture and reagents
All cell lines described were grown in high-glucose DMEM (Life Technologies) with 10% heat-inactivated fetal bovine serum (FBS) in the presence of 100 U/ml penicillin and 100 g/l streptomycin (Life Technologies). Compounds were obtained from the following companies: BFA (Sigma-Aldrich), GCA (Santa Cruz Biotechnology), MON (Enzo Life Sciences), tunicamycin (Santa Cruz Biotechnology), thapsigargin (Santa Cruz Biotechnology), doxorubicin (Sigma-Aldrich), U0126 (Sigma-Aldrich), and torin2 and MG132 (Selleck Chemicals). The ELK1-luciferase reporter construct (pELK1-Luc, cat. # LR-2061) was purchased from Signosis (USA, distributed by BioCat, Germany).
Viability assays
For viability assays, cells with diameters between 10 and 30 µm were counted using a Z2 Coulter Counter (Beckman Coulter). Cell number (typically 1500 cells per well, in 96-well plates) was adjusted to the desired concentration using fresh DMEM before cells were plated in 96-well plates (Greiner Bio One) and allowed to adhere overnight. After incubation for the desired time under the treatment conditions specified for each experiment, Cell Titer Blue viability stain (Promega) was used to quantify live cells. The fluorescent end product resurufin was measured using a Glomax Multi Detection plate reader (Promega), and all values were normalized to the appropriate vehicle control. Unless otherwise indicated in the figure legends, survival ratios were obtained after 72 h of treatment.
Gaussia luciferase assay
A549 cells stably overexpressing GLuc-Flag were seeded at a density of 5000 cells per well in 96-well assay plates and treated in fresh medium 24 h afterward with indicated concentrations of BFA, GCA, or MON for 8 or 20 h. Culture supernatant samples (50 µl) were taken at the indicated time points and transferred to a white, opaque 96-well plate. Then, 20 µl freshly-prepared Gaussia luciferase flash assay reagent (Pierce, Thermo Scientific) was added and the luminescent signal was read after a 10-s integration time.
Dual-luciferase ELK1 reporter assay
A549 cells were plated at a density of 50,000 cells per well in 12-well clusters in triplicate and reverse-transfected with 200 ng ELK1-Luc (firefly) and 4 ng Renilla luciferase per well using TransIT-LT1 as the transfection reagent (Mirus, Germany). At 48 h after transfection, cells were treated with indicated concentrations of BFA, GCA, U0126, or combinations thereof for 20 h in fresh DMEM containing 0.5% heat-inactivated fetal serum. Cells were lysed in 100 µl of provided 1X Passive Lysis Buffer. A dual-luciferase reporter assay was performed according to the provided standard protocol (Promega). In short, 50 µl of LARII buffer was added to 20 µl of cell lysates for the first luminescence readout (ELK1-Luc), followed by addition of 50 µl Stop & Glo reagent for the second readout (CMV-Renilla). For normalization of differences in transfection efficiency, the ratio derived from the luminescence values of Elk1-Luc divided by those of CMV-Renilla luciferase was calculated for every treatment condition, and ELK1 transcription factor activity was quantified relative to dimethyl sulfoxide (DMSO; vehicle).
Microarray analysis
A549 cells were treated for 8 or 20 h using 20 ng/ml BFA (∼71 nM BFA), 5 µM GCA, 10 µM MON, or the appropriate vehicle control (BFA/EtOH, MON/EtOH, GCA/DMSO). Total RNA was harvested using the RNeasy Mini Kit (Qiagen). The RNA was processed for hybridization to Affymetrix Human Gene 2.0 ST arrays using the Ambion WT Expression kit (Thermo Fisher Scientific). The resulting CEL files were analyzed for quality using Affymetrix Expression Console software and were imported into Affymetrix Transcription Analysis Console 2.0, where the data were quantile normalized using robust multiarray averaging (RMA) and baseline transformed to the median of the control samples.
The resulting entity list was subjected to a t test with Benjamini-Hochberg FDR correction. The data files have been deposited in Array Express (www.ebi.ac.uk/arrayexpress, accession number: E-MTAB-5627). GO-term and KEGG-pathway enrichment analyses were performed using the DAVID bioinformatics resource (Huang et al., 2009a, b). Transcription factor–binding motif enrichment analyses were performed using GSEA software from the Broad Institute using MSigDB, version 5.0 (Mootha et al., 2003; Subramanian et al., 2005). Venn diagrams were drawn using interactivenn (Heberle et al., 2015). Cluster and Java Tree View were used to generate the heat maps. The diagram in Figure 2B was generated using Cytoscape (Shannon, 2003) and the enrichment map plug-in (Merico et al., 2010).
Plasmids and cloning
Cloning of Flag-CREB3, Flag-γTubulin, and Flag-Rap2a was described previously (Reiling et al., 2011, 2013). For generation of Flag-tagged fusions of ELK1, GABPA, GABPB1, and ETS1, PCRs were performed with the primer sequences listed below. For cloning, the following cDNA pools were used as templates: a cDNA pool from A549 cells for ELK1; a cDNA pool from U251 cells for GABPB1 and ETS1; and a mixed cDNA pool from several cell lines for GABPA. After sequence verification, the inserts were ligated into the lentiviral vectors pLJM60 or pLJM61.
Primer sequences:
ELK1/SalI: gtcgacGATGGACCCATCTGTGACGC
ELK1/NotI: gcggccgcTCATGGCTTCTGGGGCCCTGG
GABPA/SalI: gtcgacCATGACTAAAAGAGAAGCAG
GABPB1/NotI: gcggccgcTCAATTATCCTTTTCCGTTTGC
GABPB1/SalI: GTCGACgATGTCCCTGGTAGATTTGG
GABPA/NotI: gcggccgcTTAAACAGCTTCTTTATTAGTCTG
ETS1/SalI: gtcgacAATGAGCTACTTTGTGGATTC
ETS1/NotI: gcggccgcTCACTCGTCGGCATCTGGC
Virus production and generation of stable cell lines
HEK293T were seeded at a density of 800 × 103 cells in 6-cm dishes at 24 h before transfection. Plasmids encoding ΔVPR and pCG (VSV-G envelope protein expression vector) and 1 µg of shRNA or overexpression vector were transfected into HEK293T cells using LT1 reagent transfection (Mirus) at a ratio of 3:1. At 12 h posttransfection, media was changed to GlutaMAX (Invitrogen) media plus 30% IFS + 5 mM l-glutamine. Lentiviral supernatants were collected after 48 h. Virus-containing medium was filtered to remove cellular debris, and aliquots were frozen. For lentiviral infection, cells were plated at a density of 150 × 103 cells in 6-cm dishes and allowed to adhere overnight. The culture medium was then replaced by 3 ml DMEM containing 10% IFS supplemented with 8 μg/ml polybrene (Sigma Aldrich). For shRNAs, 200 µl (A549) or 400 µl (HeLa) of viral supernatant was added to the media. For overexpression, 3 ml of polybrene-supplemented viral supernatant was added to the cells. Infected cells were selected 24 h later, using 2 μg/ml puromycin (VWR), 350 µg/ml hygromycin (VWR), and/or 1 mg/ml G418, Geneticin (VWR).
Western blotting
For immunoblotting, experiments were carried out in DMEM containing 0.5 % FBS and were processed using standard protocols. Cells were lysed in RIPA buffer containing protease and phosphatase inhibitors (Roche Applied Science), proteins were resolved by SDS–PAGE on 4–12 % NuPAGE Novex gradient Bis-Tris gels (Thermo Fisher Scientific) and transferred to polyvinylidene fluoride membranes (Immobilon). Membranes were blocked in Li-Cor Odyssey Blocking Buffer (Li-Cor). Primary antibody incubation was performed overnight at 4°C in blocking buffer containing 0.1 % Tween. Proteins were visualized using species-specific far-infrared dye coupled secondary antibodies (Li-Cor) on a Li-Cor Odyssey Sa scanner using Image Studio software (Li-Cor). The following primary antibodies were used: p84 (1:1000, GTX70220 Genetex), GAPDH (1:1000, GTX85118 Genetex), ETS1 (1:1000, #6258 Cell Signaling), cleaved PARP (1:750, #5625 Cell Signaling), β-actin (1:15000, #3700 Cell Signaling), FLAG (1:1000, #2368 Cell Signaling), GABPA (1:1000, sc-22810 Santa Cruz), ERK1/2 (1:1000, #9107 Cell Signaling), p-ERK1/2 (1:2000, #4370 Cell Signaling), MEK1/2 (1:1000, #8727 Cell Signaling), p-MEK1/2 (1:1000, #9154 Cell Signaling), p38 (1:1000, #8690 Cell Signaling), p-p38 (1:1000, #9216 Cell Signaling), JNK1/2 (1:1000, #9252 Cell Signaling), p-JNK1/2 (1:1000, #9255 Cell Signaling), AKT (1:1000, #2920 Cell Signaling), p-AKT (1:1000, #4060 Cell Signaling), GRP78 (1:1000, #3177 Cell Signaling), myc-tag (1:1000, #2276 Cell Signaling), MCL1 (1:1000, #5453 Cell Signaling), and ARF4 (1:10000, 11673-1-AP Proteintech).
qPCR
Cells were grown in 6-cm dishes, and mRNA was isolated using the RNeasy Plus Mini kit (Qiagen). One microgram of total RNA was used for the reverse transcription (RT) reaction using Oligo dT primers and Superscript III (Invitrogen Life Sciences). cDNA was diluted 1:15 after reverse transcription for use in the quantitative real-time PCR (qPCR). The qPCR was performed using QuantiNova SYBR Green Master Mix (Qiagen) in a Rotor-Gene Q (Qiagen) machine.
Primer and shRNA sequences
36B4_fwd: CAGCAAGTGGGAAGGTGTAATCC
36B4_rev: CCATTCTATCATCAACGGGTACAA
ELK1_fwd: CAGCCAGAGGTGTCTGTTACC
ELK1_rev: GAGCGCATGTACTCGTTCC
ETS1_fwd: GATAGTTGTGATCGCCTCACC
ETS1_rev: GTCCTCTGAGTCGAAGCTGTC
GABPA_fwd: TTGGCAAGTCAAGAACAACAGA
GABPA_rev: GCGCTCTTTGTACTTTGGCT
MCL1-(L)_fwd: GTGCCTTTGTGGCTAAACACT
MCL1-(L)_rev: AGTCCCGTTTTGTCCTTACGA
MCL1-(S)_fwd: GGCCTTCCAAGGATGGGTTT
MCL1-(S)_rev: ACTCCAGCAACACCTGCAAAA
BCL-X(L)_fwd: GCGTGGAAAGCGTAGACAAG
BCL-X(L)_rev: AAAAGTATCCCAGCCGCCGT
BCL-X(S)_fwd: TCCCCATGGCAGCAGTAAAG
BCL-X(S)_rev: TCCACAAAAGTATCCTGTTCAAAGC
SF3A3-fwd: GTCATGGCTAAAGAGATGCTCAC
SF3A3_rev: TCCTCCTTTCGTAATCCATCCTT
PRPF18_fwd: ACCTATGACGCTTTCTAGGCA
PRPF18_rev: TCTTATCCAAGGCTGCTTTCAAA
SRSF8_fwd: ATAGCCGGTCTCCCTACAGC
SRSF8_rev: GATCCGCCGTAGCGAGATTC
SRSF1_fwd: CCGCAGGGAACAACGATTG
SRSF1_rev: GCCGTATTTGTAGAACACGTCCT
FOSB_P1_forw: GCTGCAAGATCCCCTACGAAG
FOSB_P1_rev: ACGAAGAAGTGTACGAAGGGTT
MISSION shRNAs purchased from Sigma Aldrich:
shELK1 #1: TRCN0000237874
shELK1 #2: TRCN0000237876
shETS1 #1: TRCN0000231917
shETS1 #2: TRCN0000231919
shGABPA #1: TRCN0000235696
shGABPA #2: TRCN0000235695
Supplementary Material
Acknowledgments
We thank Cornelia Wirth for excellent technical assistance.
Abbreviations used:
- BFA
brefeldin A
- DMSO
dimethyl sulfoxide
- DNA-PK
DNA-activated protein kinase
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- FBS
fetal bovine serum
- GCA
golgicide A
- GSEA
gene set enrichment analysis
- MAPK
MAP kinase
- MON
monensin
- shRNA
short hairpin RNA
- SRF
serum response factor
- TFEB
transcription factor EB
- UPR
unfolded protein response.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0418) on November 8, 2017.
REFERENCES
- Bae J, Leo CP, Yu Hsu S, Hsueh AJW. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J Biol Chem. 2000;275:25255–25261. doi: 10.1074/jbc.M909826199. [DOI] [PubMed] [Google Scholar]
- Bingle CD, Craig RW, Swales BM, Singleton V, Zhou P, Whyte MKB. Exon skipping in Mcl-1 results in a Bcl-2 homology domain 3 only gene product that promotes cell death. J Biol Chem. 2000;275:22136–22146. doi: 10.1074/jbc.M909572199. [DOI] [PubMed] [Google Scholar]
- Boise LH, González-García M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nuñez G, Thompson CB. Bcl-X, a Bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- Boros J, Donaldson IJ, O’Donnell A, Odrowaz ZA, Zeef L, Lupien M, Meyer CA, Liu XS, Brown M, Sharrocks AD. Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res. 2009;19:1963–1973. doi: 10.1101/gr.093047.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnol S, Chambard JC. ERK and cell death: mechanisms of ERK-induced cell death—apoptosis, autophagy and senescence. FEBS J. 2010;277:2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- Chia J, Goh G, Racine V, Ng S, Kumar P, Bard F. RNAi screening reveals a large signaling network controlling the Golgi apparatus in human cells. Mol Syst Biol. 2012;8:629. doi: 10.1038/msb.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corda D, Barretta ML, Cervigni RI, Colanzi A. Golgi complex fragmentation in G2/M transition: an organelle-based cell-cycle checkpoint. IUBMB Life. 2012;64:661–670. doi: 10.1002/iub.1054. [DOI] [PubMed] [Google Scholar]
- Dinter A, Berger EG. Golgi-disturbing agents. Histochem Cell Biol. 1998;109:571–590. doi: 10.1007/s004180050256. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Hu Z, Zeng L, Lu W, Tang X, Zhang J, Li T. Golgi apparatus and neurodegenerative diseases. Int J Dev Neurosci. 2008;26:523–534. doi: 10.1016/j.ijdevneu.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Farber-Katz SE, Dippold HC, Buschman MD, Peterman MC, Xing M, Noakes CJ, Tat J, Ng MM, Rahajeng J, Cowan DM, et al. DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell. 2014;156:413–427. doi: 10.1016/j.cell.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H, Rabouille C. Signalling to and from the secretory pathway. J Cell Sci. 2011;124:171–180. doi: 10.1242/jcs.076455. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Yokotas S, Takatsukig A, Ikeharan Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem. 1988;263:18545–18552. [PubMed] [Google Scholar]
- Galluzzi L, Bravo-San Pedro JM, Kroemer G. Organelle-specific initiation of cell death. Nat Cell Biol. 2014;16:728–736. doi: 10.1038/ncb3005. [DOI] [PubMed] [Google Scholar]
- Gendarme M, Baumann J, Ignashkova TI, Lindemann RK, Reiling JH. Image-based drug screen identifies HDAC inhibitors as novel Golgi disruptors synergizing with JQ1. Mol Biol Cell. 2017;28:3756–3772. doi: 10.1091/mbc.E17-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatas NK, Stieber A, Gonatas JO. Fragmentation of the Golgi apparatus in neurodegenerative diseases and cell death. J Neurol Sci. 2006;246:21–30. doi: 10.1016/j.jns.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Halberg N, Sengelaub CA, Navrazhina K, Molina H, Uryu K, Tavazoie SF. PITPNC1 Recruits RAB1B to the Golgi network to drive malignant secretion. Cancer Cell. 2016;29:339–353. doi: 10.1016/j.ccell.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a Web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer D, Rejman Lipinski A, Machuy N, Karlas A, Wehrens A, Siedler F, Brinkmann V, Meyer TF. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature. 2009;457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- Hollander D, Donyo M, Atias N, Mekahel K, Melamed Z, Yannai S, Lev-Maor G, Shilo A, Schwartz S, Barshack I, et al. A network-based analysis of colon cancer splicing changes reveals a tumorigenesis-favoring regulatory pathway emanating from ELK1. Genome Res. 2016;26:541–553. doi: 10.1101/gr.193169.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Lempicki RA, Sherman BT. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009a;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignashkova TI, Gendarme M, Peschk K, Eggenweiler H-M, Lindemann RK, Reiling JH. Cell survival and protein secretion associated with Golgi integrity in response to Golgi stress-inducing agents. Traffic. 2017;18:530–544. doi: 10.1111/tra.12493. [DOI] [PubMed] [Google Scholar]
- Joshi G, Bekier ME, Wang Y. Golgi fragmentation in Alzheimer’s disease. Front Neurosci. 2015;9:340. doi: 10.3389/fnins.2015.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar A, Gutierrez-Hartmann A. Molecular mechanisms of ETS transcription factor-mediated tumorigenesis. Crit Rev Biochem Mol Biol. 2013;48:522–543. doi: 10.3109/10409238.2013.838202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk SJ, Cliff JM, Thomas JA, Ward TH. Biogenesis of secretory organelles during B cell differentiation. J Leukoc Biol. 2010;87:245–255. doi: 10.1189/jlb.1208774. [DOI] [PubMed] [Google Scholar]
- Knoop LL, Baker SJ. The splicing factor U1C represses EWS/FLI-mediated transactivation. J Biol Chem. 2000;275:24865–24871. doi: 10.1074/jbc.M001661200. [DOI] [PubMed] [Google Scholar]
- Knoop LL, Baker SJ. EWS/FLI alters 5′-splice site selection. J Biol Chem. 2001;276:22317–22322. doi: 10.1074/jbc.M008950200. [DOI] [PubMed] [Google Scholar]
- Laetsch TW, Liu X, Vu A, Sliozberg M, Vido M, Elci OU, Goldsmith KC, Hogarty MD. Multiple components of the spliceosome regulate Mcl1 activity in neuroblastoma. Cell Death Dis. 2014;5:e1072. doi: 10.1038/cddis.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer CE. The Golgi complex in stress and death. Front Neurosci. 2015;9:1–5. doi: 10.3389/fnins.2015.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Li H, Puertollano R. Novel roles for the MiTF/TFE family of transcription factors in organelle biogenesis, nutrient sensing, and energy homeostasis. Cell Mol Life Sci. 2014;71:2483–2497. doi: 10.1007/s00018-014-1565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast FD, Rachubinski RA, Aitchison JD. Signaling dynamics and peroxisomes. Curr Opin Cell Biol. 2015;35:131–136. doi: 10.1016/j.ceb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment Map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JL, Kim SY, Shin SW, Park JW. Regulation of brefeldin A-induced ER stress and apoptosis by mitochondrial NADP+-dependent isocitrate dehydrogenase. Biochem Biophys Res Commun. 2012;417:760–764. doi: 10.1016/j.bbrc.2011.12.030. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Xu F, Sehgal PB. Aberrant cytoplasmic sequestration of eNOS in endothelial cells after monocrotaline, hypoxia, and senescence: live-cell caveolar and cytoplasmic NO imaging. Am J Physiol Heart Circ Physiol. 2006;292:H1373–H1389. doi: 10.1152/ajpheart.00990.2006. [DOI] [PubMed] [Google Scholar]
- Odrowaz Z, Sharrocks AD. ELK1 uses different DNA binding modes to regulate functionally distinct classes of target genes. PLoS Genet. 2012a;8:e1002694. doi: 10.1371/journal.pgen.1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odrowaz Z, Sharrocks AD. The ETS transcription factors ELK1 and GABPA regulate different gene networks to control MCF10A breast epithelial cell migration. PLoS One. 2012b;7:e49892. doi: 10.1371/journal.pone.0049892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Yu J, Zhang L, Carpenter A, Zhu H, Li L, Ma D, Yuan J. A novel small molecule regulator of guanine nucleotide exchange activity of the ADP-ribosylation factor and Golgi membrane trafficking. J Biol Chem. 2008;283:31087–31096. doi: 10.1074/jbc.M806592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, et al. Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Peinado S, Ignashkova TI, van Raam BJ, Baumann J, Sennott EL, Gendarme M, Lindemann RK, Starnbach MN, Reiling JH. TRAPPC13 modulates autophagy and the response to Golgi stress. J Cell Sci. 2017;130:2251–2265. doi: 10.1242/jcs.199521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling JH, Clish CB, Carette JE, Varadarajan M, Brummelkamp TR, Sabatini DM. A haploid genetic screen identifies the major facilitator domain containing 2A (MFSD2A) transporter as a key mediator in the response to tunicamycin. Proc Natl Acad Sci USA. 2011;108:11756–11765. doi: 10.1073/pnas.1018098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling JH, Olive AJ, Sanyal S, Carette JE, Brummelkamp TR, Ploegh HL, Starnbach MN, Sabatini DM. A CREB3–ARF4 signalling pathway mediates the response to Golgi stress and susceptibility to pathogens. Nat Cell Biol. 2013;15:1473–1485. doi: 10.1038/ncb2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáenz JB, Sun WJ, Chang JW, Li J, Bursulaya B, Gray NS, Haslam DB. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat Chem Biol. 2009;5:157–165. doi: 10.1038/nchembio.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandow JJ, Dorstyn L, O’Reilly LA, Tailler M, Kumar S, Strasser A, Ekert PG. ER stress does not cause upregulation and activation of caspase-2 to initiate apoptosis. Cell Death Differ. 2014;21:475–480. doi: 10.1038/cdd.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- Selvaraj N, Kedage V, Hollenhorst PC. Comparison of MAPK specificity across the ETS transcription factor family identifies a high-affinity ERK interaction required for ERG function in prostate cells. Cell Commun Signal. 2015;13:12. doi: 10.1186/s12964-015-0089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta D, Linstedt AD. Control of organelle size: the Golgi complex. Annu Rev Cell Dev Biol. 2011;27:57–77. doi: 10.1146/annurev-cellbio-100109-104003. [DOI] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Yoshida H. TFE3, HSP47, and CREB3 pathways of the mammalian Golgi stress response. Cell Struct Funct. 2017;36:27–36. doi: 10.1247/csf.16023. [DOI] [PubMed] [Google Scholar]
- Urra H, Dufey E, Lisbona F, Rojas-Rivera D, Hetz C. When ER stress reaches a dead end. Biochim Biophys Acta. 2013;1833:3507–3517. doi: 10.1016/j.bbamcr.2013.07.024. [DOI] [PubMed] [Google Scholar]
- van der Linden L, van der Schaar HM, Lanke KHW, Neyts J, van Kuppeveld FJM. Differential effects of the putative GBF1 inhibitors Golgicide A and AG1478 on enterovirus replication. J Virol. 2010;84:7535–7542. doi: 10.1128/JVI.02684-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raam BJ, Lacina T, Lindemann RK, Reiling JH. Secretory stressors induce intracellular death receptor accumulation to control apoptosis. Cell Death Dis. 2017;8:e3069. doi: 10.1038/cddis.2017.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MJ, Kang YJ, Kim IY, Kim EH, Lee JA, Lim JH, Kwon TK, Choi KS. Monensin, a polyether ionophore antibiotic, overcomes TRAIL resistance in glioma cells via endoplasmic reticulum stress, DR5 upregulation and c-FLIP downregulation. Carcinogenesis. 2013;34:1918–1928. doi: 10.1093/carcin/bgt137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.