Abstract
Long non-coding RNAs (lncRNAs) have been established to participate in the complex network of various biological processes and play important roles in the differentiation of mesenchymal stem cells (MSCs). However, the roles of lncRNAs in the mechanisms of the osteogenic differentiation of human bone marrow-derived MSCs (HBMSCs) are poorly understood. Thus, this study aimed to investigate the effects of the lncRNA, differentiation antagonizing non-protein coding RNA (DANCR), on the proliferation and osteogenic differentiation of HBMSCs. We found that lncRNA DANCR was abnormally decreased in HBMSCs during osteogenic differentiation. DANCR knockdown induced by transfection with siRNA targeting DANCR (si-DANCR) significantly enhanced the proliferation and osteogenic differentiation of HBMSCs. By contrast, when DANCR expression was enhanced by transfection with a DANCR overexpression vector (pcDNA-DANCR), the proliferation and osteogenic differentiation of the HBMSCs were markedly inhibited. We further found that mitogen-activated protein kinase (MAPK) pathways were involved in the DANCR-mediated proliferation and osteogenic differentiation of HBMSCs. Moreover, DANCR was found to mediate the proliferation and osteogenic differentiation of HBMSCs via p38 MAPK inactivation, but not via extracellular signal-regulated protein kinase (ERK)1/2 or c-Jun N-terminal kinase (JNK) MAPKs, but. Combination treatment (pcDNA-DANCR and with the p38 specific inhibitor, SB203580) led to synergistic inhibitory effects, and these inhibitory effects were reversed by DANCR knockdown. These findings not only provide a novel interpretation for the mechanisms of the proliferation and osteogenic differentiation of HBMSCs, but also suggest that DANCR may be a novel therapeutic target for bone-destructive diseases in the future.
Keywords: human bone marrow-derived mesenchymal stem cells, long non-coding RNA, differentiation antagonizing non-protein coding RNA, osteogenic differentiation
Introduction
Bone deficiency is a socioeconomic challenge in stomatology. It not only affects the appearance of patients, but also increases the difficulty of restoring oral function (1). Currently, tissue engineering using suitable seed cells is considered to be the most effective method in the treatment of bone deficiencies. Mesenchymal stem cells (MSCs) are capable of self-renewal and can differentiate into multiple connective tissue cell types, including osteogenic, chondrogenic and adipogenic lineages. Human bone-derived marrow MSCs (HBMSCs) are associated with a high availability, low immunogenic properties and an ease of genetic manipulation; thus, they are regarded as a suitable source of cells in bone and cartilage tissue engineering (2,3). However, the molecular mechanisms of the differentiation processes of these cells involve complex pathways which remain largely unknown.
Recently, growing attention has been paid to a class of non-coding RNAs (ncRNAs), which are those RNAs that do not encode any proteins. Long non-coding RNAs (lncRNAs), which belong to a novel heterogeneous class of ncRNAs, play crucial roles in both cell fate determination and disease pathogenesis (4,5). The length of lncRNAs does not exceed 200 nt, and they can fold into complex structures to provide greater potential and versatility for both protein and target recognition (6). lncRNAs have been proposed to play crucial regulatory roles in a wide range of biological pathways and cellular processes, including chromosome inactivation (7) and differentiation (4), the reprogramming of stem cell pluripotency (8), and the modulation of invasion and apoptosis (9). Previous studies have suggested that the downregulation of the lncRNA, differentiation antagonizing non-protein coding RNA (DANCR) promotes the osteogenic differentiation of human periodontal ligament stem cells (10) and human fetal osteoblastic cells (11). However, whether lncRNA DANCR is also involved in the osteogenic differentiation of HBMSCs remains unknown.
In the present study, we obtained differential expression profiles of lncRNA DANCR in undifferentiated vs. osteogenic-differentiated HBMSCs. We found that the lncRNA DANCR expression level was significantly decreased in HBMSCs during osteogenic differentiation. DANCR knockdown abnormally enhanced the levels of alkaline phosphatase activity (ALP), the mRNA expression of osteogenic marker genes and mineralized matrix deposition in HBMSCs, whereas the overexpression of DANCR yielded opposite results. Moreover, we demonstrated that DANCR regulates the proliferation and osteogenic differentiation via the p38 mitogen-activated protein kinase (MAPK) pathway. The association of DANCR with the MAPK pathway may provide new insight into the mechanisms involved in the regulation of osteogenic differentiation.
Materials and methods
Cell culture
The HBMSC cell line, PTA-1058, was obtained from (The American Type Culture Collection, Manassas, VA, USA). The cells were cultured in Dulbecco's modified Eagle's medium (HyClone, Logan, UT, USA) supplemented with 100 U/l penicillin (Gibco Life Technologies, Carlsbad, CA, USA), 100 mg/l streptomycin (Gibco Life Technologies) and 10% FBS (Gibco Life Technologies) in a humidified atmosphere of 5% CO2 at 37°C. The confluent cells were transferred to the following passage using 0.25% trypsin for up to 3 passages and the culture medium was changed every 3 days.
Proliferation assay
Cell proliferation was measured on days 3, 5 and 7 by flow cytometry (FCM). The HBMSCs were treated with serum-free medium for 24 h, and the medium was then replaced with culture medium containing 10% FBS. The cells were harvested on days 3, 5 and 7 and then fixed with 75% ice-cold ethanol. Cell cycle fractions (G0, G1, S, and G2 M phases) were determined by FCM and the DNA content was measured using a FACScan flow cytometer (BD Biosciences, San Diego, CA, USA).
Osteogenic differentiation of HBMSCs
The HBMSCs (at passage 3) were cultured in osteogenic supplement (OS) medium containing 10 mM β-glycerophosphate, 100 nM ascorbic acid and 100 nM dexamethasone (all from Sigma-Aldrich, St. Louis, MO, USA). The cells were subjected to ALP activity assay and Alizarin Red staining. ALP activity was measured using the ALP assay kit (Jiancheng Technology Co., Ltd., Nanjing, China) according to the manufacturer's instructions. Alizarin Red staining was measured using 40 mM Alizarin Red S (pH 4.4; Jiancheng Technology Co., Ltd.) for 15 min at room temperature. Mineralized nodules were visualized using an inverted microscope (Carl Zeiss AG, Oberkochen, Germany) after rinsing with PBS and 10 images were captured for each group. SB203580 (Cell Signaling Technology Inc., Danvers, MA, USA), a highly selective inhibitor of p38 signaling, was prepared in DMSO and used in the signaling inhibition assay at the concentration of 10 mM as previously described (12). The osteogenenic differentiation was measured on days 7, 14 and 21.
Plasmid construction and cell transfection
The ectopic expression of DANCR was induced by transfection with a DANCR overexpression vector (pcDNA3.1-DANCR) using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA), with an empty pCDNA3.1 vector used as the control. Both vectors were obtained from and created by RiboBio (Guangzhou, China). The knockdown of DANCR was induced by transfection with siRNA targeting DANCR (si-DANCR; obtained from RiboBio) using Lipofectamine 2000 (Invitrogen Life Technologies), with an empty si-NC vector used as the control (RiboBio). According to the manufacturer's instructions, the fusion and transfection of the HBMSCs were performed when the cells were cultivated 0n 6-well plates using Lipofectamine 2000.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total cellular RNA was isolated from the HBMSCs using TRIzol (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the instructions recommended by the manufacturer. The RNA was reverse transcribed into cDNA using a PrimeScript RT Master Mix kit (Jiancheng Technology Co., Ltd.). Quantitative PCR was performed using a SYBR Green PCR kit (Toyobo Co., Ltd., Osaka, Japan) and the ABI 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) according to the instructions provided by the manufacturer. The primers used were as follows: human DANCR forward, 5′-GCCACTATGTAGCGGGTTTC-3′and reverse, 5′-ACCTGCGCTAAGAACTGAGG-3′; human bone gamma-carboxyglutamate protein (BGLAP) forward, 5′-CAG CGGTGCAGAGTCCAGCAAA-3′ and reverse, 5′-GATGT GGTCAGCCAACTCGTCA-3′; human collagen I (COL1) forward, 5′-GGACACAATGGATTGCAAGG-3′ and reverse, 5′-TAACCACTGCTCCACTCTGG-3′; human Runt-related transcription factor 2 (RUNX2) forward, 5′-CCGCACAACC GCACCAT-3′ and reverse, 5′-CGCTCCGGCCCACAAATCTC-3′ and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5′-GGGCTGCTTTTAACTCTGGT-3′ and reverse, 5′-GCAGGTTTTTCTAGACGG-3′. For each sample, GAPDH expression was analyzed to normalize target gene expression. Each sample was analyzed in triplicate. Relative gene expression was calculated using the 2-ΔΔCq method.
Western blot analysis
The HBMSCs were lysed in RIPA buffer containing 1 mM phenylmethane sulfonyl-fluoride according to the manufacturer's instructions. The total protein concentration was determined using a bicinchoninic acid assay kit. Protein lysates (20 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto 0.22 μm polyvinylidene difluoride membranes (Millipore Corp., Bedford, MA, USA). After blocking, the membranes were incubated overnight at 4°C with specific primary antibodies [anti-p-p38 (Thr180/Tyr182; D3F9; cat. no. 4511), p-38 (D13E1; cat. no. 8690), p-c-Jun N-terminal kinase (JNK; Thr183/Tyr185; 81E11; cat. no. 4668), JNK (cat. no. 9252), p-extracellular signal-regulated protein kinase (ERK)1/2, ERK1/2 and β-actin (13E5; cat. no. 4970) antibodies; diluted, 1/1,000; Cell Signaling Technology Inc). Following 3 washes with PBST (0.5% Tween-20 in PBS), the membranes were incubated with the relevant secondary antibodies (L27A9; cat. no. 3678) 1/1,000 diluted (Cell Signaling Technology Inc.) for 1 h at 37°C, washed and visualized with an ECL detection kit (Amersham Pharmacia Biotech, Piscataway, NJ, USA). β-actin served as an internal control.
Statistical analysis
All data are expressed as the mean ± SD and were processed using SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). Data were analyzed using one-way analysis of variance. Three independent experiments were performed. A value of P<0.05 was considered to indicate a statistically significant difference.
Results
DANCR levels are downregulated in HBMSCs during osteogenic differentiation
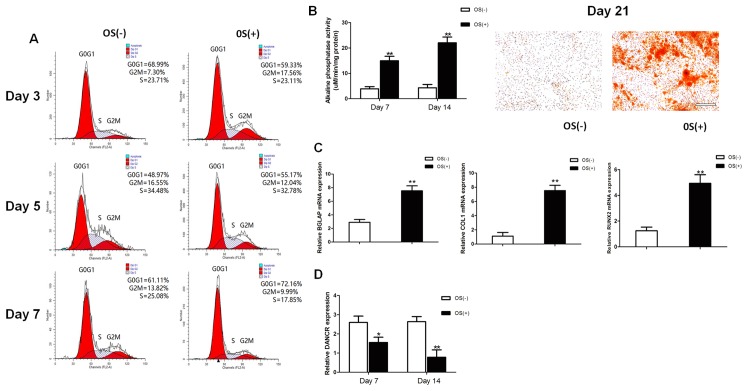
The osteogenic differentiation of HBMSCs was induced by using OS medium. Cell cycle fractions (G0, G1, S, and G2M phases) were determined by FCM on days 3, 5 and 7. The number of cells in the S phase decreased significantly in the osteogenic-differentiated HBMSCs compared to the undifferentiated HBMSCs on day 7 (Fig. 1A), while no significant differences were observed on days 3 and 5. Both ALP activity assay and Alizarin Red staining revealed that the micromass pellets had differentiated toward the osteogenic lineage following induction with OS medium, as shown by increased ALP activity and a greater amount of cells stained red (Fig. 1B). RT-qPCR detected a marked increase in the expression of osteogenic marker genes on day 14, including BGLAP, COL1 and RUNX2 (Fig. 1C). The lncRNA DANCR expression profiles during osteogenic differentiation were investigated on days 7 and 14 after osteogenic induction. The expression levels of lncRNA DANCR were significantly decreased between the undifferentiated and differentiated cells (Fig. 1D).
Figure 1.
Osteogenic differentiation of HBMSCs. (A) Cell cycle fractions (G0, G1, S, and G2 M phases) of HBMSCs were determined by flow cytometry. (B) ALP activity was measured using an ALP assay kit and mineralized matrix deposition was measured by Alizarin Red S staining. (C) The expression of BGLAP, COL1 and RUNX2 in HBMSCs was measured by RT-qPCR. (D) The expression of DANCR in the HBMSCs was measured by RT-qPCR. Scale bar, 100 μm. *P<0.05 and **P<0.01 vs. cells cultured without osteogenic supplement (OS) medium. HBMSCs, human bone marrow-derived mesenchymal stem cells; ALP, alkaline phosphatase; DANCR differentiation antagonizing non-protein coding RNA; BGLAP bone gamma-carboxyglutamate protein; COL1, collagen I; RUNX2, Runt-related transcription factor 2.
Expression of osteogenic marker genes is enhanced by the knockdown of DANCR and is suppressed by overexpression of DANCR
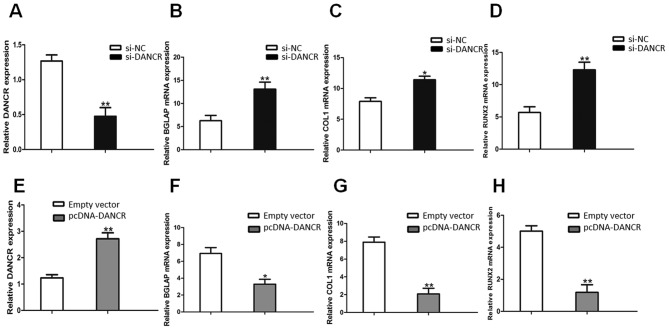
The knockdown of DANCR expression was successfully performed using RNA interference (si-NC and si-DANCR) in HBMSCs. After the HBMSCs were transfected with si-DANCR, the DANCR relative levels were significantly downregulated (Fig. 2A), compared with the cells transfected with si-NC. Following treatment with OS medium for 14 days, RT-qPCR assays revealed that the downregulation of DANCR significantly promoted the expression of osteogenic marker genes, including BGLAP, COL1 and RUNX2 (Fig. 2B, C and D). On the other hand, the DANCR relative levels in the pcDNA-DANCR-transfected HBMSCs were significantly upregulated (Fig. 2E). Compared with the cells transfected with the empty vector, the expression levels of osteogenic marker genes in the pcDNA-DANCR-transfected cells were significantly descreased following treatment with OS medium for 14 days (Fig. 2F, G and H).
Figure 2.
Effects of DANCR on osteogenic marker genes. (A) DANCR levels were assayed by RT-qPCR after the HBMSCs were transfected with si-DANCR. (B-D) The expression of BGLAP, COL1 and RUNX2 in the HBMSCs transfected with si-DANCR was measured by RT-qPCR. (E) DANCR levels were determined by RT-qPCR after the HBMSCs were transfected with pcDNA-DANCR. (F-H) The expression of BGLAP, COL1 and RUNX2 in HBMSCs transfected with pcDNA-DANCR measured by RT-qPCR. *P<0.05 and **P<0.01 vs. empty vector group. HBMSCs, human bone marrow-derived mesenchymal stem cells; DANCR differentiation antagonizing non-protein; BGLAP bone gamma-carboxyglutamate protein; COL1, collagen I; RUNX2, Runt-related transcription factor 2.
Proliferation and osteogenic differentiation of HBMSCs is promoted by the knockdown of DANCR and inhibited by the overexpression of DANCR
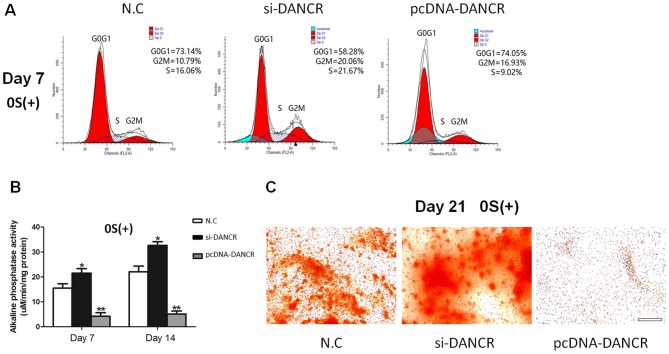
In order to determine the role of DANCR in regulating the proliferation and osteogenic differentiation of HBMSCs, the cells transfected with si-DANCR or pcDNA-DANCR were subjected to cell cycle fractions, ALP activity assay and Alizarin Red staining. We found that the knockdown of DANCR significantly increased the number of cells in the S phase in the osteogenic-differentiated HBMSCs on day 7, whereas the overexpression of DANCR inhibited cell proliferation (Fig. 3A). Moreover, higher ALP activity and a greater number of mineralized nodules were observed during osteogenic differentiation in the si-DANCR-transfected HBMSCs, as evidenced by increased ALP activity assay and Alizarin Red staining (Fig. 3B and C). These results suggested that the downregulation of DANCR promoted the proliferation and osteogenic differentiation of HBMSCs.
Figure 3.
Effects of DANCR on the proliferation and osteogenic differentiation of HBMSCs. (A) Cell cycle fractions (G0, G1, S, and G2 M phases) of HBMSCs treated with si-DANCR or pcDNA-DANCR were determined by flow cytometry. (B) ALP activity in HBMSCs transfected with si-DANCR or pcDNA-DANCR was measured using an ALP assay kit. (C) Mineralized matrix deposition of HBMSCs transfected with si-DANCR or pcDNA-DANCR was measured by Alizarin Red S staining. Scale bar, 100 μm. *P<0.05 and **P<0.01 vs. negative control (N.C). HBMSCs, human bone marrow-derived mesenchymal stem cells; DANCR differentiation antagonizing non-protein; ALP, alkaline phosphatase activity.
DANCR regulates the proliferation and osteogenic differentiation of HBMSCs via the p38 MAPK pathway
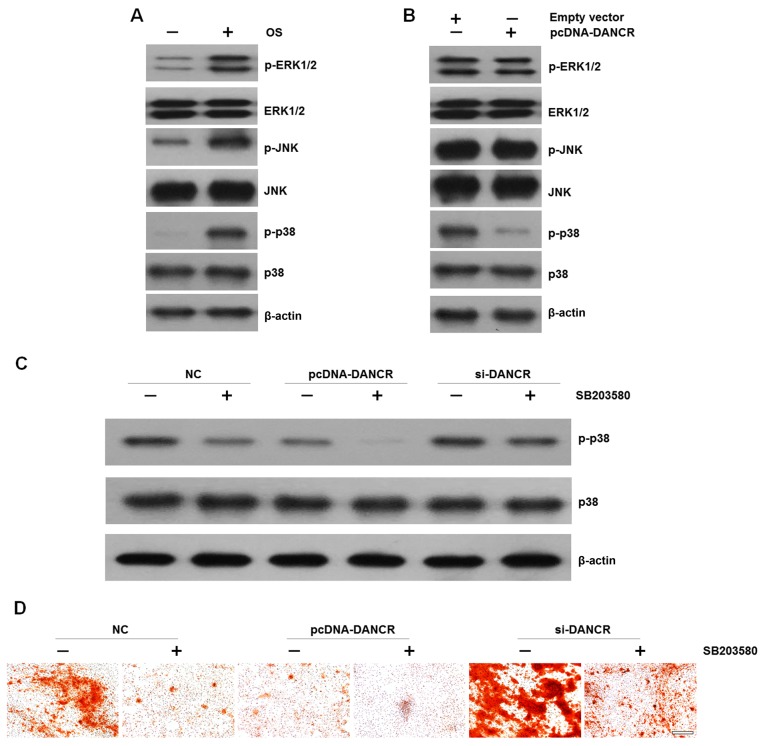
It is well known that the MAPK family regulates the differentiation, mineralization and proliferation of HBMSCs (13). Therefore, in this study, we examined the effects of DANCR on MAPK signal activation in HBMSCs. We found that cultured in OS medium enhaced the phosphorylation of ERK1/2, JNK and p38 (Fig. 4A). However, when the cells were transfected with pcDNA-DANCR, the levels of p-p38 were markedly decreased, while no significant difference was observed in the levels of p-ERK1/2 or p-JNK (Fig. 4B). In order to elucidate the effects of p38 on DANCR-related cell proliferation and osteogenic differentiation, HBMSCs that were transfected with si-DANCR or pcDNA-DANCR were treated with the specific p38 inhibitor, SB203580, which was followed by western blot analysis and Alizarin Red staining. We found that the upregulation of DANCR resulted in the inactivation of the p38 MAPK pathway. More importantly, when the p38 MAPK pathway was inactivated by the inhibitor, the inhibitory effects were reversed by DANCR knockdown (Fig. 4C). In accordance, mineralized matrix formation inhibited by the suppression of the p38 MAPK pathway was also ameliorated by DANCR knockdown (Fig. 4D)
Fig. 4.
effects of DANCR on MAPK signal activation in HBMSCs. (A) The phosphorylation of ERK1/2, JNK and p38 during osteogenic differentiation was assayed by western blot analysis. (B) The phosphorylation of ERK1/2, JNK and p38 in DANCR-overexpressing HBMSCs was assayed by western blot analysis. (C) HBMSCs transfected with nothing, pcDNA-DANCR or si-DANCR were treated with SB203580. p38 MAPK in the HBMSCs was assayed by western blot analysis. (D) BMSCs transfected with nothing, pcDNA-DANCR or si-DANCR were treated with SB203580. Mineralized matrix deposition was measured by Alizarin Red S staining. Scale bar, 100 μm. HBMSCs, human bone marrow-derived mesenchymal stem cells; DANCR differentiation antagonizing non-protein.
Discussion
Bone deficiency is an emerging medical threat in stomatology. Stomatological diseases can cause bone deficiency, which in turn can increase the difficulty of restoring oral function (14). Currently, tissue engineering using suitable seed cells that mimics the natural process has shown great potential in the treatment of bone deficiencies (15–17). Among these seed cells, MSCs present in a variety of mesenchymal tissues, have gained increasing attention. MSCs have selfrenewable capacities and multi-directional differentiation potentials (18,19). Given appropriate culture conditions, they are capable of differentiating into chondrocytes (2), adipocytes (20), endothelial cells (21), cardiomyocytes (22), osteocytes (23), as well as other cell lineages. HBMSCs have the advantages of availability, culture expansion, low immunogenicity (2) and anti-inflammatory properties (24), and thus they are widely used in tissue engineering and regenerative orthopaedics (25). The differentiation processes involve complex pathways regulated at both transcriptional and post-transcriptional levels. However, data on the precise molecular mechanisms of osteogenic differentiation remain incomprehensive.
lncRNAs are evolutionarily conserved ncRNAs that are abundantly encoded in mammalian genomes, numbering in the tens of thousands. The ratio of lncRNAs in total ncRNAs is beyond 80% but is the least well understood (26). Although initially thought to be transcriptional noise, recent evidence suggests that lncRNAs play crucial roles in gene regulatory processes, such as cellular metabolism (27), drug resistance (28) and apoptosis (29). Previous studies have suggested that some lncRNAs modulate pluripotency in embryonic stem cells (30) and play a key role in controlling the cell state (4). lncRNA DANCR, highly expressed in the epidermis and downregulated during the procedure of stem cell differentiation, is required to maintain the undifferentiated state of epidermal stem cells or osteoblasts (31). Recent studies have demonstrated that the downregulation of ANCR promotes the osteogenic differentiation of human periodontal ligament stem cells (10) and human fetal osteoblastic cells (11). Therefore, we speculated the potential role of lncRNA DANCR in the osteogenic differentiation of HBMSCs.
In the present study, we investigated the differential expression of lncRNA DANCR in undifferentiated vs. osteogenic-differentiated HBMSCs. We found that the lncRNA DANCR expression level was significantly decreased during the osteogenic differentiation of HBMSCs. By downregulating DANCR, the number of cells in the S phase, the levels of ALP activity, the mRNA expression of osteogenic marker genes and mineralized matrix deposition in the HBMSCs was abnormally increased. On the other hand, the upregulation of DANCR yielded opposite results. Thus, we confirmed that the downregulation of DANCR promoted the proliferation and osteogenic differentiation of HBMSCs.
The core purpose of the present study was to investigate the potential molecular signaling pathways engaged by DANCR. The MAPK signaling pathways, including the JNK, ERK1/2 and p38 pathways have emerged as major regulators of cellular physiology and the activation of MAPK is a crucial trigger of osteogenic differentiation (32–35). Our data indicated that the DANCR-regulated osteogenic differentiation was neither ERK1/2-dependent nor JNK-dependent, but p38-dependent, and the inhibitory effects of DANCR overexpression were more evident when introduced in combination with p38 inactivation induced by the specific inhibitor, SB203580. Moreover, we found that these inhibitory effects were reversed by DANCR knockdown.
In conclusion, the present study highlights the potential role of lncRNA DANCR in regulating HBMSC proliferation and osteogenic differentiation. We further found that the p38 MAPK pathway was involved in the precise molecular mechanisms. These findings suggested that DANCR may be both a novel potential target and regulatory factor of bone-destructive diseases. Further investigations are warranted in order to fully elucidate the complete molecular mechanisms involved in this process.
Acknowledgments
The present study was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD; 2014–37).
References
- 1.Maire P. Calibrated autologous bone grafts–their use in oral implantology. Widening - crest augmentation. Personal technic Rev Stomatol Chir Maxillofac. 1997;98(Suppl 1):27–30. In French. [PubMed] [Google Scholar]
- 2.Rastegar F, Shenaq D, Huang J, Zhang W, Zhang BQ, He BC, Chen L, Zuo GW, Luo Q, Shi Q, et al. Mesenchymal stem cells: Molecular characteristics and clinical applications. World J Stem Cells. 2010;2:67–80. doi: 10.4252/wjsc.v2.i4.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macchiarini P, Jungebluth P, Go T, Asnaghi MA, Rees LE, Cogan TA, Dodson A, Martorell J, Bellini S, Parnigotto PP, et al. Clinical transplantation of a tissue-engineered airway. Lancet. 2008;372:2023–2030. doi: 10.1016/S0140-6736(08)61598-6. [DOI] [PubMed] [Google Scholar]
- 4.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 8.Ng SY, Stanton LW. Long non-coding RNAs in stem cell pluripotency. Wiley Interdiscip Rev RNA. 2013;4:121–128. doi: 10.1002/wrna.1146. [DOI] [PubMed] [Google Scholar]
- 9.Ørom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154:1190–1193. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia Q, Jiang W, Ni L. Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch Oral Biol. 2015;60:234–241. doi: 10.1016/j.archoralbio.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432:612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 12.Hu Y, Chan E, Wang SX, Li B. Activation of p38 mitogen-activated protein kinase is required for osteoblast differentiation. Endocrinology. 2003;144:2068–2074. doi: 10.1210/en.2002-220863. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Chen B, Cao M, Sun J, Wu H, Zhao P, Xing J, Yang Y, Zhang X, Ji M, Gu N. Response of MAPK pathway to iron oxide nanoparticles in vitro treatment promotes osteogenic differentiation of hBMSCs. Biomaterials. 2016;86:11–20. doi: 10.1016/j.biomaterials.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Hertz J. Problems of maxillofacial and oral surgery. J Int Coll Surg. 1956;26:63–79. [PubMed] [Google Scholar]
- 15.Kim SH, Kim KH, Seo BM, Koo KT, Kim TI, Seol YJ, Ku Y, Rhyu IC, Chung CP, Lee YM. Alveolar bone regeneration by transplantation of periodontal ligament stem cells and bone marrow stem cells in a canine peri-implant defect model: A pilot study. J Periodontol. 2009;80:1815–1823. doi: 10.1902/jop.2009.090249. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Zhang Z, Zhao J, Zhang X, Sun X, Xia L, Chang Q, Ye D, Jiang X. Vertical alveolar ridge augmentation with beta-tricalcium phosphate and autologous osteoblasts in canine mandible. Biomaterials. 2009;30:2489–2498. doi: 10.1016/j.biomaterials.2008.12.067. [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, Zhang Z, Wang S, Sun X, Zhang X, Chen J, Kaplan DL, Jiang X. Apatite-coated silk fibroin scaffolds to healing mandibular border defects in canines. Bone. 2009;45:517–527. doi: 10.1016/j.bone.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bianco P, Robey PG. Stem cells in tissue engineering. Nature. 2001;414:118–121. doi: 10.1038/35102181. [DOI] [PubMed] [Google Scholar]
- 19.Jones E, McGonagle D. Human bone marrow mesenchymal stem cells in vivo. Rheumatology (Oxford) 2008;47:126–131. doi: 10.1093/rheumatology/kem206. [DOI] [PubMed] [Google Scholar]
- 20.Karagianni M, Brinkmann I, Kinzebach S, Grassl M, Weiss C, Bugert P, Bieback K. A comparative analysis of the adipogenic potential in human mesenchymal stromal cells from cord blood and other sources. Cytotherapy. 2013;15:76–88. doi: 10.1016/j.jcyt.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Crisan M. Transition of mesenchymal stem/stromal cells to endothelial cells. Stem Cell Res Ther. 2013;4:95. doi: 10.1186/scrt306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao JW, Zhang MR, Ji QY, Xing FJ, Meng LJ, Wang Y. The role of slingshot-1L (SSH1L) in the differentiation of human bone marrow mesenchymal stem cells into cardiomyocyte-like cells. Molecules. 2012;17:14975–14994. doi: 10.3390/molecules171214975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heino TJ, Hentunen TA. Differentiation of osteoblasts and osteocytes from mesenchymal stem cells. Curr Stem Cell Res Ther. 2008;3:131–145. doi: 10.2174/157488808784223032. [DOI] [PubMed] [Google Scholar]
- 24.Liu P, Deng Z, Han S, Liu T, Wen N, Lu W, Geng X, Huang S, Jin Y. Tissue-engineered skin containing mesenchymal stem cells improves burn wounds. Artif Organs. 2008;32:925–931. doi: 10.1111/j.1525-1594.2008.00654.x. [DOI] [PubMed] [Google Scholar]
- 25.Pećina M, Vukičević S. Tissue engineering and regenerative orthopaedics (TERO) Int Orthop. 2014;38:1757–1760. doi: 10.1007/s00264-014-2477-9. [DOI] [PubMed] [Google Scholar]
- 26.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim Biophys Acta. 2014;1843:372–386. doi: 10.1016/j.bbamcr.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 28.Jiang M, Huang O, Xie Z, Wu S, Zhang X, Shen A, Liu H, Chen X, Wu J, Lou Y, et al. A novel long non-coding RNA-ARA: Adriamycin resistance-associated. Biochem Pharmacol. 2014;87:254–283. doi: 10.1016/j.bcp.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 29.Wu W, Zhang S, Li X, Xue M, Cao S, Chen W. Ets-2 regulates cell apoptosis via the Akt pathway, through the regulation of urothelial cancer associated 1, a long non-coding RNA, in bladder cancer cells. PLoS One. 2013;8:e73920. doi: 10.1371/journal.pone.0073920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu N, Feng C, Jiang Y, Miao Q, Liu H. Regulative Effect of Mir-205 on Osteogenic Differentiation of Bone Mesenchymal Stem Cells (BMSCs): Possible Role of SATB2/Runx2 and ERK/MAPK Pathway. Int J Mol Sci. 2015;16:10491–10506. doi: 10.3390/ijms160510491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herbert BA, Valerio MS, Gaestel M, Kirkwood KL. Sexual Dimorphism in MAPK-Activated Protein Kinase-2 (MK2) Regulation of RANKL-Induced Osteoclastogenesis in Osteoclast Progenitor Subpopulations. PLoS One. 2015;10:e0125387. doi: 10.1371/journal.pone.0125387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin L, Tang B, Deng B, Mohan C, Wu T, Peng A. Extracellular regulated protein kinases play a key role via bone morphogenetic protein 4 in high phosphate-induced endothelial cell apoptosis. Life Sci. 2015;131:37–43. doi: 10.1016/j.lfs.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Ki YW, Park JH, Lee JE, Shin IC, Koh HC. JNK and p38 MAPK regulate oxidative stress and the inflammatory response in chlorpyrifos-induced apoptosis. Toxicol Lett. 2013;218:235–245. doi: 10.1016/j.toxlet.2013.02.003. [DOI] [PubMed] [Google Scholar]