Abstract
In May 2012, the first authenticated cases of active chikungunya virus infection were detected in Champasak Province, Southern Laos. Analysis of series of human samples and mosquito specimens collected during the outbreak and over the year that followed the emergence enabled the drawing up of a map of the progression of CHIKV and the establishment of a full genetic characterization of the virus.
Introduction
Chikungunya virus (CHIKV; Togaviridae, genus alphavirus) was first identified in Tanzania in the 1950s. Less than a decade later, an outbreak was recorded in Thailand, initially considered as a possible consequence of the introduction of CHIKV from Africa [1; 2].
Genetic variability studies of CHIKV strains evidenced three different genotypes, referred to as Western African, Asian, and Eastern/Central/Southern African (ECSA) genotypes. While only a few complete genome sequences of Asian CHIKV isolates were obtained before 2005, phylogenetic analyses supported the contention that Asian isolates had significantly diversified to form a specific Asian genotype present in Asia since at least the eighteenth century [2]. Historical descriptions of clinical syndromes and epidemiologic reports also support the probable presence of CHIKV in Asia during past centuries [2, 3]. Unlike Western African and Asian genotypes that spread poorly out of their regions of origin, the ECSA genotype has dispersed widely since 2006, from the Indian Ocean islands to Asia and Western countries [4, 5, 6]. Several recent studies in Asia over the last ten years evidenced that the ECSA genotype tends to supplant the Asian genotypes [7, 8, 9]. However, Asian genotypes were maintained, at least in Indonesia, and have recently spread out into Southern Pacific territories, the Caribbean islands, and the continental Americas [9, 10, 11].
Follow up on CHIKV emergence events in different countries offered opportunities to study the evolution of the virus in new environmental conditions [12, 13, 14, 15]. The extension of Aedes species into new territories during inter-epidemic periods favored the emergence or re-emergence of CHIKV [4, 5, 6, 10]. Such events have been investigated at the phylogenetic level and reveal micro-evolutionary processes with both common and specific signatures within the viral sequences [6, 12, 13]. Among the markers of genetic viral microevolution, adaptive mutation to specific vector species or possibly associated with viral virulence have been reported in structural and/or non-structural genes.
Since that time, numbers of outbreaks have been recorded in urban centers throughout the Indochinese peninsula, while there is very little evidence to support a possible maintenance of the virus in sylvatic cycles [16, 17, 18]. More recently, the emergence of CHIKV was reported in China and Singapore [19].
Detection and isolation of CHIKV from mosquitoes is increasingly reported during epidemics [20, 21 22, 23]. Virological surveillance in vector populations provides valuable information for vector control monitoring and for the assessment of the co-circulation of other Aedes-borne viruses such as dengue virus (DENV).
Despite evidence of a frequent circulation of CHIKV in Southeast Asia, only indirect serological data were available in the Lao PDR. The presence of CHIKV or a closely related virus has been mentioned in the past, but relies only on serological investigations [1, 24]. The risk of exposure for populations living in rural or remote mountainous areas has been estimated to be very low [25]. Other alphaviruses, members of the Semliki Forest virus antigenic complex such as Me Tri or Getah, have been reported in neighboring countries, raising the question of the specificity of the antibodies detected in the former studies [26, 27]. A seroprevalence study conducted in Vientiane City detected no positivity, suggesting that the population, at least in this urban setting, was mostly naïve to CHIKV infection [24].
Based on the report of the National Center for Laboratory and Epidemiology (NCLE), a dengue-like syndrome outbreak that occurred from May to September 2012 was linked to CHIKV, as suggested by detection of the viral genome in some suspected cases [24]. A recent published study highlighted the co-circulation of CHIKV and DENV and the occurrence of CHIKV-DENV coinfection in Champasak Province, Southern Laos [28]. On the other hand, some confirmed cases were reported in Phreah Vihear, Northern Cambodia, close to the Lao border, in late 2011, and some scarce molecular information based only on partial envelope gene sequencing suggested a link between Lao and other Southeast Asian CHIKV strains [24, 28, 29]. Furthermore, no geographic extension of CHIKV from the original outbreak area has yet been determined.
We performed multidisciplinary studies from the early beginning of the emergence of CHIKV in Champasak Province in summer 2012 up to December 2013 when the last case was reported. We describe here the general context of CHIKV emergence and spread and provide genetic information to help understand the origin and evolution of the CHIKV virus in the southern provinces of the Lao PDR.
Materials and methods
Human samples
Human samples from Champasak Province were collected on different occasions. In early August 2012, grouped cases of dengue-like syndromes were reported by the district health services in two villages of Moonlapamok, Champasak Province, located at the Laos–Cambodia border [24]. Human cases were recorded by the district health authorities using a case definition adapted from the WHO recommendation used for dengue surveillance (i.e. acute fever ≥38.5°C with at least one companion symptom from the list referred to in the WHO 2009 classification of dengue patients) [30]. Patients matching this case definition in Champasak Province were sampled after informed consent by venous puncture within seven days of fever onset and submitted to CHIKV screening. Some samples were first screened for the presence of anti-CHIKV antibodies and viral genome at the NCLE in Vientiane Capital [24]. Some of these samples (n = 16) were then transferred to the Institut Pasteur du Laos for confirmation and viral isolation assays as described elsewhere [31]. An additional series of 51 CHIKV suspected cases were identified and sampled during a joint field mission involving WHO, NCLE, and IPL staff which was set up in Moonlapamock and Khong Districts from July to September 2012.
In 2013, volunteers sampled during a seroprevalence study held in the districts of Moonlapamock and Khong (see below) and presenting a serological profile compatible with an acute phase of a CHIKV infection (n = 34) were screened for the presence of CHIKV genome sequences. Clinicians from provinces located north of Champasak (i.e. Sekong, Savannakhet, Salavan, Attapeu) were informed of the risk of CHIKV circulation and encouraged to collect early blood samples for a combined CHIKV–DENV investigation. A total of 475 cases matching the CHIKV case definition were collected through this investigative network from June to December 2013.
Dengue-like cases collected by the NCLE (n = 47), negative for active dengue infection markers, were screened for CHIKV genome by RT-PCR at IPL as described by Pastorino et al. [31].
Seroprevalence study in Champasak Province
A seroprevalence study was carried out from March 18th to April 11th 2013 in Moonlapamock and Khong Districts, Champasak Province. Data from the National Institute of Statistics were used to make a random selection of villages and determine the size of the sampled population. The whole populations in Moonlapamock and Khong Districts were 33,111 and 84,449 inhabitants respectively. The minimum sample size has been determined for an expected precision of 5% and a statistical accuracy of 80% using the OPEN EPI program. With an estimated prevalence of 50% in a total population of 670,122 inhabitants living in the area studied, the minimum sample size was 384 volunteers. The EPI WHO program was used to calculate the representative number of villages and the required number of volunteers per village. To be significant, it was necessary to include at least 30 villages with a minimum sample of 13 people in each village.
Microsoft Excel software was used to proceed with a random selection of the villages in the two targeted districts. Among the selected villages, four were drawn twice. Thus the targeted number of volunteers was increased to 26 in these villages. At the end of the process, 22 villages were selected in Khong District and four villages in Moonlapamok District. A standardized questionnaire was administered by field workers to collect data on demographics, past symptoms compatible with a CHIKV infection during the previous year, and treatments. A blood sample (5 ml) was collected after informed consent had been obtained. Anti-chikungunya IgM and IgG were detected by means of in-house ELISA methods described elsewhere [6, 32].
Ethical statements
The studies protocol and surveillance project were approved by the National Ethics Committee for Health Research of the Ministry of Health of the Lao PDR. All public hospitals’ management committees approved the studies and obtained the agreement of the Ministry of Health for participating in the protocol.
All adult volunteers provided written informed consent. A parent or a legal guardian of any child included in the study signed a consent form on their behalf.
Chikungunya vector(s) investigation
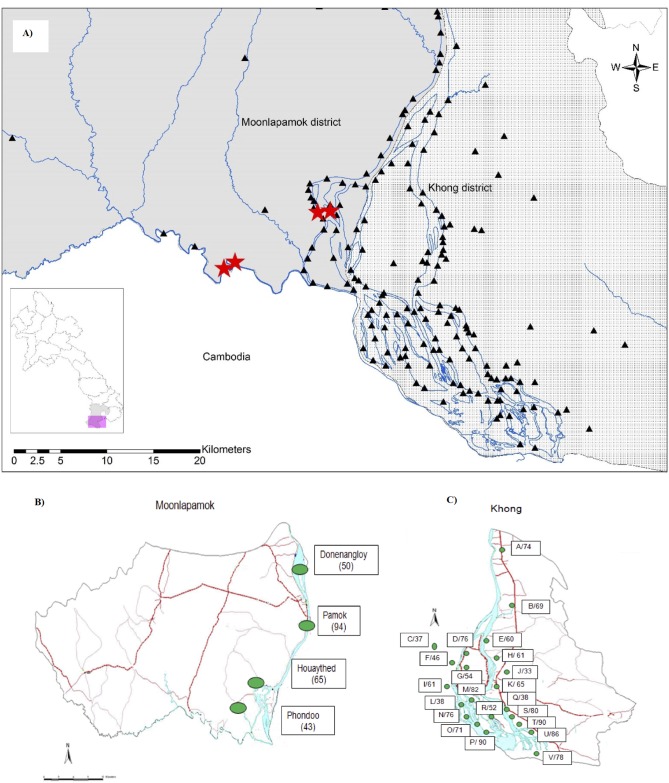
Entomological surveys were carried out in August 2012 in three villages of Moonlapamok District and one village of Khong District, where suspected and confirmed chikungunya cases were reported (Fig 1A). The houses and neighborhoods of clinically ill patients were selected for larval and adult mosquito sampling. For larval collection, all water-holding containers in and around a house were checked for the presence of immature mosquitoes. Mosquito larvae and pupae were collected using standard 250 ml dippers or sieves and pipettes. The main characteristics of larvae breeding sites (type of container, container function, and maximum capacity) were recorded. The larvae and pupae were counted and reared to adult to improve morphological identification. The House Index (i.e. percentage of houses with Aedes breeding sites), Container Index (percentage of containers positive for the presence of Aedes larvae), and Breteau Index (positive containers per 100 houses) were calculated.
Fig 1. Chikungunya virus study sites in 2012–2013.
1A) Entomologic surveillance sites in September 2012. Black triangles represent villages in Moonlapamok and Khong Districts. Red stars represent villages where mosquito larvae were sampled. 1B) Chikungunya virus IgM seroprevalence in villages in Moonlapamok District. 1C) Chikungunya virus IgM seroprevalence in villages in Khong District. Letters correspond to the villages’ code and numbers to the recorded seroprevalence level.
Adult mosquitoes resting inside a house were collected using sweep nets and aspirators. CDC light traps were set up from 3:30–4:30 pm to 7:30–8:30 am, both inside and outside houses. The mosquito adults (both those that emerged from larvae or nymphs and those collected as adults) were identified morphologically using the keys from Thailand and Vietnam and pooled for virus detection (by species, sex, and method of capture) [33, 34]. Mosquito pools were stored in liquid nitrogen and sent to the Institut Pasteur du Laos in Vientiane Capital.
Analysis of mosquito samples
Adults or imagoes emerged from larvae were identified and sorted by species and sex. All specimens were dissected individually. Abdomens, wings, and legs of up to ten specimens were grouped in pools for rapid screening. Head-thorax segments were kept frozen at −80°C. Tissue pools were suspended in 400 μl of cold PBS and crushed for 1 min at full speed (25 oscillation/s) in a TissueLyser homogenizer (Qiagen) in the presence of LysingMatrix E beads (MP Biomedicals). Residual tissue fragments were pelleted by spinning the tubes at 10,000g for 5 minutes. One half of the supernatants (200 μl) were used for total nucleic acid extraction using Nucleospin Viral RNA kits (Macherey Nagel) according to manufacturer’s instructions. The rest of the tissue lysates were kept frozen at −80°C for viral isolation assays. Samples were screened for the presence of CHIKV sequences by the real-time RT-PCR method [31].
Head-thorax segments from positive pools were investigated individually by the same procedure to determine the effective number of infected mosquitoes.
Chikungunya virus isolation
Human samples positive by RT-PCR were inoculated on Vero E6 cells with 100μl of each serum after filtration through a 0.22 μm membrane. Cultures were monitored by daily observation for the presence of cytopathic effect (CPE). Supernatants from cultures displaying a CPE were tested by RT-PCR, generally between Day 3 to Day 5 post-infection. At this stage, CPE generally reached at least 70% of the cell monolayers.
Supernatants of positive pools and head-thorax segments were inoculated to Vero E6 cells. Sub-confluent Vero E6 cells monolayers prepared in 25 cm2 flasks were inoculated by 200 μl of supernatant diluted 5 times in DMEM medium after filtration on 0.22μm membranes (Sartorius). The inoculum was removed after 2 hours’ incubation at 37°C and replaced by 5 ml of fresh DMEM completed with 2.5% fetal calf serum. Cultures were monitored using real-time RT-PCR [31].
Sequence analysis
Viral genomic RNA extraction was carried out from human plasma or from CHIKV primary isolates (passage 1) in Vero E6 cells supernatant using NucleoSpin II RNA kits (Macherey Nagel) according to the manufacturer’s instructions. Sequencing of the E2-6K-E1 region (2,771 nt) or the entire viral genome was performed using primers designed to obtain 700 bp RT-PCR amplicons [6, 12,]. Amplicons generated presented an overlap of 100 bp between contiguous fragments. RT-PCR was performed using SuperScript One-Step RT-PCR with Platinum Taq (Invitrogen) with primers targeting the E2-6K-E1 region or the entire genome [12]. RT-PCR fragments were purified by ultrafiltration. Sequencing reactions were performed using the BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems). Sequence chromatograms from both strands were obtained on automated sequence analyzer ABI3730XL (Applied Biosystems). For sequence analysis, contig assembly and sequence alignment were performed using the BioNumerics version 6.5 program (Applied-Maths, Saint-Martens-Latem, Belgium).
Phylogenetic analysis
For phylogenetic analysis of the E2-6K-E1 region or complete genome, maximum-likelihood trees were constructed using MEGA version 6 (www.megasoftware.net), with the Kimura-2 parameter corrections of multiple substitutions. Reliability of nodes was assessed by bootstrap resampling with 5,000 replicates. A sequence of Ivory Coast (West Africa lineage) isolate (HM045818) was used as an outgroup to root the tree. Nucleotide sequences of the E2-6K-E1 region and the complete genome of isolates used in this study were submitted to EMBL-EBI and their accession numbers are shown in Table 1.
Table 1. List of Lao CHIKV strains from Champasak Province genetically characterized in this study (with GenBank accession numbers for Lao E2-6K-E1 and complete genome generated sequences).
| Isolate ID | District | Village | Date | Acc. Nbr E2-6K-E1 | Acc. Nbr Genome |
|---|---|---|---|---|---|
| H2013-005-16 | Khong | Khone Noy | 28/03/2013 | LN901359 | MF076568 |
| H2013-007-16 | Khong | Khone Noy | 28/03/2013 | LN901362 | MF076569 |
| H2013-009-16 | Khong | Khone Noy | 28/03/2013 | LN901356 | MF076570 |
| H2013-120 | Khong | Loparkchok | 26/03/2013 | LN901357 | |
| H2013-015 | Khong | Huaphiman | 29/03/2013 | LN901351 | |
| H2013-016 | Khong | Huaphiman | 26/03/2013 | LN901352 | |
| H2013-017 | Khong | Huaphiman | 30/03/2013 | LN901353 | |
| H2013-024 | Khong | Huaphiman | 30/03/2013 | LN901344 | |
| H2012-033 | Khong | Donkao | 31/08/2012 | LN901366 | MF076576 |
| H2012-037 | Khong | Donkao | 31/08/2012 | LN901046 | |
| H2013-198 | Khong | Loparkchok | 26/03/2013 | LN901360 | |
| H2013-209 | Khong | Loparkchok | 26/03/2013 | LN901358 | |
| H2013-213 | Khong | Loparkchok | 26/03/2013 | LN901361 | |
| M2012-006P* | Khong | Donkao | 31/08/2012 | LN901364 | MF076577 |
| M2012-5866T* | Khong | Donkao | 31/08/2012 | LN901365 | |
| M2012-5917T* | Khong | Donkao | 31/08/2012 | LN901372 | |
| H2013-019-16 | Mounlapamok | Luangso | 12/03/2013 | LN901373 | MF076572 |
| H2013-020-16 | Mounlapamok | Luangso | 12/03/2013 | LN901374 | |
| H2013-021-16 | Mounlapamok | Luangso | 12/03/2013 | LN901342 | |
| H2013-002 | Mounlapamok | Mai | 30/03/2013 | LN901343 | |
| H2013-003 | Mounlapamok | Mai | 31/03/2013 | LN901345 | |
| H2013-005 | Mounlapamok | Mai | 31/03/2013 | LN901375 | |
| H2013-006 | Mounlapamok | Mai | 30/03/2013 | LN901376 | |
| H2013-007 | Mounlapamok | Mai | 30/03/2013 | LN901377 | |
| H2013-008 | Mounlapamok | Mai | 31/03/2013 | LN901378 | |
| H2013-009 | Mounlapamok | Mai | 30/03/2013 | LN901379 | |
| H2013-010 | Mounlapamok | Mai | 01/04/2013 | LN901380 | |
| H2013-011 | Mounlapamok | Mai | 31/03/2013 | LN901381 | |
| H2013-012 | Mounlapamok | Mai | 31/03/2013 | LN901382 | |
| H2013-013 | Mounlapamok | Mai | 31/03/2013 | LN901383 | |
| H2013-030 | Mounlapamok | Done Nang Loi | 31/03/2013 | LN901349 | |
| H2012-001 | Mounlapamok | Kanleuang | 19/07/2012 | LN901368 | |
| H2012-003 | Mounlapamok | Kanleuang | 20/07/2012 | LN901367 | |
| H2012-018 | Mounlapamok | Nadee | 07/08/2012 | LN901369 | |
| H2012-019 | Mounlapamok | Salaosong | 07/08/2012 | LN901370 | MF076573 |
| H2012-021 | Mounlapamok | Huiko | 16/08/2012 | LN901347 | MF076574 |
| H2012-022 | Mounlapamok | Nadee | 16/08/2012 | LN901341 | |
| H2012-028 | Mounlapamok | Nadee | 16/08/2012 | LN901371 | MF076575 |
| H2012-029 | Mounlapamok | Nadee | 16/08/2001 | LN901354 | |
| H2013-445 | Pakse | Songse | 25/12/2013 | LN901048 | |
| H2013-011-16 | Pathoumphone | Bangliengnai | 28/03/2013 | LN901363 | |
| H2013-013-16 | Pathoumphone | Bangliengnai | 28/03/2013 | LN901355 | MF076571 |
| H2013-025-16 | Soukhoumma | Soukhoummatai | 18/03/2013 | LN901350 |
* Mosquito isolates (P = Pool and T = Thorax Head).
Results
Detection of Chikungunya virus markers in acute human cases
During the initial investigation of the outbreak from May to July 2012, 52 suspected cases were investigated by the NCLE. A direct diagnosis by RT-PCR could be established for 16 patients (31%) [24]. From these 16 samples referred to the Institut Pasteur du Laos for viral culture assays, 14 CHIKV isolates were obtained.
After the initial alert, a joint field mission involving WHO, NCLE, and IPL staff was set up in the Moonlapamock and Khong Districts. During this period, 51 human suspected cases with clinical profiles compatible with a recent CHIKV infection were recorded in 10 villages located in Moonlapamock and Khong Districts between July 19th and September 10th 2012 (Table 2). All samples were investigated for the presence of anti-CHIKV antibodies. Serological analysis evidenced recent exposure profile (i.e. presence of anti-CHIKV IgM) in 21 of the samples tested negative by RT-PCR. Only one patient was also found positive for anti-CHIKV IgG. Altogether in this initial series, 37 suspected cases (72.5%) displayed a marker of recent chikungunya infection confirming its etiological role in the Champasak Province outbreak.
Table 2. Virologic markers of CHIKV infection recorded in suspected cases recorded during the initial field investigation in August 2012.
| Villages (n = 51) | District | Date of collection | IFI (IgM/IgG) | RT-PCR |
|---|---|---|---|---|
| Kanleuang (6) | Moonlapamok | 19/07/2012 | 3 | 2 |
| Nadee (13) | Moonlapamok | 07–16/08/2012 | 4 | 4 |
| Salaosong (1) | Moonlapamok | 07/08/2012 | 0 | 1 |
| Houayko (3) | Moonlapamok | 16–17/08/2012 | 0 | 1 |
| Donkao (6) | Khong | 31/08/2012 | 3 | 2 |
| Thakang (7) | Moonlapamok | 24–26/08/2012 | 6† | 0 |
| Done hed (4) | Khong | 01/09/2012 | 3 | 0 |
| Thangsadam (7) | Khong | 06/09/2012 | 2 | 4 |
| Veunkham (3) | Khong | 06–07/09/2012 | 0 | 2 |
| Nafang (1) | Khong | 10/09/2012 | 0 | 0 |
† 1 case was found positive for IgM and IgG.
In 2013, a major dengue epidemic hit all the provinces of the Lao PDR. Differential diagnosis to discriminate dengue from possible chikungunya infection was applied on samples collected in southern Lao provinces. A total of 485 human samples were tested for the presence of CHIKV genome by real time RT-PCR of which 33 (6.8%) were found positive.
From an independent series of 47 samples of dengue suspected cases recorded in Champasak Province, all found negative for any direct markers of dengue infection by the NCLE, the CHIKV genome was detected in 19 samples (40.4%).
Among the 568 samples collected for the seroprevalence study, 34 (6.0%) showed a serological profile compatible with an acute CHIKV infection, of which two (5.9%) were CHIKV positive by real-time RT-PCR and yielded a viral isolate in culture.
Altogether, a total of 69 CHIKV isolates were obtained from these different series.
Chikungunya virus seroprevalence in Champasak Province
A total of 568 volunteers were recruited in 26 villages, of which 4 were located in Moonlapamock District and 22 in Khong District of Champasak Province. Of the volunteers, 158 were male (sex ratio M/F: 1/ 2.6), aged 45.5 ± 17.6 (range 3–87 years) and mostly farmers (78.2%). The serological survey revealed that all villages randomly selected had been affected by CHIKV with IgM/IgG seroprevalence levels ranging from 43% to 94% in the four villages selected in Moonlapamok District (Fig 1B) and from 33% to 90% in the 22 villages in Khong District (Fig 1C). No significant differences have been found between the two districts.
Clinical profile of CHIKV infections
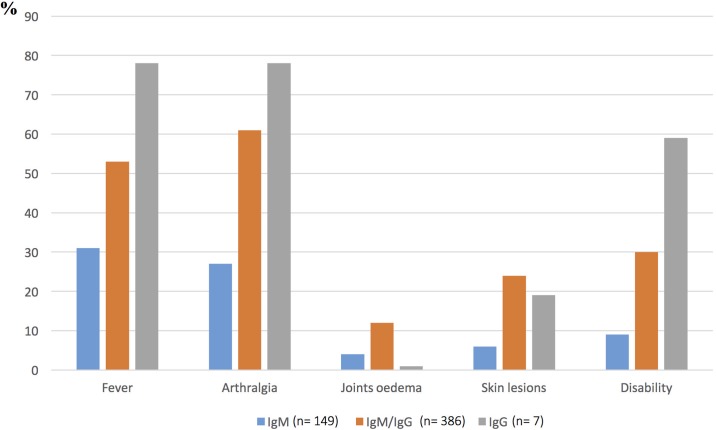
Volunteers were classified into three different groups based on their serological profiles as follows: (i) group 1, presence of anti-CHIKV IgM suggesting recent or possible acute infection stage; (ii) group 2, presence of both IgM and IgG suggestive of a possible convalescent phase; and (iii) group 3, presence of anti-CHIKV IgG only, corresponding to past infection. Among the volunteers positive for at least one immunoglobulin isotype (n = 542), 42.7% declared no clinical history compatible with a CHIKV infection. Most of the participants who experienced symptoms reported an acute fever, often accompanied by severe arthralgia affecting small joints. In summary, the main clinical symptoms recorded in our series were fever, arthralgia, and skin rash. Fig 2 summarizes the main symptoms reported by the participants displaying different anti-CHIKV serologic profiles. Disability, mainly associated with severe recurrent arthralgia, was declared by nearly 60% of the volunteers with isolated IgG.
Fig 2. Main symptoms recorded declared by the volunteers during the retrospective seroprevalence study in 2013.
Of the participants classified in group 1 (IgM+; n = 149), only 35.1% declared clinical signs and symptoms such as fever (33.1%) and arthralgia (29.1%). Joint pain was localized equally in fingers (18.9%) and/or wrists (18.9%) and/or hands (18.9%). Lower limb pain affected feet (21%) and ankles (16.2%). Swelling of joints was reported by 3.4% of participants and joint pain when walking in 7.4% of cases. Other main symptoms reported were myalgia (12.8%), back pain (15.5%), headache (21%), eye pain (7.4%), and vomiting (1.2%). Skin signs were reported by 6.8% of group 1 volunteers of whom 97% declared the occurrence of a rash. In this group of 149 volunteers, 18 (12.1%) reported a prolonged general weakness leading to work disablement for 16 (10.7%) of them.
Clinical signs and symptoms were reported by 65.5% of participants classified in group 2 (i.e. recent infection suggested by positive IgM and IgG; n = 386). Fever and arthralgia were reported by 56.2% and 63.5% respectively of the volunteers. Joint pain was located mainly in the fingers (51%), wrists (51.3%), and hands (48.4%). Lower limb pain affected feet (51.6%) and ankles (45.9%) and swelling of joints was reported by 16.1% of the participants. A significant statistical association was found between this serological profile and headache (34%), asthenia (27.7%), skin rash (26%), back pain (27.7%), myalgias (24.6%), joint pain when walking (24.1%), and eye pain (17.6%). Stoppage of work was declared by 123 volunteers (31.9%). In this group, 19 subjects (4.9%) experienced from 1 to 10 recurrences of arthralgia for a period that never exceeded one month after the acute phase.
Within the last group (past infections IgG+; n = 7), no episode of recurrent arthralgia was reported; the only symptoms statistically associated with this serological profile were ocular pain (80%) and asthenia (60%).
Questionnaire analysis revealed limited population mobility. Only one volunteer (0.7%) reported visiting Cambodia in the neighboring province of Phreah Vihear within the two weeks that preceded the symptom onset, a delay compatible with the CHIKV incubation phase. The possibility of an imported infection, mostly from Cambodia, could be considered for 20 participants (5.2%) as they declared having trading activities in Cambodia but without precise information on dates of stay.
Entomologic investigation
In the four villages investigated where suspected or confirmed cases were previously reported, high household (81%), container (48%), and Breteau (156) indices were found. Larval breeding sites were found in buckets (1%), drums (2%), tires (7%), discarded household waste (3%), pots (2%), and water jars (85%). Among the different kinds of water containers, water jars used for storing rainwater, with a median volume capacity of 100 liters (range: 2–300 liters) were mostly found positive for Ae. aegypti larvae, accounting for 84% (3819/4552) of the total larvae collection. As shown in Table 3, the two main vectors of CHIKV were recorded in the different villages. Ae. aegypti was clearly predominant, representing a mean of 82% of the total population captured (mostly from larvae collection). Among adult mosquitoes, Ae. aegypti represented 25% of the total population captured by different methods. The CHIKV genome could be detected by RT-PCR in 1 pool of 10 mosquitoes among the 2003 tested. The pools grouped Aedes aegypti females captured in Donekhao Village, Moonlapamock District. In order to determine the real number of infected mosquitoes per pool, head-thorax segments preserved were separately analyzed. By this approach, evidence of the CHIKV genome was only found in two mosquitoes within the positive pool. Viral isolation attempted on Vero E6 cells was successful for both the positive specimens.
Table 3. Distribution of Chikungunya virus vectors and mosquito species composition from adult and larval collections in villages visited during the initial field investigation in August 2012 (see Fig 1A).
| Collection methods/Mosquito species | Khong District | Moonlapamok District | Total | Percentage (%) | ||
|---|---|---|---|---|---|---|
| Donkao Village | Kanleung Village | Nadi Village | Thakang Village | |||
| Adult collection | ||||||
| CDC light traps | ||||||
| Ae. aegypti | Na | Na | 4 | 0 | 4 | 0.50 |
| Ae. albopictus | Na | Na | 2 | 2 | 4 | 0.50 |
| Ae. vittatus | Na | Na | 0 | 1 | 1 | 0.12 |
| Other speciesb | Na | Na | 189 | 115 | 304 | 37.91 |
| Total | 195 | 118 | 313 | 39.03 | ||
| Sweep nets | ||||||
| Ae. aegypti | 46a | Na | 131 | 17 | 194 | 24.19 |
| Ae. albopictus | 0 | Na | 0 | 9 | 9 | 1.12 |
| Ae. species | 0 | Na | 0 | 9 | 9 | 1.12 |
| Other speciesb | 96 | Na | 152 | 29 | 277 | 34.54 |
| Total | 142 | 283 | 64 | 489 | 60.97 | |
| Total adult collection | 142 | 478 | 182 | 802 | 100 | |
| Larvae collection | ||||||
| Ae. aegyptic | 132 | 1 | 875 | 76 | 1084 | 80.84 |
| Ae. albopictusc | 37 | 2 | 60 | 68 | 167 | 12.45 |
| Ae. vittatusc | 0 | 0 | 0 | 6 | 6 | 0.45 |
| Other speciesb | 82 | 0 | 2 | 0 | 84 | 6.26 |
| Total larvae collection | 251 | 3 | 937 | 150 | 1341 | 100 |
| Total | 393 | 3 | 1415 | 332 | 2143 | |
Na: Not applicable (since trapping was not conducted).
a Two pools of females were positive for CHIKV: One pool of 10 non-engorged females and one pool of one engorged female.
b Other mosquito genera including Culex species, Anopheles species, Armigeres species, and Mansonia were also analyzed by RT-PCR for CHIKV but all were negative (132 pools of 286 mosquitoes).
c Species identified after larvae metamorphosis
In order to document a possible vertical transmission of CHIKV in Aedes species, the same procedure was applied to reared mosquitoes and adult males but this approach provided no positive identification.
All mosquito pools were also checked for the presence of DENV. Two pools of Ae aegypti adult females were found positive for the presence of DENV genome. After individual testing, only one Aedes mosquito was found infected in each pool. Typing of DENV by real-time RT-PCR revealed that both isolates belonged to serotype 3. No DENV–CHIKV co-infection could be evidenced in these samples.
Sequence analysis
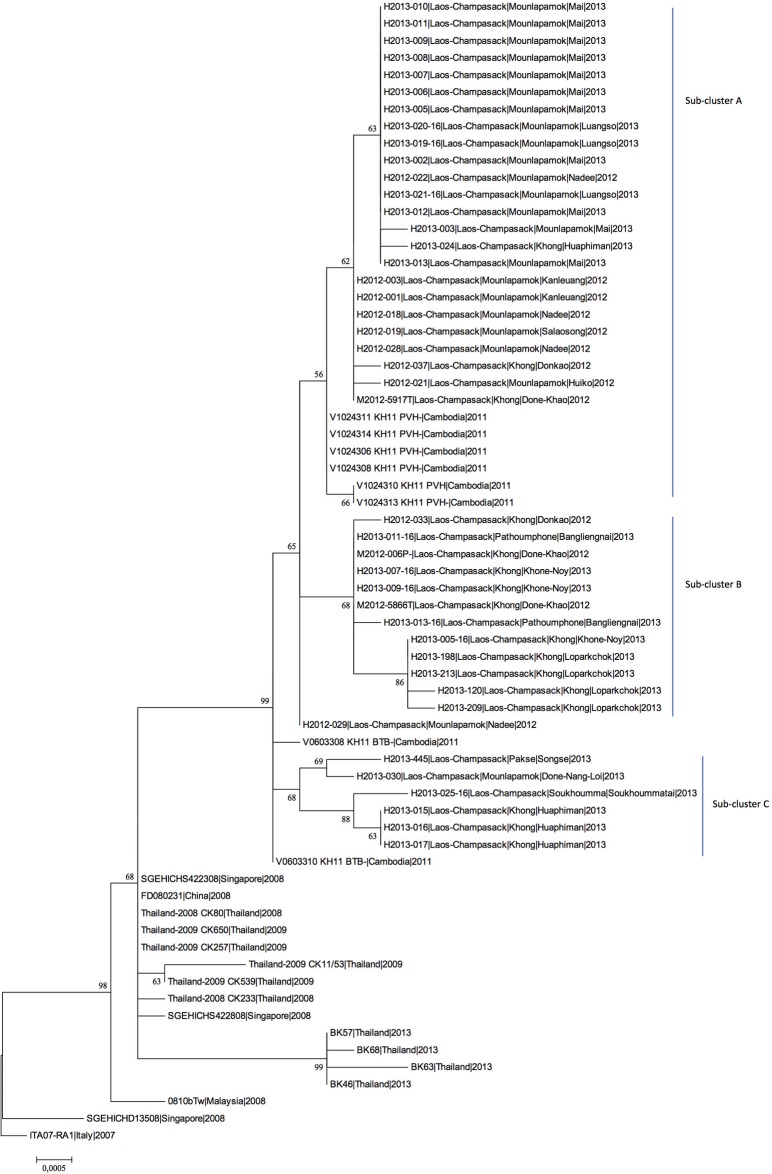
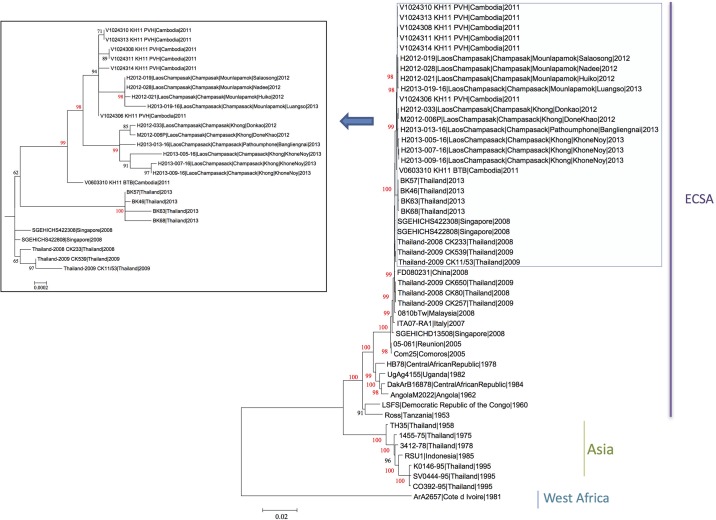
A total of 69 CHIKV isolates could be obtained among the different series of human and mosquito samples. Of them, 40 human and 3 mosquito isolates (1 from a pool, 2 from individual specimens of the positive pool) were selected for partial or complete genome sequencing. These isolates, obtained from specimens collected between July 2012 and December 2013, were analyzed by sequencing the E2-6K-E1 region (Fig 3). The full genome sequencing has been determined for ten of them (5 from 2012 and 5 from 2013). The phylogenetic tree based on full open reading frames (Fig 4) which identifies the three main phylogroups of CHIKV (West African, East/Central/South African (ECSA) and Asian) showed that all the Lao isolates belonged to the ECSA phylogroup. In this phylogroup, since the major outbreak in the Indian Ocean in 2005, the Indian Ocean lineage (IOL) has spread and diverged in two sub-lineages, Indian Ocean basin and Asian. All the Lao isolates felt in the Asian sub-lineage.
Fig 3. Phylogenetic tree of CHIKV strains based on E2-6K-E1 gene sequences, constructed using the maximum likelihood method.
Fig 4. Phylogenetic relationships of CHIKV strains from the Lao PDR based on complete genomes.
Phylogenetic tree constructed using the maximum likelihood method.
The phylogenetic tree based on 43 Lao E2-6K-E1 sequences, compared with isolates from neighboring countries in Southeast Asia from the same period (Cambodia, China, and Thailand) revealed a cluster between the southern Lao and the Cambodian isolates alone from 2011, well supported by bootstrap value. Interestingly, a mean identity of 99.8% was found between the Lao isolates and the two clusters of strains seen so far in Cambodian provinces (i.e. Battambang, Western Cambodia, and Phreah Vihear, Northern Cambodia near the Lao border) [29]. This analysis also suggests that the Lao isolates could form three different sub-clusters named A, B, and C (Fig 3) without clear correlation with the date and place of collection. This discrimination within the Lao strains, only supported by low bootstrap values when analysis was limited to the E2-6K-E1 segments, could be confirmed by the full genome sequences for the subclusters A and B (bootstraps >98%) for whom full sequence could be established (Fig 4). However, sub-cluster A only comprised the Phreah Vihear (Northern Cambodia) isolates, close to the Lao isolates. The sub-clusters A, B, and C displayed respectively 99.92%, 99.92%, and 99.84% nucleotide identity to each other and harbored specific genetic signatures (Table 4; S1 Table). All these proposed sub-clusters presented the E1-A226V mutation.
Table 4. Genetic signatures displayed by the CHIKV Lao isolates over the studied period (August 2012–December 2013).
| nsp1 | nsp2 | nsp3 | nsp4 | C | E2 | 6K | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Key | Country | Date | 249 | 455 | 130 | 503 | 539* | 679 | 330 | 455 | 463 | 76 | 179 | 438 | 604 | 24 | 93 | 252* | 60 | |
| Sub-cluster A | H2012-019 | Laos | 2012 | Q | L | H | I | S | H | S | P | H | I | Y | V | V | T | A | Q | S |
| H2012-028 | Laos | 2012 | Q | L | H | I | S | H | S | P | H | I | Y | V | V | T | A | Q | S | |
| H2012-021 | Laos | 2012 | Q | L | H | I | S | H | S | P | H | I | Y | V | V | T | A | Q | S | |
| H2013-019-16 | Laos | 2013 | Q | P | H | V | S | H | S | P | R | I | Y | V | V | T | A | Q | S | |
| V1024310_KH11_PVH | Cambodia | 2011 | P | L | H | I | S | H | S | P | H | I | H | V | I | T | A | Q | S | |
| V1024313_KH11_PVH | Cambodia | 2011 | P | L | H | I | S | H | S | P | H | I | H | V | I | T | A | Q | S | |
| V1024306_KH11_PVH | Cambodia | 2011 | P | L | H | I | S | H | S | P | H | I | H | V | I | T | A | Q | S | |
| V1024308_KH11_PVH | Cambodia | 2011 | P | L | H | I | S | H | S | P | H | I | H | V | I | T | A | Q | S | |
| V1024311_KH11_PVH | Cambodia | 2011 | P | L | H | I | S | H | S | P | H | I | H | V | I | T | A | Q | S | |
| V1024314_KH11_PVH | Cambodia | 2011 | P | L | H | I | S | H | S | P | H | I | H | V | I | T | A | Q | S | |
| V0603310_KH11_BTB | Cambodia | 2011 | P | L | H | I | S | Y | S | P | H | I | H | V | I | T | A | Q | S | |
| Sub-cluster B | H2012-033 | Laos | 2012 | P | L | Y | I | S | Y | F | P | H | I | H | V | I | T | V | Q | S |
| M2012-006 | Laos | 2012 | P | L | Y | I | S | Y | F | P | H | I | H | V | I | T | V | Q | S | |
| H2013-013-16 | Laos | 2013 | P | L | Y | I | S | Y | F | S | H | I | H | V | I | T | V | Q | S | |
| H2013-007-16 | Laos | 2013 | P | L | Y | I | S | Y | F | P | H | V | H | D | I | A | V | Q | S | |
| H2013-009-16 | Laos | 2013 | P | L | Y | I | S | Y | F | P | H | V | H | V | I | A | V | Q | S | |
| H2013-005-16 | Laos | 2013 | P | L | Y | I | S | Y | F | P | H | I | H | V | I | A | V | Q | N | |
| BK46 | Thailand | 2013 | P | L | H | I | S | Y | S | P | H | I | H | V | I | T | A | Q | S | |
| BK57 | Thailand | 2013 | P | L | H | I | S | Y | S | P | H | I | H | V | I | T | A | Q | S | |
| BK63 | Thailand | 2013 | P | L | H | I | S | Y | S | P | H | I | H | V | I | T | A | Q | S | |
| BK68 | Thailand | 2013 | P | L | H | I | S | Y | S | P | H | I | H | V | I | T | A | Q | S | |
| sl1 | sl1 | |||||||||||||||||||
(*) identifies amino acid residues specific of CHIKV sub-lineage 1.
The consensus sequence obtained from the CHIKV isolated from the mosquito pool (M2012-006P) fell into sub-cluster B and was 100% identical to the subset of human Lao isolates belonging to sub-cluster B.
The E2-6K-E1 sequences of the two isolates obtained from the head-thorax specimens from the positive pool were also established. One isolate (M2012-5866T) was identical to the consensus sequence of the pool (sub-cluster B), but surprisingly, the second one (M2012-5917T) felt into sub-cluster A. The envelope genes sequence of the M2012-5917T isolate shared 99.86% nucleotide identity with the two other mosquito sequences.
Sequencing of the full coding region of the genome was performed for Lao suggested sub-clusters A and B but not in sub-cluster C for which no virus had been isolated from human or mosquito samples. Therefore, a full genome analysis could be achieved for 10 isolates (5 from 2012 and 5 from 2013) in order to better define the position of Lao isolates among the CHIKV lineages from Southeast Asia. The phylogenetic tree based on complete ORF sequences (11,160 nucleotides) revealed the same overall structure as the E2-6K-E1 tree, and at least confirmed the sub-clusters A and B (with strong bootstrap values), with the presence of Lao-Cambodian sub-clusters. Overall homogeneity rates between Lao and Cambodian isolates were 99.84% at the nucleotide and 99.83% at the amino acid levels. These isolates harbored the two mutations nsp2-L539S and E2-K252Q which defined the sub-lineage 1 (sl1) within the IOL [35]. All six Lao isolates from sub-cluster B displayed one specific marker in the structural region C-A93V, and three of the six harbored C-T24A, of which one (isolate H2013-005-16) displayed an additional 6k-S60N substitution, suggesting this sub-cluster was likely in the process of diversification. Isolates within sub-cluster B displayed common signatures in non-structural regions (nsp2-H130Y and nsp3-S330F), with additional markers for three isolates: (i) nsp3-P455S, (ii) nsp4-I76V, and (iii) nsp4-V438D (Table 4; S1 Table).
Interestingly, the four isolates of sub-cluster A shared a common marker (nsp2-Y679H) with all Cambodian isolates. Of these four isolates, none displayed any amino-acid substitution in the structural polyprotein, but only presented some silent mutations in the E2-6K-E1 (6k-nt9860) (4/4), E2-nt9161 (1/4), and E1-nt10931 (1/4) regions (Table 4; S1 Table). In the non-structural regions, four specific markers were recorded in all isolates (i.e. nsp1-P249Q, nsp2-Y679H, nsp4-H179Y, and nsp4-I604V). One isolate harbored additional markers (i.e. nsp1-L455P, nsp2-I503V, nsp3-H463R).
As shown in Fig 3, the Lao isolates grouped in a sub-lineage “Asia” within the ECSA genotype. Close relationships were found with the Cambodian strains. Moreover, the Lao isolates were closely related (99.6% homology) to isolates from Thailand in the same period (2013). Finally, links were found with Chinese (2008: 99.4% homology) and Malaysian (2008: 99.4% homology) strains. Interestingly, one isolate (H2013-013-16) from Pathoumphone Village (in the north of Champasak Province) matching with sub-cluster B displayed some specific markers, such as the amino-acid mutation nsp3-P455S. Moreover, this isolate presented an unambiguous quasi-species at E1-10787 nt position (50%C/50%G analyzed by Sanger sequencing), attesting to the ongoing evolution.
Discussion
The circulation of CHIKV in the Lao PDR has been suspected for decades based on serological studies but despite the presence of wild and urban vectors, CHIKV had never been isolated in this country [1, 25]. However, the weakness of virology laboratories limited the ability to perform an etiologic diagnosis and may have contributed to confusion between CHIKV and other arboviral infections such as dengue fever. Moreover, the circulation in Southeast Asia of different members of the Semliki Forest virus antigenic complex has been clearly established [26, 27]. Indeed, the presence of anti-CHIKV antibodies reported by different authors may reflect these cryptic (misdiagnosed) infections. However, the low overall rate of seroprevalence in the Lao population leaves the way open for CHIKV to spread in the human population. This threat is confirmed by the fact that most of the patients included in our study displayed markers of recent infection (i.e. positive RT-PCR; presence of IgM). These data evidenced that, despite the official declaration of the end of the epidemic in September 2012, the virus transmission was maintained at a low level in 2013. It is worth noting that the sudden re-emergence of DENV serotype 3 at the beginning of a major outbreak countrywide may have hidden the persistence of CHIKV as a consequence of clinical confusion and/or lack of laboratory diagnosis resources. Previous descriptions of DENV–CHIKV co-infection support these assumptions [28].
Circulation of CHIKV was proven in the Lao PDR in 2012 by the detection of the virus genome in human samples but the real extent of this emergence and especially the phylogenetic position of the virus remained unknown [24]. A large-scale seroprevalence study has been organized to evaluate its true incidence.
Anti-CHIKV antibodies were detected in some groups in the Vientiane City population in the 1960s, but neither the prevalence nor the specificity of these antibodies were specified [25]. A second seroprevalence study performed in Vientiane City in the late 1970s recorded an anti-CHIKV antibody rate of 30% in the general population [36]. Over nearly 40 years, only one more study was conducted in the capital but none of the samples tested (n = 200) were found positive for anti-CHIKV IgG [24]. These results suggest that changes in CHIKV transmission may have occurred in the capital city but could also be linked to the methods used for antibody investigation.
The high proportion of recent infection profiles (isolated IgM: 26.1%) in our study may be related to a low anti-CHIKV immunity level in Lao people. Since the phase of viremia may overlap the rise of IgM antibodies, RT-PCR was tentatively used to detect the presence of CHIKV genome in the plasma of the corresponding cases. Of the 34 samples analyzed, two were found positive by RT-PCR and virus isolation, demonstrating the continued presence of CHIKV several months after the official declaration of the end of the outbreak. Moreover, the combined CHIKV–DENV surveillance in 2013 evidenced the persistence of CHIKV transmission until December 2013.
Among the volunteers included in our study, 42.7% did not report any symptoms suggestive of a CHIKV infection. This rate of asymptomatic infection appears dramatically higher compared to previously published data. A study published in 2008 reported that among the 568 participants, only 3.2% had asymptomatic infections [37]. Among other studies recently reviewed, the maximum asymptomatic rate recorded so far reached 27.7% [38]. However, more recently, an unexpected high rate of CHIKV symptomatic infections (82%) has been recorded in a prospective cohort [39]. In our study, asymptomatic forms were nearly three times higher compared to most of the previous studies but remain lower compared to the last observation in the Philippines. Some bias may explain this unexpected rate. First, a language barrier linked to a specific local dialect and the low educational level of the population may have an impact on the quality of data collection. For example, the difference between symptoms and symptom history may have been misunderstood. Possible cross-reactivity of antibodies with closely related viruses such as Me Tri or Getah with different clinical presentations may have contributed to increasing the rate of asymptomatic cases. And finally, as suggested by Yoon and colleagues, the evaluation of the subclinical CHIKV infection rate could be influenced by the study protocol (prospective vs retrospective) [39].
Severe recurrent arthralgia is observed in at least 30% and up to 93% of patients with acute CHIKV infection, whereas in our series only 7.5% of participants reported such episodes [36, 40]. This low rate of recurrent arthralgia may be due to the short delay between the CHIKV emergence and our seroprevalence study. Up to 60% of the participants displaying IgG declared post-acute phase disabilities from 15 days to 6 months following their CHIKV infection, suggesting that, as reported elsewhere, CHIKV may carry serious economic consequences [41, 42]. However, a direct link between CHIKV and these disabilities could not be established with certainty but it could be explained, at least in part, by the harshness of work, as farming is the predominant occupational activity of the population. Thus, a long-term follow up of this exposed population is needed to determine the actual frequency of post-CHIKV arthralgia and evaluate its degree of severity.
In the two districts, the recorded seroprevalence rates ranged from 43% to 94%, revealing the magnitude of the outbreak missed by the preliminary investigations performed in 2012 [24]. These seroprevalence rates give an accurate picture of the herd immunity that may serve as a barrier against future re-emergence (or reintroduction) of CHIKV in the studied area. However, this interpretation should be moderated. Indeed, the higher prevalence observed in females seems to reflect the temporary emigration of males for occupational purposes outside Champasak Province. Thus a substantial proportion of the natives from this region still remained susceptible to CHIKV. Moreover, as no further cases have been diagnosed since December 2013, the level of anti-CHIKV herd immunity in the Lao provinces could be assumed to be very low or non-existent.
Entomologic investigations performed in the two districts in summer 2012 evidenced the presence of the two main recognized vectors of CHIKV, i.e. Aedes aegypti and Aedes albopictus, with a clear predominance of Ae. aegypti (82%). However, no correlation could be established between registered entomologic indexes and the anti-CHIKV seroprevalence rates. The vector control campaign implemented a few days before the beginning of the field mission may explain the low vector population densities recorded in the villages. Moreover, only a few villages could be investigated. The disequilibrium observed between the two Aedes species vector of CHIKV could also in part be linked to environmental conditions particularly favorable to the development of Ae. aegypti in the villages where a high infestation rate of jars used for water storage was found. Among many mosquito specimens collected, only Ae. aegypti yielded positive detection and culture of CHIKV. Comparison of complete viral genome sequences highlighted a strong identity between human and mosquito CHIKV isolates corroborating the vector role of this mosquito species. This does not exclude the possible participation of Ae. albopictus, as both human and mosquito viral isolates displayed genetic markers of adaptation to Ae. albopictus, including the E1A226V signature [12, 13]. However, this adaptation process did not occur during the epidemic in Laos as the more closely related 2010–2011 CHIKV isolates from Cambodia already harbored the E1A226V substitution [29]. Aedes albopictus has been associated with numbers of recent CHIKV epidemics linked to A226V CHIKV in Southeast Asia whereas Ae. aegypti has only been involved in wild-type virus transmission [10, 43]. Our findings highlight the potential role of Ae. aegypti as a vector of ECSA A226V CHIKV in a rural area in the Indochinese peninsula.
As observed in neighboring countries (i.e. Cambodia and Thailand) since the epidemic of 2005 in the Indian Ocean, the CHIKV isolates that circulated in Laos also belonged to the ECSA phylogroup. Interestingly, we could detect and characterize CHIKV isolates from mosquitoes, and the analysis evidenced congruence with the Lao CHIKV human isolates in the same period. More precisely, our analysis shows that all Lao isolates harbor the two specific mutations of the sub-lineage sl1, confirming the filiation of Lao isolates with the Cambodian-Thai cluster and the maintenance of this sub-lineage in this Asian area.
The Lao PDR is surrounded by five countries where chikungunya infection has been reported. There are therefore several possible sources of introduction. Regarding the phylogenetic analysis, the more probable source of CHIKV introduction into the Lao PDR is Cambodia. Sequence comparison allowed the drawing of a clear link between the Lao strains and the Cambodian strains circulating in 2011. Intriguingly, the co-circulation in Laos of different variants previously identified in two remote provinces of Cambodia (i.e. Battambang and Phreah Vihear) raises questions about the introduction pathways of CHIKV. A multiple introduction can be considered, but it cannot be demonstrated due to the low number of strains isolated in Cambodia in 2011 [29]. Another explanation could be that different CHIKV variants co-circulated in Cambodia in the provinces affected by the epidemic, but, as only a few strains were sequenced, only the predominant variant was detected. A recent study of intra-patient genetic diversity confirmed that different genetic variants may co-infect a single individual [44].
Conversely, the possibility of a silent circulation of CHIKV of unknown origin in Laos, before their diffusion to Cambodia, is also a tenable assumption.
Our analysis, including a comparison between isolates from Thailand and Laos in 2013, showed that an introduction from Thailand is highly unlikely.
CHIKV and DENV cocirculation has been evidenced during different outbreaks in Africa and Asia. Moreover, in France, a non-endemic country, the co-emergence of both viruses was reported in 2010 [6, 45]. Occurrences of coinfection in humans and mosquitoes have been reported for decades [23, 28, 46, 47, 48]. CHIKV and DENV co-infection raised concerns that the severity of symptoms may be exacerbated, but such a negative effect has not yet been reported [48].
Recently published data, including our own, provide a general view of the emergence of CHIKV in Laos and shed light on the country’s complex situation [24, 28]. Reinforcement of laboratory diagnostic capacities at the national level are needed, as demonstrated by the high rates of DENV–CHIKV co-infection recorded in a study [28]. The absence of new declared CHIKV cases since December 2013, the limited spread of the virus in the country, and the low level of herd immunity leave the country vulnerable to a large-scale epidemic. As CHIKV still actively circulates in Southeast Asia, it represents, with other viruses such as Zika, a permanent threat for Laos and justifies the maintenance and extension of surveillance systems for arboviral infections.
Supporting information
(XLSX)
Acknowledgments
These studies were funded by the International Division of Institut Pasteur, Paris (PTR-408; ACIP-A16/2011). We thank Ruairí Ó hEithir for his prompt help in manuscript revision.
Data Availability
All sequence data (reference strains; primary isolates) are available on GenBank. Accession numbers are provided in the core of the manuscript. Sequences are publicly accessible. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the International Division of Institut Pasteur, Paris, France (MG). The funder had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Halstead SB, Udomsakdi S, Scanlon JE, Rohitayodhin S. Dengue and chikungunya virus infection in man in Thailand, 1962–1964. V. Epidemiologic observations outside Bangkok. Am J Trop Med Hyg. 1969. November;18(6):1022–1033. [DOI] [PubMed] [Google Scholar]
- 2.Ng LC, Hapuarachchi HC. Tracing the path of Chikungunya virus—evolution and adaptation. Infect Genet Evol. 2010. October;10(7):876–885. doi: 10.1016/j.meegid.2010.07.012 [DOI] [PubMed] [Google Scholar]
- 3.Carey DE. Chikungunya and dengue: a case of mistaken identity? J Hist Med Allied Sci. 1971. July;26(3):243–262. [DOI] [PubMed] [Google Scholar]
- 4.Angelini R, Finarelli AC, Angelini P, Po C, Petropulacos K, Macini P, et al. An outbreak of chikungunya fever in the province of Ravenna, Italy. Euro Surveill. 2007. September 6;12(9):E070906.1 [DOI] [PubMed] [Google Scholar]
- 5.Sreekumar E, Issac A, Nair S, Hariharan R, Janki MB, Arathy DS, et al. Genetic characterization of 2006–2008 isolates of Chikungunya virus from Kerala, South India, by whole genome sequence analysis. Virus Genes. 2010. February;40(1):14–27. doi: 10.1007/s11262-009-0411-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grandadam M, Caro V, Plumet S, Thiberge JM, Souarès Y, Failloux AB, et al. Chikungunya virus, southeastern France. Emerg Infect Dis. 2011. May;17(5):910–913. doi: 10.3201/eid1705.101873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sam IC, Loong SK, Michael JC, Chua CL, Wan Sulaiman WY, Vythilingam I, et al. Genotypic and phenotypic characterization of Chikungunya virus of different genotypes from Malaysia. PLoS One. 2012;7(11):e50476 doi: 10.1371/journal.pone.0050476 Epub 2012 Nov 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffey LL, Forrester N, Tsetsarkin K, Vasilakis N, Weaver SC. Factors shaping the adaptive landscape for arboviruses: implications for the emergence of disease. Future Microbiol. 2013. February;8(2):155–176. doi: 10.2217/fmb.12.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont-Rouzeyrol M, Caro V, Guillaumot L, Vazeille M, D’Ortenzio E, Thiberge JM, et al. Chikungunya virus and the mosquito vector Aedes aegypti in New Caledonia (South Pacific Region). Vector Borne Zoonotic Dis. 2012. December;12(12):1036–1041. doi: 10.1089/vbz.2011.0937 [DOI] [PubMed] [Google Scholar]
- 10.Zeller H, Van Bortel W, Sudre B. Chikungunya: Its History in Africa and Asia and Its Spread to New Regions in 2013–2014. J Infect Dis. 2016. December 15;214(suppl 5):S436–S440. doi: 10.1093/infdis/jiw391 [DOI] [PubMed] [Google Scholar]
- 11.Gallian P, Leparc-Goffart I, Richard P, Maire F, Flusin O, Djoudi R, et al. Epidemiology of Chikungunya Virus Outbreaks in Guadeloupe and Martinique, 2014: An Observational Study in Volunteer Blood Donors. PLoS Negl Trop Dis. 2017. January 12;11(1):e0005254 doi: 10.1371/journal.pntd.0005254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006. July;3(7):e263 doi: 10.1371/journal.pmed.0030263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsetsarkin KA, McGee CE, Volk SM, Vanlandingham DL, Weaver SC, Higgs S. Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. aegypti mosquitoes. PLoS One. 2009. August 31;4(8):e6835 doi: 10.1371/journal.pone.0006835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volk SM, Chen R, Tsetsarkin KA, Adams AP, Garcia TI, Sall AA, et al. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol. 2010. July;84(13):6497–6504. doi: 10.1128/JVI.01603-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stapleford KA, Moratorio G, Henningsson R, Chen R, Matheus S, Enfissi A, et al. Whole-Genome Sequencing Analysis from the Chikungunya Virus Caribbean Outbreak Reveals Novel Evolutionary Genomic Elements. PLoS Negl Trop Dis. 2016. January 25;10(1):e0004402 doi: 10.1371/journal.pntd.0004402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chastel C. Human infections in Cambodia by the chikungunya virus or a closely related agent. 3. Epidemiology. Bull Soc Patho Ex Filiales. 1964. Jan-Feb;57:65–82. French [PubMed] [Google Scholar]
- 17.Apandi Y, Nazni WA, Noor Azleen AZ, Vythilingam I, Noorazian MY, Azahari AH, et al. The first isolation of chikungunya virus from nonhuman primates in Malaysia. J Gen Mol Virol. 2009;1:35–39. [Google Scholar]
- 18.Sam IC, Chua CL, Rovie-Ryan JJ, Fu JY, Tong C, Sitam FT, et al. Chikungunya Virus in Macaques, Malaysia. Emerg Infect Dis. 2015. September;21(9):1683–1685. doi: 10.3201/eid2109.150439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leo YS, Chow AL, Tan LK, Lye DC, Lin L, Ng LC. Chikungunya outbreak, Singapore, 2008. Emerg Infect Dis. 2009. May;15(5):836–837. doi: 10.3201/eid1505.081390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bessaud M, Peyrefitte CN, Pastorino BA, Tock F, Merle O, Colpart JJ, et al. Chikungunya virus strains, Reunion Island outbreak. Emerg Infect Dis. 2006. October;12(10):1604–1606. doi: 10.3201/eid1210.060596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delatte H, Paupy C, Dehecq JS, Thiria J, Failloux AB, Fontenille D. Aedes albopictus, vector of chikungunya and dengue viruses in Reunion Island: biology and control. Parasite. 2008. March;15(1):3–13. doi: 10.1051/parasite/2008151003 [DOI] [PubMed] [Google Scholar]
- 22.Niyas KP, Abraham R, Unnikrishnan RN, Mathew T, Nair S, Manakkadan A, et al. Molecular characterization of Chikungunya virus isolates from clinical samples and adult Aedes albopictus mosquitoes emerged from larvae from Kerala, South India. Virol J. 2010. August 13;7:189 doi: 10.1186/1743-422X-7-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caron M, Paupy C, Grard G, Becquart P, Mombo I, Nso BB, et al. Recent introduction and rapid dissemination of Chikungunya virus and Dengue virus serotype 2 associated with human and mosquito coinfections in Gabon, central Africa. Clin Infect Dis. 2012. September;55(6):e45–53. doi: 10.1093/cid/cis530 [DOI] [PubMed] [Google Scholar]
- 24.Soulaphy C, Souliphone P, Phanthavong K, Phonekeo D, Phimmasine S, Khamphaphongphane B, et al. Emergence of chikungunya in Moonlapamok and Khong Districts, Champasak Province, the Lao People’s Democratic Republic, May to September 2012. Western Pac Surveill Response J. 2013. March; 18;4(1):46–50. doi: 10.5365/WPSAR.2012.3.4.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halstead SB. Mosquito-borne haemorrhagic fevers of South and South-East Asia. Bull World Health Organ. 1966;35(1):3–15. [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin DE. Neuronal cell death in alphavirus encephalomyelitis. Curr Top Microbiol Immunol. 2005;289:57–77. [DOI] [PubMed] [Google Scholar]
- 27.Tan le V, Ha do Q, Hien VM, van der Hoek L, Farrar J, de Jong MD. Me Tri virus: a Semliki Forest virus strain from Vietnam? J Gen Virol. 2008. September;89(Pt 9):2132–2135. doi: 10.1099/vir.0.2008/002121-0 [DOI] [PubMed] [Google Scholar]
- 28.Phommanivong V, Kanda S, Shimono T, Lamaningao P, Darcy AW, Mishima N, et al. Co-circulation of the dengue with chikungunya virus during the 2013 outbreak in the southern part of Lao PDR. Trop Med Health. 2016. August 4;44:24 doi: 10.1186/s41182-016-0020-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duong V, Andries AC, Ngan C, Sok T, Richner B, Asgari-Jirhandeh N, et al. Reemergence of Chikungunya virus in Cambodia. Emerg Infect Dis. 2012. December;18(12):2066–2069. doi: 10.3201/eid1812.120471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michael B, Deen J, Buchy P, Gubler D, Harris E, Hombach J. World Health Organization dengue guidelines for diagnosis, treatment, prevention, and control (new edition) 2009 Dengue. Guidelines for diagnosis, treatment prevention and control. Geneva, WHO/HTM/NTD/DEN; / 2009. [Google Scholar]
- 31.Pastorino B, Bessaud M, Grandadam M, Murri S, Tolou HJ, Peyrefitte CN. Development of a TaqMan RT-PCR assay without RNA extraction step for the detection and quantification of African Chikungunya viruses. J Virol Methods. 2005. March;124(1–2):65–71. doi: 10.1016/j.jviromet.2004.11.002 [DOI] [PubMed] [Google Scholar]
- 32.Prat CM, Flusin O, Panella A, Tenebray B, Lanciotti R, Leparc-Goffart I. Evaluation of commercially available serologic diagnostic tests for chikungunya virus. Emerg Infect Dis. 2014. December;20(12):2129–2132. doi: 10.3201/eid2012.141269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stojanovich CJ, Scott HG. Illustrated Key to Aedes Mosquitoes of Vietnam. U. S. Department of Health, Education and Welfare, Public Health Service, Communicable Disease Center; 1965. [Google Scholar]
- 34.Rattanarithikul R, Harbach RE, Harrison BA, Panthusiri P, Coleman RE, Richardson JH. 2010. Illustrated keys to the mosquitoes of Thailand. VI. Tribe Aedini. Southeast Asian J Trop Med Public Health 41 Suppl 1: 1–225. [PubMed] [Google Scholar]
- 35.Tsetsarkin KA, Chen R, Yun R, Rossi SL, Plante KS, Guerbois M, et al. Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat Commun. 2014. June 16;5:4084 doi: 10.1038/ncomms5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanamitsu M, Taniguchi K, Urasawa S, Ogata T, Wada Y, Wada Y, Saroso JS. Geographic distribution of arbovirus antibodies in indigenous human populations in the Indo-Australian archipelago. AM J Trop Med Hyg. 1979;28(2):351–363. [DOI] [PubMed] [Google Scholar]
- 37.Queyriaux B, Simon F, Grandadam M, Michel R, Tolou H, Boutin JP. Clinical burden of chikungunya virus infection. Lancet Infect Dis. 2008. January;8(1):2–3. doi: 10.1016/S1473-3099(07)70294-3 [DOI] [PubMed] [Google Scholar]
- 38.Thiberville SD, Moyen N, Dupuis-Maguiraga L, Nougairede A, Gould EA, Roques P, et al. Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy. Antiviral Res. 2013. September;99(3):345–370. doi: 10.1016/j.antiviral.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon IK, Alera MT, Lago CB, Tac-An IA, Villa D, Fernandez S, et al. High rate of subclinical chikungunya virus infection and association of neutralizing antibody with protection in a prospective cohort in the Philippines. PLoS Negl Trop Dis. 2015. May 7;9(5):e0003764 doi: 10.1371/journal.pntd.0003764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol. 2010. July;8(7):491–500. doi: 10.1038/nrmicro2368 [DOI] [PubMed] [Google Scholar]
- 41.Soumahoro MK, Boelle PY, Gaüzere BA, Atsou K, Pelat C, Lambert B, et al. The Chikungunya epidemic on La Réunion Island in 2005–2006: a cost-of-illness study. PLoS Negl Trop Dis. 2011. June;5(6):e1197 doi: 10.1371/journal.pntd.0001197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vijayakumar K, George B, Anish TS, Rajasi RS, Teena MJ, Sujina CM. Economic impact of chikungunya epidemic: out-of-pocket health expenditures during the 2007 outbreak in Kerala, India. Southeast Asian J Trop Med Public Health. 2013. January;44(1):54–61. [PubMed] [Google Scholar]
- 43.Ng LC, Tan LK, Tan CH, Tan SS, Hapuarachchi HC, Pok KY, et al. Entomologic and virologic investigation of Chikungunya, Singapore. Emerg Infect Dis. 2009; 15:1243–1249. doi: 10.3201/eid1508.081486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stapleford KA, Moratorio G, Henningsson R, Chen R, Matheus S, Enfissi A, et al. Whole-Genome Sequencing Analysis from the Chikungunya Virus Caribbean Outbreak Reveals Novel Evolutionary Genomic Elements. PLoS Negl Trop Dis. 2016. January 25;10(1):e0004402 doi: 10.1371/journal.pntd.0004402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.La Ruche G, Souarès Y, Armengaud A, Peloux-Petiot F, Delaunay P, Desprès P, et al. First two autochthonous dengue virus infections in metropolitan France, September 2010. Euro Surveill. 2010. September 30;15(39):19676 [PubMed] [Google Scholar]
- 46.Myers RM, Carey DE. Concurrent isolation from patient of two arboviruses, Chikungunya and dengue type 2. Science. 1967. September 15;157(3794):130. [DOI] [PubMed] [Google Scholar]
- 47.Leroy EM, Nkoghe D, Ollomo B, Nze-Nkogue C, Becquart P, Grard G, et al. Concurrent chikungunya and dengue virus infections during simultaneous outbreaks, Gabon, 2007. Emerg Infect Dis. 2009. April;15(4):591–593. doi: 10.3201/eid1504.080664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furuya-Kanamori L, Liang S, Milinovich G, Soares Magalhaes RJ, Clements AC, Hu W, et al. Co-distribution and co-infection of chikungunya and dengue viruses. BMC Infect Dis. 2016. March 3;16:84 doi: 10.1186/s12879-016-1417-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All sequence data (reference strains; primary isolates) are available on GenBank. Accession numbers are provided in the core of the manuscript. Sequences are publicly accessible. All other relevant data are within the paper and its Supporting Information files.