Abstract
Transforming growth factor β1 (TGFβ1) is a cytokine with multiple functions. TGFβ1 significantly induces migration and invasion of liver cancer cells. However, the molecular mechanisms underlying this effect remain unclear. Epithelial-to-mesenchymal transition (EMT) is crucial for the development of invasion and metastasis in human cancers. The aim of the present study was to determine whether TGFβ1-induced EMT promoted migration and invasion in HepG2 cells. The underlying mechanism and the effect of EMT on HepG2 cells were also investigated. The results demonstrated that TGFβ1 may induce EMT to promote migration and invasion of HepG2 cells, and this effect depends on activation of the Janus kinase/signal transducer and activator of transcription 3 (JAK/STAT3) signaling pathway. JAK/STAT3 signaling is involved in human malignancies, including lung cancer, and is implicated in cell transformation, tumorigenicity, EMT and metastasis. In the present study, TGFβ1 also activated JAK/STAT3 signaling in HepG2 cells and promoted Twist expression, but these events were abolished by treatment with the STAT3 inhibitor AG490. Additionally, Twist siRNA blocked TGFβ1-induced EMT. Thus, TGFβ1 was shown to induce EMT, thereby promoting the migration and invasion of HepG2 cells via JAK/STAT3/Twist signaling.
Keywords: epithelial-to-mesenchymal transition, transforming growth factor β1, signal transducer and activator of transcription 3
Introduction
Liver cancer is one of the most common malignant tumors worldwide, and its incidence is the second highest in China. Liver cancer cannot be easily detected at an early stage due to the lack of distinct symptoms and the scarcity of clinically specific markers for serodiagnosis. Therefore, the majority of the patients are diagnosed at advanced or late stages, resulting in distant metastasis and a low 5-year survival rate (1). Thus, the metastasis and invasion of liver cancer must be clinically investigated to prevent progression of this disease and improve its prognosis.
The invasion and metastasis of malignant tumors are regulated and controlled by various factors and mechanisms. Epithelial-to-mesenchymal transition (EMT) is a key mechanism participating in the invasion and metastasis of solid cancers, such as colon, lung and pancreatic cancer (2–4). However, the association between EMT and the onset and progression of liver cancer has not been fully elucidated. In this context, Lee et al (5) and Giannelli et al (6) previously reported that EMT is involved in the invasion and metastasis of liver cancer cells.
A number of studies reported that transforming growth factor β1 (TGFβ1) is a cytokine with multiple functions that promotes EMT (7,8). The activation abnormalities in the signal transducer and activator of transcription 3 (STAT3) signaling pathway are associated with tumor onset and progression (9). The activation of this pathway is regulated and controlled by the upstream factor Janus kinase (JAK). The activation of JAK/STAT3 signaling may directly affects EMT and promotes the invasion and metastasis of tumor cells in lung cancer and ovarian tumors (10). However, whether the EMT mediated by the JAK/STAT3 signaling pathway promotes TGFβ1-induced invasion and metastasis of liver cancer cells has not been clearly determined.
The present study investigated the human liver cancer line HepG2, in which invasion and metastasis were induced by TGFβ1. The role of JAK/STAT3 signaling in mediating the involvement of EMT in the invasion and metastasis of HepG2 cells induced by TGFβ1 was also determined. Experiments were performed to confirm whether Twist is a target of STAT3. Overall, the aim of this study was to provide new experimental evidence and potential targets for preventing the invasion and metastasis of liver cancer cells.
Materials and methods
Cell culture
The liver cancer cell line HepG2 was purchased from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). HepG2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-high glucose containing trypsin (cat no. SH30022.01B) supplemented with 10% fetal bovine serum (FBS; cat no. SH30084.03) (both from HyClone, Logan, UT, USA), 100 U/ml penicillin (cat no. ST488-1; Beyotime Institute of Biotechnology, Shanghai, China) and 100 U/ml streptomycin (cat no. ST488-2; Beyotime Institute of Biotechnology) at 37°C under 95% air and 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from the tissue samples using TRIzol® reagent (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer's instructions. Subsequently, cDNA was synthesized using a TaqMan Reverse Transcription Reagents kit (Thermo Fisher Scientific), according to the manufacturer's protocol. The relative expression levels of mRNA were determined using a Power SYBR-Green PCR Master Mix kit (Thermo Fisher Scientific) and normalized to GAPDH. RT-PCR was performed using the Applied Biosystems 7500 Fast Dx Real-Time PCR instrument (cat no. 4425757; Thermo Fisher Scientific) and the following gene-specific primers (Sangon Biotech Co., Ltd., Shanghai, China): GAPDH: Sense, 5′-TGCCATCAACGACCCCTTCA-3′ and antisense, 5′-TGACCTTGCCCACAGCCTTG-3′; E-cadherin: Sense, 5′-AGCTATCCTTGCACCTCAGC-3′ and antisense, 5′-CCCAGGAGTTTGAG-3′; N-cadherin: Sense, 5′-TCCTGCTCACCACCACTACTT-3′ and antisense, 5′-CTGACAATGACCCCACAGC-3′; Smad: Sense, 5′-ATAAGCAACCGCCTGAACAT-3′ and anti-sense, 5′-TTACCTGCCTCCTGAAGACC-3′; Twist: Sense, 5′-GCTGATTGGCACGACCTCT-3′ and antisense, 5′-CACCATCCTCACACCTCTGC-3′; and vimentin: Sense, 5′-CCAAACTTTTCCTCCCTGAACC-3′ and antisense, 5′-GTGATGCTGAGAAGTTTCGTTGA-3′. A control siRNA specific for the red fluorescent protein, 5′-CCACTACCTGAGCACCCAG-3′, was used as the negative control (sc-37007; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). All primers were designed using the National Center for Biotechnology Information Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). PCR was performed under the following conditions: Denaturation at 50°C for 2 min, followed by 38 cycles at 95°C for 15 sec and 60°C for 1 min. Gene expression was normalized to internal controls and fold changes were calculated using the relative quantification method (2−ΔΔCq) (11).
Western blot analysis
Cells were washed 3 times with ice-cold PBS and then incubated on ice with 250 μl RIPA buffer (cat no. P0013; Beyotime Institute of Biotechnology) with 2.5 μl phenylmethylsulfonyl fluoride (cat no. ST506-2; Beyotime Institute of Biotechnology) for 15–30 min. The cells were collected and centrifuged at 13,000 × g for 10 min at 4°C. The protein concentrations of the cell lysates were measured in duplicate using a bicinchoninic acid protein assay kit (cat no. 23227; Thermo Fisher Scientific). The protein lysates and 6X loading buffer were mixed at a ratio of 4:1 and then boiled for 5 min at 100°C. Equal amounts of total protein were separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (cat no. FFP39; Beyotime Institute of Biotechnology). The membranes were immunoblotted with monoclonal mouse β-actin (1:1,000; cat. no. sc-8432), goat anti-human p-STAT3 (1:1,000; cat. no. sc-21876), monoclonal mouse anti-human JAK (1:400; cat. no. sc-376996), monoclonal mouse anti-human STAT3 (1:400; cat no. sc-293151), p-JAK (1:400; cat. no. sc-16773), E-cadherin (1:500; cat. no. sc-21791), N-cadherin (1:400; cat. no. sc-393933) and vimentin (1:400; cat. no. sc-373717) antibodies (all from Santa Cruz Biotechnology, Inc.) at 4°C overnight. All antibodies were diluted with 0.5% bovine serum albumin. Following incubation, the corresponding secondary antibody conjugated with peroxidase and enhanced chemiluminescence reagents (Beyo ECL Plus; cat. no. P0018; Beyotime Institute of Biotechnology) were applied, and the blot was visualized (cat. no. 121-2550; Beijing Liuyi Biotechnology Co., Ltd., Beijing, China). The amount of total protein was semiquantified as ratio to β-actin on each gel.
Scattering assay
Scattering assay was performed as previously described (7). HepG2 cells (3×105/ml) were seeded into each well of a 24-well plate (cat. no. 662102; Greiner Bio-One GmbH, Frickenhausen, Germany), and incubated overnight at 37°C in an atmosphere of 5% CO2. The cells were pretreated with 10 μM TGFβ1 for 48 h at 37°C for 48 h in 95% air and 5% CO2. Representative images were captured at a magnification of ×20 using the Eclipse TE2000-U inverted microscope (Nikon Corporation, Tokyo, Japan).
Invasion and migration assay
The invasion assay was performed using Transwell 24-well plates with 8-μm pore polycarbonate membranes (BD Biosciences, Franklin Lakes, NJ, USA). Briefly, the upper side of the membranes was coated with Matrigel (20 μg/well) and the membranes were then air-dried for 1 h at 37°C. The lower side of the membranes was coated with 5 μg fibronectin, and the treated or untreated HepG2 cells (2×105) in 200 μl of DMEM medium with 2.5% FBS were placed in the upper chamber. The lower chamber was filled with DMEM medium with 10% FBS as the chemoattractant. The invasion chamber was incubated for 8 h at 37°C and 5% CO2. The cells on the upper surface of the membrane were removed by gentle scrubbing with a cotton swab. The membranes were fixed in a stationary liquid of 95% ethanol and 5% acetic acid for 30 min and stained with crystal violet. The number of cells on the lower surface of the membrane in 5 random visual fields (magnification, ×200) was then counted using an Eclipse TE2000-U inverted microscope. Each assay was performed in triplicate.
Wound healing assay
The wound healing assay was performed as previously described: HepG2 cells (3×105/ml) were seeded into a 6-well plate (cat. no. 657160; Greiner Bio-One GmbH) in serum-containing medium, and incubated at 37°C in an atmosphere of 5% CO2 in order to form a confluent monolayer. The monolayer was scratched using a sterile plastic pipette tip (cat. no. CLS4860; Sigma-Aldrich, Merck KGaA, St. Louis, MO, USA), and washed with PBS to remove cell debris. Subsequently, fresh medium was added, and 10 μM TGFβ1 or 0.1 ml DMSO was added to each well. The scratched mono-layer was incubated at 37°C in an atmosphere of 5% CO2 for 48 h. Wound closure was measured in 6 random high-power fields at a magnification of ×200, using Image-Pro®Express software, version 6 (Media Cybernetics, Inc., Rockville, MD, USA) and an Eclipse TE2000-U inverted microscope (11).
Statistical analysis
Data were analyzed using SPSS software, version 1.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism software, version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Analysis of variance was conducted followed by the Student's t-test. The data are presented as mean ± standard deviation. P<0.05 was considered to indicate statistically significant differences.
Results
TGFβ1 induces migration and invasion of HepG2 cells
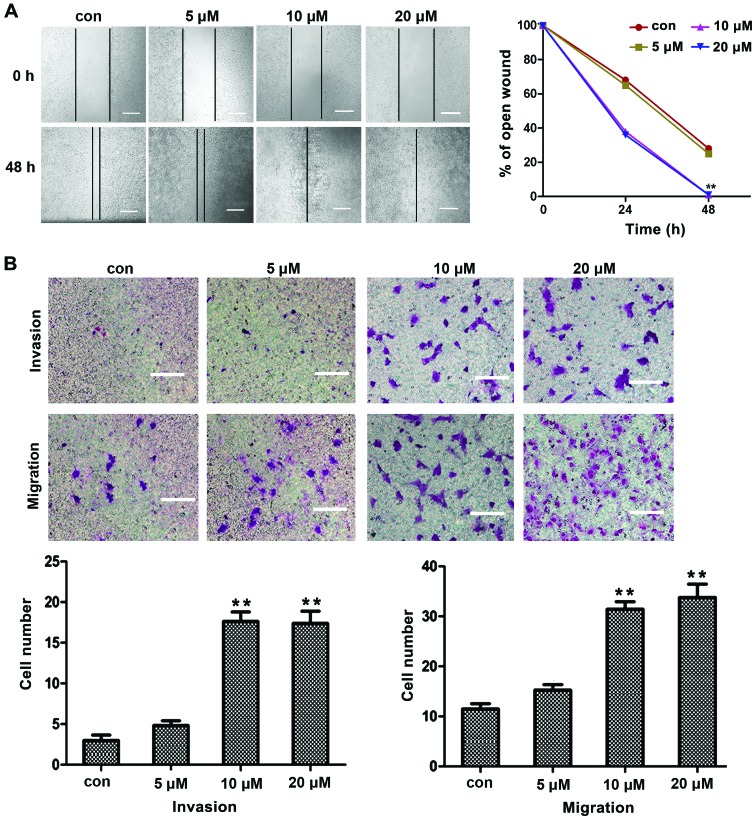
To determine the migration and invasion induced by TGFβ1, liver cancer HepG2 cells were treated with different concentrations of TGFβ1 for 48 h, and the migration and invasion of cancer cells were assessed by wound closure assays and Matrigel Transwell chamber invasion assays. The effects of TGFβ1 were observed at concentrations as low as 5 μM. As the TGFβ1 concentration increased, the migration and invasion of HepG2 cells also increased in a concentration-dependent manner, with the most prominent effects observed at a concentration of 10 μM (Fig. 1A and B). These results indicated that TGFβ1 induced HepG2 cell migration and invasion in a concentration-dependent manner. Hence, the concentration of 10 μM was selected for all further mechanistic studies.
Figure 1.
Migration and invasion induced by TGFβ1 in HepG2 cells. (A) In wound-healing assay, HepG2 cells were treated with TGFβ1 at different concentrations (5, 10 and 20 μM) for 48 h, and TGFβ1-induced cell motility was determined by measuring wound closure at a magnification of ×200 with Image-Pro® Express software. Scale bar, 10 μm. (B) For the migration and invasion assays, HepG2 cells were treated with TGFβ1 at different concentrations (5, 10 and 20 μM) for 48 h, and the cells were visualized by staining with toluidine blue and counted in 6 random high-power fields at a magnification of ×200 using Image-Pro® Express software. Scale bar, 5 μm. The experiments were performed in triplicate. Data are presented as mean ± standard deviation. **P<0.01 vs. control group. TGFβ1, transforming growth factor β1; con, control.
TGFβ1 induces EMT in HepG2 cells
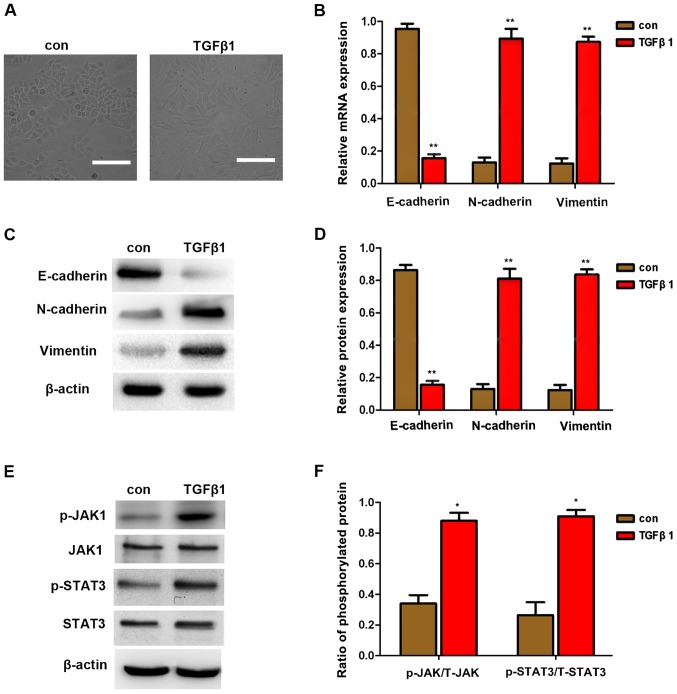
TGFβ1 is a factor that promotes EMT in cancer cells, as previously reported (7,8). EMT is an important mechanism of cancer cell invasion and metastasis. The downregulation of E-cadherin expression and the upregulation of vimentin and N-cadherin expression are considered to be markers of EMT. In the present study, TGFβ1 also induced cell scattering (Fig. 2A), indicating that TGFβ1 induces EMT, thereby increasing the migration and invasion of HepG2 cells. To further investigate whether EMT is involved in TGFβ1-induced scattering, migration and invasion of HepG2 cells, the expression of E-cadherin, vimentin and N-cadherin were first detected by qPCR and western blot analysis. As shown in Fig. 2B and C, the expression of vimentin and N-cadherin was upregulated, whereas that of E-cadherin was downregulated following treatment with 10 μM TGFβ1. These results demonstrated that TGFβ1-induced EMT promoted the migration and invasion of HepG2 cells.
Figure 2.
EMT induced by TGFβ1 in HepG2 cells. (A) TGFβ1 induced HepG2 cell scattering. Cells were incubated with 10 μM TGFβ1 for 48 h. Representative images were captured at a magnification of ×200 using an Eclipse TE2000-U inverted microscope. Scale bar, 10 μm. (B–D) qPCR and western blot analysis of EMT markers in HepG2 cells after treatment with 10 μM TGFβ1 for 48 h. Data are presented as mean ± standard deviation of 3 independent experiments. *P<0.05 vs. control. (E and F) Western blot analysis revealed that TGFβ1 promoted the expression of JAK, p-JAK, STAT3, and p-STAT3. Data are presented as mean ± standard deviation of three independent experiments. *P<0.05 vs. control. TGFβ1, transforming growth factor β1; con, control; EMT, epithelial-to-mesenchymal transition; qPCR, quantitative polymerase chain reaction; JAK, Janus kinase; STAT3, signal transducer and activator of transcription 3.
Moreover, JAK/STAT3 protein expression was detected by western blot analysis and it was observed that TGFβ1 stimulated the expression of p-JAK and p-STAT3. This finding indicates that TGFβ1-induced EMT may be activated by JAK/STAT3 signaling.
JAK/STAT3 signaling is involved in TGFβ1-induced EMT to increase migration and invasion of HepG2 cells
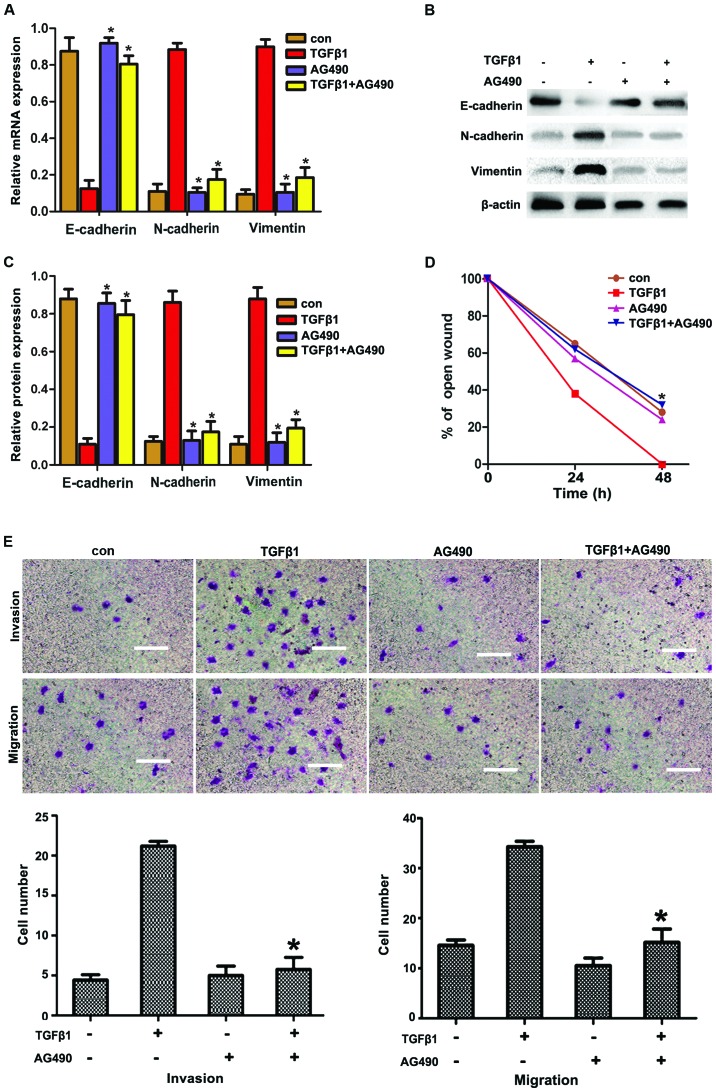
STAT3 is the key transcription factor regulating cell proliferation and survival. STAT3 may be activated by oncostatin M, interferons, interleukin-6 (IL-6) and epidermal growth factor (EGF). It was recently reported that TGFβ1 induced JAK/STAT3 signaling to increase migration and invasion in lung carcinoma cells. Based on these reports, we hypothesized that JAK/STAT3 signaling may be involved in TGFβ1-induced EMT to increase the migration and invasion in HepG2 cells. To confirm this hypothesis, HepG2 cells were incubated with the STAT3 inhibitor AG490 prior to treatment with TGFβ1. As shown in Fig. 3A and C, AG490 significantly suppressed the TGFβ1-induced upregulation of N-cadherin and vimentin expression and the downregulation of E-cadherin expression, compared with the TGFβ1 group. Furthermore, AG490 treatment significantly reduced the number of TGFβ1-induced invasive and migratory cells (Fig. 3E and F), which was consistent with the results obtained from metastasized cell-wound closure (Fig. 3D).
Figure 3.
AG490 inhibits TGFβ1-induced EMT to increase migration and invasion via JAK/STAT3 signaling in HepG2 cells (A–C). Effects of TGFβ1 and AG490 on EMT-related protein expression in HepG2 cells. After the cells were treated with TGFβ1, AG490 and TGFβ1 + AG490 for 48 h, reverse transcription-quantitative polymerase chain reaction and western blot analysis of EMT markers in HepG2 cells were performed. Data are presented as mean ± standard deviation of three independent experiments. *P<0.05 vs. TGFβ1 group. (D) Wound-healing assay suggested that TGFβ1 markedly promoted cell motility. The promoting effects of TGFβ1 were abolished by AG490 treatment in HepG2 cells. The experiments were performed in triplicate. Data are presented as mean ± standard deviation. *P<0.05 vs. TGFβ1 group. (E) AG490 inhibited TGFβ1 to promote migration and invasion in HepG2 cells. In migration and invasion assays, cells were treated with TGFβ1, AG490 and TGFβ1 + AG490 for 48 h. The promoting effects of TGFβ1 were abolished by AG490 treatment in the HepG2 cells. Cells were visualized by staining with toluidine blue and counted in six random high-power fields at a magnification of ×200 using Image-Pro® Express software. Scale bar, 10 μm. The experiments were performed in triplicate. Data are presented as mean ± standard deviation. *P<0.05 vs. TGFβ1 group. TGFβ1, transforming growth factor β1; con, control; EMT, epithelial-to-mesenchymal transition; JAK, Janus kinase; STAT3, signal transducer and activator of transcription 3.
Twist is involved in TGFβ1-induced EMT depending on STAT3
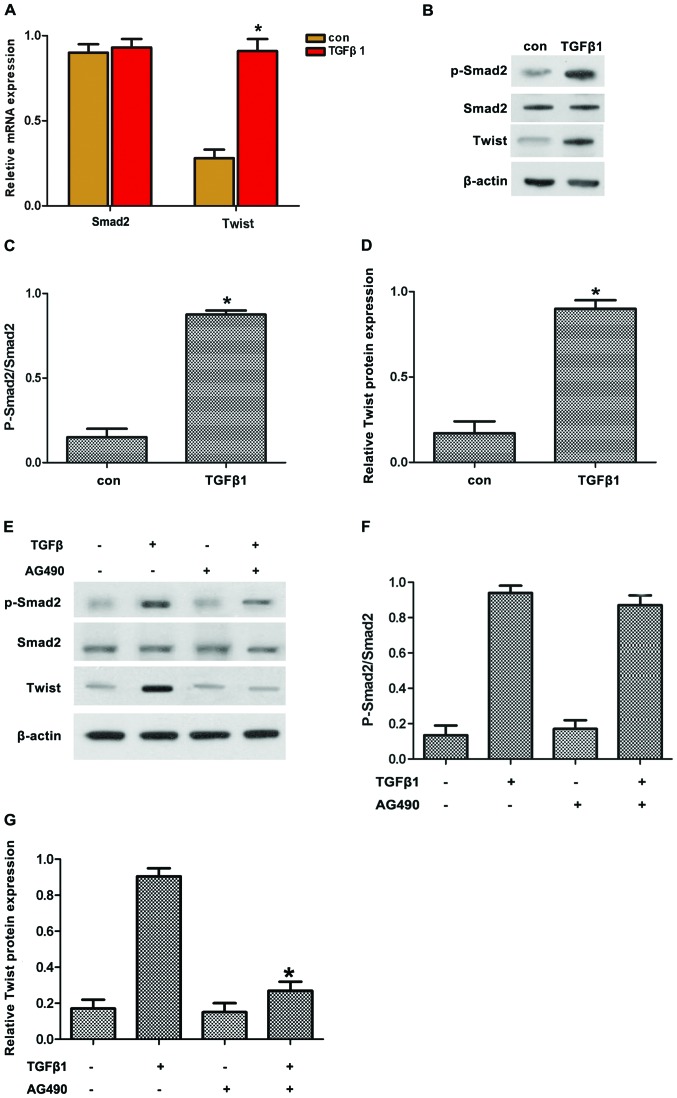
Smad2 and Twist are important factors regulating EMT through TGFβ1. Thus, we hypothesized that TGFβ1 induced EMT by upregulating Twist expression via JAK/STAT3 signaling. To confirm this hypothesis, the expression of pSmad2 and Twist in HepG2 cells treated with TGFβ1 was first detected. The results revealed that TGFβ1 induced the protein expression of pSmad2 and Twist (Fig. 4A–D). By contrast, AG490 treatment reversed the TGFβ1-induced protein expression of pSmad2 and Twist (Fig. 4E–G). Along these lines, TGFβ1 induced the protein expression of pSmad2 and Twist in accordance with the activated JAK/STAT3 signaling.
Figure 4.
(A–D) AG490 inhibited TGFβ1-induced expression of Twist and p-Smad2 in HepG2 cells. The effects of TGFβ1 on the expression of p-Smad2 and Twist in HepG2 cells were investigated by incubating HepG2 cells with 10 μM TGFβ1 for 48 h, followed by reverse transcription-quantitative polymerase chain reaction and western blot analysis of Smad2 and Twist expression. (E–G) The expression of p-Smad2 was analyzed by western blot analysis; AG490 inhibited the TGFβ1-induced expression of Twist and p-Smad2 in HepG2 cells. The cells were pretreated with AG490 for 2 h and then incubated with or without TGFβ1 (10 μM) for 48 h, followed by western blot analysis of p-Smad2, Smad2 and Twist expression. The experiments were performed in triplicate. Data are presented as mean ± standard deviation. *P<0.05 vs. TGFβ1 group. TGFβ1, transforming growth factor β1; EMT, epithelial-to-mesenchymal transition.
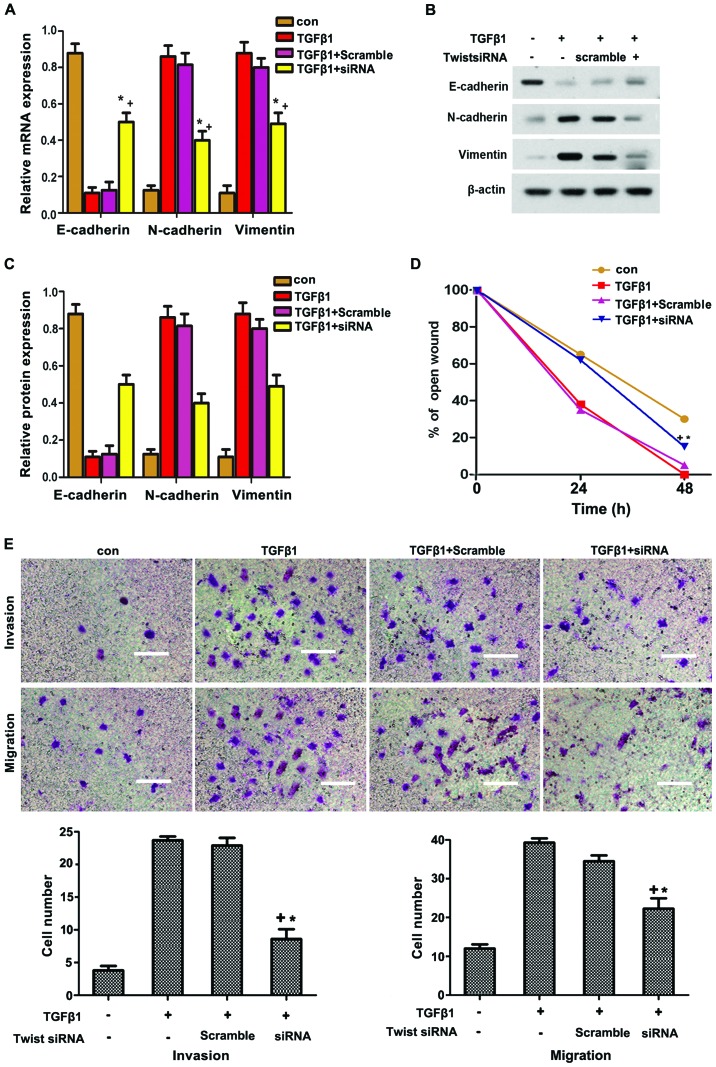
To further determine whether Twist participated in TGFβ1-induced EMT, HepG2 cells were transfected with siRNA of Twist. As shown in Fig. 5A–C, Twist knockdown significantly suppressed the TGFβ1-induced expression of N-cadherin and vimentin, but reversed the TGFβ1-inhibited expression of E-cadherin. The TGFβ1-induced migration and invasion in HepG2 cells were also reversed through Twist knockdown. Overall, these data strongly suggest that Twist participates in TGFβ1-induced EMT to increase the migration and invasion of HepG2 cells via JAK/STAT3 signaling.
Figure 5.
(A–C) Twist siRNA blocked the TGFβ1-induced EMT-mediated increase in the migration and invasion of HepG2 cells. Twist siRNA blocked the TGFβ1-induced EMT-related protein expression. HepG2 cells were pretreated with Twist siRNA for 12 h and then incubated with or without TGFβ1 (10 μM) for 48 h. E-cadherin, N-cadherin and vimentin proteins were analyzed by western blotting. (D) Wound-healing assay revealed that TGFβ1 markedly promoted cell motility. The promoting effects of TGFβ1 were abolished by Twist siRNA treatment in HepG2 cells. (E) Twist siRNA blocked TGFβ1-induced migration and invasion in HepG2 cells. HepG2 Cells were pretreated with Twist siRNA for 12 h and then incubated with or without TGFβ1 (10 μM) for 48 h. Cells were visualized by staining with toluidine blue and counted in six random high-power fields at a magnification of ×200 by using Image-Pro® Express software. Scale bar, 10 μm. The experiments were performed in triplicate. Data are presented as mean ± standard deviation. *P<0.05 vs. TGFβ1 group. TGFβ1, transforming growth factor β1; con, control; EMT, epithelial-to-mesenchymal transition.
Discussion
Invasion and metastasis are the leading causes of death from liver cancer (12). The onset and progression of liver cancer are regulated and controlled by various factors, among which EMT is the key mechanism promoting invasion and metastasis (13). A number of studies demonstrated that molecular markers are altered during EMT in cancer cells; for example, E-cadherin (an epithelial marker) and ZO-1 (a closely connected protein) are downregulated, whereas the levels of molecular markers derived from interstitial cells, including vimentin and N-cadherin, are upregulated. Hence, adhesions among tumor cells are reduced, thereby increasing the invasion and metastasis of cancer cells (14–16). The occurrence of EMT is affected by various factors. TGFβ1 is a key factor that induces and participates in the entire process of EMT (17–19). In the present study, TGFβ1 was found to upregulate vimentin and downregulate E-cadherin expression. Moreover, TGFβ1 induced scattering, invasion and metastasis of HepG2 cells. These results demonstrated that TGFβ1-induced EMT, thereby promoting the invasion and metastasis of HepG2 cells.
STAT3 is a signal transduction and transcription activator. Abnormal regulation of the STAT3 signaling pathway is associated with tumor occurrence and development (20). Following activation by cytokines or growth factors, the activated JAK may collect STAT3 monomers to produce homologous or heterogonous dimers; subsequently, nuclei and specific DNA sequences regulate the transcription of target genes (21). The abnormal expression and activation of STAT3 in various tumor tissues and cell lines (including liver cancer cells) are associated with the invasion and metastasis of tumor cells (21). EMT is the first focus in studies investigating the invasion and metastasis of tumor cells. The activation of the STAT3 signaling pathway is associated with EMT, invasion and metastasis of tumors. Colomiere et al (22) demonstrated that the JAK̸STAT3 pathway is aberrantly activated in ovarian cancer tissues. Furthermore, EMT in ovarian cancer cells may be induced by EGF or IL-6 (23,24). These results indicated that the action of EGF or IL-6 relies on the activation of JAK̸STAT3 signaling; EMT induced by EGF or IL-6 may be significantly inhibited by treatment with the JAK̸STAT3 pathway inhibitor AG490, and the invasion and metastasis of ovarian cancer cells may be reduced. Xiong et al (25) also reported that AG490 significantly suppressed STAT3 activation; consequently, AG490 treatment upregulated E-cadherin expression but reduced the invasion of tumor cells in colorectal cancer. In the present study, the JAK/STAT3 pathway was activated when the TGFβ1-induced EMT promoted the migration and invasion of HepG2 cells, whereas AG490 reversed these effects. Overall, the results demonstrated that TGFβ1-induced EMT was inhibited, thereby confirming the involvement of the JAK/STAT3 pathway in EMT induction.
Lee et al (7) reported that JAK̸STAT3 pathway activation promotes the expression of Twist and, thus, reduces the EMT of breast cancer cells. Cheng et al (17) also reported that the activated STAT3 may directly bind to the STAT3 binding site of the Twist promoter in breast cancer cells. These studies support that STAT3 activation may regulate the expression of Twist, a key transcription factor regulating EMT. In the present study, Twist was downstream from the JAK/STAT3 pathway in HepG2 cells; thus, AG490 treatment was applied for 2 h prior to incubation with TGFβ1. Our results demonstrated that the expression of Twist was inhibited by AG490. Overall, TGFβ1 induced the expression of Twist in accordance with the JAK/STAT3 pathway activation. Furthermore, we observed that Twist participated in the TGFβ1-induced EMT that promoted invasion and metastasis in HepG2 cells. This finding is consistent with the report of Liu et al (26). Thus, these results indicated that TGFβ1 upregulated the expression of Twist via the JAK/STAT3 pathway, thereby promoting the invasion and metastasis of HepG2 cells.
The findings of the present study verified the biological functions of TGFβ1 in liver cancer HepG2 cells and provided evidence that the TGFβ1-induced EMT promoted the invasion and metastasis of HepG2 cells in vitro. It was further demonstrated that these actions may be mediated via the JAK/STAT3/Twist signaling pathway. In conclusion, TGFβ1 appears to be involved in the progression of liver cancer and represents a potential molecular target for the treatment of this disease.
Acknowledgments
This study was supported by the Natural Science Foundation of China (grant no. 81600342), the Medical Foundation of Hui Zhou (grant no. 2015y134); the Medical Research Foundation of Guangdong Province (grant no. A2015620); and the Graduate Student Research Innovation Project of Hunan Province (grant no. CX2013B396).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Huang HC, Hu CH, Tang MC, Wang WS, Chen PM, Su Y. Thymosin beta4 triggers an epithelial-mesenchymal transition in colorectal carcinoma by upregulating integrin-linked kinase. Oncogene. 2007;26:2781–2790. doi: 10.1038/sj.onc.1210078. [DOI] [PubMed] [Google Scholar]
- 3.Gjerdrum C, Tiron C, Høiby T, Stefansson I, Haugen H, Sandal T, Collett K, Li S, McCormack E, Gjertsen BT, et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc Natl Acad Sci USA. 2010;107:1124–1129. doi: 10.1073/pnas.0909333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70:2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee TK, Poon RT, Yuen AP, Ling MT, Kwok WK, Wang XH, Wong YC, Guan XY, Man K, Chau KL, et al. Twist overexpression correlates with hepatocellular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clin Cancer Res. 2006;12:5369–5376. doi: 10.1158/1078-0432.CCR-05-2722. [DOI] [PubMed] [Google Scholar]
- 6.Giannelli G, Fransvea E, Bergamini C, Marinosci F, Antonaci S. Laminin-5 chains are expressed differentially in metastatic and nonmetastatic hepatocellular carcinoma. Clin Cancer Res. 2003;9:3684–3691. [PubMed] [Google Scholar]
- 7.Lee J, Choi JH, Joo CK. TGF-β1 regulates cell fate during epithelial-mesenchymal transition by upregulating survivin. Cell Death Dis. 2013;4:e714. doi: 10.1038/cddis.2013.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Connor JW, Gomez EW. Cell adhesion and shape regulate TGF-beta1-induced epithelial-myofibroblast transition via MRTF-A signaling. PLoS One. 2013;8:e83188. doi: 10.1371/journal.pone.0083188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wendt MK, Balanis N, Carlin CR, Schiemann WP. STAT3 and epithelial-mesenchymal transitions in carcinomas. JAK-STAT. 2014;3:e28975. doi: 10.4161/jkst.28975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamran MZ, Patil P, Gude RP. Role of STAT3 in cancer metastasis and translational advances. Biomed Res Int. 2013;421821 doi: 10.1155/2013/421821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hills CE, Squires PE. The role of TGF-β and epithelial-to mesenchymal transition in diabetic nephropathy. Cytokine Growth Factor Rev. 2011;22:131–139. doi: 10.1016/j.cytogfr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Książkiewicz M, Markiewicz A, Zaczek AJ. Epithelial-mesenchymal transition: A hallmark in metastasis formation linking circulating tumor cells and cancer stem cells. Pathobiology. 2012;79:195–208. doi: 10.1159/000337106. [DOI] [PubMed] [Google Scholar]
- 13.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological irnpact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y, Sarkissyan M, Vadgama JV. Epithelial-Mesenchymal Transition and Breast Cancer. J Clin Med. 2016;5:2–18. doi: 10.3390/jcm5020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brivio S, Cadamuro M, Fabris L, Strazzabosco M. Epithelial-to-mesenchymal transition and cancer invasiveness: What can we learn from cholangiocarcinoma. J Clin Med. 2015;4:2028–2041. doi: 10.3390/jcm4121958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nalluri SM, O'Connor JW, Gomez EW. Cytoskeletal signaling in TGFβ-induced epithelial-mesenchymal transition. Cytoskeleton. 2015;72:557–569. doi: 10.1002/cm.21263. [DOI] [PubMed] [Google Scholar]
- 17.Cheng GZ, Zhang WZ, Sun M, Wang Q, Coppola D, Mansour M, Xu LM, Costanzo C, Cheng JQ, Wang LH. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J Biol Chem. 2008;283:14665–14673. doi: 10.1074/jbc.M707429200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Sun WY, Wu JJ, Gu YJ, Wei W. Decreased expression of the type III TGF-β receptor enhances metastasis and invasion in hepatocellullar carcinoma progression. Oncol Rep. 2016;35:2373–2381. doi: 10.3892/or.2016.4615. [DOI] [PubMed] [Google Scholar]
- 19.Yeh YH, Wang SW, Yeh YC, Hsiao HF, Li TK. Rhapontigenin inhibits TGF-β-mediated epithelial mesenchymal transition via the I3K/AKT/mTOR pathway and is not associated with HIF-1α degradation. Oncol Rep. 2016;35:2887–2895. doi: 10.3892/or.2016.4664. [DOI] [PubMed] [Google Scholar]
- 20.Piccirillo R, Giavazzi R. Inactivating STAT3: Bad for tumor, good for muscle. Cell Cycle. 2015;14:939–940. doi: 10.1080/15384101.2015.1010979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wake MS, Watson CJ. STAT3 the oncogene - still eluding therapy. FEBS J. 2015;282:2600–2611. doi: 10.1111/febs.13285. [DOI] [PubMed] [Google Scholar]
- 22.Colomiere M, Ward AC, Riley C, Trenerry MK, Cameron-Smith D, Findlay J, Ackland L, Ahmed N. Cross talk of signals between EGFR and IL-6R through JAK2/STAT3 mediate epithelial-mesenchymal transition in ovarian carcinomas. Br J Cancer. 2009;100:134–144. doi: 10.1038/sj.bjc.6604794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li CJ, Li YC, Zhang DR, Pan JH. Signal transducers and activators of transcription 3 function in lung cancer. J Cancer Res Ther. 2013;9(Suppl 2):S67–S73. doi: 10.4103/0973-1482.119100. [DOI] [PubMed] [Google Scholar]
- 24.Liu A, Zhao F, Wang J, Zhao Y, Luo Z, Gao Y, Shi J. Regulation of TRPM7 function by IL-6 through the JAK2 STAT3 signaling pathway. PLoS One. 2016;11:e0152120–e0152132. doi: 10.1371/journal.pone.0152120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong H, Zhang ZG, Tian XQ, Sun DF, Liang QC, Zhang YJ, Lu R, Chen YX, Fang JY. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10:287–297. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu RY, Zeng Y, Lei Z, Wang L, Yang H, Liu Z, Zhao J, Zhang HT. JAK/STAT3 signaling is required for TGF-β-induced epithelial-mesenchymal transition in lung cancer cells. Int J Oncol. 2014;44:1643–1651. doi: 10.3892/ijo.2014.2310. [DOI] [PubMed] [Google Scholar]