Abstract
Auxin controls a myriad of plant developmental processes and plant response to environmental conditions. Precise trafficking of auxin transporters is essential for auxin homeostasis in plants. Here, we report characterization of Arabidopsis CTL1, which controls seedling growth and apical hook development by regulating intracellular trafficking of PIN-type auxin transporters. The CTL1 gene encodes a choline transporter-like protein with an expression pattern highly correlated with auxin distribution and is enriched in shoot and root apical meristems, lateral root primordia, the vascular system, and the concave side of the apical hook. The choline transporter-like 1 (CTL1) protein is localized to the trans-Golgi network (TGN), prevacuolar compartment (PVC), and plasma membrane (PM). Disruption of CTL1 gene expression alters the trafficking of 2 auxin efflux transporters—Arabidopsis PM-located auxin efflux transporter PIN-formed 1 (PIN1) and Arabidopsis PM-located auxin efflux transporter PIN-formed 3 (PIN3)—to the PM, thereby affecting auxin distribution and plant growth and development. We further found that phospholipids, sphingolipids, and other membrane lipids were significantly altered in the ctl1 mutant, linking CTL1 function to lipid homeostasis. We propose that CTL1 regulates protein sorting from the TGN to the PM through its function in lipid homeostasis.
Author summary
Auxin, a plant hormone, controls many aspects of plant growth and development. The precise transport and distribution of auxin hold the key to its function. A number of transport proteins are known to be involved in auxin translocation, and the PIN proteins, which are an integral part of membranes in plants, play a pivotal role in this process. Several PIN proteins are localized in the plasma membrane to mediate auxin efflux from cells, but their regulation is not well known. In this report, we analyze the role of a choline transport protein, choline transporter-like 1 (CTL1), and find that it controls the trafficking of Arabidopsis PM-located auxin efflux transporter PIN-formed 1 (PIN1) and Arabidopsis PM-located auxin efflux transporter PIN-formed 3 (PIN3) to the plasma membrane, thereby regulating auxin distribution during plant growth and development. In addition, we show that CTL1 has a role in lipid homeostasis in the membrane; thus, these findings provide a mechanistic link between choline transport, lipid homeostasis, and vesicle trafficking in plants. We conclude that CTL1 is a new factor in secretory protein sorting and that this finding contributes to the understanding of not only auxin distribution in plants but also protein trafficking in general.
Introduction
Dynamic endomembrane trafficking delivers proteins and other cargo molecules to a variety of organelles and thus controls almost all aspects of plant development and physiology, including gravitropism, epidermis differentiation, guard cell movement, cell wall remodeling, defense against pathogens, and hormone signaling [1]. In this context, exocytosis and endocytosis determine the abundance and dynamics of signaling receptors and transporters at the plasma membrane (PM), which in turn afford cells the ability to respond to extracellular stimuli. For example, PM-located auxin transporters PIN-formed (PINs) and AUXIN RESISTANT 1 (AUX1) have been shown to continuously cycle between PM and endosomal compartments, resulting in dynamic changes in localization and abundance at the PM, where they function to control auxin gradients between cells. Many components have been shown to affect the trafficking and PM abundance of auxin transporters. These include the nucleotide exchange factor for ADP-ribosylation-factor-type GTPases (ARF-GEFs) [2–4], retromer proteins [5], adaptor protein complex 3 (AP-3) [6, 7], endosomal sorting complexes [8, 9], the trans-Golgi network/early endosome (TGN/EE) protein ECHIDNA [10], and several protein kinases [11–13]. The trafficking regulators safeguard the polarity and dynamics of auxin transporters, making possible the spatiotemporal variations in auxin distribution that act as an instructive signal for a wide variety of cellular events, ranging from polarity establishment during the earliest phases of plant embryogenesis to other morphogenetic events, including root development and the formation of the apical hook during seedling development [2–5, 10–14]. Because of the dynamics of these auxin transporters in the endomembrane system, they can be used as cargo proteins to study the regulation of the membrane trafficking process in cell biology. In genetic analysis, the mutations in genes encoding these transporters are often trackable because the mutants often show severe phenotypic changes in plant growth and development.
In a screen for mutants defective in phloem development, Dettmer et al. [15] identified choline transporter-like 1 (CTL1) to be essential for sieve tube formation and thus overall plant development. The CTL1 gene encodes a protein with multiple transmembrane domains that has been shown to mediate choline transport [15]. However, the cellular mechanism for CTL1 function in plant development remains unknown. In tracking down the mechanism of CTL1 function, we discovered that CTL1 is closely associated with auxin signaling in the control of plant developmental processes. We further found that disruption of CTL1 function impaired endomembrane trafficking of several auxin transporters and probably some other unidentified cargo proteins as well. CTL1 acts at both the secretory vesicles (SVs) and the clathrin-coated vesicles (CCVs) of the TGN to regulate the trafficking of PIN1 and PIN3, which may explain the phenotypes of the ctl1 mutant in seedling growth and apical hook development. As a choline transporter, CTL1 plays a crucial role in the homeostasis of membrane lipids, including phospholipids and sphingolipids. As lipid compositions of cell membranes influence vesicular transport, CTL1 function provides the mechanistic link between choline transport, lipid remodeling, and vesicular trafficking of PM proteins. We conclude that CTL1 is a previously unrecognized regulator of endomembrane trafficking in plants.
Results
A null mutant lacking CTL1 displays a cell elongation defect
A significant portion of plant genomes encode proteins with multiple transmembrane domains that may function as transporters. However, a large majority of these open reading frames (ORFs) are designated as “proteins of unknown function.” We took a comprehensive reverse genetic approach to identify the function of these proteins by screening Arabidopsis SALK transfer deoxyribonucleic acid (T-DNA) insertion lines disrupting the expression of corresponding genes. One of the lines, SALK_065853C in Columbia-0 (Col-0) background, was recovered for its severely stunted growth phenotype. This line contains a T-DNA insertion in the At3g15380 gene, previously named CHOLINE-TRANSPORTER-LIKE 1 (CTL1) for its biochemical function as a choline transporter and its function in sieve tube formation [15].
The insertion in CTL1 occurred in the first intron (281 bp from the start codon) to generate a transcriptional knockout (S1A–S1C Fig). We backcrossed the ctl1 mutant with the wild type for 2 generations to segregate other possible T-DNA insertions and found that the isolated homozygous ctl1 mutant lacking other insertions consistently showed the typical ctl1 phenotype observed with the original SALK line. We further confirmed that the phenotype of the ctl1 mutant resulted from lack of CTL1 function by complementation using a genomic fragment of the CTL1 gene and a CTL1- enhanced green fluorescent protein (EGFP) fusion construct driven by the CTL1 promoter, respectively (S1D Fig and S2A Fig). The transgenic plants expressing CTL1 genomic fragment or CTL1-EGFP fusion protein in the ctl1 mutant background displayed a similar growth phenotype to that of the wild-type plants (Fig 1A, S1E Fig and S2D Fig), supporting the conclusion that disruption of the CTL1 gene rendered the dwarf phenotype of the mutant plants.
Fig 1. The CTL1-knockout mutant has a defect in cell elongation.
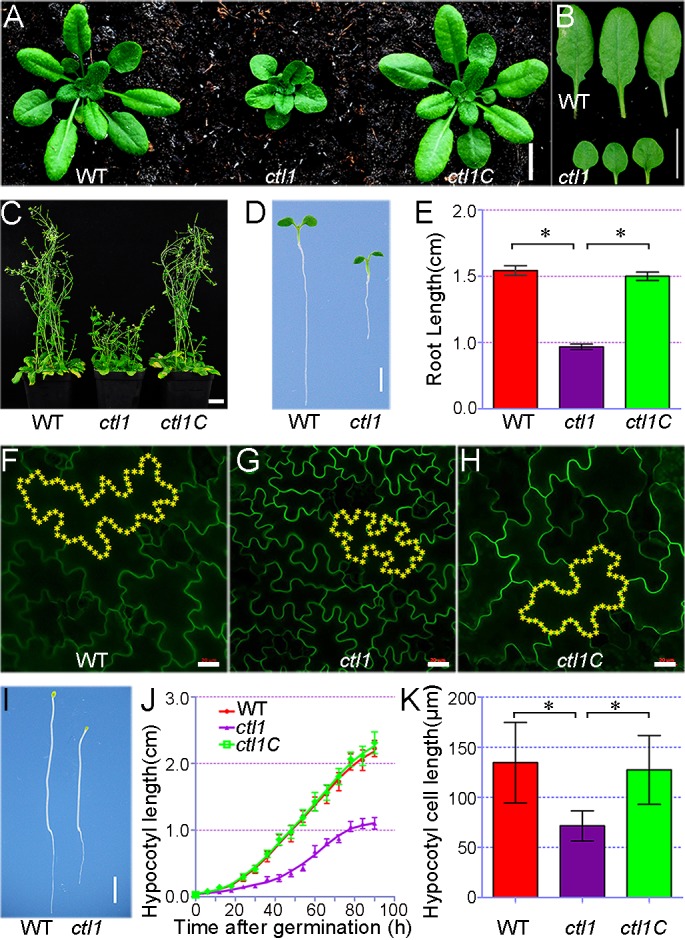
(A) The growth phenotype of wild-type (WT), ctl1 mutant, and complementation lines transformed with At3g15380 genomic DNA (ctl1C). After being grown on half-strength Murashige and Skoog (MS) medium for 7 days, the seedlings were transferred to the soil and grown for 23 days before photographs were taken. Bar = 1 cm. (B) The rosette leaves from 30-day-old ctl1 mutant as compared to those of the WT. Bar = 1 cm. (C) The dwarfed phenotype of 2-month-old adult ctl1 plants compared to the WT and ctl1C. Bar = 2 cm. (D) Five-day-old WT and ctl1 mutant grown on half-strength MS medium. Bar = 3 mm. (E) The root length of 5-day-old WT, ctl1, and ctl1C. Three independent experiments were performed. Data are mean ± SD. n = 6 for each genotype. Significant differences are indicated as *P < 0.05 (Student t test). (F–H) Comparison of epidermal cells in the cotyledon of 5-day-old WT (F), ctl1 (G), and ctl1C seedlings (H). Representative cells were highlighted with yellow asterisks. Scale bars in panels F–H are 20 μm. (I) Growth phenotype of 5-day-old etiolated WT and ctl1 seedlings. Bar = 2 mm. (J) Hypocotyl length of etiolated WT, ctl1, and ctl1C seedlings. Data are mean ± SD. n = 5 for each genotype. (K) The hypocotyl cell length of 5-day-old etiolated WT, ctl1, and ctl1C seedlings. Data are mean ± SD, and significant differences are indicated as *P < 0.05 (n = 30, Student t test). The raw data for panels E, J, and K can be found in S1 Data.
We examined the ctl1 phenotype in detail and found that 30-day-old ctl1 seedlings grown in the soil displayed smaller, round, and dark green epinastic leaves with short petioles (Fig 1A and 1B and S1F–S1H Fig) and that 2-month-old adult ctl1 mutants exhibited a dwarfed phenotype (Fig 1C). To examine root growth, we planted seeds on half-strength Murashige and Skoog (MS) agar medium and found that 5-day-old ctl1 seedlings had shorter roots as compared to the wild type (Fig 1D and 1E), consistent with previous observations [15].
We further examined the cell size in both wild-type and mutant plants and found that the stunted growth phenotype in the ctl1 mutant resulted from a defect in cell elongation. To determine cell size, we expressed AUX1-GFP in wild-type, ctl1, and complemented lines (ctl1C) so that the cell periphery lit up with the GFP for confocal imaging. The images collected from confocal laser scanning microscopy revealed that the size of leaf epidermal cells in ctl1 was smaller than that of those in the wild type and ctl1C (Fig 1F–1H). Typically, the cell size in the ctl1 mutant plants was only 49% of the wild type (S1 Table). As hypocotyl cells elongate quickly without cell division, hypocotyls grown in the dark are excellent models for examining cell elongation [16]. We thus analyzed the hypocotyl length of etiolated seedlings and found that the hypocotyls of ctl1 mutant seedlings elongated more slowly compared with the wild type and ctl1C (Fig 1I and 1J). Moreover, the epidermal cell length of ctl1 hypocotyls was only about half of that of the wild type (Fig 1K). Taken together, these results demonstrate that CTL1 is an essential regulator of cell elongation in Arabidopsis. This finding explains an overall dwarf phenotype of the ctl1 mutant seedlings beyond the defect in phloem development observed earlier [15].
CTL1 is localized to the PM, TGN, and prevacuolar compartment (PVC)
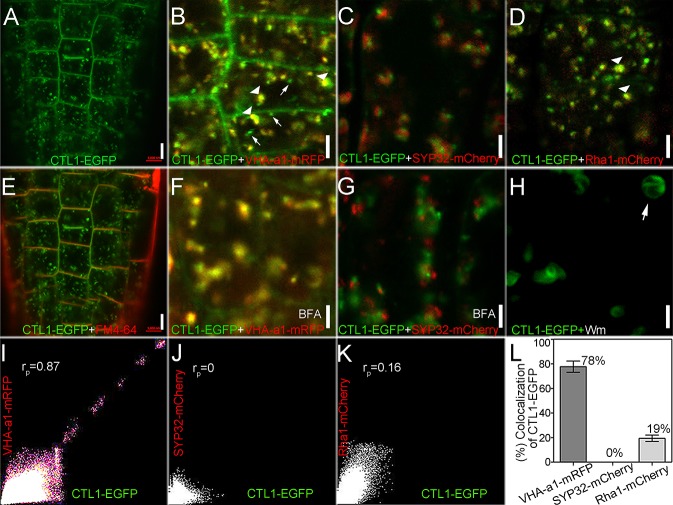
To further study the cellular mechanism of CTL1 function, we determined the subcellular localization of CTL1 in root cortex cells of Arabidopsis seedlings by using a CTL1-EGFP transgenic line in the ctl1 mutant background. Real-time quantitative reverse transcription PCR (qRT-PCR) assay showed that this transgenic line had levels of CTL1 transcript that were similar to those in the wild-type plants (S2B Fig). Furthermore, the CTL1-EGFP construct in this transgenic line complemented the dwarfed phenotype of the ctl1 mutant (S2D Fig), indicating that this line expressed functional CTL1-EGFP fusion proteins in the same compartments as the native CTL1 protein. Therefore, we used this line to track down the subcellular location of CTL1-EGFP protein using confocal microscopy. The fluorescence signal generated by CTL1-EGFP fusion appeared at the PM and also displayed a punctate pattern in the cytosol (Fig 2A and S2E Fig). To identify intracellular punctate structures at which CTL1-EGFP is localized, we crossed this CTL1-EGFP transgenic line with the transgenic lines that stably express fluorescent markers for several organelles. We found that most of the punctate signals of CTL1-EGFP colocalized with vacuolar proton ATPase subunit VHA-a isoform 1 monomer red fluorescent protein (VHA-a1-mRFP) (Fig 2B), a TGN/EE marker [17]. The linear Pearson correlation coefficient (rp) for the colocalization was 0.87 (Fig 2I). We did not observe colocalization of CTL1-EGFP fluorescence signals with Syntaxin of plants 32 (SYP32)-mCherry (rp = 0) (Fig 2C and 2J), the Golgi apparatus marker [18]. In addition, some CTL1-EGFP fluorescence signals overlapped with those of Arabidopsis Rab homolog F2A (Rha1)-mCherry (rp = 0.16) (Fig 2D and 2K), the PVC marker [18]. We conducted more experiments to further examine the subcellular localization of CTL1. First, we stained root tips with FM4-64 for a short time to label the PM [19] and found that the CTL1-EGFP fluorescence colocalized with the FM4-64 signal (Fig 2E). Secondly, we treated root tips with fungal toxin brefeldin A (BFA), which blocks trafficking from the TGN to the PM, leading to the formation of enlarged endosomal compartments referred to as BFA bodies [20]. We detected both CTL1-EGFP and VHA-a1-mRFP fluorescence in the BFA bodies (Fig 2F). However, the CTL1-EGFP and SYP32-mCherry signals did not overlap in the presence of BFA (Fig 2G). Finally, we treated root tips with wortmannin (Wm), a drug that preferentially triggers the swelling of PVC but has little effect on the morphology of the Golgi apparatus and the TGN/EE [21]. After 1 hour of treatment with 33 μM Wm, the morphology of some organelles labeled by CTL1-EGFP became swollen and showed a PVC-specific ring-like structure (Fig 2H), indicating that CTL1 is also sorted to the PVC.
Fig 2. Subcellular localization of choline transporter-like 1 (CTL1).
(A) CTL1- enhanced green fluorescent protein (EGFP) signals in root cortex cells of CTL1-EGFP transgenic plants expressing CTL1-EGFP in the ctl1 mutant background. Bar = 5 μm. (B–D) Comparison of CTL1-EGFP (green) signal with subcellular markers tagged with monomer red fluorescent protein (mRFP) or mCherry (red). Bar = 2 μm. (B) CTL1-EGFP is largely colocalized with the trans-Golgi network/early endosome (TGN/EE) marker, vacuolar proton ATPase subunit VHA-a isoform 1 (VHA-a1)-mRFP. Arrowheads and arrows indicate overlapping and nonoverlapping signals, respectively. (C) CTL1-EGFP is localized to distinct compartments from Syntaxin of plants 32 (SYP32)-mCherry. (D) Partial colocalization of CTL1-EGFP with Arabidopsis Rab homolog F2A (Rha1)-mCherry. Arrowheads indicate overlapping signals. (E) Root cortex cells labeled with FM4-64 (red) and CTL1-EGFP (green). The image was captured within 5 minutes of incubation with 4 μM FM4-64. Bars = 5 μm. (F) Colocalization of CTL1-EGFP and VHA-a1-mRFP in root cortex cells treated with brefeldin A (BFA). (G) CTL1-EGFP and SYP32-mCherry are not colocalized in BFA-treated cells. The images in (F) and (G) were captured after the roots were treated with 100 μM BFA for 60 minutes. Bars = 2 μm. (H) CTL1-EGFP signals in root cells after they were treated with 33 μM wortmannin (Wm) for 60 minutes. White arrow shows the ring-like structure. Bars = 2 μm. (I–K) Colocalization analysis of CTL1-EGFP with VHA-a1-mRFP (I), SYP32-mCherry (J), or Rha1-mCherry (K) using the ImageJ software with the colocalization finder plugin. The linear Pearson correlation coefficient (rp) indicates the percentage of signal overlap, and an rp value of 1.0 represents 100% colocalization. (L) The colocalization percentage of CTL1-EGFP with VHA-a1-mRFP, SYP32-mCherry, or Rha1-mCherry. Data are mean ± SD. Six images acquired from 3 roots were used. The raw data for panel L can be found in S1 Data.
We also quantified the colocalization of CTL1-EGFP with various organelle markers in root tip cells of 4-day-old seedlings. This analysis revealed a large proportion (78% ± 4.6%) of colocalization between CTL1-EGFP and the TGN marker VHA-a1-mRFP, a smaller proportion (19% ± 2.6%) of colocalization between CTL1-EGFP and the PVC marker Rha1-mCherry, and no colocalization with SYP32-mCherry (Fig 2L). The TGN is subdivided into 2 structure forms, SVs and CCVs [22]. We also detected that CTL1-EGFP colocalizes with mCherry-clathrin light chain 2 (CLC2) (S3 Fig). As VHA-a1 and CLC2 reside in the SVs and the CCVs, respectively [22], colocalization with both VHA-a1-mRFP and mCherry-CLC2 illustrated that CTL1 is localized to both subdomains of the TGN.
Postembryonic expression of CTL1
We used the CTL1-EGFP transgenic line to study the expression pattern of CTL1, since the expression of CTL1-EGFP was driven by the CTL1 promoter region (S2 Fig). The GFP fluorescence was ubiquitously distributed in the meristem and elongation zones but restricted to vascular tissues in the mature zone of roots (Fig 3A). Cross sections through the root meristem zone displayed the distribution of CTL1-EGFP signal in all cell layers (Fig 3B and 3C). Furthermore, GFP was also highly expressed in the newly initiated lateral roots (Fig 3D and 3E).
Fig 3. Expression pattern of CTL1 gene reporters in Arabidopsis seedlings.
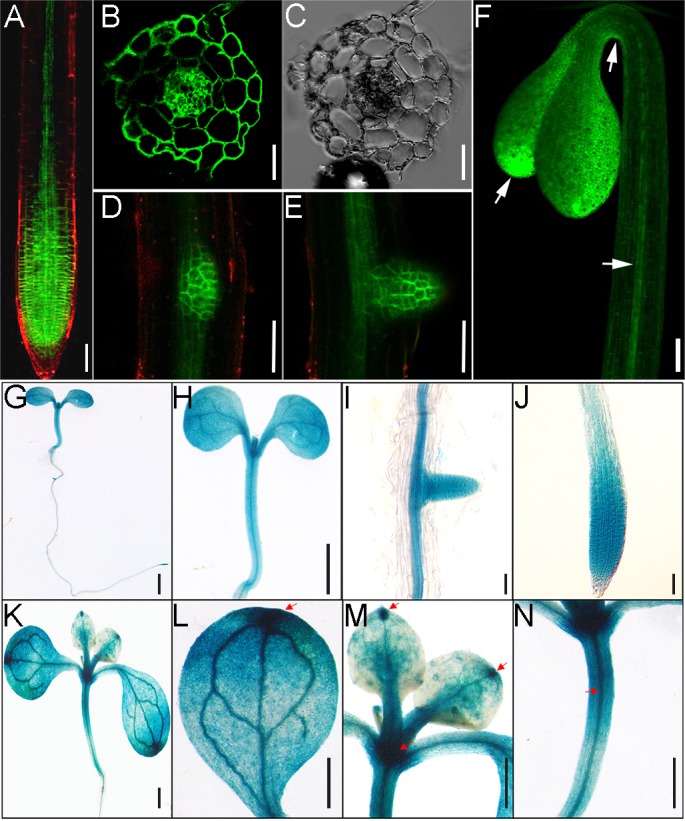
(A–F) Green fluorescence signals in transgenic plants harboring the CTL1-EGFP construct. Choline transporter-like 1 enhanced green fluorescent protein (CTL1-EGFP) signals were detected in the primary root (A), lateral root primordium (D), and emerged lateral root (E) of 5-day-old seedlings (Bars = 50 μm). The transection view of CTL1-GFP at the GFP channel (B) and the differential interference contrast (DIC) channel (C) shows GFP signal in all cell types in the meristem region of a primary root from a 5-day-old seedling. Bars = 25 μm. (F) CTL1-EGFP signals in a 3-day-old etiolated seedling. Arrows indicate the sites of strong expression. Bars = 1 mm. (G–N) Histochemical analysis of CTL1 promoter–β-glucuronidase (GUS) expression in transgenic plants. (G–J) GUS staining of a 5-day-old seedling (G), showing expression in shoot (H), emerging lateral root (I), and primary root (J). (K–N) GUS staining of a 7-day-old seedling (K), showing expression in cotyledon (L), the first pair of true leaves (M), and the hypocotyl (N). The scale bars in (I) and (J) are 100 μm, and other bars are 1 mm.
In dark-grown seedlings, CTL1 was expressed in the whole cotyledon with enrichment at the blade tips, the vasculature of the hypocotyl, and the concave side of the apical hook (Fig 3F). When grown on the half-strength MS medium under dark conditions, Arabidopsis apical hook development proceeds through 3 phases: the hook formation phase, the maintenance phase, and the opening phase [10, 23, 24]. CTL1-EGFP signals appeared at both the convex and the concave sides of the hook, with the maximum in the concave side (S4G and S4H Fig), and the expression at the concave side gradually disappeared during the opening phase (S4A–S4F and S4J Fig), suggesting that CTL1 function may be associated with apical hook development.
To further confirm the expression pattern of CTL1, we generated transgenic plants harboring the CTL1pro:CTL1:GUS construct in which the EGFP reporter was replaced by the β-glucuronidase (GUS) reporter. In the apical hook region, CTL1-GUS shows the same expression pattern as the CTL1-EGFP in the concave side of the apical hook (S4I Fig). In the roots of 5-day-old seedlings, GUS activity was detected in the similar tissues labeled by EGFP signals (Fig 3G–3J). In shoots of 7-day-old seedlings, the expression pattern of CTL1 was ubiquitous, but a relatively higher level of GUS activity was detected in the apical meristem, at the tips of cotyledon and true leaves, and in vascular tissues (Fig 3K–3N), largely overlapping with the GFP signals detected earlier (Fig 3F).
CTL1 loss-of-function mutant shows defects associated with auxin
Phenotypic analyses of the ctl1 mutant in detail provided insights into CTL1 function in growth and development. The ctl1 mutant showed reduced primary root growth, whereas the lateral roots reached approximately the same length as the primary root, apparently resulting from lack of apical dominance [15]. In addition, we found that the ctl1 showed a severe defect in apical hook development, consistent with high levels of expression of CTL1 in the apical hook (Fig 3F and S4 Fig). In particular, etiolated ctl1 seedlings never formed the fully closed apical hook, and the hook quickly opened without an apparent maintenance phase (Fig 4A and 4B).
Fig 4. The ctl1 mutant shows auxin-related phenotypic changes.
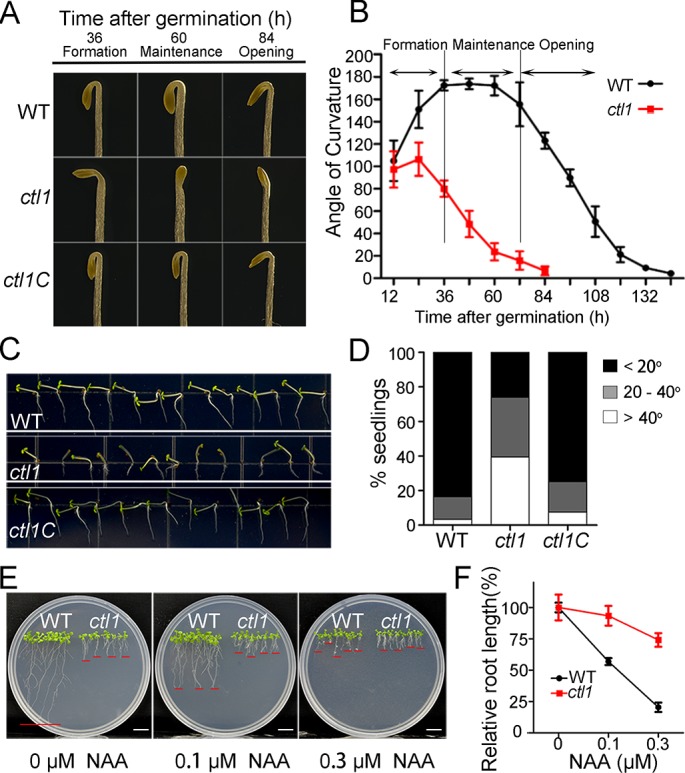
(A) Representative images of the apical hooks of the wild type (WT), ctl1, and the ctl1 complementation line (ctl1C) at 36, 60, and 84 hours after germination in the dark. (B) Kinetic analysis of apical hook angles of etiolated seedlings of WT, ctl1, and ctl1C during apical hook development. Data are mean ± SD of 4 replicate experiments. (C) Representative images of phototropic growth of WT, ctl1, and ctl1C seedlings. After being vertically grown on half-strength Murashige and Skoog (MS) medium in the dark for 2 days, the seedlings were exposed to unilateral light for 2 days before the photographs were taken. (D) Quantitation of the phenotypes shown in panel C. The bending angles of the hypocotyls towards the horizontal direction were measured using the ImageJ software. The percentages of seedlings in each of 3 categories (<20°, 20–40°, and >40°) were calculated. WT, n = 116; ctl1, n = 119; and ctl1C, n = 119. (E) Growth phenotype of 10-day-old WT and ctl1 mutant plants grown on half-strength MS medium containing 0, 0.1, or 0.3 μM 1-naphthylacetic acid (NAA). Bars = 1 cm. (F) The ctl1 mutant shows reduced NAA sensitivity. Relative root length (%) indicates shortening of the primary roots as a result of NAA in the medium. Data are mean ± SD. Three independent experiments were performed. n = 6 for each experiment. The raw data for panels B, D, and F can be found in S1 Data.
ctl1 defects in cell elongation (described earlier) and defects in primary and lateral root growth all point to relevance with the auxin response. Furthermore, apical hook development is the result of differential cell elongation mediated by uneven auxin distribution [23]. These auxin-related phenotypes of ctl1 prompted us to investigate the connections between CTL1 and auxin. We went on exploring whether there are other auxin-related phenotypes altered in the ctl1 mutant. One process we studied was phototropism, a well-characterized light response mediated by asymmetric auxin distribution in plants [4]. As shown in Fig 4, ctl1 had a reduced hypocotyl phototropic response to unilateral light as compared with the wild type and the complemented line (Fig 4C and 4D). In addition, we also found that ctl1 seedlings were less sensitive to high levels of 1-naphthylacetic acid (NAA) that inhibit root elongation. Compared to the wild type, ctl1 roots were resistant to auxin inhibition over a range of auxin concentrations (0, 0.1, and 0.3 μM) (Fig 4E and 4F). We also tested the sensitivity to salicylic acid (SA) and found that both the wild type (WT) and the ctl1 mutant were sensitive to SA (S5 Fig), unlike the situation with auxin. All these findings support the hypothesis that CTL1 is involved in auxin-related processes.
Auxin distribution is altered in the ctl1 mutant
To study auxin distribution in the ctl1 mutant, we crossed it with a transgenic line harboring DR5::GUS, a reporter of auxin levels [5]. In the ctl1 mutant background, the DIRECT REPEAT5 (DR5)-driven GUS expression was greatly enhanced in the cotyledons, especially at the tips, but was dramatically reduced in the vascular tissues of the hypocotyls and roots, as compared to the pattern of DR5::GUS in the WT background (Fig 5A–5H and S6A Fig). In the roots, the DR5::GFP signal in the WT and the ctl1 mutant showed a similar distribution pattern, with a condensed signal in the primary root tips. However, after being treated with NAA (0.1, 0.3, 0.5, and 1 μM) for 12 hours, the DR5::GFP signal was enhanced in the meristem and elongation zones of WT roots with increasing NAA concentrations, but the level of this response was reduced in ctl1, suggesting reduced transport or reduced response of auxin (Fig 5I and S6B Fig). Moreover, a maximum DR5::GFP signal was detected in the concave side of the apical hooks of the WT, but this was largely diminished in the mutant (Fig 5J and S6C Fig).
Fig 5. The ctl1 mutant shows altered auxin distribution pattern.
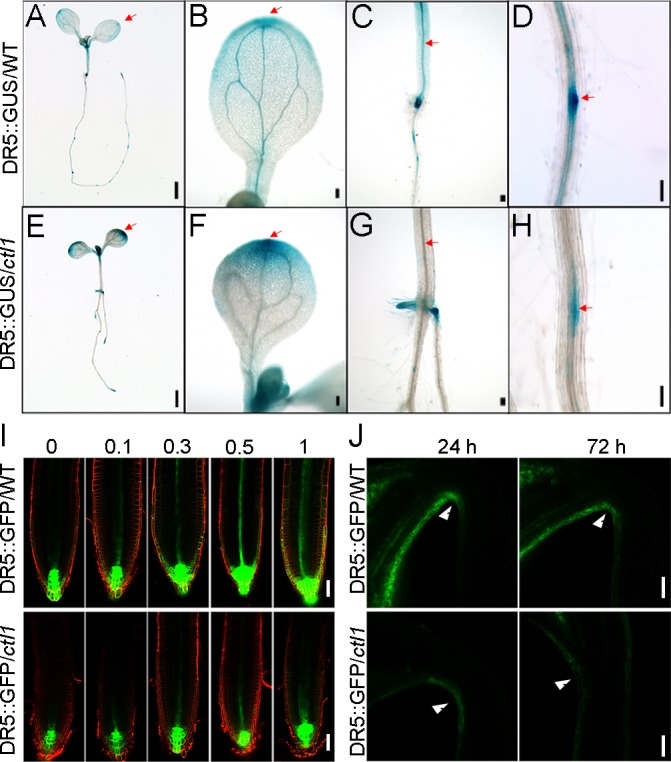
(A–H) DR5::GUS expression pattern in 7-day-old wild type (DR5::GUS/WT) (A–D) and ctl1 mutant (DR5::GUS/ctl1) (E–H). The red arrows point to the areas where signals are different between DR5::GUS/WT and DR5::GUS/ctl1. Scale bars in panel A and panel E are 1 mm, and those in panels B–D and F–H are 100 μm. (I) Expression pattern of DR5::GFP in the wild type (DR5::GFP/WT) (upper panel) and the ctl1 mutant (DR5::GFP/ctl1) (lower panel) treated with 0, 0.1, 0.3, 0.5, or 1 μM 1-naphthylacetic acid (NAA). Scale bars are 50 μm. (J) DR5::GFP signals in the concave side of the apical hook of the wild type (WT) and the ctl1 mutant at 1 and 3 days after germination. The red arrows show the sites where green fluorescent protein (GFP) signals were accumulated in the WT but not in the mutant. Scale bars are 20 μm. DR5, DIRECT REPEAT5; GUS, β-glucuronidase.
CTL1 controls the abundance of PIN1 and PIN3 in the root and the apical hook
We showed that disruption of CTL1 function appears to affect auxin transport and distribution (Figs 4 and 5). Together with the finding of CTL1 subcellular localization to the PM and endomembranes (Fig 2), we speculated that CTL1 may regulate the trafficking of PM-localized proteins so that distribution of auxin transporters may be altered in the ctl1 mutant. In order to test this hypothesis, we examined the expression and localization of several auxin transporters in the ctl1 mutant by using auxin transporter-GFP markers. We crossed the ctl1 mutant with transgenic lines containing PIN1pro:PIN1:GFP, PIN2pro:PIN2:GFP, PIN3pro:PIN3:GFP, AUX1pro:AUX1:GFP, or LAX3pro:LAX3:GFP and monitored the GFP signals by confocal microscopy. We found a dramatically reduced accumulation of PIN1-GFP and PIN3-GFP in the ctl1 mutant as compared to the WT. More specifically, the abundance of PIN1-GFP in the mutant was about half (56% ± 14.2%) of the WT, and for PIN3 it was only a quarter (26% ± 19.3%) of the WT (Fig 6A, 6C and 6F). However, other auxin transporters, including PIN2, AUX1, and Like AUX1 protein 3 (LAX3), were not affected by the CTL1 mutation (Fig 6B and 6D–6F). We also found that the brassinosteroid receptor BRI1 showed no difference in the WT and the ctl1 mutant (S8 Fig). Because PIN-mediated auxin efflux also plays a major role during apical hook development [24, 25], we also examined GFP signal in the apical hook and again found reduced levels of PIN1-GFP and PIN3-GFP, but not of AUX1-GFP, in the ctl1 mutant as compared to the WT (S7 Fig). The loss of PIN3 can cause an enhanced DR5 signal in the cotyledons of Arabidopsis [25]. As we also detected an enhanced DR5 signal in the cotyledons of ctl1 mutant, we proposed that reduced abundance of PIN1 and PIN3 may lead to changed auxin transport and distribution (shown by DR5::GUS/GFP lines earlier) that in turn causes changes in auxin-related phenotypes in the ctl1 mutant.
Fig 6. The ctl1 mutant shows reduced levels of Arabidopsis PM-located auxin efflux transporter PIN-formed 1 (PIN1) and Arabidopsis PM-located auxin efflux transporter PIN-formed 3 (PIN3).
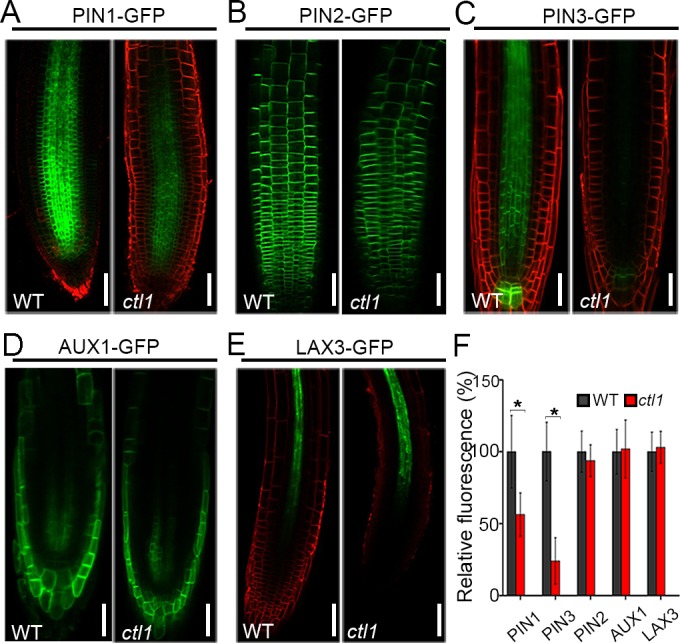
The green fluorescent protein (GFP) signals indicate the abundance of several auxin transporters, including PIN1pro:PIN1:GFP (A), PIN2pro:PIN2:GFP (B), PIN3pro:PIN3:GFP (C), AUX1pro:AUX1:GFP (D), and LAX3pro:LAX3:GFP (E), in 4-day-old primary root of the wild type (WT) and the ctl1 mutant. The red signals are from FM4-64 staining. Scale bars are 50 μm. (F) Relative abundance of auxin transporters (PIN1, PIN2, PIN3, AUXIN RESISTANT 1 [AUX1], and Like AUX1 protein 3 [LAX3]) in ctl1 mutant is shown as % of the WT. Data are mean ± SD from 91, 64, 44, 39, and 40 cells in 5 roots of the transgenic lines expressing PIN1-GFP, PIN3-GFP, PIN2-GFP, AUX1-GFP, and LAX3-GFP, respectively. Three independent experiments were performed. Asterisks indicate a significant difference (*P < 0.05, Student t test). The raw data for panel F can be found in S1 Data.
To determine whether the reduced abundance of PIN1 and PIN3 proteins in the ctl1 mutant results from a decrease in the transcript level, we examined the RNA levels of auxin transporter genes in the WT and the ctl1 mutant. The qRT-PCR analysis showed that the mRNA levels of PIN1 and PIN3 in ctl1 were slightly higher than those in the WT (S9 Fig), suggesting that down-regulation of PIN1 and PIN3 proteins in ctl1 occurs at the post-transcriptional level.
CTL1 regulates trafficking of PIN1 and PIN3 to the PM
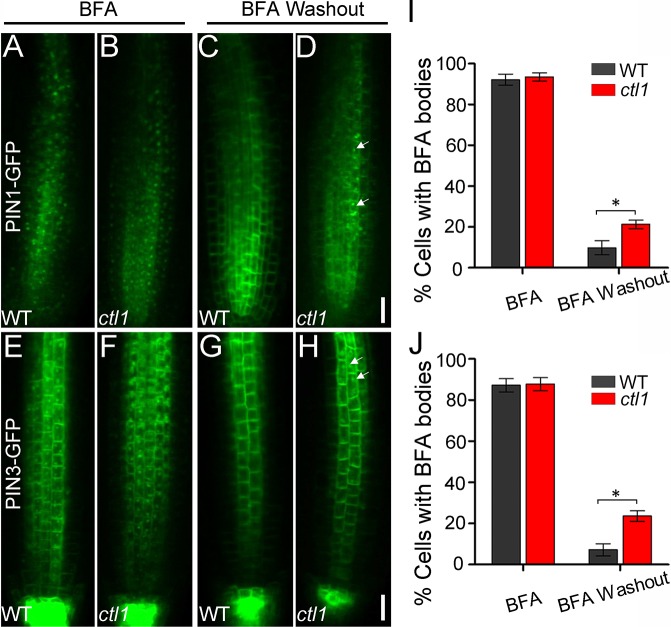
The reduced abundance of PIN1 and PIN3 in the ctl1 mutant, together with CTL1 localization to the TGN, suggests that CTL1 protein may affect the recruitment of PINs from the TGN to the PM. In the study of protein trafficking, BFA is often used to block protein sorting from the TGN to the PM and to induce protein internalization from the PM to the TGN. Thus, BFA treatment of cells results in the formation of large endosomes called BFA bodies that contain PM-destined proteins. These BFA bodies are rapidly disintegrated, and the proteins are targeted back to the PM again after BFA washout [26–28]. Therefore, BFA washout has been widely used to measure the speed of trafficking from the TGN back to the PM [26–29]. We analyzed the trafficking of PIN1-GFP and PIN3-GFP using a similar approach. Upon treatment of WT and ctl1 roots with 50 μM BFA for 60 minutes, similar percentages of cells with BFA bodies were detected in the WT and the ctl1 mutant (Fig 7A, 7B and 7I). However, after BFA washout for 90 minutes, we detected 9.7% and 21.6% of root cells containing PIN1-GFP-decorated BFA bodies in the WT and ctl1, respectively (Fig 7C, 7D and 7I). Similarly, about 7.6% and 23.5% of cells contained PIN3-GFP-labeled BFA bodies in the WT and ctl1, respectively (Fig 7E–7H and 7J). We also did the BFA washout experiment using the PIN2-GFP/WT and PIN2-GFP/ctl1 transgenic lines and found no difference between the WT and the ctl1 mutant (S10 Fig). These data indicate that loss of CTL1 causes a delay of trafficking in PIN1 and PIN3 proteins, which may in turn lead to the auxin-related developmental defects we observed in the ctl1 mutant plants.
Fig 7. Choline transporter-like 1 (CTL1) affects Arabidopsis PM-located auxin efflux transporter PIN-formed 1 (PIN1) and Arabidopsis PM-located auxin efflux transporter PIN-formed 3 (PIN3) recycling.
(A–D) PIN1-green fluorescent protein (GFP) localization in wild-type (WT) (A and C) and ctl1 roots (B and D) treated with brefeldin A (BFA) for 60 minutes before (A and B) or after (C and D) washout for 90 minutes. Insets at the lower right corner of panels A–D show enlarged views of PIN1-GFP labeled BFA bodies (from areas indicated by arrows). Scale bars are 20 μm. (I) Percentage of cells with PIN1-GFP labeled BFA bodies before and after BFA washout in the WT and the ctl1 mutant. Data are mean ± SD. Three independent experiments were performed. Five roots were measured for each experiment (Student t test, *P < 0.05). (E–H) PIN3-GFP localization in WT (E and G) and ctl1 (F and H) roots treated with BFA as shown for PIN1-GFP. Scale bars are 20 μm. (J) Percentage of cells with PIN3-GFP labeled BFA bodies before and after BFA washout in the WT and the ctl1 mutant. Data are mean ± SD. Three independent experiments were performed. Five roots were used for each experiment (Student t test, *P < 0.05). The raw data for panels I and J can be found in S1 Data.
CTL1 plays a vital role in the homeostasis of membrane lipids in Arabidopsis
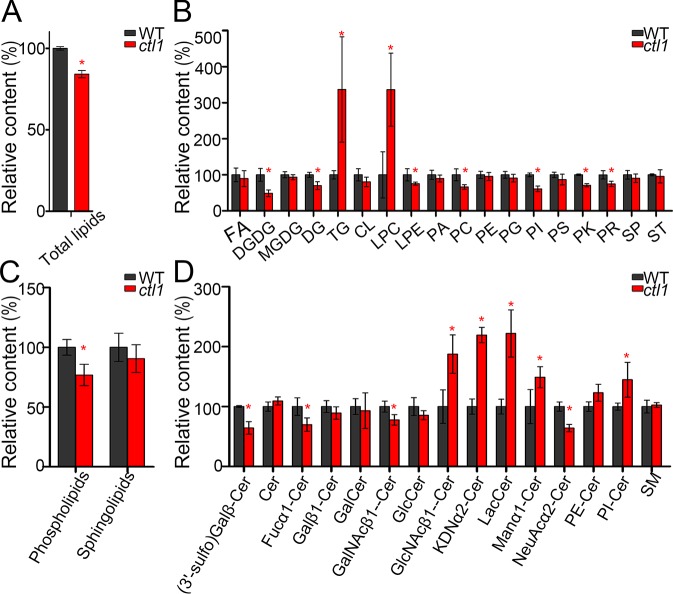
It has been reported that sphingolipids are involved in the trafficking of auxin transporters [30–32]. Choline is predominantly utilized for the synthesis of essential lipid components of the cell membranes, including sphingolipid [33]. A defect in choline transport may impair the homeostasis of these lipids, thereby affecting vesicle trafficking. To test if CTL1 functions in lipid homeostasis, we first monitored the lipid content in situ in the WT and mutant seedlings using Nile Red staining [32, 34]. The results showed that both polar and nonpolar lipids were reduced in the apical hook region of the ctl1 mutant as compared to the WT (S11 Fig). To provide a more detailed analysis of lipid content, we used ultra-performance liquid chromatography coupled with mass spectrometry (UPLC-MS) to survey the lipid profiles of WT and ctl1 plants. In the ctl1 mutant, the total lipids were reduced to about 84% of the WT level (Fig 8A). In further analysis, we found that the glycerophospholipids (lysophosphatidylethanolamine, phosphatidylcholine, and phosphatidylinositol) in the ctl1 mutant were reduced (Fig 8B). The content of total glycerophospholipids in ctl1 mutant was about 77% of the level in the WT (Fig 8C). Although the total sphingolipid contents were similar between the WT and the ctl1 mutant (Fig 8C), we found that the content of some specific types of sphingolipids were significantly altered in the ctl1 mutant (Fig 8D). In particular, the abundance of sphingolipids containing very-long-acyl-chain fatty acids (C > 18 carbons) was altered in the ctl1 mutant (S12 Fig), consistent with the earlier finding [31] that these sphingolipids are required for the targeting of specific auxin carriers to the PM.
Fig 8. The lipidomics analysis of wild type and ctl1.
(A) The relative content of total lipids in the wild type (WT) and the ctl1 mutant. (B) The changes in the subclasses of lipids in the ctl1 mutant. Fatty acyls (FAs), digalactosyldiacylglycerol (DGMG), monogalactosyldiacylglycerol (MGDG), diacylglycerol (DG), triacylglycerol (TG), lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidylserine (PS), polyketides (PKs), prenol lipids (PRs), sphingolipids (SPs), and sterol lipids (STs). (C) The relative intensity of total glycerophospholipids and SPs in the WT and the ctl1 mutant. (D) Some subclasses of SPs are significantly altered in the ctl1 mutant. These include a group of ceramides (Cer) such as Fucα1-Cer: Fucα1-2GalNAcβ1-4(NeuAcα2-8NeuAcα2–3) Galβ1-4Glcβ-Cer; Galβ1-Cer: Galβ1-3GalNAcβ1-3Galα1-3Galβ1-4Glcβ-Cer; GalNAcβ1-Cer: GalNAcβ1-3Galα1-3Galα1-3Galα1-3Galα1-4Galβ1-4Glcβ-Cer; GlcNAcβ1-Cer: GlcNAcβ1-4Manβ1-4Glcβ-Cer; KDNα2-Cer: KDNα2-6Galβ1-4GlcNAcβ1-3Galβ1-4Glcβ-Cer; Manα1-Cer: Manα1-3Manβ1-4Glcβ-Cer; and NeuAcα2-Cer: NeuAcα2-8NeuAcα2-3Galβ1-4Glcβ-Cer. The classification of these SPs was from the method provided by the website LIPID Metabolites and Pathways Strategy(LIPID MAPS) (http://www.lipidmaps.org/). Data are mean ± SD. Four independent experiments were performed. Red asterisks indicate a significant difference (*P < 0.05, Student t test). The raw data for panels A–D can be found in S1 Data.
To link the sphingolipids metabolism to CTL1 function further, we used the sphingolipid synthesis inhibitor fumonisin B1 (FB1) to treat the WT seedlings and found that this treatment induced apical hook defects as observed in the ctl1 mutant (S13 Fig). Taken together, CTL1, as a choline transport protein, plays a role in homeostasis of lipids, including sphingolipids. The disturbed homeostasis of sphingolipids and other lipids in ctl1 mutant may affect the delivery of PIN1, PIN3, and other cargos from the TGN to the PM.
Discussion
CTL1 is essential for auxin-regulated plant development
Auxin is the most investigated phytohormone and plays a pivotal role in virtually every aspect of plant growth and development, and such function often depends on its differential distribution within plant tissues [35]. At the whole plant level, auxin controls organogenesis, apical dominance, lateral root emergence, apical hook development and tropism, vascular development, and many other developmental processes [23, 36–39]. At the cellular level, auxin regulates cell elongation, cell division, and cell differentiation [40].
We showed that Arabidopsis CTL1 plays a key role in a number of auxin-regulated processes, including cell elongation, apical hook development, root morphology, and hypocotyl phototropic response to unilateral light in Arabidopsis. CTL1 expression corresponds to the auxin maxima in the shoot apical meristems, the tips of leaves, the lateral root primordia, the vascular system, and the concave side of apical hook. The dynamic changes in the CTL1 expression pattern strongly resemble the changes in auxin distribution during apical hook development [24]. Moreover, CTL1 is required for maintaining a normal pattern of DR5 expression, a marker for auxin distribution. Loss of function of CTL1 results in enhanced DR5 signals in the tips of cotyledons and reduced signals in the vascular system of hypocotyls, roots, and the concave side of the apical hook. The auxin-related phenotypes and altered auxin distribution observed in the ctl1 mutant provide insights on the cellular and physiological processes that underpin the role of CTL1 in plant development.
CTL1 affects the trafficking of auxin transporters to the PM
Auxin action depends on its differential distribution in plant tissues through a process called polar auxin transport [41]. A number of PM auxin transporters, such as PINs, AUX1, LIKE AUX1 (LAX), and subfamily B of the ATP-binding cassette proteins (ABCB), contribute to the influx and efflux of auxin and subsequently determine the distribution of auxin among different tissues and cells. Therefore, auxin homeostasis requires accurate subcellular targeting of these auxin transporters through secretory trafficking processes. In plant cells, the TGN is a central traffic hub responsible for protein trafficking in both biosynthetic and endocytic pathways. The endocytosed cargos from the PM were received by the TGN, and also through the TGN, biosynthetic cargos were sorted to the PM, cell wall, cell plate, PVC, tonoplast, and vacuole [42]. Protein composition and localization at the PM are regulated by exocytosis and endocytosis in response to developmental and environmental cues.
Here we provide strong evidence that CTL1 is located at multiple subcellular sites associated with secretory trafficking, including the TGN, PVC, and PM. As an endosomal protein, CTL1 may contribute to the trafficking of proteins from the TGN to the PM, especially those PM-localized auxin transporters, as supported by the finding that the abundance of some PIN-type auxin transporters was reduced in the ctl1 mutant, which displays a number of auxin-related phenotypes. In particular, CTL1 is required for the efficient trafficking of PIN1 and PIN3 from the TGN to the PM. As a consequence of losing CTL1 function, the abundance of PIN1 and PIN3 is reduced, and auxin distribution is altered in the root and the apical hook. Further, the CTL1 expression pattern largely overlaps with those of PIN1 and PIN3 in roots and apical hooks [43] [44]. Loss of function of PIN1 or PIN3 causes defects in apical hook development, primary and lateral root growth, and response to unilateral light, which are also the defects found in the ctl1 mutant [4, 45, 46]. Together, results on expression pattern, CTL1 contribution to PIN1 and PIN3 trafficking, and similar mutant phenotypes all support the conclusion that CTL1 regulates the trafficking of PIN1 and PIN3, thereby controlling auxin-related plant developmental processes.
The mechanistic insights of CTL1 function in plant cells
CTL1 has been recently identified as a choline transporter required for sieve plate development [15], although the cellular mechanism underlying CTL1 function is not clear. Our discovery that CTL1 functions in promoting the trafficking of PIN1 and PIN3 may provide a mechanism for CTL1 function in auxin-related developmental processes, including previously described sieve plate biogenesis. In eukaryotic cells, lipid-dependent protein sorting is a major delivery mechanism for cargo proteins from the TGN to cell surface [47]. However, the mechanism by which lipids control vesicular trafficking remains unclear. As membrane lipids provide the microenvironment for protein actions, each membrane system consists of different domains with a specific composition of lipids, such as the sphingolipid-rich microdomains and lipid rafts. Recent studies also reveal various TGN subdomains with different sphingolipid and sterol compositions [48]. Choline is one of the precursors for the synthesis of membrane lipids, including phospholipids and sphingolipids. It is conceivable that CTL1, as a choline transport protein, would play a role in choline delivery to various cell membranes and thus modulate lipid composition of multiple organelles in plant cells. The defect in the homeostasis of membrane lipids in the ctl1 mutant would cause the trafficking defects of some cargo proteins such as PIN1 and PIN3.
Although we cannot detect any differences between the WT and the ctl1 mutant on the morphology, size, and number of the compartments labelled by different fluorescence-tagged organelle markers using confocal microscopy (S14 Fig), there might be other defects on the endomembrane system in ctl1 that we were not able to identify. It remains unclear whether CTL1 functions as a transporter-like protein that prefers choline over other substrates [15]. Further investigations will be required to determine whether CTL1 transports choline specifically or can also transport other substrates. CTL1 is localized to different cellular compartments, including the TGN, PM, and PVC, and its trafficking is sensitive to BFA, suggesting that CTL1 itself is a cargo protein that goes through the secretory pathway. We found that CTL1 affects the trafficking of some special cargo proteins such as PIN1 and PIN3, but not PIN2. This indicates that CTL1 is not required for the proper function of global trafficking machinery; instead, it affects subgroups of proteins. Further survey on the cargo proteins that are affected by CTL1 and identification of proteins that interact with CTL1 will help further understanding of the mechanisms of CTL1 function.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Col-0 was used in this study. Arabidopsis WT, the T-DNA insertion line (SALK_065853C), and the transgenic lines expressing SYP32-mCherry (CS781677) or Rha1-mCherry (CS781672) were obtained from the Arabidopsis Biological Resource Center. The transgenic lines expressing VHA-a1-mRFP [17], DR5::GFP [49], DR5::GUS [50], PIN1pro:PIN1:GFP [51], PIN2pro:PIN2:GFP [52], PIN3pro:PIN3:GFP [52], mCherry-CLC2 [53], AUX1pro:AUX1:GFP [24], and BRI1pro:BRI1:GFP [29] have been described previously. Homozygous individuals were screened by PCR using the primers described in S2 Table. The surface-sterilized Arabidopsis seeds were plated on half-strength MS medium containing 1% (w/v) sucrose. The pH of the medium was adjusted to 5.7 and solidified using 1% (w/v) agar. For soil culture, 7-day-old Arabidopsis seedlings grown on half-strength MS medium were transferred to nutrient-rich soil (Pindstrup Mosebrug, Denmark). The seedlings were grown in a greenhouse under 150 μmol/m2/s light intensity with a 16-hour light/8-hour dark photoperiod at 22°C, unless otherwise indicated. For dark culture, the seeds were planted on half-strength MS and then transferred to white light for 6 hours to enhance germination; subsequently, the plates were wrapped with 2 layers of aluminum foil. For the unilateral light response experiment, seedlings were grown in darkness for 2 days and then exposed to unilateral white light (150 μmol/m2/s) for another 2 days, according to the published procedure [54].
Pharmacological treatments
The seedlings were incubated in liquid half-strength MS medium containing a final concentration of 50 μM BFA (Sigma-Aldrich) or 33 μM wortmannin (Sigma-Aldrich) for 60 minutes, unless described otherwise in the figure legends. Labeling with FM4-64 (Molecular Probes) was carried out as described [29]. For BFA washouts, the seedlings were treated with 50 μM BFA for 1 hour, rinsed 3 times with water, and then incubated in half-strength MS liquid medium for 90 minutes before the imaging procedure.
Root length, hypocotyl length, and apical hook curvature measurements
Images of individual seedlings were acquired using the OLYMPUS stereoscopic microscope equipped with a DP27 camera. The root length, hypocotyl length, and hook angle were then measured using the ImageJ software. The bending angles of the apical hook were scored as previously described [24]. For each time-lapse experiment, 6 seedlings were analyzed.
Generation of plasmid constructs and transgenic Arabidopsis lines
For genetic complementation, a 6,107-bp genomic DNA fragment containing a 1,918-bp promoter region, a 3,117-bp coding region, and a 1,072-bp terminator region after the stop codon of the CTL1 gene were amplified from WT genomic DNA and then cloned into the binary vector pCAMBIA-1300. To generate the CTL1pro:CTL1:EGFP construct, we fused the EGFP and NOS terminator sequence with the 5,032-bp genomic DNA of CTL1, including the 1,918-bp promotor region and the 3,114-bp coding region, and then cloned this recombinant DNA into the binary vector pCAMBIA-1300. A 10-alanine (Ala) linker sequence (GCT GCT GCC GCT GCC GCT GCG GCA GCG GCC) was also inserted between CTL1 and EGFP. To generate the CTL1pro:CTL1:GUS construct, we fused the GUS and NOS terminator sequence to the CTL1 gene as described for the CTL1pro:CTL1:EGFP construct. The organization of these 2 constructs is shown in S1D and S2A Figs, respectively. To generate the LAX3pro:LAX3:EGFP construct, a 3,007-bp genomic DNA of LAX3 including the promotor region and the coding region was fused with the EGFP and NOS terminator sequence and then cloned into pCAMBIA1300. The constructs were introduced into the Agrobacterium tumefaciens GV3101 strain for transformation into Arabidopsis by the floral dipping method [55]. The primers used for plasmids constructions are listed in S2 Table.
Transgenic lines expressing various markers including DR5::GFP, DR5::GUS, PIN1pro:PIN1:GFP, PIN2pro:PIN2:GFP, PIN3pro:PIN3:GFP, AUX1pro:AUX1:GFP, LAX3pro:LAX3:GFP, BRI1pro:BRI1:GFP, VHA-a1-mRFP, SYP32-mCherry, and Rha1-mCherry were individually crossed into the ctl1 mutant background. Homozygous ctl1 plants harboring various markers were isolated from F2 populations by scoring the specific phenotype of the mutant. The F3 and later generations were used for analyses. For colocalization analysis, transgenic lines expressing VHA-a1-mRFP, SYP32-mCherry, Rha1-mCherry, and mCherry-CLC2 were individually crossed with transgenic lines expressing CTL1pro:CTL1: EGFP.
Confocal microscopy analysis
Imaging was performed on an LSM-710 confocal microscope (Zeiss) equipped with an argon/krypton laser. For the quantitative fluorescence intensity, confocal pictures were acquired using strictly identical acquisition parameters (laser power, photomultiplier, offset, zoom factor, and resolution) among the experimental seedlings. The excitation wave lengths for the GFP, FM4-64, mRFP, and mCherry signals were 488, 514, 543, and 587 nm, respectively. For colocalization analysis, the Colocalization Finder plugin of ImageJ was used. The linear Pearson correlation coefficient (rp) was used to indicate the extent of colocalization, with the value of +1.0 as complete colocalization. Detection of nonpolar lipids (excitation at 514 nm, emission at 520–560 nm) and polar lipids (excitation at 534 nm, emission at 600–700 nm) after Nile red staining was performed according to the published protocol [56].
Histochemical GUS analysis
The staining was conducted according to the published protocol [57], with slight modifications. Briefly, transformed Arabidopsis seedlings were treated with 90% (vol/vol) acetone on ice for 30 minutes and then incubated in GUS staining solution (0.5 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide, 100 mM Na3PO4, 10 mM EDTA, 0.1 mM K3[Fe(CN)6], 5 mM K4[Fe(CN)6] and 0.1% [vol/vol] Triton X-100, pH 7.0) at 37°C in darkness for 12 hours. After being sufficiently decolorized with 75% (vol/vol) ethanol, the plant tissues were photographed with an Olympus SZX12 microscope equipped with a camera.
Quantitative real-time PCR analysis
Total RNA was extracted from plant samples using TRIzol reagent (Invitrogen), followed by synthesis of Poly (dT) complementary DNA using the M-MLV Reverse Transcriptase (Promega). qRT-PCR was performed using the SYBR Green I Master kit (Roche Diagnostics) according to the manufacturer’s instructions on a CFX Connect Real-Time System (Bio-Rad). All individual reactions were done in triplicate. The primers used for qRT-PCR are listed in S2 Table.
Lipidomics analysis by UPLC-MS
Plant lipids from 5-day-old WT and ctl1 mutant seedlings were extracted according to the published method [58], and the extracts were directly subjected to LC-MS analysis. The lipidomic data were recorded on an ESI-QTOF/MS (Xevo G2-S Q-TOF, Waters) coupled with a UPLC (ACQUITY UPLC I-Class system, Waters). Parameters for mass spectrometry were as follows: scan range, m/z 50–1,500; ion source, ESI; loop time, 0.2 seconds; cone voltages, 25 KV; capillary voltage, 3 KV; collision energy, 15–60 V; source temperature, 120°C; desolvation temperature, 400°C; desolvation gas flow, 500 L/h; and cone gas flow, 25 L/h. Raw data were imported into the commercial software Progenesis QI (Version 2.3) for data processing, including peak picking and acquiring compounds’ associated information such as m/z, retention time, and intensity. Next, data filtering was performed to delete low-quality data. R project was also applied in further data processing and statistical analysis.
Supporting information
(XLSX)
(XLSX)
(A) Schematic representation of the CTL1 gene. The T-DNA insertion site is located in the first intron (281-bp downstream of the ATG start codon) and is indicated by a triangle. Solid boxes and lines indicate the exons and introns, respectively. (B) Confirmation of the T-DNA insertion in the ctl1 mutant by PCR using the forward primer (F) and reverse primer (R) of CTL1 paired to the T-DNA border primer LB1. (C) Reverse transcription PCR (RT-PCR) analysis of CTL1 mRNA from 10-day-old wild type (WT), ctl1, and ctl1 complemented with a genomic fragment of the CTL1 gene (ctl1C). ACTIN2 mRNA was analyzed as a loading control. (D) Schematic model of the T-DNA region of vector pCAMBIA-1300 containing a genomic fragment of the CTL1 gene. (E) The growth phenotype of 10-day-old WT, ctl1, and ctl1C. (F–H) Quantitative analysis of the petiole length of rosette leaves (F), the ratio of leaf length to width (G), and the rosette diameter (H) in 30-day-old WT, ctl1 and ctl1C grown in the soil. Data are mean ± SD, and significant differences are indicated as ***P < 0.01 (n = 5, Student t test). The raw data for panels F–H can be found in S1 Data.
(TIF)
(A) Schematic model of the transfer deoxyribonucleic acid (T-DNA) region of vector pCAMBIA-1300 containing a genomic fragment of the choline transporter-like 1 (CTL1) gene fused with enhanced green fluorescent protein (EGFP). A 10-alanine (Ala) linker sequence was inserted between CTL1 and EGFP. (B) Real-time quantitative reverse transcription PCR (qRT-PCR) analysis of CTL1 mRNA levels in 10-day-old wild type (WT), ctl1, and lines expressing CTL1pro:CTL1:EGFP in the ctl1 mutant background. Relative expression levels were calculated as the ratio of CTL1 in various seedlings to that in WT. Data are mean ± SD (n = 3). Fifteen CTL1pro:CTL1:EGFP/ctl1 lines were selected for qRT-PCR analysis, and one of them, #11, with CTL1 levels similar to those of the WT, was used for further studies. (C) RT-PCR analysis of CTL1 mRNA in 10-day-old ctl1 mutant and CTL1pro:CTL1:EGFP-transformed line (CTL1pro:CTL1:EGFP/ctl1 #11). ACTIN2 mRNA was analyzed as a loading control. (D) Growth phenotype of 5-day-old WT, ctl1, and CTL1pro:CTL1:EGFP/ctl1 #11 grown on half-strength Murashige and Skoog (MS) solid medium. Bar = 2 mm. (E) Fluorescence signals in the primary root of a 4-day-old CTL1pro:CTL1:EGFP/ctl1 #11 seedling grown on half-strength MS solid medium. Bar = 50 μm. The raw data for panel B can be found in S1 Data.
(TIF)
A representative confocal image of CTL1-EGFP signals (A), mCherry-CLC2 signals (B), and a merge of the 2 signals (C). Arrows indicated the overlapping signals. Bars = 2 μm. (D), Colocalization analysis of CTL1-EGFP with mCherry-CLC2 using the ImageJ software with the colocalization finder plugin.
(TIF)
(A–E) The fluorescence signals in the apical hook region of etiolated seedlings at 12 (A), 24 (B), 48 (C), 72 (D), 96 (E), and 120 (F) hours after germination. Arrows indicate the signals of CTL1-EGFP in the concave side of the apical hook. Bars = 100 μm. (G) The fluorescence intensity of pseudocolor images (blue-yellow-red palette) of CTL1pro:CTL1:EGFP at the apical hook of an etiolated seedling at 1 day after germination (DAG). The arrow indicates a maximum signal of CTL1 at the concave side of the apical hook. Bar = 50 μm. (H) The cortex view of fluorescence signals of CTL1pro:CTL1:EGFP in the hook region of an etiolated seedling at 1 DAG. The arrow indicates a strong signal in the concave side. Bar = 20 μm. (I) The β-glucuronidase (GUS) staining result of CTL1pro:CTL1:GUS in the apical hook region of etiolated seedling at 1 DAG. Bar = 100 μm. (J) The relative intensity of the fluorescence signals at the concave and the convex side of the apical hook. The fluorescence intensity of the concave side of 1 DAG seedling was set as 100%. The other relative values were the ratio of the fluorescence intensity against the concave signal at 1 DAG. Data are mean ± SD (n = 15). The raw data for panel J can be found in S1 Data.
(TIF)
(A–E) Growth phenotype of 10-day-old wild-type (WT) and ctl1 mutant plants grown on half-strength Murashige and Skoog (MS) medium containing 0, 10, 30, or 50 μM SA. Bars = 1 cm. (F) Relative root length (%) indicates shortening of the primary roots as a result of SA in the medium. Data are mean ± SD. Three independent experiments were performed. n = 10 for each experiment. The raw data for panel F can be found in S1 Data.
(TIF)
(A) Relative abundance of DR5–β-glucuronidase (DR5-GUS) signals in the cotyledon tip, hypocotyl, and root parts of the WT and the ctl1 mutants. Data are mean ± SD calculated from 5 seedlings (Student t test, *P < 0.05). (B) Relative intensity of DR5-green fluorescent protein (DR5-GFP) signals in the root tips of the WT and ctl1 mutants when treated with 0, 0.1, 0.3, 0.5, or 1 μM 1-naphthylacetic acid (NAA). Data are mean ± SD calculated from 5 seedlings. (C) Relative intensity of DR5-GFP signals in the concave sides of the apical hook of the WT and the ctl1 mutant at 1 and 3 days after germination. Data are mean ± SD calculated from 5 seedlings (Student t test, *P < 0.05). The raw data for panels A–C can be found in S1 Data.
(TIF)
Fluorescence signals in the apical section of the wild type (WT) and the ctl1 mutant indicate levels of various auxin transporters, including PIN1pro:PIN1:GFP (A and B), PIN3pro:PIN3:GFP (C and D), and AUX1pro:AUX1:GFP (E and F). Green fluorescent protein (GFP) signals in the etiolated seedlings (A–F) were examined on the first day after germination. Scale bars are 50 μm. (G) Relative fluorescence intensity of auxin transporters (PIN1, PIN3, and AUXIN RESISTANT 1 [AUX1]) in the ctl1 mutant as compared to that in the WT (set as 100%). Data are mean ± SD calculated from 40 cells of 5 etiolated seedlings (Student t test, *P < 0.05). The raw data for panel G can be found in S1 Data.
(TIF)
The GFP signals indicate the abundance of BRI1-GFP in 4-day-old primary root of the wild type (WT) (A) and the ctl1 mutant (B). (C) Relative abundance of BRI1-GFP in the ctl1 mutant is shown as % of the WT. Data are mean ± SD from 63 cells in 5 roots of the transgenic lines expressing BRI1-GFP in the WT versus mutant background. Three independent experiments were performed. The raw data for panel C can be found in S1 Data.
(TIF)
Total RNA was extracted from 5-day-old wild type (WT) and ctl1 mutant seedlings. The relative level of the WT was set as 1.0, and mutant levels were ratios against the WT level. Data are mean ± SD (n = 3). The raw data for this figure can be found in S1 Data.
(TIF)
(A–D) PIN2-green fluorescent protein (GFP) localization in wild-type (WT) (A and C) and ctl1 roots (B and D) treated with brefeldin A (BFA) for 60 minutes before (A and B) or after (C and D) washout for 90 minutes. (E) Percentage of cells with PIN2-GFP labeled BFA bodies before and after BFA washout in the WT and the ctl1 mutant. Data are mean ± SD. Three independent experiments were performed. Five roots were used for each experiment (Student t test, *P < 0.05). The raw data for panel E can be found in S1 Data.
(TIF)
(A) Nile red staining of nonpolar lipid and polar lipid in the apical hook region of the wild type (WT) and the ctl1 mutant. (Scale bars = 50 μm.) (B) Quantification of signal intensity of nonpolar and polar lipids as in panel A. Data are mean ± SD from 40 cells in 5 etiolated seedlings. The raw data for panel B can be found in S1 Data.
(TIF)
(A) Ceramide (cer); (B) Galβ1-3GalNAcβ1-3Galα1-3Galβ1-4Glcβ-Cer (Galβ1-Cer); (C) (3'-sulfo) Galβ-Cer; (D) Fucα1-2GalNAcβ1-4(NeuAcα2-8NeuAcα2–3) Galβ1-4Glcβ-Cer (Fucα1-Cer); (E) GalNAcβ1-3Galα1-3Galα1-3Galα1-3Galα1-4Galβ1-4Glcβ-Cer (GalNAcβ1-Cer); (F) Gal-Cer; (G) Manα1-3Manβ1-4Glcβ-Cer (Manα1-Cer); (H) NeuAcα2-8NeuAcα2-3Galβ1-4Glcβ-Cer (NeuAcα2-Cer); (I) GlcNAcβ1-4Manβ1-4Glcβ-Cer (d18:1/16:0), KDNα2-6Galβ1-4GlcNAcβ1-3Galβ1-4Glcβ-Cer (d18:1/26:0) and LacCer (d18:0/24:1); (J) Glc-Cer; (K) phosphatidylethanolamine (PE)-Cer; (L) phosphatidylinositol (PI)-Cer; and (M) sphingomyelins (SM). Data are mean ± SD. Four independent experiments were performed. Red asterisks indicate a significant difference (*P < 0.05, Student t test). The raw data can be found in S1 Data.
(TIF)
(A) Representative images of apical hooks of the wild type (WT) at 36, 60, and 84 hours after germination in the dark. Seedlings were grown on half-strength Murashige and Skoog (MS) medium containing 1 μM FB1. As FB1 was dissolved in the dimethyl sulfoxide (DMSO), the same volume of DMSO was added to the control. (B) Angles of hook curvature calculated from multiple experiments as shown in panel A. Data are mean ± SD, and 3 independent experiments were performed, with similar findings (Student t test, *P < 0.05). The raw data for panel B can be found in S1 Data.
(TIF)
(A and B) VHA-a1-mRFP labeled trans-Golgi network (TGN) in wild type (WT) (A) and ctl1 (B). (C and D) SYP32-mCherry labeled Golgi apparatus in WT (C) and ctl1 (D). (E and F) Rha1-mCherry labeled prevacuolar compartment (PVC) in WT (E) and ctl1 (F). Bars = 2 μm. (G and H) The statistical analysis of organelle size (G) and intensity (H) in WT and ctl1. The organelle size and intensity were measured using the ImageJ software. Data are mean ± SD, and the significant difference was analyzed by Student t test. Six images from 3 roots were used. The raw data for panels G and H can be found in S1 Data.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
We thank Drs. Zhaojun Ding, Weimin Shi, Zhi Hong and Feng Yu for providing plant materials and Drs. Ye Lu and Buhong Gao for assistance in laser confocal analysis.
Abbreviations
- ABCB
subfamily B of ATP-binding cassete transporters
- AP-3
adaptor protein complex 3
- ARF-GEF
ADP-ribosylation factor-type GTPase
- AUX1
AUXIN RESISTANT 1
- BFA
brefeldin A
- BRI1
brassinosteroid receptor
- CCV
clathrin-coated vesicle of the TGN
- CLC2
clathrin light chain 2
- Col-0
Columbia-0
- CTL1
choline transporter-like 1
- DR5
DIRECT REPEAT5
- EGFP
enhanced green fluorescent protein
- FB1
sphingolipid synthesis inhibitor fumonisin B1
- GUS
β-glucuronidase
- LAX
LIKE AUX1
- LAX3
Like AUX1 protein 3
- mRFP
monomer red fluorescent protein
- MS
Murashige and Skoog
- NAA
1-naphthylacetic acid
- ORF
open reading frame
- PIN1
Arabidopsis PM-located auxin efflux transporter PIN-formed 1
- PIN2
Arabidopsis PM-located auxin efflux transporter PIN-formed 2
- PIN3
Arabidopsis PM-located auxin efflux transporter PIN-formed 3
- PM
plasma membrane
- PVC
prevacuolar compartment
- qRT-PCR
real-time quantitative reverse transcription PCR
- Rha1
Arabidopsis Rab homolog F2A
- SA
salicylic acid
- SV
secretory vesicle of the TGN
- SYP32
Syntaxin of plants 32
- T-DNA
transfer deoxyribonucleic acid
- TGN/EE
trans-Golgi network/early endosome
- UPLC-MS
ultra-performance liquid chromatography coupled with mass spectrometry
- VHA-a1
vacuolar proton ATPase subunit VHA-a isoform 1
- Wm
wortmannin
- WT
wild type
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
National Natural Science Foundation of China Grants (grant number 31770267). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The National Science Foundation grant (grant number 1339239). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Fundamental Research Funds for the Central Universities (grant number 020814380062). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Otegui MS, Spitzer C. Endosomal functions in plants. Traffic. 2008; 9(10):1589–98. doi: 10.1111/j.1600-0854.2008.00787.x . [DOI] [PubMed] [Google Scholar]
- 2.Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, et al. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science.1999; 286(5438):316–8. . [DOI] [PubMed] [Google Scholar]
- 3.Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112(2):219–30. . [DOI] [PubMed] [Google Scholar]
- 4.Ding Z, Galvan-Ampudia CS, Demarsy E, Langowski L, Kleine-Vehn J, Fan Y, et al. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol. 2011; 13(4):447–52. doi: 10.1038/ncb2208 . [DOI] [PubMed] [Google Scholar]
- 5.Jaillais Y, Santambrogio M, Rozier F, Fobis-Loisy I, Miege C, Gaude T. The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell. 2007;130(6):1057–70. doi: 10.1016/j.cell.2007.08.040 . [DOI] [PubMed] [Google Scholar]
- 6.Feraru E, Paciorek T, Feraru MI, Zwiewka M, De Groodt R, De Rycke R, et al. The AP-3 beta adaptin mediates the biogenesis and function of lytic vacuoles in Arabidopsis. Plant Cell. 2010; 22(8):2812–24. doi: 10.1105/tpc.110.075424 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zwiewka M, Feraru E, Moller B, Hwang I, Feraru MI, Kleine-Vehn J, et al. The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 2011;21(12):1711–22. doi: 10.1038/cr.2011.99 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao C, Luo M, Zhao Q, Yang R, Cui Y, Zeng Y, et al. A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr Biol. 2014;24(21):2556–63. doi: 10.1016/j.cub.2014.09.014 . [DOI] [PubMed] [Google Scholar]
- 9.Spitzer C, Reyes FC, Buono R, Sliwinski MK, Haas TJ, Otegui MS. The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell. 2009;21(3):749–66. doi: 10.1105/tpc.108.064865 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutte Y, Jonsson K, McFarlane HE, Johnson E, Gendre D, Swarup R, et al. ECHIDNA-mediated post-Golgi trafficking of auxin carriers for differential cell elongation. Proc Natl Acad Sci USA. 2013;110(40):16259–64. doi: 10.1073/pnas.1309057110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130(6):1044–56. doi: 10.1016/j.cell.2007.07.033 . [DOI] [PubMed] [Google Scholar]
- 12.Rigo G, Ayaydin F, Tietz O, Zsigmond L, Kovacs H, Pay A, et al. Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell. 2013;25(5):1592–608. doi: 10.1105/tpc.113.110452 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhonukshe P, Huang F, Galvan-Ampudia CS, Mahonen AP, Kleine-Vehn J, Xu J, et al. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development. 2010;137(19):3245–55. doi: 10.1242/dev.052456 . [DOI] [PubMed] [Google Scholar]
- 14.Willige BC, Ogiso-Tanaka E, Zourelidou M, Schwechheimer C. WAG2 represses apical hook opening downstream from gibberellin and PHYTOCHROME INTERACTING FACTOR 5. Development. 2012;139(21):4020–8. doi: 10.1242/dev.081240 . [DOI] [PubMed] [Google Scholar]
- 15.Dettmer J, Ursache R, Campilho A, Miyashima S, Belevich I, O'Regan S, et al. CHOLINE TRANSPORTER-LIKE1 is required for sieve plate development to mediate long-distance cell-to-cell communication. Nat Commun. 2014;5:4276 doi: 10.1038/ncomms5276 . [DOI] [PubMed] [Google Scholar]
- 16.Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Hofte H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997;114(1):295–305. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18(3):715–30. doi: 10.1105/tpc.105.037978 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geldner N, Denervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 2009;59(1):169–78. doi: 10.1111/j.1365-313X.2009.03851.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer-Parton S, Parton RM, Hickey PC, Dijksterhuis J, Atkinson HA, Read ND. Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J Microsc. 2000;198(Pt 3):246–59. . [DOI] [PubMed] [Google Scholar]
- 20.Contento AL, Bassham DC. Structure and function of endosomes in plant cells. J Cell Sci. 2012;125(Pt 15):3511–8. doi: 10.1242/jcs.093559 . [DOI] [PubMed] [Google Scholar]
- 21.Tse YC, Mo B, Hillmer S, Zhao M, Lo SW, Robinson DG, et al. Identification of Multivesicular Bodies as Prevacuolar Compartments in Nicotiana tabacum BY-2 Cells. Plant Cell. 2004;16(3):672–93. doi: 10.1105/tpc.019703 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA. Electron tomography of RabA4b- and PI-4Kbeta1-labeled trans Golgi network compartments in Arabidopsis. Traffic. 2011;12(3):313–29. doi: 10.1111/j.1600-0854.2010.01146.x . [DOI] [PubMed] [Google Scholar]
- 23.Abbas M, Alabadi D, Blazquez MA. Differential growth at the apical hook: all roads lead to auxin. Front Plant Sci. 2013;4:441 doi: 10.3389/fpls.2013.00441 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandenbussche F, Petrasek J, Zadnikova P, Hoyerova K, Pesek B, Raz V, et al. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development. 2010. February;137(4):597–606. doi: 10.1242/dev.040790 . [DOI] [PubMed] [Google Scholar]
- 25.Zadnikova P, Petrasek J, Marhavy P, Raz V, Vandenbussche F, Ding Z, et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development. 2010;137(4):607–17. doi: 10.1242/dev.041277 . [DOI] [PubMed] [Google Scholar]
- 26.Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413(6854):425–8. doi: 10.1038/35096571 . [DOI] [PubMed] [Google Scholar]
- 27.Zhang C, Brown MQ, van de Ven W, Zhang ZM, Wu B, Young MC, et al. Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc Natl Acad Sci USA. 2017;113(1):E41–50. doi: 10.1073/pnas.1521248112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang JB, Liu H, Chen M, Li X, Wang M, Yang Y, et al. ROP3 GTPase contributes to polar auxin transport and auxin responses and is important for embryogenesis and seedling growth in Arabidopsis. Plant Cell. 2014;26(9):3501–18. doi: 10.1105/tpc.114.127902 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo Y, Scholl S, Doering A, Zhang Y, Irani NG, Di Rubbo S, et al. V-ATPase activity in the TGN/EE is required for exocytosis and recycling in Arabidopsis. Nat Plants. 2015;1:15094 doi: 10.1038/nplants.2015.94 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Richter GL, Wang X, Mlodzinska E, Carraro N, Ma G, et al. Sterols and sphingolipids differentially function in trafficking of the Arabidopsis ABCB19 auxin transporter. Plant J. 2013;74(1):37–47. doi: 10.1111/tpj.12103 . [DOI] [PubMed] [Google Scholar]
- 31.Markham JE, Molino D, Gissot L, Bellec Y, Hematy K, Marion J, et al. Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell. 2011;23(6):2362–78. doi: 10.1105/tpc.110.080473 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li R, Sun R, Hicks GR, Raikhel NV. Arabidopsis ribosomal proteins control vacuole trafficking and developmental programs through the regulation of lipid metabolism. Proc Natl Acad Sci USA. 2015;112(1):E89–98. doi: 10.1073/pnas.1422656112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeisel SH, Da Costa KA, Franklin PD, Alexander EA, Lamont JT, Sheard NF, et al. Choline, an essential nutrient for humans. FASEB J. 1991;5(7):2093–8. . [PubMed] [Google Scholar]
- 34.Diaz G, Melis M, Batetta B, Angius F, Falchi AM. Hydrophobic characterization of intracellular lipids in situ by Nile Red red/yellow emission ratio. Micron. 2008;39(7):819–24. doi: 10.1016/j.micron.2008.01.001 . [DOI] [PubMed] [Google Scholar]
- 35.Benjamins R, Scheres B. Auxin: the looping star in plant development. Annu Rev Plant Biol. 2008;59:443–65. doi: 10.1146/annurev.arplant.58.032806.103805 . [DOI] [PubMed] [Google Scholar]
- 36.Scarpella E, Barkoulas M, Tsiantis M. Control of leaf and vein development by auxin. Cold Spring Harb Perspect Biol. 2010;2(1):a001511 doi: 10.1101/cshperspect.a001511 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karampelias M, Neyt P, De Groeve S, Aesaert S, Coussens G, Rolcik J, et al. ROTUNDA3 function in plant development by phosphatase 2A-mediated regulation of auxin transporter recycling. Proc Natl Acad Sci USA. 2016;113(10):2768–73. doi: 10.1073/pnas.1501343112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rakusova H, Fendrych M, Friml J. Intracellular trafficking and PIN-mediated cell polarity during tropic responses in plants. Curr Opin Plant Biol. 2015;23:116–23. doi: 10.1016/j.pbi.2014.12.002 . [DOI] [PubMed] [Google Scholar]
- 39.Lucas WJ, Groover A, Lichtenberger R, Furuta K, Yadav SR, Helariutta Y, et al. The plant vascular system: evolution, development and functions. J Integr Plant Biol. 2013;55(4):294–388. doi: 10.1111/jipb.12041 . [DOI] [PubMed] [Google Scholar]
- 40.Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136(6):1005–16. doi: 10.1016/j.cell.2009.03.001 . [DOI] [PubMed] [Google Scholar]
- 41.Petrasek J, Friml J. Auxin transport routes in plant development. Development 2009;136(16):2675–88. doi: 10.1242/dev.030353 . [DOI] [PubMed] [Google Scholar]
- 42.Reyes FC, Buono R, Otegui MS. Plant endosomal trafficking pathways. Curr Opin Plant Biol. 2011;14(6):666–73. doi: 10.1016/j.pbi.2011.07.009 . [DOI] [PubMed] [Google Scholar]
- 43.Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell. 2007;19(1):131–47. doi: 10.1105/tpc.106.040782 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2002;415(6873):806–9. doi: 10.1038/415806a . [DOI] [PubMed] [Google Scholar]
- 45.Blakeslee JJ, Bandyopadhyay A, Peer WA, Makam SN, Murphy AS. Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol. 2004;134(1):28–31. doi: 10.1104/pp.103.031690 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peret B, De Rybel B, Casimiro I, Benkova E, Swarup R, Laplaze L, et al. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 2009;14(7):399–408. doi: 10.1016/j.tplants.2009.05.002 . [DOI] [PubMed] [Google Scholar]
- 47.Surma MA, Klose C, Simons K. Lipid-dependent protein sorting at the trans-Golgi network. Biochimica et biophysica acta. 2012;1821(8):1059–67. doi: 10.1016/j.bbalip.2011.12.008 . [DOI] [PubMed] [Google Scholar]
- 48.Wattelet-Boyer V, Brocard L, Jonsson K, Esnay N, Joubes J, Domergue F, et al. Enrichment of hydroxylated C24- and C26-acyl-chain sphingolipids mediates PIN2 apical sorting at trans-Golgi network subdomains. Nat Commun. 2016;7:12788 doi: 10.1038/ncomms12788 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ottenschlager I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, et al. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA. 2003;100(5):2987–91. doi: 10.1073/pnas.0437936100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997. November;9(11):1963–71. doi: 10.1105/tpc.9.11.1963 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115(5):591–602. . [DOI] [PubMed] [Google Scholar]
- 52.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433(7021):39–44. doi: 10.1038/nature03184 . [DOI] [PubMed] [Google Scholar]
- 53.Li X, Wang X, Yang Y, Li R, He Q, Fang X, et al. Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell. 2011;23(10):3780–97. doi: 10.1105/tpc.111.091454 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang KX, Xu HH, Yuan TT, Zhang L, Lu YT. Blue-light-induced PIN3 polarization for root negative phototropic response in Arabidopsis. Plant J. 2013;76(2):308–21. doi: 10.1111/tpj.12298 . [DOI] [PubMed] [Google Scholar]
- 55.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–43. . [DOI] [PubMed] [Google Scholar]
- 56.Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. Journal Cell Biol. 1985;100(3):965–73. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geldner N, Richter S, Vieten A, Marquardt S, Torres-Ruiz RA, Mayer U, et al. Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development. 2004;131(2):389–400. doi: 10.1242/dev.00926 . [DOI] [PubMed] [Google Scholar]
- 58.Okazaki Y, Otsuki H, Narisawa T, Kobayashi M, Sawai S, Kamide Y, et al. A new class of plant lipid is essential for protection against phosphorus depletion. Nature Commun. 2013;4:1510 doi: 10.1038/ncomms2512 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(A) Schematic representation of the CTL1 gene. The T-DNA insertion site is located in the first intron (281-bp downstream of the ATG start codon) and is indicated by a triangle. Solid boxes and lines indicate the exons and introns, respectively. (B) Confirmation of the T-DNA insertion in the ctl1 mutant by PCR using the forward primer (F) and reverse primer (R) of CTL1 paired to the T-DNA border primer LB1. (C) Reverse transcription PCR (RT-PCR) analysis of CTL1 mRNA from 10-day-old wild type (WT), ctl1, and ctl1 complemented with a genomic fragment of the CTL1 gene (ctl1C). ACTIN2 mRNA was analyzed as a loading control. (D) Schematic model of the T-DNA region of vector pCAMBIA-1300 containing a genomic fragment of the CTL1 gene. (E) The growth phenotype of 10-day-old WT, ctl1, and ctl1C. (F–H) Quantitative analysis of the petiole length of rosette leaves (F), the ratio of leaf length to width (G), and the rosette diameter (H) in 30-day-old WT, ctl1 and ctl1C grown in the soil. Data are mean ± SD, and significant differences are indicated as ***P < 0.01 (n = 5, Student t test). The raw data for panels F–H can be found in S1 Data.
(TIF)
(A) Schematic model of the transfer deoxyribonucleic acid (T-DNA) region of vector pCAMBIA-1300 containing a genomic fragment of the choline transporter-like 1 (CTL1) gene fused with enhanced green fluorescent protein (EGFP). A 10-alanine (Ala) linker sequence was inserted between CTL1 and EGFP. (B) Real-time quantitative reverse transcription PCR (qRT-PCR) analysis of CTL1 mRNA levels in 10-day-old wild type (WT), ctl1, and lines expressing CTL1pro:CTL1:EGFP in the ctl1 mutant background. Relative expression levels were calculated as the ratio of CTL1 in various seedlings to that in WT. Data are mean ± SD (n = 3). Fifteen CTL1pro:CTL1:EGFP/ctl1 lines were selected for qRT-PCR analysis, and one of them, #11, with CTL1 levels similar to those of the WT, was used for further studies. (C) RT-PCR analysis of CTL1 mRNA in 10-day-old ctl1 mutant and CTL1pro:CTL1:EGFP-transformed line (CTL1pro:CTL1:EGFP/ctl1 #11). ACTIN2 mRNA was analyzed as a loading control. (D) Growth phenotype of 5-day-old WT, ctl1, and CTL1pro:CTL1:EGFP/ctl1 #11 grown on half-strength Murashige and Skoog (MS) solid medium. Bar = 2 mm. (E) Fluorescence signals in the primary root of a 4-day-old CTL1pro:CTL1:EGFP/ctl1 #11 seedling grown on half-strength MS solid medium. Bar = 50 μm. The raw data for panel B can be found in S1 Data.
(TIF)
A representative confocal image of CTL1-EGFP signals (A), mCherry-CLC2 signals (B), and a merge of the 2 signals (C). Arrows indicated the overlapping signals. Bars = 2 μm. (D), Colocalization analysis of CTL1-EGFP with mCherry-CLC2 using the ImageJ software with the colocalization finder plugin.
(TIF)
(A–E) The fluorescence signals in the apical hook region of etiolated seedlings at 12 (A), 24 (B), 48 (C), 72 (D), 96 (E), and 120 (F) hours after germination. Arrows indicate the signals of CTL1-EGFP in the concave side of the apical hook. Bars = 100 μm. (G) The fluorescence intensity of pseudocolor images (blue-yellow-red palette) of CTL1pro:CTL1:EGFP at the apical hook of an etiolated seedling at 1 day after germination (DAG). The arrow indicates a maximum signal of CTL1 at the concave side of the apical hook. Bar = 50 μm. (H) The cortex view of fluorescence signals of CTL1pro:CTL1:EGFP in the hook region of an etiolated seedling at 1 DAG. The arrow indicates a strong signal in the concave side. Bar = 20 μm. (I) The β-glucuronidase (GUS) staining result of CTL1pro:CTL1:GUS in the apical hook region of etiolated seedling at 1 DAG. Bar = 100 μm. (J) The relative intensity of the fluorescence signals at the concave and the convex side of the apical hook. The fluorescence intensity of the concave side of 1 DAG seedling was set as 100%. The other relative values were the ratio of the fluorescence intensity against the concave signal at 1 DAG. Data are mean ± SD (n = 15). The raw data for panel J can be found in S1 Data.
(TIF)
(A–E) Growth phenotype of 10-day-old wild-type (WT) and ctl1 mutant plants grown on half-strength Murashige and Skoog (MS) medium containing 0, 10, 30, or 50 μM SA. Bars = 1 cm. (F) Relative root length (%) indicates shortening of the primary roots as a result of SA in the medium. Data are mean ± SD. Three independent experiments were performed. n = 10 for each experiment. The raw data for panel F can be found in S1 Data.
(TIF)
(A) Relative abundance of DR5–β-glucuronidase (DR5-GUS) signals in the cotyledon tip, hypocotyl, and root parts of the WT and the ctl1 mutants. Data are mean ± SD calculated from 5 seedlings (Student t test, *P < 0.05). (B) Relative intensity of DR5-green fluorescent protein (DR5-GFP) signals in the root tips of the WT and ctl1 mutants when treated with 0, 0.1, 0.3, 0.5, or 1 μM 1-naphthylacetic acid (NAA). Data are mean ± SD calculated from 5 seedlings. (C) Relative intensity of DR5-GFP signals in the concave sides of the apical hook of the WT and the ctl1 mutant at 1 and 3 days after germination. Data are mean ± SD calculated from 5 seedlings (Student t test, *P < 0.05). The raw data for panels A–C can be found in S1 Data.
(TIF)
Fluorescence signals in the apical section of the wild type (WT) and the ctl1 mutant indicate levels of various auxin transporters, including PIN1pro:PIN1:GFP (A and B), PIN3pro:PIN3:GFP (C and D), and AUX1pro:AUX1:GFP (E and F). Green fluorescent protein (GFP) signals in the etiolated seedlings (A–F) were examined on the first day after germination. Scale bars are 50 μm. (G) Relative fluorescence intensity of auxin transporters (PIN1, PIN3, and AUXIN RESISTANT 1 [AUX1]) in the ctl1 mutant as compared to that in the WT (set as 100%). Data are mean ± SD calculated from 40 cells of 5 etiolated seedlings (Student t test, *P < 0.05). The raw data for panel G can be found in S1 Data.
(TIF)
The GFP signals indicate the abundance of BRI1-GFP in 4-day-old primary root of the wild type (WT) (A) and the ctl1 mutant (B). (C) Relative abundance of BRI1-GFP in the ctl1 mutant is shown as % of the WT. Data are mean ± SD from 63 cells in 5 roots of the transgenic lines expressing BRI1-GFP in the WT versus mutant background. Three independent experiments were performed. The raw data for panel C can be found in S1 Data.
(TIF)
Total RNA was extracted from 5-day-old wild type (WT) and ctl1 mutant seedlings. The relative level of the WT was set as 1.0, and mutant levels were ratios against the WT level. Data are mean ± SD (n = 3). The raw data for this figure can be found in S1 Data.
(TIF)
(A–D) PIN2-green fluorescent protein (GFP) localization in wild-type (WT) (A and C) and ctl1 roots (B and D) treated with brefeldin A (BFA) for 60 minutes before (A and B) or after (C and D) washout for 90 minutes. (E) Percentage of cells with PIN2-GFP labeled BFA bodies before and after BFA washout in the WT and the ctl1 mutant. Data are mean ± SD. Three independent experiments were performed. Five roots were used for each experiment (Student t test, *P < 0.05). The raw data for panel E can be found in S1 Data.
(TIF)
(A) Nile red staining of nonpolar lipid and polar lipid in the apical hook region of the wild type (WT) and the ctl1 mutant. (Scale bars = 50 μm.) (B) Quantification of signal intensity of nonpolar and polar lipids as in panel A. Data are mean ± SD from 40 cells in 5 etiolated seedlings. The raw data for panel B can be found in S1 Data.
(TIF)
(A) Ceramide (cer); (B) Galβ1-3GalNAcβ1-3Galα1-3Galβ1-4Glcβ-Cer (Galβ1-Cer); (C) (3'-sulfo) Galβ-Cer; (D) Fucα1-2GalNAcβ1-4(NeuAcα2-8NeuAcα2–3) Galβ1-4Glcβ-Cer (Fucα1-Cer); (E) GalNAcβ1-3Galα1-3Galα1-3Galα1-3Galα1-4Galβ1-4Glcβ-Cer (GalNAcβ1-Cer); (F) Gal-Cer; (G) Manα1-3Manβ1-4Glcβ-Cer (Manα1-Cer); (H) NeuAcα2-8NeuAcα2-3Galβ1-4Glcβ-Cer (NeuAcα2-Cer); (I) GlcNAcβ1-4Manβ1-4Glcβ-Cer (d18:1/16:0), KDNα2-6Galβ1-4GlcNAcβ1-3Galβ1-4Glcβ-Cer (d18:1/26:0) and LacCer (d18:0/24:1); (J) Glc-Cer; (K) phosphatidylethanolamine (PE)-Cer; (L) phosphatidylinositol (PI)-Cer; and (M) sphingomyelins (SM). Data are mean ± SD. Four independent experiments were performed. Red asterisks indicate a significant difference (*P < 0.05, Student t test). The raw data can be found in S1 Data.
(TIF)
(A) Representative images of apical hooks of the wild type (WT) at 36, 60, and 84 hours after germination in the dark. Seedlings were grown on half-strength Murashige and Skoog (MS) medium containing 1 μM FB1. As FB1 was dissolved in the dimethyl sulfoxide (DMSO), the same volume of DMSO was added to the control. (B) Angles of hook curvature calculated from multiple experiments as shown in panel A. Data are mean ± SD, and 3 independent experiments were performed, with similar findings (Student t test, *P < 0.05). The raw data for panel B can be found in S1 Data.
(TIF)
(A and B) VHA-a1-mRFP labeled trans-Golgi network (TGN) in wild type (WT) (A) and ctl1 (B). (C and D) SYP32-mCherry labeled Golgi apparatus in WT (C) and ctl1 (D). (E and F) Rha1-mCherry labeled prevacuolar compartment (PVC) in WT (E) and ctl1 (F). Bars = 2 μm. (G and H) The statistical analysis of organelle size (G) and intensity (H) in WT and ctl1. The organelle size and intensity were measured using the ImageJ software. Data are mean ± SD, and the significant difference was analyzed by Student t test. Six images from 3 roots were used. The raw data for panels G and H can be found in S1 Data.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.