Abstract
Inflammation and excessive proliferation of vascular smooth muscle cells (VSMCs) have key roles in various vascular disorders, including restenosis, atherosclerosis and pulmonary artery hypertension. However, the underlying mechanism remains unclear. The present study investigated the role of nuclear factor-κB (NF-κB) and microRNA (miRNA) in the regulation of VSMC proliferation under inflammatory conditions. It was demonstrated that miR-17 stimulated the proliferation of VSMCs, enhanced cell cycle G1/S transition, and increased levels of proliferating cell nuclear antigen and E2F1. By directly targeting the retinoblastoma (RB) protein mRNA-3′ untranslated region, miR-17 suppressed the expression of RB. Activation of NF-κB p65 resulted in increased miR-17 expression in VSMCs, whereas inactivation of NF-κB p65 resulted in decreased expression of miR-17 in VSMCs. NF-κB p65 signalling directly regulates miR-17 promoter activity. NF-κB p65 activation also suppressed RB expression, which was abrogated by miR-17 inhibitor. Taken together, the present results indicated that VSMC proliferation is regulated by activation of the NF-κB p65/miR-17/RB pathway. As NF-κB p65 signalling is activated in and is a master regulator of the inflammatory response, the present findings may provide a mechanism for the excessive proliferation of VSMCs under inflammation during vascular disorders and may identify novel targets for the treatment of vascular diseases.
Keywords: microRNA-17, retinoblastoma protein, nuclear factor-κB transcription factor, vascular smooth muscle cells, cell proliferation
Introduction
Excess proliferation of vascular smooth muscle cells (VSMCs) results in vascular remodelling and serves a key role in several vascular disorders, including restenosis (1,2), atherosclerosis (3) and pulmonary artery hypertension (4). However, the molecular mechanism underlying VSMC proliferation remains unclear.
Inflammation is a key response to the damage in vascular diseases (5–8). Clinical studies have demonstrated that inflammation is a marker for predicting internal carotid artery restenosis following eversion endarterectomy (9). The transcription factor nuclear factor-κB (NF-κB) is activated in and is a master regulator of inflammatory responses (10). It has been reported that NF-κB serves a key role in restenosis; NF-κB decoy may suppress restenosis following percutaneous coronary intervention (11) and attenuates in-stent restenosis in hypercholesterolemic rabbits (12). However, the mechanism by which VSMC proliferation is regulated by NF-κB during inflammation remains unclear.
MicroRNA (miRNA) are a type of non-coding RNA that regulate cellular functions (13). Studies have indicated that miRNA may have roles in vascular restenosis (14,15). miRNA-21 mediates thrombospondin-1-induced VSMC migration and proliferation (16) and serves a role in the proliferation of platelet-derived growth factor-induced human aortic vascular smooth muscle cells (17). miR-21 has been applied in attempts to treat restenosis (18,19). miR-17 belongs to the miR-17-92 cluster (20,21). Studies have revealed that the miR-17-92 cluster promotes VSMC proliferation in a murine model (22) and mediates inhibition of VSMC proliferation via bone morphogenetic protein receptor type II (BMPR2) (23). The Smad3/miR-17-92/BMPR2 pathway (23) and downregulation of the miR-17-92 cluster by thrombospondin-1 (24) serve important roles in the regulation of VSMC proliferation and function. However, it remains unknown how miRNA mediate the regulation of VSMC proliferation during inflammation through activation of NF-κB.
The present study investigated the molecular mechanism responsible for VSMC proliferation under inflammation. It was demonstrated that overexpression of miR-17 stimulated VSMC proliferation, enhanced cell cycle G1/S transition and increased levels of E2F1 and proliferating cell nuclear antigen (PCNA). miR-17 targeted retinoblastoma (RB) protein, which is a key regulator of cell cycle progression and proliferation (25,26). p65 controlled the expression of miR-17 and suppressed RB expression, which was abrogated by miR-17 inhibitor. The present data indicated that activation of the NF-κB p65/miR-17/RB pathway resulted in VSMC proliferation. This finding may identify the mechanism responsible for the excess proliferation of VSMCs under inflammation, and miR-17 may be a novel target for controlling several vascular disorders, including restenosis, atherosclerosis, and pulmonary artery hypertension.
Materials and methods
Cell culture and treatment
The VSMC (T/G HA-VSMC) and 293T cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). All types of cells were cultured on poly-(L-lysine)-coated plates/format discs (Sumitomo Bakelite Co., Ltd., Tokyo, Japan) in Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml penicillin and 100 mg/ml streptomycin in a humidified atmosphere with 5% CO2 at 37°C.
Bioinformatics analyses
To examine which cell cycle-regulating factor is a potential target of miR-17, cell cycle-regulating factors were surveyed for potential sites targeted by miR-17 using RNAhybrid (bibiserv.cebitec.uni-bielefeld.de/rnahybrid/), miRBase (mirbase.org/) and TargetScan (targetscan.org/vert_71/). Transcription factor prediction was performed using RNAhybrid. The potential target was selected based on conservation of the binding region and the strength of the predicted interaction.
Transfection and reporter luciferase activity assay
VSMCs and 293T cells were seeded on 6-well plates at a density of 6×105 cells/cm2 and cultured overnight in a humidified atmosphere with 5% CO2 at 37°C. Cells were then transfected with 20 nM miR-17 mimic or 20 nM inhibitor or 20 nM control miRNA (Guangzhou RiboBio Co., Ltd., Guangzhou, China) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After 24 h, cells were collected for western blotting and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analyses.
To construct the pGL3-RB-3′ untranslated region (3′UTR), the full length 3′UTR of the human RB mRNA was cloned into the pGL3-control vector (Promega Corp., Madison, WI, USA). To construct the miR-17 promoter reporter genes, the DNA fragment upstream of the transcription start site of the miR-17 gene was inserted into the pGL3-basic vector (Promega Corp.). For luciferase reporter assays, 293T cells were transiently transfected with miR-17 mimics and Renilla-luciferase reporter plasmids (Promega Corp.) containing the pGL3-RB-3′UTR (with the binding region: AGGCGCUU) or pGL3-RB-3′UTR MUT (with the mutant binding region: UUCGUGAA) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, cells were collected and reporter gene activity was determined using the dual-luciferase assay system (Invitrogen; Thermo Fisher Scientific, Inc.). Renilla luciferase activity was used for normalization of transfection efficiency. Reporter luciferase activity assay was also used to assess the function of NF-κB p65 regulating miR-17 promoter activity. There were three NF-κB p65 binding sites in the regulatory sequence of the miR-17 gene.
MTT assay
Cells were seeded on 96-well plates at a density of 3×103 cells/well and allowed to grow in the growth medium for 24 h in a humidified atmosphere with 5% CO2 at 37°C. Following transfection with miR-17 mimic or inhibitor and control for 48 h, cells were incubated with 5 mg/ml MTT for 4 h. Subsequently, dimethyl sulfoxide (100 µl/well) was added. The absorbance at 570 nm was then measured using an enzyme-linked immunosorbent assay (ELISA) reader (Autobio Diagnostics Co., Ltd., Zhengzhou, China). Experiments were repeated at least three times.
RT-qPCR analysis
Total RNA was isolated from experimental cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A cDNA library was generated through reverse transcription using 5X reaction buffer, RNase-free water, dNTPs, enzyme inhibitor, MMLV reverse transcriptase (Takara Bio, Inc., Otsu, Japan) and specific RT primers of miRNA (Guangzhou RiboBio Co., Ltd.). The RT conditions were as follows: 42°C for 60 min followed by 95°C for 5 min, then immediate cooling to 4°C. The expression levels of mature miRNA were determined using ExiLENT SYBR®-Green master mix (Takara Bio, Inc.). U6 small nuclear RNA was used as an internal control. Primers of miR-17 and U6 were purchased from Guangzhou RiboBio Co., Ltd. The qPCR conditions used were as follows: 95°C 10 min for polymerase activation/denaturation, 95°C for 10 sec and 60°C for 1 min with ramp rate 1.6°C/sec for 40 amplification cycles. To determine the relative levels of RB transcripts, RT-qPCR was performed under the following conditions, with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an endogenous control: RT conditions of 25°C for 10 min, 50°C 15 min, 85°C for 5 min, followed by immediate cooling to 4°C; qPCR conditions of 95°C for 10 min for polymerase activation/denaturation, 95°C for 10 sec and 60°C for 1 min for 40 amplification cycles. An ABI PRISM 7500 Fast Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to amplify and detect the specific products. The following PCR primers were used: RB forward, 5′-AAGGAGACAAGTTCGCATGT-3′ and reverse, 5′-GCCGGTAATTGTCGTAGTTT-3′; and GAPDH forward, 5′-CCATGTTCGTCATGGGTGTGAACCA-3′ and reverse, 5′-GCCAGTAGAGGCAGGGATGATGTTC-3′. The fold change for each miRNA and RB transcript relative to the control was calculated using the 2−ΔΔCq method (27).
Western blotting
Nuclear extracts were isolated from human VSMCs using a Nuclear and Cytoplasmic Protein Extraction kit (Nanjing Keygen Biotech Co., Ltd., Nanjing, China). For western blot analysis, VSMCs were homogenized in radioimmunoprecipitation assay buffer and nM phenylmethylsulfonyl fluoride (Beyotime Institute of Biotechnology, Haimen, China). The protein concentration was determined according to a bicinchoninic assay (BCA) standard curve using a BCA Protein αssay kit (Beyotime Institute of Biotechnology). Proteins (30–50 µg) from each cell lysate were mixed (4:1) with 5X sample buffer (Beyotime Institute of Biotechnology). A total of 30 µl protein and buffer mixture were loaded and resolved using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred onto polyvinylidene difluoride membranes by electroblotting. Membranes were blocked with QuickBlock (Beyotime Institute of Biotechnology) at room temperature for 15 min and then incubated with an appropriate dilution of p65, RB, PCNA, E2F1, H3 and GAPDH antibodies (1:1,000–1:2,000) overnight at 4°C. The membranes were then incubated with secondary antibodies conjugated to horseradish peroxidase at room temperature for 1 h. Antibody details are presented in Table I. Detection was performed using an enhanced chemiluminescence kit (cat. no. 32209; Thermo Fisher Scientific, Inc.). Proteins were detected with Chemi Genius 2 via GeneSnap software 7.02 (Syngene, Frederick, MD, USA), and relative intensities of the protein bands were analysed using ImageJ 1.47 software (National Institutes of Health, Bethesda, MD, USA).
Table I.
Primary and secondary antibodies used in western blotting.
| Type of antibody | Antibody | Supplier details | Cat. no. |
|---|---|---|---|
| Primary | p65 | Cell Signaling Technology, Inc., | 8242 |
| RB | Danvers, MA, USA | Ab181616 | |
| PCNA | Abcam, Cambridge, MA, USA | Ab92552 | |
| E2F1 | Ab179445 | ||
| H3 | Cell Signaling Technology, Inc., | 4499 | |
| GAPDH | Danvers, MA, USA | Ab8245 | |
| Secondary | Goat anti-rabbit IgG H&L (HRP) | Abcam, Cambridge, MA, USA | Ab6721 |
| Rabbit anti-mouse IgG H&L (HRP) | Abcam, Cambridge, MA, USA | Ab6728 |
RB, retinoblastoma; PCNA, proliferating cell nuclear antigen; IgG, immunoglobulin G; HRP, horseradish peroxidase; GAPDH, glyceraldehyde 3-phosphate dehydrogenas.
Flow cytometric analysis
All experimental cells were harvested by trypsinization, washed in ice-cold phosphate-buffered saline (PBS) three times, and fixed in 75% ice-cold ethanol in PBS overnight. Before staining, the cells were centrifuged (200 × g for 5 min at 4°C), and then resuspended in ice-cold PBS containing RNase A (0.2 mg/ml). Cells were incubated at 37°C for 30 min and then stained for 10 min at room temperature with 400 µl propidium iodide (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). Finally, samples of 10,000 cells were analysed on a FACSCalibur cytometer and data collected using CellQuest 3.1 software (BD Biosciences, San Jose, CA, USA).
Statistical analysis
Data were analysed using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA). Results were presented as mean ± standard deviation. Two-tailed Student's t-tests were used for comparing data between two groups. One-way analysis of variance was used to compare differences among groups, followed by least significant difference post hoc tests. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-17 directly targets the RB gene
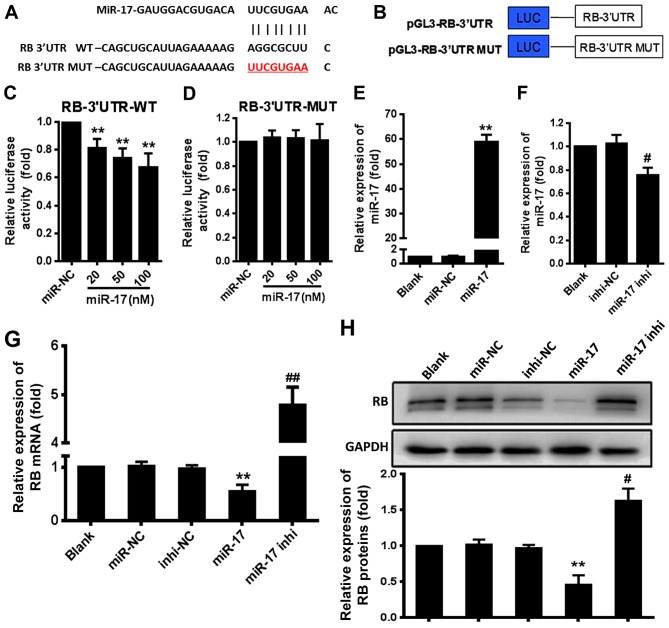
The bioinformatics analyses indicated a potential site for miR-17 in the RB mRNA-3′UTR (Fig. 1A). To confirm that this site was an actual target of miR-17, 293T cells were co-transfected with luciferase reporter plasmid constructs that contained the RB mRNA-3′UTR or RB mRNA-3′UTR mutant (MUT) (Fig. 1B) and miR-17 mimic (miR-17) or inhibitor (miR-17 inhi), and the luciferase activity was determined. The results demonstrated that miR-17 significantly suppressed the expression of the luciferase reporter gene directed by the wild-type (WT) RB mRNA-3′UTR compared with the expression in the miR-negative control (NC) group (P<0.01) (Fig. 1C). However, miR-17 did not significantly suppress the expression of the luciferase reporter gene directed by RB mRNA-3′UTR MUT compared with the miR-NC (Fig. 1D). Overexpression of miR-17 mimic resulted in a significant increase in the miR-17 expression level compared with the expression level induced by miR-NC (P<0.01) (Fig. 1E), and overexpression of miR-17 inhi significantly decreased the endogenous miR-17 level in VSMCs compared with the level induced by inhi-NC (P<0.05) (Fig. 1F). The level of RB mRNA in VSMCs was significantly decreased by treatment with miR-17 mimic compared with the levels in the miR-NC group (P<0.01) and significantly increased by treatment with miR-17 inhibitor compared with the levels in the inhi-NC group (P<0.01) (Fig. 1G). This finding was consistent with that of western blotting (Fig. 1H). These results indicated that miR-17 suppressed RB expression by directly targeting the RB mRNA-3′UTR in VSMCs.
Figure 1.
miR-17 directly targets the RB gene. (A) The target sites and free energy of binding between miR-17 and RB mRNA-3′UTR predicted by RNA hybrid and miRBase targets. (B) Schematic for the luciferase reporter plasmid constructs that contained the RB mRNA-3′UTR or MUT RB mRNA-3′UTR. (C) miR-17 suppressed expression of the luciferase reporter gene directed by the RB mRNA-3′UTR WT. (D) miR-17 did not suppress expression of the luciferase reporter gene directed by the RB mRNA-3′UTR MUT. (E) The levels of pri-miR-17 in miR-17 mimic-transfected cells. (F) The levels of miR-17 in miR-17 inhi transfected cells. (G) Expression of RB protein in VSMCs was decreased by overexpression of miR-17 mimic but not by overexpression of miR-17 inhi. The expression of RB protein was determined using an immunocytochemistry assay. (H) Expression of RB protein in VSMCs was decreased by overexpression of miR-17 mimic but not by overexpression of miR-17 inhi. The relative expression levels were quantitated using densitometry. Representative results were obtained from three repeats. *P<0.05 and **P<0.01 vs. miR-NC; #P<0.05 and ##P<0.01 vs. inhi-NC. miR, microRNA; RB, retinoblastoma; UTR, untrans-lated region; MUT, mutants; WT, wild-type; inhi, inhibitor; NC, negative control.
miR-17 stimulates VSMC proliferation and enhances cell cycle G1/S transition
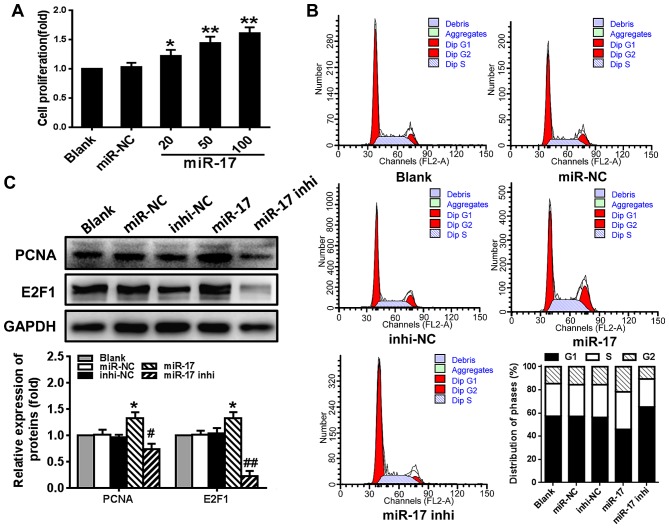
To determine whether miR-17 serves a role in regulating VSMC proliferation, the effect of miR-17 on cell viability, cell cycle progression and the expression of cell cycle-regulating factors PCNA and E2F1 was examined. The results demonstrated that VSMC proliferation was significantly stimulated by miR-17 mimic in a dose-dependent manner compared with the proliferation level in the miR-NC group (P<0.05) (Fig. 2A). Compared with the miR-NC group, overexpression of miR-17 (miR-17 group) significantly increased the number of cells that entered the S phase and decreased the number of the cells in the G1 phase, whereas downregulating the expression of miR-17 (miR-17 inhi group) markedly decreased the number of cells that entered S phase and increased the number of cells in the G1 phase (Fig. 2B). The levels of PCNA and E2F1 protein were significantly increased in VSMCs transfected with miR-17 mimic compared with the levels in those transfected with miR-NC (P<0.05). The miR-17 inhibitor had the opposite effect on the levels of these proteins, and significantly reduced the expression levels compared with the levels in those transfected with inhi-NC (P<0.05) (Fig. 2C). These results suggested that overexpression of miR-17 stimulated VSMC proliferation and enhanced cell cycle progression through promoting G1/S transition by increasing the levels of PCNA and E2F1.
Figure 2.
miR-17 stimulates proliferation of VSMCs. (A) miR-17 mimic stimulated proliferation of VSMCs. VSMCs were transfected with miR-17 mimic, and cell proliferation was determined using MTT assay. (B) Regulation of cell cycle progression in VSMCs by miR-17. The proportions of cells in each cycle were examined using flow cytometry. (C) The expression levels of cell cycle-regulating factors PCNA and E2F1 were regulated by miR-17. The protein levels were determined using western blotting. Representative results were obtained from three repeats. *P<0.05 and **P<0.01 vs. miR-NC; #P<0.05 and ##P<0.01 vs. inhi-NC. miR, microRNA; VSMCs, vascular smooth muscle cells; inhi, inhibitor; NC, negative control; PCNA, proliferating cell nuclear antigen.
NF-κB p65 subunit induces miR-17 transcription by binding its promoter elements
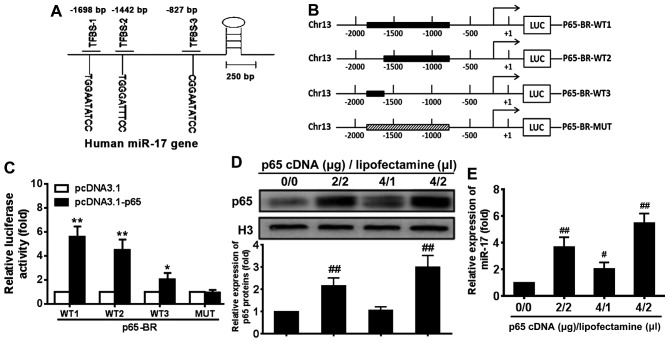
Previous studies have demonstrated that exogenous miR-17 mimic and inhibitor regulated RB protein level and VSMC proliferation (28,29). To further investigate the endogenous mechanism of regulation of miR-17, the potential binding sites for p65 in the upstream regulatory region of miR-17 were investigated and three potential p65 binding sites were identified around −1698, −1442 and −827 bp (relative to the transcription start site) in the regulatory sequence of the miR-17 gene (Fig. 3A). To determine whether these sites mediated the regulation of miR-17 expression by p65, luciferase reporter genes driven by the upstream regulatory DNA of miR-17 that contained all or some of the potential p65 binding sites were constructed (Fig. 3B). These constructs and p65cDNA WT expression plasmid or p65cDNA loss-of-function MUT expression plasmid were co-transfected into 293T cells, and the reporter luciferase activity was determined. The results demonstrated that the expression of the reporter genes containing p65 binding sites was significantly upregulated by pcDNA3.1-p65 compared with the levels induced by pcDNA3.1 in 293T cells (P<0.05) (Fig. 3C). The magnitude of the upregulation was related to the number of potential p65 binding sites contained in the reporter constructs.
Figure 3.
NF-κB p65 regulates miR-17 promoter activity. (A) NF-κB p65 binding sites in the regulatory sequence of the miR-17 gene. (B) Schematic for reporter luciferase genes driven by DNA fragments in the regulatory sequence of the miR-17 gene. WT1, WT2 and WT3 represent three different binding sites. (C) Expression of reporter genes p65-BR-Luc-WT1/2/3 was regulated by p65 but not p65-BR-MUT in 293T cells. 293T cells were transfected with the indicated plasmids and incubated for 48 h. Luciferase activity was assayed after cells were collected following incubation. (D) p65 protein expression in VSMCs transfected with different amounts of p65 cDNA and Lipofectamine. (E) Expression of miR-17 in VSMCs transfected with different amounts of p65 cDNA and Lipofectamine. The relative expression levels were quantitated using densitometry. Representative results were obtained from three repeats. *P<0.05 and **P<0.01 vs. pcDNA3.1; #P<0.05 and ##P<0.01 vs. 0 µg p65 cDNA/0 µl Lipofectamine. NF-κB, nuclear factor-κB; miR, microRNA; WT, wild-type; MUT, mutant; VSMCs, vascular smooth muscle cells.
Following transfection with different amounts of p65 cDNA and Lipofectamine, the level of p65 protein was upregulated compared with the level in the control group (Fig. 3D). To determine whether NF-κB p65 signalling regulates miR-17 expression, the miR-17 levels in VSMCs transfected with p65 cDNA were examined (Fig. 3E). The miR-17 expression level was also significantly increased in VSMCs transfected with p65 cDNA in a dose-dependent manner compared with 0 µg p65 cDNA/0 µl Lipofectamine group (P<0.05) (Fig. 3E). These results indicated that activation of NF-κB p65 signal-ling induced an increase in miR-17 levels in VSMCs.
NF-κB/p65 activation decreases RB level through miR-17 in VSMCs
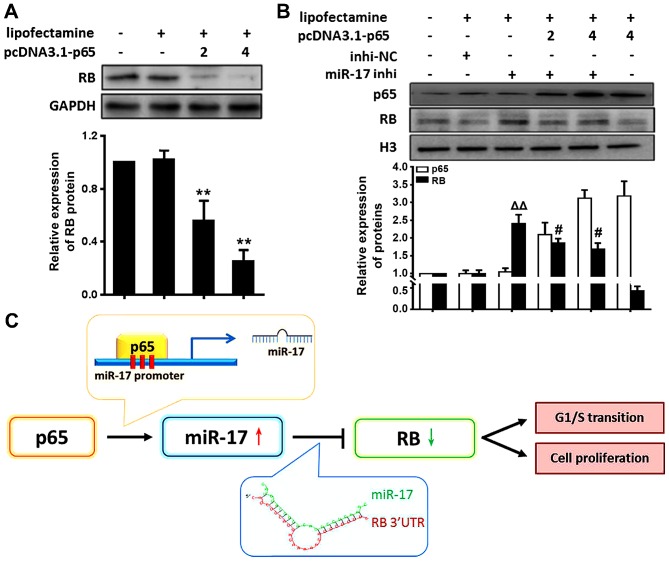
The present study demonstrated that the expression of miR-17 is directly regulated by NF-κB p65 activation and that miR-17 suppressed the RB levels by direct targeting. These data suggested that NF-κB p65 signalling decreased the RB levels in VSMCs through induction of miR-17 expression. To determine whether RB expression was actually regulated by NF-κB p65, RB expression in VSMCs transfected with p65 cDNA was examined. The results demonstrated that RB levels were significantly decreased in VSMCs treated with p65 cDNA compared with the levels in those treated with Lipofectamine only (P<0.01) (Fig. 4A). RB levels were decreased in VSMCs by activating NF-κB p65. These results suggested that activation of NF-κB p65 reduced RB levels.
Figure 4.
Activation of NF-κB p65 suppresses RB levels through miR-17 in VSMCs. (A) The RB protein level was decreased in cells treated with different amounts of p65 cDNA (2 or 4 µg/ml). (B) VSMCs were treated with p65 cDNA and miR-17 inhi. VSMCs were transfected with indicated plasmids and treated with the indicated chemicals. After incubation for 24 h, cells were collected for determination of protein expression. The relative expression levels were quantitated using densitometry. (C) A schematic model for the NF-κB/miR-17/RB pathway and its effects on VSMC proliferation. NF-κB p65 is activated under inflammation, and in turn it activates the expression of miR-17 at the transcription level. miR-17 directly targets RB, which stimulates VSMC proliferation and enhances cell cycle G1/S transition. Representative results were obtained from three repeats. **P<0.01 vs. Lipofectamine only; △△P<0.01 vs. RB level in inhi-NC group; #P<0.05 vs. RB level in miR-17 inhi group. NF-κB, nuclear factor-κB; miR, microRNA; inhi, inhibitor; NC, negative control; RB, retinoblastoma; VSMCs, vascular smooth muscle cells.
In order to further verify whether p65 activation inhibited RB expression through miR-17, the p65 and RB protein levels in VSMCs following treatment with miR-17 inhi and p65 cDNA were examined. The results revealed that RB protein levels were significantly increased in VSMCs following treatment with miR-17 inhi (miR-17 inhi vs. inhi-NC, P<0.01); however, this trend was weakened following overexpression of p65 (miR-17 inhi vs. miR-17 inhi + pcDNA3.1-p65, P<0.05) (Fig. 4B). These results suggested that decreases in miR-17 levels elevated RB levels suppressed by NF-κB p65 activation, supporting a model that activation of NF-κB p65 reduced RB levels through miR-17 in VSMCs under inflammation, as represented by the schematic in Fig. 4C.
Discussion
Excess proliferation of VSMCs and inflammation have a key role in several vascular disorders; however, the underlying mechanism remains unclear. The present study demonstrated that miR-17 was the most highly induced miRNA in proliferating VSMCs and overexpression of miR-17 stimulated VSMC proliferation, enhanced cell cycle G1/S transition and increased levels of E2F1 and PCNA. NF-κB p65 signalling activated the expression of miR-17, and miR-17 targeted RB, which is a key regulator of cell cycle progression and proliferation (25,26). p65 activation resulted in suppression of RB expression, which was abrogated by miR-17 inhibitor. These data suggest that NF-κB p65/miR-17/RB pathway activation leads to the proliferation of VSMCs. These findings may indicate a novel mechanism for restenosis and suggest a novel target for restenosis treatment or prevention.
Transcription factor E2F1 activates the expression of genes that promote DNA synthesis during cell cycle progression (30,31). PCNA is important for DNA synthesis and DNA repair (32). It is well known that RB is a negative regulator of E2F function in the regulation of cell cycle progression and proliferation (33). Activation of E2F1, E2F2 and E2F3a promotes cell cycle progression and proliferation by stimulating the expression of S phase genes (25,26,34,35). In the present study, miR-17 suppressed RB expression by directly targeting the RB mRNA-3′UTR. Overexpression of miR-17 decreased RB levels, releasing E2F1, which in turn promoted the transition from G1 to S phase of the cell cycle. Consistently, a previous study indicated that E2F3 promotes VSMC proliferation (36). How miR-17 increases the levels of E2F1 and PCNA requires further investigation.
It has been demonstrated that p65 disrupts the physical interaction between the activator E2Fs and the histone acetyltransferase cofactor transactivation/transformation-domain associated protein, resulting in a reduction in E2F-responsive gene expression in normal human fibroblasts. This leads to inhibition of cell cycle progression in an RB-independent manner (37). In the present study, it was indicated that NF-κB p65 signalling directly activated the expression of miR-17, which directly targeted RB expression in VSMCs. Therefore, NF-κB p65 signalling may regulate cell cycle progression and proliferation through RB-dependent and -independent pathways.
Interplay between NF-κB and miRNA has important roles in cell growth and proliferation (13,38). As a transcription factor, NF-κB regulates the expression of miR-143, which enhances hepatocellular carcinoma metastasis by repressing fibronectin expression (39). In the present study, it was demonstrated that miR-17 levels were increased by activation of NF-κB p65 signalling and decreased by inactivation of NF-κB p65 signalling in VSMCs. NF-κB p65 signalling directly regulates the miR-17 promoter activity, likely through potential p65 binding sites in the upstream regulatory region of the miR-17 gene. These data indicated that miR-17 expression is directly regulated by NF-κB p65 signalling. The present study also indicated that activation of p65 decreases the levels of RB, which is a direct target of miR-17. Furthermore, suppression of RB expression by activation of p65 was abrogated by miR-17 inhibitor in the present study. These findings support that p65 may regulate gene expression, and cell growth and proliferation by modulating the expression levels of miRNA, such as miR-17.
Taken together, the results of the present study suggest that miR-17 expression is highly induced in proliferating VSMCs. Overexpression of miR-17 stimulates the proliferation of VSMCs, likely via enhancing cell cycle G1/S transition by increasing the levels of E2F1 and PCNA. p65 activates the expression of miR-17 at the transcription level, and miR-17 directly targets RB, a key regulator of cell cycle progression and proliferation, through interaction with E2Fs. NF-κB p65 signalling actually decreases the RB level in VSMCs, which is abrogated by miR-17 inhibitor. These data support that proliferation of VSMCs is regulated by activation of the NF-κB p65/miR-17/RB pathway. The transcription factor NF-κB is activated in and is a master regulator of the inflammatory response, which is a characteristic of restenosis. The present findings may provide insight into the mechanism responsible for the excessive proliferation of VSMCs under inflammation during restenosis, adding to the regulation of proliferation of VSMCs by the Smad3/miR-17-92/BMPR2 pathway (23) and thrombospondin-1-mediated downregulation of the miR-17-92 cluster (24) in pathophysiological conditions, including restenosis, atherosclerosis and pulmonary artery hypertension. miR-17 may be a potential novel target for the treatment of restenosis.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (grant nos. 81170295 and 81000136), Liaoning Provincial Science and Technology Department (grant no. 2013225086) and Liaoning Provincial Special Program of Medical Science (grant no. LNCCC-C03-2015).
References
- 1.Bauters C, Isner JM. The biology of restenosis. Prog Cardiovasc Dis. 1997;40:107–116. doi: 10.1016/S0033-0620(97)80003-5. [DOI] [PubMed] [Google Scholar]
- 2.Yahagi K, Otsuka F, Sakakura K, Sanchez OD, Kutys R, Ladich E, Kolodgie FD, Virmani R, Joner M. J Cardiovasc Surg. Vol. 55. Torino: 2014. Pathophysiology of superficial femoral artery in-stent restenosis; pp. 307–323. [PubMed] [Google Scholar]
- 3.McNamara CA, Sarembock IJ, Bachhuber BG, Stouffer GA, Ragosta M, Barry W, Gimple LW, Powers ER, Owens GK. Thrombin and vascular smooth muscle cell proliferation: Implications for atherosclerosis and restenosis. Semin Thromb Hemost. 1996;22:139–144. doi: 10.1055/s-2007-999001. [DOI] [PubMed] [Google Scholar]
- 4.Sheikh AQ, Lighthouse JK, Greif DM. Recapitulation of developing artery muscularization in pulmonary hypertension. Cell Reports. 2014;6:809–817. doi: 10.1016/j.celrep.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Satoh K, Satoh T, Kikuchi N, Omura J, Kurosawa R, Suzuki K, Sugimura K, Aoki T, Nochioka K, Tatebe S, et al. Basigin mediates pulmonary hypertension by promoting inflammation and vascular smooth muscle cell proliferation. Circ Res. 2014;115:738–750. doi: 10.1161/CIRCRESAHA.115.304563. [DOI] [PubMed] [Google Scholar]
- 6.Donners MM, Daemen MJ, Cleutjens KB, Heeneman S. Inflammation and restenosis: Implications for therapy. Ann Med. 2003;35:523–531. doi: 10.1080/07853890310014876. [DOI] [PubMed] [Google Scholar]
- 7.Drachman DE, Simon DI. Inflammation as a mechanism and therapeutic target for in-stent restenosis. Curr Atheroscler Rep. 2005;7:44–49. doi: 10.1007/s11883-005-0074-5. [DOI] [PubMed] [Google Scholar]
- 8.Schillinger M, Minar E. Restenosis after percutaneous angioplasty: The role of vascular inflammation. Vasc Health Risk Manag. 2005;1:73–78. doi: 10.2147/vhrm.1.1.73.58932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaskovic S, Isenovic ER, Radak D. Inflammation as a marker for the prediction of internal carotid artery restenosis following eversion endarterectomy - evidence from clinical studies. Angiology. 2011;62:535–542. doi: 10.1177/0003319710398010. [DOI] [PubMed] [Google Scholar]
- 10.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, Lee R, Tatevossian RG, Phoenix TN, Thiruvenkatam R, et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki J, Tezuka D, Morishita R, Isobe M. An initial case of suppressed restenosis with nuclear factor-kappa B decoy transfection after percutaneous coronary intervention. J Gene Med. 2009;11:89–91. doi: 10.1002/jgm.1266. [DOI] [PubMed] [Google Scholar]
- 12.Ohtani K, Egashira K, Nakano K, Zhao G, Funakoshi K, Ihara Y, Kimura S, Tominaga R, Morishita R, Sunagawa K. Stent-based local delivery of nuclear factor-kappaB decoy attenuates in-stent restenosis in hypercholesterolemic rabbits. Circulation. 2006;114:2773–2779. doi: 10.1161/CIRCULATIONAHA.105.582254. [DOI] [PubMed] [Google Scholar]
- 13.Beitzinger M, Meister G. Preview. MicroRNAs: From decay to decoy. Cell. 2010;140:612–614. doi: 10.1016/j.cell.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 14.Gareri C, De Rosa S, Indolfi C. MicroRNAs for restenosis and thrombosis after vascular injury. Circ Res. 2016;118:1170–1184. doi: 10.1161/CIRCRESAHA.115.308237. [DOI] [PubMed] [Google Scholar]
- 15.Lv J, Wang L, Zhang J, Lin R, Wang L, Sun W, Wu H, Xin S. Long noncoding RNA H19-derived miR-675 aggravates restenosis by targeting PTEN. Biochem Biophys Res Commun: Jan. 4:2017. doi: 10.1016/j.bbrc.2017.01.011. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 16.Stein JJ, Iwuchukwu C, Maier KG, Gahtan V. Thrombospondin-1-induced vascular smooth muscle cell migration and proliferation are functionally dependent on microRNA-21. Surgery. 2014;155:228–233. doi: 10.1016/j.surg.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Yan L, Zhang W, Hu N, Chen W, Wang H, Kang M, Ou H. MicroRNA-21 inhibits platelet-derived growth factor-induced human aortic vascular smooth muscle cell proliferation and migration through targeting activator protein-1. Am J Transl Res. 2014;6:507–516. [PMC free article] [PubMed] [Google Scholar]
- 18.Muthiah M, Islam MA, Cho CS, Hwang JE, Chung IJ, Park IK. Substrate-mediated delivery of microRNA-145 through a polysorbitol-based osmotically active transporter suppresses smooth muscle cell proliferation: Implications for restenosis treatment. J Biomed Nanotechnol. 2014;10:571–579. doi: 10.1166/jbn.2014.1737. [DOI] [PubMed] [Google Scholar]
- 19.Santulli G, Wronska A, Uryu K, Diacovo TG, Gao M, Marx SO, Kitajewski J, Chilton JM, Akat KM, Tuschl T, et al. A selective microRNA-based strategy inhibits restenosis while preserving endothelial function. J Clin Invest. 2014;124:4102–4114. doi: 10.1172/JCI76069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Wu J, Yang C, Fan P, Balazs L, Jiao Y, Lu M, Gu W, Li C, Pfeffer LM, et al. DiGeorge syndrome critical region 8 (DGCR8) protein-mediated microRNA biogenesis is essential for vascular smooth muscle cell development in mice. J Biol Chem. 2012;287:19018–19028. doi: 10.1074/jbc.M112.351791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo T, Cui S, Bian C, Yu X. Crosstalk between TGF-β/Smad3 and BMP/BMPR2 signaling pathways via miR-17-92 cluster in carotid artery restenosis. Mol Cell Biochem. 2014;389:169–176. doi: 10.1007/s11010-013-1938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maier KG, Ruhle B, Stein JJ, Gentile KL, Middleton FA, Gahtan V. Thrombospondin-1 differentially regulates microRNAs in vascular smooth muscle cells. Mol Cell Biochem. 2016;412:111–117. doi: 10.1007/s11010-015-2614-9. [DOI] [PubMed] [Google Scholar]
- 25.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 26.Burke JR, Hura GL, Rubin SM. Structures of inactive retinoblastoma protein reveal multiple mechanisms for cell cycle control. Genes Dev. 2012;26:1156–1166. doi: 10.1101/gad.189837.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Shi J, Bei Y, Kong X, Liu X, Lei Z, Xu T, Wang H, Xuan Q, Chen P, Xu J, et al. miR-17-3p contributes to exercise-induced cardiac growth and protects against myocardial ischemia-reperfusion injury. Theranostics. 2017;7:664–676. doi: 10.7150/thno.15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen T, Zhou Q, Tang H, Bozkanat M, Yuan JX, Raj JU, Zhou G. miR-17/20 controls prolyl hydroxylase 2 (HD2)/hypoxia-inducible factor 1 (HIF1) to regulate pulmonary artery smooth muscle cell proliferation. J Am Heart Assoc. 2016;5:e004510. doi: 10.1161/JAHA.116.004510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blais A, Dynlacht BD. Hitting their targets: An emerging picture of E2F and cell cycle control. Curr Opin Genet Dev. 2004;14:527–532. doi: 10.1016/j.gde.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Wong JV, Dong P, Nevins JR, Mathey-Prevot B, You L. Network calisthenics: Control of E2F dynamics in cell cycle entry. Cell Cycle. 2011;10:3086–3094. doi: 10.4161/cc.10.18.17350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, Vermeulen W. Nuclear dynamics of PCNA in DNA replication and repair. Mol Cell Biol. 2005;25:9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer M, Müller GA. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit Rev Biochem Mol Biol. 2017 Aug 11; doi: 10.1080/10409238.2017.1360836. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 34.Korenjak M, Brehm A. E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr Opin Genet Dev. 2005;15:520–527. doi: 10.1016/j.gde.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Schaal C, Pillai S, Chellappan SP. The Rb-E2F transcriptional regulatory pathway in tumor angiogenesis and metastasis. Adv Cancer Res. 2014;121:147–182. doi: 10.1016/B978-0-12-800249-0.00004-4. [DOI] [PubMed] [Google Scholar]
- 36.Giangrande PH, Zhang J, Tanner A, Eckhart AD, Rempel RE, Andrechek ER, Layzer JM, Keys JR, Hagen PO, Nevins JR, et al. Distinct roles of E2F proteins in vascular smooth muscle cell proliferation and intimal hyperplasia. Proc Natl Acad Sci USA. 2007;104:12988–12993. doi: 10.1073/pnas.0704754104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Araki K, Kawauchi K, Tanaka N. IKK/NF-kappaB signaling pathway inhibits cell-cycle progression by a novel Rb-independent suppression system for E2F transcription factors. Oncogene. 2008;27:5696–5705. doi: 10.1038/onc.2008.184. [DOI] [PubMed] [Google Scholar]
- 38.Kravtsova-Ivantsiv Y, Shomer I, Cohen-Kaplan V, Snijder B, Superti-Furga G, Gonen H, Sommer T, Ziv T, Admon A, Naroditsky I, et al. KPC1-mediated ubiquitination and proteasomal processing of NF-κB1 105 to 50 restricts tumor growth. Cell. 2015;161:333–347. doi: 10.1016/j.cell.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–499. doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]