Abstract
Acute coronary syndrome (ACS) is a common disease with high mortality and morbidity rates. The methylation status of blood DNA may serve as a potential early diagnosis and prevention biomarker for numerous diseases. The present study was designed to explore novel genome-wide aberrant DNA methylation patterns associated with ACS. The Infinium HumanMethylation450 assay was used to examine genome-wide DNA methylation profiles in 3 pairs of ACS and control group samples. Epigenome-wide DNA methylation, genomic distribution, Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed. The results were confirmed using methylation-specific polymerase chain reaction (MSP) and Sequenom MassARRAY analyses in ACS, stable coronary artery disease (SCAD) and control samples. A total of 11,342 differentially methylated (DM) 5′-C-phosphate-G-3′ (CpG) sites were identified, including 8,865 hypomethylated and 2,477 hypermethylated CpG sites in the ACS group compared with the control samples. They varied in frequency across genomic compartments, but were particularly notable in gene bodies and shores. The results of GO term and KEGG pathway enrichment analyses revealed that the methylated genes were associated with certain biological processes and pathways. Despite the considerable variability in methylation data, the candidate selected possessed significant methylation alteration in mothers against decapentaplegic homolog 3 (SMAD3) transcription start site 155 (Chr1:67356838-Chr1:67356942). MSP analysis from 81 ACS samples, 74 SCAD samples and 53 healthy samples, and Sequenom MassARRAY analysis, confirmed that differential CpG methylation of SMAD3 was significantly corrected with the reference results of the HumanMethylation450 array. The data identified an ACS-specific DNA methylation profile with a large number of novel DM CpG sites, some of which may serve as candidate markers for the early diagnosis of ACS.
Keywords: DNA methylation, genome-wide, alterations, acute coronary syndrome, Infinium HumanMethylation450 assay
Introduction
Acute coronary syndrome (ACS) is a type of cardiovascular disease (CVD) that occurs as a result of an acute obstruction in the coronary arteries, and it represents a leading cause of mortality and disability worldwide (1–3). A previous epidemiological survey revealed that ~50% of cardiovascular mortalities are due to ACS (4). Therefore, it is necessary to clarify the underlying biological mechanisms of ACS to enable early diagnosis and treatment of the disease.
The traditional diagnosis of ACS is achieved by a combination of electrocardiogram (ECG) results and the detection of specific serum markers (5). However, the sensitivity of ECGs is generally <50% and the application of serum markers, including creatine kinase isoenzyme MB (CK-MB), cardiac troponin (cTn)I and cTnT, also has limitations (6). Despite the high specificity of cTnT and cTnI, they may only be detected 6–12 h following myocardial ischemia, which is insufficient for early diagnosis and treatment of ACS prior to myocardial injury (7). The development of novel, sensitive and accurate ACS biomarkers are urgently required to enable the early diagnosis and treatment of ACS.
Epigenetics are defined as stable, heritable alterations in gene expression, which are not due to changes in the DNA sequence (8). The role of epigenetics has mainly been evaluated in relation to cancer (9), but selected previous studies have explored epigenetic involvement in CVD development and progression (10–13). DNA methylation-induced genetic repression is a major type of epigenetic modification (14). DNA methylation occurs at the C5 position of cytosine by the covalent modification of a methyl group, which is most commonly found in 5′-C-phosphate-G-3′ (CpG) dinucleotides (15). Alterations in genome-wide and site-specific DNA methylation have been described in CVD, although their specific functions are still largely uninvestigated (16,17). Altered DNA methylation (hyper methylation or hypomethylation) is one of the early events in the pathogenesis of ACS (18,19). Hypermethylation of specific promoter regions of crucial genes, such as transcription factor genes, can repress the expression of these genes and alter biological functions, which in turn increases the risk of ACS (20). Furthermore, it has been reported that numerous risk factors, including hypertension (21,22), diabetes (23–26), atherosclerosis (27–30) and smoking (31,32), associated with the development of ACS may cause epigenetic alterations. These previous studies suggested that the detection of DNA methylation alterations may be a promising tool for early ACS prediction and diagnosis. However, systematic analyses of the changes to genome-wide DNA methylation in patients with ACS are still lacking. In the present study, the genome-wide aberrant DNA methylation patterns predominantly associated with ACS were explored. Additionally, the differentially methylated (DM) regions associated with ACS pathogenesis were characterized, and novel DNA methylation biomarkers potentially beneficial to the early diagnosis of ACS were identified.
Materials and methods
Study subjects
The present study recruited 81 patients with ACS, 74 patients with stable coronary artery disease (SCAD) and 53 control subjects. The patients with CAD in the present study were admitted to the Department of Cardiology of Jinling Hospital (Nanjing, China) between January, 2014 and December, 2016, who were undergoing clinically necessary coronary angiography. Angiograms of all patients with CAD revealed ≥50% stenosis in a minimum of one coronary artery. The 81 patients with ACS included patients with acute myocardial infarction and unstable angina, with Braunwald classification II or III who exhibited a positive cardiac biomarker result (cTnI ≥0.090 ng/ml or cTnT ≥0.014 ng/ml), acute ischemic-type chest pain (lasting for >15 min, duration from symptom onset to emergency admission within 72 h) and characteristic ECGs. The 74 patients with SCAD had a normal ECG and documented normal left ventricular contractility, with the exception of possible minor nonspecific ST-T features, and they had a ≥1 year history without any cardiac events or procedures suggestive of ACS. The exclusion criteria for the patients with CAD included mild disease identified on angiography (a stenosis of 10–50% of the luminal diameter in all three coronary arteries), prior coronary revascularization and the presence of renal disease. The 53 control subjects were recruited to the study from routine health examinations at Jinling Hospital over the same period. The control patients had a normal physical examination, ECG and laboratory tests, and were did not show signs of other diseases, including hyperlipidemia, hypertension and diabetes mellitus, or any clinically evident signs of atherosclerosis. The clinicopathological features of all the patients and controls are listed in Table I. The present study was approved by the Ethics Committee of Jinling Hospital and all the recruited subjects provided written informed consent prior to the study.
Table I.
Clinical and biochemical characteristics of the participants included in the present study.
| Variable | Group
|
||
|---|---|---|---|
| Control (n=53) | SCAD (n=74) | ACS (n=81) | |
| Age, years (mean ± range) | 64.68±8.18 | 68.99±13.62 | 72.72±12.93 |
| Sex, n=m/f | 34/19 | 47/27 | 42/39 |
| History of hypertension, n (%) | 0 (0) | 58 (78) | 61 (75) |
| History of diabetes, n (%) | 0 (0) | 15 (20) | 37 (46) |
| TC, mmol/l | 4.43±0.47 | 4.16±0.95 | 4.50±0.73 |
| TG, mmol/l | 1.01±0.33 | 1.48±0.97a | 1.66±0.57b |
| HDL-C, mmol/l | 1.39±0.26 | 1.07±0.30b | 0.94±0.16b |
| LDL-C, mmol/l | 2.70±0.58 | 2.74±0.52 | 2.80±0.33 |
| Cardiac troponin I, ng/ml | – | 0.03±0.01 | 6.61±17.31c |
| Cardiac troponin T, ng/ml | – | 0.015±0.03 | 0.758±1.95d |
| Creatine kinase-MB, U/l | – | 10.24±7.44 | 45.57±80.27d |
P<0.05 and
P<0.001 vs. the control group;
P<0.05 and
P<0.01 vs. the SCAD group. HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride; SCAD, stable coronary artery disease; ACS, acute coronary syndrome; m, male; f, female.
Blood sampling
Blood samples were collected from the patients with 81 ACS and 74 patients with SCAD in tubes containing EDTA immediately following their hospitalization. The 53 control samples were drawn from the elbow vein of a paired healthy individual on an empty stomach. The samples were stored at −80°C prior to analysis.
DNA isolation and sodium bisulfite modification
Genomic DNA was isolated from 100 µl blood using the DNeasy Blood and Tissue kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's protocol. The quality and quantity of the extracted genomic DNA was analyzed with a NanoDrop™ 2000 Spectrophotometer. Bisulfite modification of 1 μg DNA was conducted using an EpiTect Bisulfite kit (Qiagen, Inc.) according to the manufacturer's protocol.
Infinium HumanMethylation450 assay
A total of 3 patients with ACS were randomly selected and paired with 3 control subjects for the Infinium HumanMethylation450 assay (Illumina, Inc., San Diego, CA, USA). The Infinium HumanMethylation450 assay was performed according to the manufacturer's standard protocol. Processed methylation chips were scanned using an iScan reader (Illumina, Inc.). Paired control samples were processed on the same chip and all samples were processed at the same time to avoid chip-to-chip variation. The assays were conducted in the Beijing Genomics Institute (Beijing, China).
Methylation-specific semi-quantitative polymerase chain reaction (MSP) analysis
In the present study, MSP was developed for the detection of mothers against decapentaplegic homolog 3 (SMAD3; Chr1:67356838-Chr1:67356942) methylation in 81 patients with ACS, 74 patients with SCAD and 53 healthy individuals using two pairs of primers, one was specific for the methylated alleles while the other was specific for the unmethylated alleles. Primer sequences are listed in Table II. The thermocycling conditions were as follows: Complete denaturation of the DNA at 95°C for 5 min; 35 cycles of denaturation at 95°C for 30 sec, annealing at 58°C for 30 sec, extension at 72°C for 45 sec; and a final extension at 72°C for 10 min. The MSP products were assessed by electrophoresis in 2% agarose gels with 0.5 μg/ml ethidium bromide and analyzed using a UVC1-1100 SmartView Pro 1100 Imaging system with version 1.0.0.2 software (Major Science, Saratoga, CA, USA). Images demonstrating the different methylation patterns were captured.
Table II.
Primer sequences for the methylation analysis of mothers against decapentaplegic homolog 3 (Chr1:67356838-Chr1:67356942).
| Method | Primer | Sequence (5′-3′) | Amplicon size (bp) |
|---|---|---|---|
| MSP | UF | TTTAGATATTGGTTAGTTGGTTGT | 141 |
| UR | ACACATTAAAAAACTCCACAACTT | ||
| MF | AGATATCGGTTAGTCGGTTGC | 141 | |
| MR | CATTAAAAAACTCCGCGACTT | ||
| Sequenom | F | AGGAAGAGAGGTTTTGTAATTTTTTATTTATTTGAGGG | 352 |
| MassARRAY | R | CAGTAATACGACTCACTATAGGGAGAAGGCTATTTTATTTCCAAAACAATAAACTCACA |
UF, unmethylated forward; UR, unmethylated reverse; MF, methylated forward; MR, methylated reverse; F, forward; R, reverse.
Sequenom MassARRAY
Primer pairs were designed using EpiDesigner (epidesigner.com) and are presented in Table II. MassArray analysis was performed in triplicate using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY Analyzer 4; Sequenom, San Diego, CA, USA) according to manufacturer's protocol in 8 ACS patients, 8 SCAD patients and 8 healthy individuals randomly from each group. The experiments were conducted at the Beijing Genomics Institute.
Differential methylation analysis
The Infinium HumanMethylation450 assay data were normalized using internal controls (normalization control probe pairs, which were designed to target the same region within housekeeping genes and had no underlying CpG sites in the probe) with GenomeStudio software (version 2011.1; Illumina, Inc.). For quality control, methylation measures of P>0.05 and samples with a CpG coverage <95% were removed. The methylation levels of CpG sites were calculated as the β score [β score = intensity of the methylated allele/(intensity of the unmethylated allele + intensity of the methylated allele + 100)]. Limma software (version 3.24.15; bioconductor. org/packages/release/bioc/html/limma.html) was used to perform paired t-tests followed by Benjamini and Hochberg correction for multiple testing in order to identify DM CpG sites in the ACS group vs. the control group. Sites with a Benjamini and Hochberg-corrected value of P<0.05 and a β score ≥0.1 were considered to be significantly statistically different. A volcano plot was used to display the mean DNA methylation differences for the DM CpG sites. Hierarchical clustering of the methylation data was performed when the β score was ≥0.2.
Functional annotation of DM CpG sites
The enrichment analysis of the methylated regions for Gene Ontology (GO) (33) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) (34) pathways was conducted using the Genomic Regions Enrichment of Annotations Tool (35). Binomial statistics with false discovery rate (FDR) correction were used to identify significantly enriched ontologies. The most significant and replicable corresponding genes were selected to be candidate markers for ASC detection. These genes were chosen from the results of the differential methylation tests at the site, gene region and gene island region levels, and had a β score difference between the ACS and control samples of >0.3.
Statistical analysis
Statistical analyses were performed using SPSS software (version 20.0; SPSS, Inc., Chicago, IL, USA). Normally distributed values are expressed as the mean ± standard deviation. A one-sample Kolmogorov-Smirnov test was used to evaluate the normality of distribution of the variables. The significance of differences between variants among groups was analyzed by one-way analysis of variance, and the differences between groups were subsequently determined using a post hoc Fisher's least significant difference test. The differences in cTnI and CK-MB between two groups were analyzed using the Student's t-test. The Kruskal-Wallis H test was performed for non-parametric data. P<0.05 was considered to indicate a statistically significant difference.
Results
Normalization of methylation data
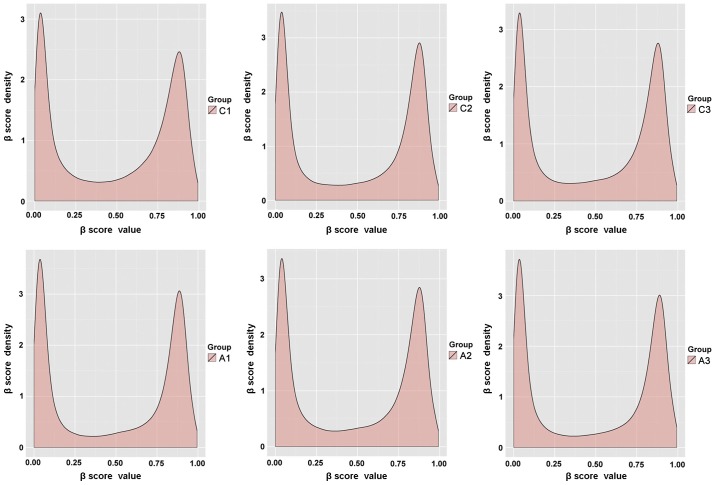
For the methylated sites identified by Illumina HumanMethylation450 assay, the original data of the 3 patients with ACS and 3 paired controls were normalized to the internal controls to eliminate the possibility of system error, and the β scores were subsequently calculated. The β scores were generated based on the intensity of the methylated and unmethylated probes, and were used as an indicator of the degree of methylation at each CpG site. Subsequently, a built-in detection score filter was used prior to further calculations. This only left values with significantly higher mean signal intensities from multiple probes for a given CpG locus compared with those of the negative control in the same set of chip data. The average number of loci detected for the three pairs of ACS and control samples was 484,352 covering 42,343 genes (Table III). The density plots of the β score for the 6 samples exhibited a bimodal distribution (Fig. 1). These results indicate that the hybridization and amplification conditions were uniform for all samples.
Table III.
DM CpG sites and genes in patients with acute coronary syndrome compared with the control group.
| CpG site methylation | No. of CpG sites | No. of genes |
|---|---|---|
| Loci detected | 484,352 | 42,343 |
| DM | 11,342 | 5,073 |
| Hypermethylated | 2,477 | 1,746 |
| Hypomethylated | 8,865 | 5,071 |
CpG, 5′-C-phosphate-G-3′; DM, differentially methylated.
Figure 1.
Density plots of the distribution of the β scores. The β score distribution for the three pairs of samples following normalization, as visualized in density plots.
Epigenome-wide DNA methylation analysis
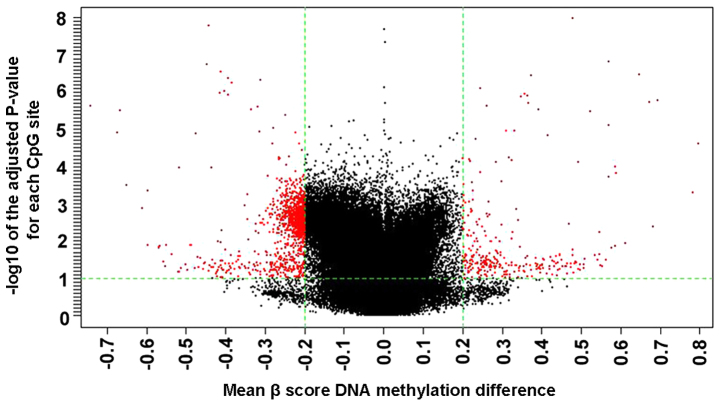
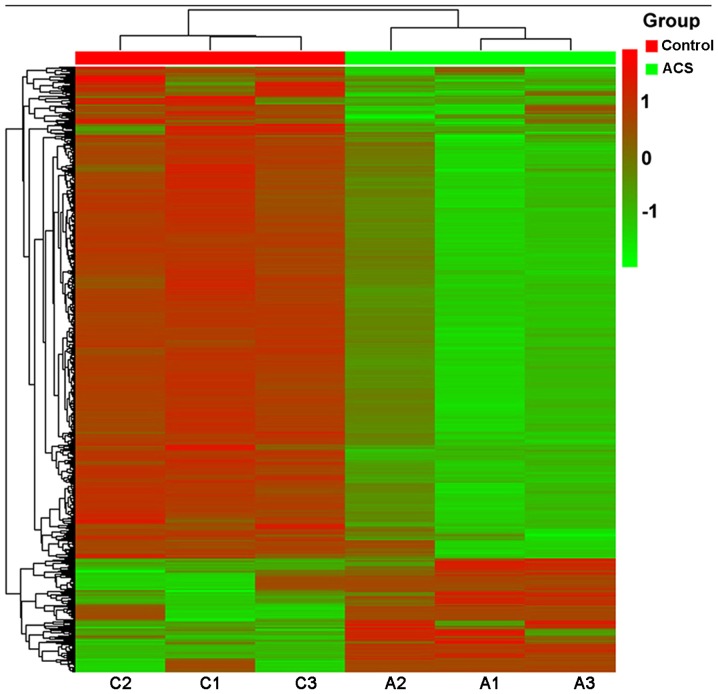
Using Benjamini and Hochberg correction for multiple comparisons, a total of 11,342 (2.34%) DM CpG sites covering 5,073 genes were identified in patients with ACS compared with the control patients (Table III). Among the 11,342 DM CpG sites, 8,865 (78.2%) covering 5,071 genes were significantly hypomethylated, while 2,477 (21.8%) covering 1,746 genes were significantly hypermethylated in the ACS samples (Table III). A volcano plot of the data exhibited the mean DNA methylation differences for all CpG sites (Fig. 2). The frequencies of all significant CpG sites between the ACS and control groups were analyzed (Table IV). A total of 9.04% (224) of the hypermethylated CpG sites and 11.8% (982) of the hypomethylated CpG sites had an absolute ACS/control group methylation difference ≥20%. Fig. 3 presents a hierarchical cluster analysis of the 1,206 CpG sites that distinguished the ACS samples from the control samples.
Figure 2.
Volcano plot of the significant differential DNA methylation analysis. All 11,342 sites among the 3 paired acute coronary syndrome and control samples were presented in the volcano plot. Red markers indicate the 11,342 differentially methylated CpG sites between the ACS and the control samples. CpG, 5′-C-phosphate-G-3′; ACS, acute coronary syndrome.
Table IV.
Frequency distribution of all significantly differentially methylated CpG sites between acute coronary syndrome and control samples by methylation status.
| ACS vs. control group methylation β score difference (%) | Hypermethylated CpG sites, n (%) | Cumulative (%) | Hypomethylated CpG sites, n (%) | Cumulative (%) |
|---|---|---|---|---|
| ≥50 | 24 (0.97) | 0.97 | 16 (0.18) | 0.18 |
| ≥40–<50 | 32 (1.29) | 2.26 | 22 (0.25) | 0.43 |
| ≥30–<40 | 35 (1.41) | 3.67 | 40 (0.45) | 0.88 |
| ≥20–<30 | 133 (5.37) | 9.04 | 904 (10.20) | 11.08 |
| ≥10–<20 | 2,253 (90.96) | 100 | 7,883 (88.92) | 100 |
| Total | 2,477 | – | 8,865 | – |
CpG, 5′-C-phosphate-G-3′; ACS, acute coronary syndrome.
Figure 3.
Hierarchical cluster analysis of the significantly DM CpG sites. A hierarchical cluster analysis was performed on the 1,206 significantly differentially methylated CpG sites. DM, differentially methylated; ACS, acute coronary syndrome; CpG, 5′-C-phosphate-G-3′.
Genomic distribution of DM CpG sites
Significant DM CpG sites were analyzed based on their region and island region level throughout the genome (Table V). There were 6 gene-based regions, including the transcription start site (TSS)1500, TSS200 (within 200 base pairs of a TSS), 5′-untranslated region (5′-UTR), 1stExon, body and 3′-UTR. In addition, there were 5 CpG island-based regions, including the island, N-Shore, S-Shore, N-Shelf and S-Shelf sites. The gene and island regions were used to calculate the methylation index. All the given region definitions were based on the original Illumina methylation annotation for the Human Methylation450 BeadChip array. The gene-based variant of the region-level test resulted in the identification of 4,809 significant DM CpG sites, which covered 2,188 genes. This number contained all unique genes, which resulted from six different, separately analyzed gene categories: TSS1500 (349 genes), TSS200 (182 genes), 5′-UTR (246 genes), 1stExon (31 genes), body (1,250 genes) and 3′-UTR (130 genes). Similarly, the CpG island-based variant of the region-level test resulted in 1,224 DM CpG sites located in islands, and 1,593 and 1,247 DM CpG sites distributed inside the N-Shore and S-Shore, respectively, while 751 and 685 DM CpG sites were located in the N-Shelf and S-Shelf, respectively. In total, 5,500 unique DM CpG sites covering 2,322 genes were identified among the five CpG-related regions.
Table V.
Statistics of DM CpG sites identified by site, region and island region level.
| Test | Region | No. of DM CpG sites | No. of genes |
|---|---|---|---|
| CpG site-level | Whole genome | 11,342 | 5,073 |
| Region-level gene based | TSS1500 | 861 | 349 |
| TSS200 | 421 | 182 | |
| 5′-UTR | 515 | 246 | |
| 1st Exon | 73 | 31 | |
| Body | 2,662 | 1,250 | |
| 3′-UTR | 277 | 130 | |
| Region-level island based | Island | 1224 | 462 |
| N-Shore | 1593 | 644 | |
| S-Shore | 1247 | 513 | |
| N-Shelf | 751 | 372 | |
| S-Shelf | 685 | 331 |
UTR, untranslated region; CpG, 5′-C-phosphate-G-3′; DM, differentially methylated; TSS, transcription start site; N, north; S, south.
The genomic regions of the significantly hypermethylated or hypomethylated CpG sites were distributed similarly (Table VI). The largest portion of hypermethylated CpGs (60.36%) were located in the body of the genes and subsequently decreased in other categories (15.17% in TSS1500, 6.88% in TSS200, 10.01% in the 5′-UTR, 1.72% in the 1stExon and 5.86% in the 3′-UTR). In addition, over half (54.06%) of the significantly hypomethylated CpG sites were located in the body, while the rest were distributed in the TSS1500 (18.16%), TSS200 (9.24%), 5′-UTR (10.89%), 1stExon (1.47%) and 3′-UTR (5.73%) regions. Furthermore, the distribution analysis of DM CpG sites in island regions revealed that 18.5% (190) of the significantly hypermethylated CpG sites were in CpG island regions, with 47.81% (491) in CpG shores and 33.69% (346) in shelf regions. As for the significantly hypomethylated CpG sites, 23.12% (1,034) were located in islands, while the majority were identified in shore (52.52%) or shelf (24.36%) regions (Table VII), suggesting they serve a potential role in genomic instability.
Table VI.
Distribution of the genomic region of significantly differentially methylated CpG sites in the acute coronary syndrome compared with the control samples.
| Genomic region of CpG sites | All CpG sites, n (%) | Hypermethylated CpG sites, n (%) | Hypomethylated CpG sites, n (%) |
|---|---|---|---|
| TSS1500 | 861 (17.90) | 150 (15.17) | 711 (18.61) |
| TSS200 | 421 (8.75) | 68 (6.88) | 353 (9.24) |
| 5′-UTR | 515 (10.71) | 99 (10.01) | 416 (10.89) |
| 1st Exon | 73 (1.52) | 17 (1.72) | 56 (1.47) |
| Body | 2,662 (55.36) | 597 (60.36) | 2,065 (54.06) |
| 3′-UTR | 277 (5.76) | 58 (5.86) | 219 (5.73) |
| Total | 4,809 | 989 | 3,820 |
CpG, 5′-C-phosphate-G-3′; UTR, untranslated region; TSS, transcription start site.
Table VII.
Distribution of island region of significantly differentially methylated GpG sites in ACS vs. control samples.
| Region level island-based GpG sites | All GpG sites | Hypermethylated CpG sites, no. (%) | Hypomethylated CpG sites, no. (%) |
|---|---|---|---|
| Island | 1,224 (22.25) | 190 (18.50) | 1,034 (23.12) |
| N-Shore | 1,593 (28.96) | 304 (29.60) | 1,289 (28.82) |
| S-Shore | 1,247 (22.68) | 187 (18.21) | 1,060 (23.70) |
| N-Shelf | 751 (13.65) | 166 (16.16) | 585 (13.08) |
| S-Shelf | 685 (12.46) | 180 (17.53) | 505 (11.28) |
| Total | 5,500 | 1,027 | 4,473 |
CpG, 5′-C-phosphate-G-3′; N, north; S, south.
GO term and KEGG pathway enrichment analyses of the DM CpG sites
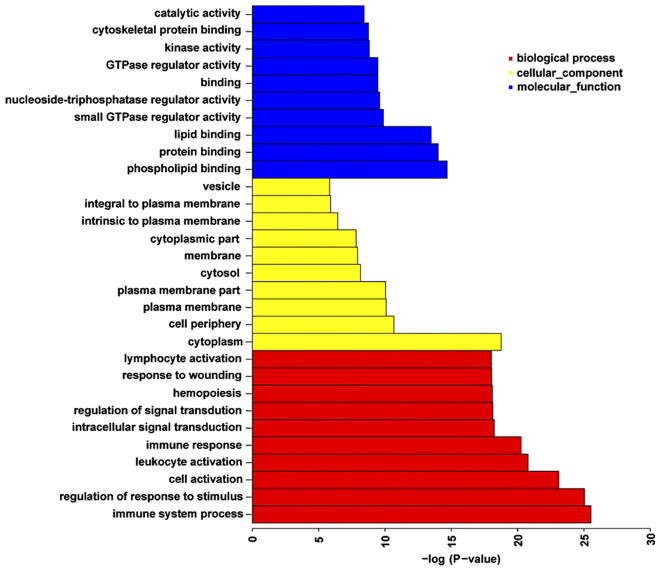
To identify the functions and pathways possibly influenced by the genes nearest the DM CpG sites in patients with ACS compared with the controls, GO term and KEGG pathway enrichment analyses were performed (Fig. 4). There were 767 functional categories of DM genes (FDR <0.05), the largest portion of the functional terms comprised of biological processes (603), while the rest were cellular components (76) or molecular functions (88). When the biological processes were used for categorization, it was observed that the most enriched groups included immune system processes, the regulation of responses to stimuli and cell activation. Categorization by cellular components indicated that proteins encoded by the analyzed genes were mainly associated with the cytoplasm. The molecular function category demonstrated significant enrichment for phospholipid, protein and lipid binding activities. Fig. 4 presents the top 10 terms of the three GO categories ranked by statistical significance.
Figure 4.
Top 10 terms in each GO category ranked according to their statistical significance. The top terms within the biological process, cellular component and molecular function categories are presented. GO, Gene Ontology.
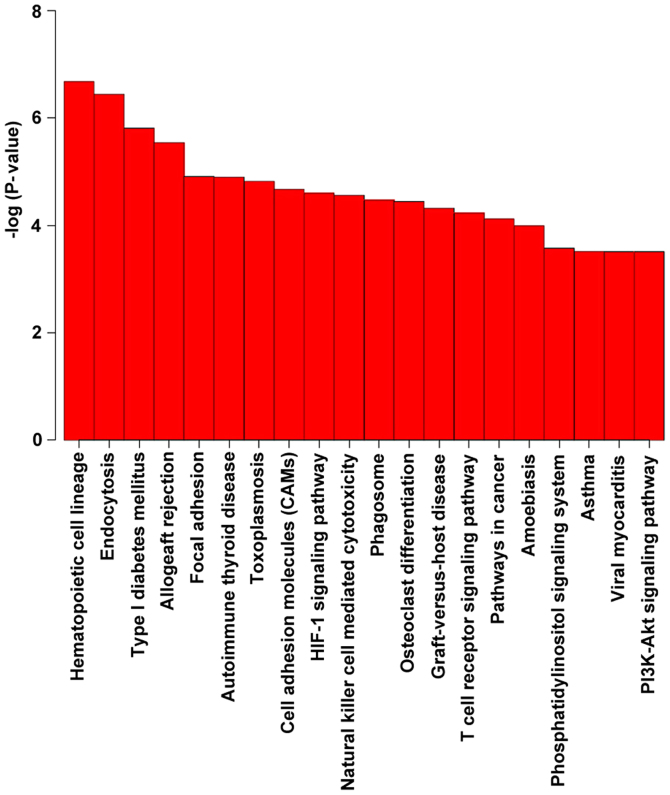
KEGG pathway analysis revealed a significant enrichment of genes associated with the hematopoietic cell lineage and endocytosis (Table VIII and Fig. 5). Among them, the immune system, and the transport and catabolism pathways attracted were ranked highest in terms of significance.
Table VIII.
KEGG pathway enrichment analysis of the hypermethylated and hypomethylated 5′-C-phosphate-G-3′ sites between the acute coronary syndrome and control samples by methylation status.
| KEGG ID no. | Pathway name | Pathway class | No. of genes in pathway | P-value | FDR |
|---|---|---|---|---|---|
| hsa04640 | Hematopoietic cell lineage | Immune system | 43 | 2.13×10−7 | 4.72×10−5 |
| hsa04144 | Endocytosis | Transport and catabolism | 83 | 3.63×10−7 | 4.72×10−5 |
| hsa04940 | Type I diabetes mellitus | Endocrine and metabolic diseases | 25 | 1.55×10−7 | 1.35×10−4 |
| hsa05330 | Allograft rejection | Immune diseases | 22 | 2.90×10−6 | 1.89×10−4 |
| hsa04510 | Focal adhesion | Cell communication | 80 | 1.24×10−5 | 5.51×10−4 |
| hsa05320 | Autoimmune thyroid disease | Immune diseases | 24 | 1.27×10−5 | 5.51×10−4 |
| hsa05145 | Toxoplasmosis | Infectious diseases: Parasitic | 50 | 1.51×10−5 | 5.63×10−4 |
| hsa04514 | Cell adhesion molecules | Signaling molecules and interaction | 58 | 2.12×10−5 | 6.90×10−4 |
| hsa04066 | HIF-1 signaling pathway | Signal transduction | 47 | 2.54×10−5 | 7.17×10−4 |
| hsa04650 | Natural killer cell mediated cytotoxicity | Immune system | 52 | 2.76×10−5 | 7.17×10−4 |
| hsa04145 | Phagosome | Transport and catabolism | 60 | 3.41×10−5 | 7.83×10−4 |
| hsa04380 | Osteoclast differentiation | Development | 54 | 3.61×10−5 | 7.83×10−4 |
| hsa05332 | Graft vs. host disease | Immune diseases | 21 | 4.81×10−5 | 9.63×10−4 |
| hsa04660 | T cell receptor signaling pathway | Immune system | 46 | 5.94×10−5 | 1.10×10−3 |
| hsa05200 | Pathways in cancer | Cancers: Overview | 113 | 7.67×10−5 | 1.33×10−3 |
| hsa05146 | Amoebiasis | Infectious diseases: Parasitic | 46 | 1.03×10−4 | 1.67×10−3 |
| hsa04070 | Phosphatidylinositol signaling system | Signal transduction | 35 | 2.66×10−4 | 4.03×10−3 |
| hsa05310 | Asthma | Immune diseases | 16 | 3.06×10−4 | 4.03×10−3 |
| hsa05416 | Viral myocarditis | Cardiovascular diseases | 31 | 3.10×10−4 | 4.03×10−3 |
| hsa04151 | PI3K-Akt signaling pathway | Signal transduction | 112 | 3.10×10−4 | 4.03×10−3 |
| hsa05221 | Acute myeloid leukemia | Cancers: Specific types | 27 | 3.34×10−4 | 4.14×10−3 |
| hsa05212 | Pancreatic cancer | Cancers: Specific types | 30 | 4.07×10−4 | 4.82×10−3 |
| hsa04670 | Leukocyte transendothelial migration | Immune system | 46 | 5.73×10−4 | 6.28×10−3 |
| hsa04630 | Jak-STAT signaling pathway | Signal transduction | 53 | 5.80×10−4 | 6.28×10−3 |
| hsa05166 | HTLV-I infection | Infectious diseases: Viral | 89 | 7.26×10−4 | 7.55×10−3 |
| hsa05032 | Morphine addiction | Substance dependence | 38 | 8.19×10−4 | 8.01×10−3 |
| hsa05223 | Non-small cell lung cancer | Cancers: Specific types | 25 | 8.32×10−4 | 8.01×10−3 |
| hsa04062 | Chemokine signaling pathway | Immune system | 67 | 9.16×10−4 | 8.32×10−3 |
| hsa04150 | mTOR signaling pathway | Signal transduction | 27 | 9.29×10−4 | 8.32×10−3 |
| hsa04662 | B cell receptor signaling pathway | Immune system | 32 | 1.06×10−3 | 9.16×10−3 |
| hsa04915 | Estrogen signaling pathway | Endocrine system | 39 | 1.30×10−3 | 1.05×10−2 |
| hsa04612 | Antigen processing and presentation | Immune system | 29 | 1.34×10−3 | 1.05×10−2 |
| hsa04664 | Fc epsilon RI signaling pathwayc | Immune system | 30 | 1.36×10−3 | 1.05×10−2 |
| hsa04060 | Cytokine-cytokine receptor interaction | Signaling molecules and interaction | 83 | 1.37×10−3 | 1.05×10−2 |
| hsa05222 | Small cell lung cancer | Cancers: Specific types | 35 | 1.75×10−3 | 1.30×10−2 |
| hsa00562 | Inositol phosphate metabolism | Carbohydrate metabolism | 26 | 2.97×10−3 | 2.15×10−2 |
| hsa05140 | Leishmaniasisc | Infectious diseases: Parasitic | 28 | 3.90×10−3 | 2.74×10−2 |
| hsa04666 | Fcγ R-mediated phagocytosis | Immune system | 35 | 5.30×10−3 | 3.62×10−2 |
| hsa04360 | Axon guidance | Development | 47 | 5.58×10−3 | 3.72×10−2 |
| hsa04725 | Cholinergic synapse | Nervous system | 41 | 7.36×10−3 | 4.78×10−2 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate.
Figure 5.
Top 20 Kyoto Encyclopedia of Genes and Genomes pathways ranked according to their statistical significance.
Selection of significant genes as possible diagnostic markers
The selection criteria used to select potential diagnostic marker genes included CpG sites that were methylated in the site, gene and gene island region, and a β score difference between the ACS and control samples of >0.3. Following filtering, a list of 19 hypermethylated and 17 hypomethylated CpG sites were selected that matched these criteria (Table IX). These methylated CpG sites were mapped to 37 known genes.
Table IX.
List of the top 19 hypermethylated and 17 hypomethylated corresponding genes selected as possible diagnostic markers.
| A, Hypermethylated corresponding genes | ||||
|---|---|---|---|---|
|
| ||||
| CpG site | ACS vs. control group methylation β score difference | Gene affected | P-value | FDR |
| cg03951394 | 0.31859 | DEAF1 | 4.61×10−2 | 0.43464 |
| cg11478607 | 0.33696 | GSTT1 | 1.43×10−3 | 0.24422 |
| cg01387905 | 0.34720 | ADAMTS2 | 1.35×10−6 | 0.03278 |
| cg13829625 | 0.35709 | SIN3B | 1.13×10−6 | 0.03160 |
| cg18424635 | 0.35947 | HLA-DRB5 | 2.83×10−2 | 0.39069 |
| cg26951705 | 0.38792 | ZNF787 | 4.81×10−2 | 0.43861 |
| cg00243527 | 0.39404 | LIF | 4.99×10−2 | 0.44222 |
| cg23782583 | 0.40248 | JAKMIP3 | 4.53×10−2 | 0.43270 |
| cg00248861 | 0.40500 | WHSC2 | 3.01×10−2 | 0.39564 |
| cg17005068 | 0.41609 | GSTT1 | 1.49×10−5 | 0.14324 |
| cg10662395 | 0.46909 | HCN2 | 3.48×10−3 | 0.24464 |
| cg19235645 | 0.47402 | FAM101A | 4.16×10−2 | 0.42548 |
| cg21829038 | 0.48534 | LRRK1 | 3.54×10−2 | 0.41016 |
| cg17713488 | 0.49270 | COL5A3 | 7.66×10−5 | 0.24422 |
| cg14594459 | 0.50806 | PKHD1L1 | 4.34×10−2 | 0.42957 |
| cg18443741 | 0.79680 | PYROXD1 | 2.48×10−5 | 0.18747 |
| cg19577958 | 0.54856 | SERPINB9 | 4.46×10−2 | 0.43166 |
| cg04234412 | 0.64833 | LOC391322 | 3.49×10−7 | 0.01914 |
| cg16836675 | 0.68159 | TOM1L1 | 4.18×10−3 | 0.24951 |
|
| ||||
| B, Hypomethylated corresponding genes | ||||
|
| ||||
| cg20592836 | −0.59591 | TP53INP2 | 1.33×10−2 | 0.32577 |
| cg03549208 | −0.55680 | ICA1L | 3.62×10−2 | 0.41253 |
| cg27141807 | −0.53542 | SPSB4 | 2.27×10−2 | 0.37233 |
| cg24851651 | −0.48666 | CCS | 1.31×10−2 | 0.32488 |
| cg06180910 | −0.43609 | GSTT1 | 1.11×10−4 | 0.24422 |
| cg10864200 | −0.41318 | PCGF3 | 1.12×10−2 | 0.31160 |
| cg24875593 | −0.40991 | PDXK | 5.26×10−3 | 0.26054 |
| cg04613734 | −0.39972 | TRPS1 | 4.07×10−2 | 0.42315 |
| cg19961153 | −0.38234 | ALPPL2 | 4.02×10−2 | 0.42219 |
| cg05141159 | −0.37248 | DEXI | 3.71×10−2 | 0.41476 |
| cg06049774 | −0.34306 | PLAC9 | 9.33×10−3 | 0.29687 |
| cg23231631 | −0.33541 | GABRB1 | 3.02×10−6 | 0.05052 |
| cg23731272 | −0.32429 | SMAD3 | 5.37×10−3 | 0.26137 |
| cg08253809 | −0.32072 | C14orf119 | 3.87×10−2 | 0.41857 |
| cg01655658 | −0.31142 | HLA-L | 4.82×10−7 | 0.02123 |
| cg13120108 | −0.31114 | PRKCH | 4.08×10−2 | 0.42353 |
| cg20433858 | −0.30584 | OR2L13 | 3.36×10−3 | 0.24464 |
CpG, 5′-C-phosphate-G-3′; ACS, acute coronary syndrome; FDR, false discovery rate.
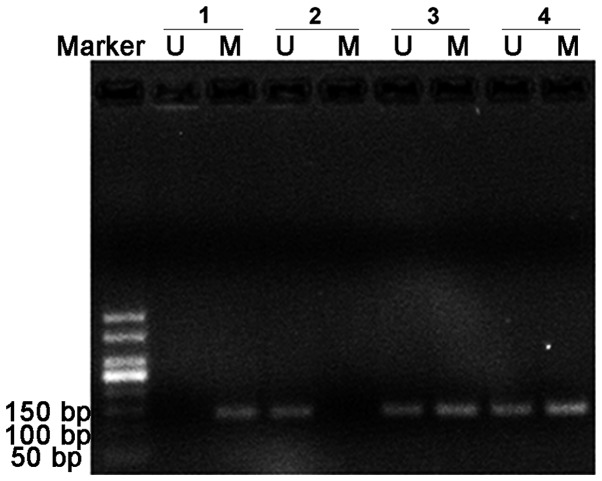
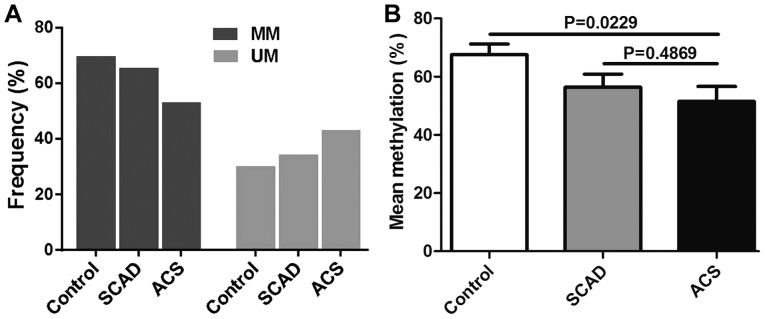
CpG methylation validation
The methylation state of the representative TSS1500 region (SMAD3; Chr1:67356838-Chr1:67356942) on the basis of DM magnitude was detected by MSP (Figs. 6 and 7A) and Sequenom MassARRAY (Fig. 7B). These results were compared with the results from the HumanMethylation450 array. The frequency of semi-methylated sites (ummethylated/methylated status) in patients with ACS was notably higher than that in the patients with SCAD and the healthy donors (Fig. 7A). The mean methylation level was identified as being significantly lower in patients with ACS compared with the SCAD and healthy control groups (Fig. 7B).
Figure 6.
Electrophoresis of the TSS1500 region of mothers against decapentaplegic homolog 3 (Chr1:67356838-Chr1:67356942) following methylation-specific polymerase chain reaction analysis. The M primers and U primers amplified the 141 bp products. M, methylated; U, unmethylated.
Figure 7.
Representative measurements of the SMAD3 TSS1500 methylation status. (A) The TSS1500 methylation frequency of SMAD3 in 81 patients with ACS, 74 patients with SCAD and 53 healthy individuals was measured using methylation-specific semi-quantitative polymerase chain reaction analysis. (B) The DNA methylation level of SMAD3 TSS1500 using Sequenom MassARRAY in 8 paired ACS, SCAD and healthy individuals was measured. SMAD3, mothers against decapentaplegic homolog 3; MM, fully methylated; MU, methylated/unmethylated or semi-methylated; ACS, acute coronary syndrome; SCAD, stable coronary artery disease.
Discussion
To the best of our knowledge, the present study presents the first genome-wide DNA methylation profiling of 3 paired ACS and normal human blood samples, using a high-throughput DNA methylation microarray covering >450,000 CpG sites on the human genome. The DM CpGs identified were enriched in genes associated with immune system processes, the regulation of responses to stimuli and cell activation, which suggests that they are of potential mechanistic and therapeutic relevance.
The impact of epigenetic mechanisms in cardiovascular pathophysiology is emerging as a key participant in the interface between genotype and phenotype variability. Several previous studies have focused on the role of DNA methylation in the development of atherosclerosis and CVD. Hiltunen et al (27) reported that a global hypomethylation of DNA was present in advanced human, mouse and rabbit atherosclerotic lesions. Notably, the results of Kim et al (36) indicated that elevated peripheral blood leukocyte DNA methylation was positively associated with the prevalence of CVD predisposing conditions and obesity in Chinese Singaporeans. A recent study also revealed that lower global DNA methylation is associated with higher cardiovascular risk in postmenopausal women (37). These studies led to the use of DNA methylation as a novel biomarker for CAD. However, the majority of their results were obtained from animal experiments or gene-specific DNA methylation, which lack of the systematic analysis of genome-wide changes to DNA methylation in patients with ACS. Zaina et al (38) identified an atherosclerosis-specific DNA methylation profile that highlighted the contribution of different genes and pathways to the disorder; however, this study did not take into account the characteristics of acute onset in patients with atherosclerotic CVD.
The present study analyzed the changes to genome-wide DNA methylation in patients with ACS using the Illumina HumanMethylation450 assay. A total of 11,342 CpG sites across the whole genome demonstrated statistically significant DM in the ACS samples compared with the control samples. The majority of the DM CpG sites were hypomethylated (8,865; 78.2%) rather than hypermethylated (2,477; 21.8%), which was consistent with the results of a previous methylation study in atherosclerotic lesions and CAD (28). In addition, global hypomethylation, particularly in repeated sequences, serves an important role in increasing chromosomal instability and may lead to ACS (39). This suggests that alteration of the DNA methylation profile, particularly hypomethylation, is an important process in the pathogenesis of ACS. Sharma et al (40) proposed that genomic DNA methylation in patients with CAD is significantly altered, as higher genomic DNA methylation changes were observed in patients with CAD than in controls. Kim et al (36) examined the association between peripheral blood leukocyte global genomic DNA methylation and the prevalence of CVD (myocardial infarction and stroke) or its risk factors (hypertension and diabetes) to verify the value of DNA methylation as a CVD risk biomarker. However, Kim et al (36) only compared differences in the overall methylation status between normal subjects and patients with coronary heart disease; further details on the differences in DNA methylation sites were not mentioned.
The genomic distribution of significant CpG sites revealed that significantly DM CpG sites were more frequently located at the body (55.3%) and shore (51.6%) sites. It has been previously reported that methylation at the CpG island shore regions regulates promoter activity (41), and that differential DNA methylation in gene bodies and intergenic loci indicates more complex mechanistic implications, as they are considered to promote transcriptional elongation, and to regulate gene splicing and enhancer activities (42).
The GO term and KEGG pathway enrichment analyses results demonstrated a significant enrichment of the corresponding methylated genes associated with the immune system processes, regulation of responses to stimuli and cell activation, which are all associated with the hematopoietic cell lineage and endocytosis signaling pathways. The proteins encoded by the corresponding methylated genes were mainly associated with the cytoplasm, and were enriched in the molecular functions of phospholipid, protein and lipid binding activities. These results are highly consistent with data from a recent genome-wide systematic pathway analysis of body mass index, which provided insights into the genetics of CVD and its risk factors (43). The results of the present study revealed a significant enrichment to immune system processes in the ACS samples, which may be due to the altered maturation status and possible immune exhaustion of immune cells in the peripheral blood of patients with ACS (44). Additionally, the neutrophil/lymphocyte ratio (NLR) has been identified to be independently associated with the severity and 3-year outcomes of CAD: Patients with higher NLR values had more advanced obstructive CAD and a poorer prognosis (45). CVD is often accompanied by disorders of lipid metabolism, and previous studies have demonstrated that elevated concentrations of oxidized lipoprotein were associated with the presence and severity of ACS (46,47). In the present study, the network analysis for GO term and KEGG pathway enrichment of the DM CpG sites revealed significant enrichment change in patients with ACS, which indicated that the methylation alterations at these sites were biologically meaningful.
In the present study, 19 hypermethylated genes (DEAF1, GSTT1, ADAMTS2, SIN3B, HLA-DRB5, ZNF787, LIF, JAKMIP3, WHSC2, HCN2, FAM101A, LRRK1, COL5A3, PKHD1L1, PYROXD1, SERPINB9, LOC391322 and TOM1L1) and 17 hypomethylated genes (TP53INP2, ICA1L, SPSB4, CCS, GSTT1, PCGF3, PDXK, TRPS1, ALPPL2, DEXI, PLAC9, GABRB1, SMAD3, C14orf119, HLA-L, PRKCH and OR2L13) demonstrated a potential association with ACS. These genes may thus represent potential novel candidate biomarkers for the early diagnosis of ACS. Three DM genes were identified that have been deeply studied in CVD: SMAD3, SERPINB9 and PRKCH. SMAD3, a key contributor to the transforming growth factor-β pathway signaling, has been reported to serve an essential role in CAD (48–51). Meta-analyses of genome-wide association studies have identified that an intronic single nucleotide polymorphism (SNP) in SMAD3 is associated with protection from CAD (52). SERPINB9 is most commonly known for its role in the control of granzyme B, which can be applicable for prevention of ACS and atherosclerotic plaque formation in patients with left ventricular remodeling following acute myocardial infarction (53). However, SERPINB9 also serves a role in suppressing interleukin-1β maturation through inhibiting caspase-1 (54). Genome-wide association and molecular mechanism studies have revealed that the SNPs rs2230500 (55) and rs2230500 (56,57) in PKRCH were significantly associated with the risk of ischemic stroke. However, whether SMAD3, SERPINB9 or PRKCH methylation is associated with the pathogenesis of ACS is currently unknown. The validation experiments performed for SMAD3 and TSS1500 (Chr1:67356838-Chr1:67356942) in the present study indicated that the methylation degree was negatively associated with the pathogenesis of ACS, and this was significantly corrected by the reference results of the Illumina HumanMethylation450 assay.
In conclusion, the present study characterized the genome-wide DNA methylation patterns occurring in ACS, and identified significantly differentiated CpG sites and genes that were correlated with ACS. Additionally, GO term and KEGG pathway enrichment analyses provided information on the processes and pathways that the methylated CpG sites and the participating genes affected. These results may help clarify the role of aberrant methylation in ACS pathogenesis, and provide the basis for future research into potentially abnormally methylated genes as biomarkers for the prevention and early diagnosis of ACS.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (grant nos. 81271904, 81401742, 81572073 and 81572074), the Special-Funded Program on National Key Scientific Instruments and Equipment Development of China (grant no. 2012YQ030261) and Jinling Hospital Foundation (grant no. 2017060).
Glossary
Abbreviations
- ACS
acute coronary syndrome
- SCAD
stable coronary artery disease
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DM
differentially methylated
- MSP
methylation-specific polymerase chain reaction
- ECG
electrocardiogram
- CK-MB
creatine kinase isoenzyme MB
- cTn
cardiac troponin
- CVD
cardiovascular disease
- TSS
transcription start site
- UTR
untranslated region
- CAD
coronary artery disease
References
- 1.Falk E, Nakano M, Bentzon JF, Finn AV, Virmani R. Update on acute coronary syndromes: The pathologists' view. Eur Heart J. 2013;34:719–728. doi: 10.1093/eurheartj/ehs411. [DOI] [PubMed] [Google Scholar]
- 2.Corcoran D, Grant P, Berry C. Risk stratification in non-ST elevation acute coronary syndromes: Risk scores, biomarkers and clinical judgment. Int J Cardiol Heart Vasc. 2015;8:131–137. doi: 10.1016/j.ijcha.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Gara PT, Kushner FG, Ascheim DD, Casey DE, Jr, MK, de Lemos JA, Ettinger SM, Fang JC, Fesmire FM, Franklin BA, et al. American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2013;127:e362–e425. doi: 10.1161/CIR.0b013e3182742c84. [DOI] [PubMed] [Google Scholar]
- 4.Redfors B, Råmunddal T, Angerås O, Dworeck C, Haraldsson I, Ioanes D, Petursson P, Libungan B, Odenstedt J, Stewart J, et al. Angiographic findings and survival in patients undergoing coronary angiography due to sudden cardiac arrest in western Sweden. Resuscitation. 2015;90:13–20. doi: 10.1016/j.resuscitation.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 5.Hofer TP, Frankenberger M, Mages J, Lang R, Meyer P, Hoffmann R, Colige A, Ziegler-Heitbrock L. Tissue-specific induction of ADAMTS2 in monocytes and macrophages by glucocorticoids. J Mol Med (Berl) 2008;86:323–332. doi: 10.1007/s00109-007-0284-0. [DOI] [PubMed] [Google Scholar]
- 6.Tomson T, Surges R, Delamont R, Haywood S, Hesdorffer DC. Who to target in sudden unexpected death in epilepsy prevention and how? Risk factors, biomarkers, and intervention study designs. Epilepsia. 2016;57(Suppl 1):4–16. doi: 10.1111/epi.13234. [DOI] [PubMed] [Google Scholar]
- 7.Mosleh W, Abdel-Qadir H, Farkouh M. Biomarkers in the emergency workup of chest pain: Uses, limitations, and future. Cleve Clin J Med. 2013;80:589–598. doi: 10.3949/ccjm.80a.12112. [DOI] [PubMed] [Google Scholar]
- 8.Wu SS, Lin X, Yuan LQ, Liao EY. The role of epigenetics in arterial calcification. BioMed Res Int. 2015;2015:320849. doi: 10.1155/2015/320849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heyn H, Esteller M. DNA methylation profiling in the clinic: Applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 10.Handy DE, Castro R, Loscalzo J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation. 2011;123:2145–2156. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace RG, Twomey LC, Custaud MA, Moyna N, Cummins PM, Mangone M, Murphy RP. Potential diagnostic and prognostic biomarkers of epigenetic drift within the cardiovascular compartment. BioMed Res Int. 2016;2016:2465763. doi: 10.1155/2016/2465763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loscalzo J, Handy DE. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease (2013 Grover Conference series) Pulm Circ. 2014;4:169–174. doi: 10.1086/675979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turgeon PJ, Sukumar AN, Marsden PA. Epigenetics of cardiovascular disease - A new 'Beat' in coronary artery disease. Med Epigenet. 2014;2:37–52. doi: 10.1159/000360766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandi V, Vadakedath S. Effect of DNA methylation in various diseases and the probable protective role of nutrition: A mini-review. Cureus. 2015;7:e309. doi: 10.7759/cureus.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razin A, Webb C, Szyf M, Yisraeli J, Rosenthal A, Naveh-Many T, Sciaky-Gallili N, Cedar H. Variations in DNA methylation during mouse cell differentiation in vivo and in vitro. Proc Natl Acad Sci USA. 1984;81:2275–2279. doi: 10.1073/pnas.81.8.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Afzali M, Nakhaee A, Tabatabaei SP, Tirgar-Fakheri K, Hashemi M. Aberrant promoter methylation profile of Niemann-pick type C1 gene in cardiovascular disease. Iran Biomed J. 2013;17:77–83. doi: 10.6091/ibj.11432.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nazarenko MS, Markov AV, Lebedev IN, Freidin MB, Sleptcov AA, Koroleva IA, Frolov AV, Popov VA, Barbarash OL, Puzyrev VP. A comparison of genome-wide DNA methylation patterns between different vascular tissues from patients with coronary heart disease. PLoS One. 2015;10:e0122601. doi: 10.1371/journal.pone.0122601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Liu Y, Strickland FM, Richardson B. Age-dependent decreases in DNA methyltransferase levels and low transmethylation micronutrient levels synergize to promote overexpression of genes implicated in autoimmunity and acute coronary syndromes. Exp Gerontol. 2010;45:312–322. doi: 10.1016/j.exger.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JM, Stewart R, Kang HJ, Bae KY, Kim SW, Shin IS, Hong YJ, Ahn Y, Jeong MH, Yoon JS. BDNF methylation and depressive disorder in acute coronary syndrome: The K-DEPACS and EsDEPACS studies. Psychoneuroendocrinology. 2015;62:159–165. doi: 10.1016/j.psyneuen.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Lü CX, Xu RD, Cao M, Wang G, Yan FQ, Shang SS, Wu XF, Ruan L, Quan XQ, Zhang CT. FOXP3 demethylation as a means of identifying quantitative defects in regulatory T cells in acute coronary syndrome. Atherosclerosis. 2013;229:263–270. doi: 10.1016/j.atherosclerosis.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Zhang LN, Liu PP, Wang L, Yuan F, Xu L, Xin Y, Fei LJ, Zhong QL, Huang Y, Xu L, et al. Lower ADD1 gene promoter DNA methylation increases the risk of essential hypertension. PLoS One. 2013;8:e63455. doi: 10.1371/journal.pone.0063455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JD, Lee A, Choi J, Park Y, Kang H, Chang W, Lee MS, Kim J. Epigenetic modulation as a therapeutic approach for pulmonary arterial hypertension. Exp Mol Med. 2015;47:e175. doi: 10.1038/emm.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Zheng Y, Hou C, Yang L, Li X, Lin J, Huang G, Lu Q, Wang CY, Zhou Z. DNA methylation impairs TLR9 induced Foxp3 expression by attenuating IRF-7 binding activity in fulminant type 1 diabetes. J Autoimmun. 2013;41:50–59. doi: 10.1016/j.jaut.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Pasquier J, Hoarau-Véchot J, Fakhro K, Rafii A, Abi Khalil C. Epigenetics and cardiovascular disease in diabetes. Curr Diab Rep. 2015;15:108. doi: 10.1007/s11892-015-0677-3. [DOI] [PubMed] [Google Scholar]
- 25.Babu M, Durga Devi T, Mäkinen P, Kaikkonen M, Lesch HP, Junttila S, Laiho A, Ghimire B, Gyenesei A, Ylä-Herttuala S. Differential promoter methylation of macrophage genes is associated with impaired vascular growth in ischemic muscles of hyperlipidemic and type 2 diabetic mice: Genome-wide promoter methylation study. Circ Res. 2015;117:289–299. doi: 10.1161/CIRCRESAHA.115.306424. [DOI] [PubMed] [Google Scholar]
- 26.Yip L, Fuhlbrigge R, Taylor C, Creusot RJ, Nishikawa-Matsumura T, Whiting CC, Schartner JM, Akter R, von Herrath M, Fathman CG. Inflammation and hyperglycemia mediate Deaf1 splicing in the pancreatic lymph nodes via distinct pathways during type 1 diabetes. Diabetes. 2015;64:604–617. doi: 10.2337/db14-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiltunen MO, Turunen MP, Häkkinen TP, Rutanen J, Hedman M, Mäkinen K, Turunen AM, Aalto-Setälä K, Ylä-Herttuala S. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc Med. 2002;7:5–11. doi: 10.1191/1358863x02vm418oa. [DOI] [PubMed] [Google Scholar]
- 28.Castillo-Díaz SA, Garay-Sevilla ME, Hernández-González MA, Solís-Martínez MO, Zaina S. Extensive demethylation of normally hypermethylated CpG islands occurs in human atherosclerotic arteries. Int J Mol Med. 2010;26:691–700. doi: 10.3892/ijmm_00000515. [DOI] [PubMed] [Google Scholar]
- 29.Jiang YZ, Manduchi E, Stoeckert CJ, Jr, Davies PF. Arterial endothelial methylome: Differential DNA methylation in atherosusceptible disturbed flow regions in vivo. BMC Genomics. 2015;16:506. doi: 10.1186/s12864-015-1656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 31.Virani S, Rentschler KM, Nishijo M, Ruangyuttikarn W, Swaddiwudhipong W, Basu N, Rozek LS. DNA methylation is differentially associated with environmental cadmium exposure based on sex and smoking status. Chemosphere. 2016;145:284–290. doi: 10.1016/j.chemosphere.2015.10.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu X, Li J, Deng S, Yu K, Liu X, Deng Q, Sun H, Zhang X, He M, Guo H, et al. Genome-wide analysis of DNA methylation and cigarette smoking in a chinese population. Environ Health Perspect. 2016;124:966–973. doi: 10.1289/ehp.1509834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The Gene Ontology Consortium: Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5:e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramos RB, Fabris V, Lecke SB, Maturana MA, Spritzer PM. Association between global leukocyte DNA methylation and cardiovascular risk in postmenopausal women. BMC Med Genet. 2016;17:71. doi: 10.1186/s12881-016-0335-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaina S, Heyn H, Carmona FJ, Varol N, Sayols S, Condom E, Ramírez-Ruz J, Gomez A, Gonçalves I, Moran S, et al. DNA methylation map of human atherosclerosis. Circ Cardiovasc Genet. 2014;7:692–700. doi: 10.1161/CIRCGENETICS.113.000441. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Chen L, Du L, Li M. Cage the firefly luciferin! - a strategy for developing bioluminescent probes. Chem Soc Rev. 2013;42:662–676. doi: 10.1039/C2CS35249D. [DOI] [PubMed] [Google Scholar]
- 40.Sharma P, Kumar J, Garg G, Kumar A, Patowary A, Karthikeyan G, Ramakrishnan L, Brahmachari V, Sengupta S. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol. 2008;27:357–365. doi: 10.1089/dna.2007.0694. [DOI] [PubMed] [Google Scholar]
- 41.Bockmühl Y, Patchev AV, Madejska A, Hoffmann A, Sousa JC, Sousa N, Holsboer F, Almeida OF, Spengler D. Methylation at the CpG island shore region upregulates Nr3c1 promoter activity after early-life stress. Epigenetics. 2015;10:247–257. doi: 10.1080/15592294.2015.1017199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 43.Zhao X, Gu J, Li M, Xi J, Sun W, Song G, Liu G. Pathway analysis of body mass index genome-wide association study highlights risk pathways in cardiovascular disease. Sci Rep. 2015;5:13025. doi: 10.1038/srep13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zidar DA, Mudd JC, Juchnowski S, Lopes JP, Sparks S, Park SS, Ishikawa M, Osborne R, Washam JB, Chan C, et al. Altered maturation status and possible immune exhaustion of CD8 T lymphocytes in the peripheral blood of patients presenting with acute coronary syndromes. Arterioscler Thromb Vasc Biol. 2016;36:389–397. doi: 10.1161/ATVBAHA.115.306112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arbel Y, Finkelstein A, Halkin A, Birati EY, Revivo M, Zuzut M, Shevach A, Berliner S, Herz I, Keren G, et al. Neutrophil/lymphocyte ratio is related to the severity of coronary artery disease and clinical outcome in patients undergoing angiography. Atherosclerosis. 2012;225:456–460. doi: 10.1016/j.atherosclerosis.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Wang JJ, Zhang CN, Meng Y, Han AZ, Gong JB, Li K. Elevated concentrations of oxidized lipoprotein(a) are associated with the presence and severity of acute coronary syndromes. Clin Chim Acta. 2009;408:79–82. doi: 10.1016/j.cca.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 47.Wang JJ, Han AZ, Meng Y, Gong JB, Zhang CN, Li K, Liu YX. Measurement of oxidized lipoprotein (a) in patients with acute coronary syndromes and stable coronary artery disease by 2 ELISAs: Using different capture antibody against oxidized lipoprotein (a) or oxidized LDL. Clin Biochem. 2010;43:571–575. doi: 10.1016/j.clinbiochem.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 48.Lee AM, Shimizu C, Oharaseki T, Takahashi K, Daniels LB, Kahn A, Adamson R, Dembitsky W, Gordon JB, Burns JC. Role of TGF-β signaling in remodeling of noncoronary artery aneurysms in kawasaki disease. Pediatr Dev Pathol. 2015;18:310–317. doi: 10.2350/14-12-1588-OA.1. [DOI] [PubMed] [Google Scholar]
- 49.Turner AW, Nikpay M, Silva A, Lau P, Martinuk A, Linseman TA, Soubeyrand S, McPherson R. Functional interaction between COL4A1/COL4A2 and SMAD3 risk loci for coronary artery disease. Atherosclerosis. 2015;242:543–552. doi: 10.1016/j.atherosclerosis.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Peng Q, Deng Y, Yang X, Leng X, Yang Y, Liu H. Genetic variants of ADAM17 are implicated in the pathological process of Kawasaki disease and secondary coronary artery lesions via the TGF-β/SMAD3 signaling pathway. Eur J Pediatr. 2016;175:705–713. doi: 10.1007/s00431-016-2696-8. [DOI] [PubMed] [Google Scholar]
- 51.Lusis AJ. Cardiovascular disease genes come together. Atherosclerosis. 2015;242:630–631. doi: 10.1016/j.atherosclerosis.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 52.Turner AW, Martinuk A, Silva A, Lau P, Nikpay M, Eriksson P, Folkersen L, Perisic L, Hedin U, Soubeyrand S, et al. Functional analysis of a novel genome-wide association study signal in SMAD3 that confers protection from coronary artery disease. Arterioscler Thromb Vasc Biol. 2016;36:972–983. doi: 10.1161/ATVBAHA.116.307294. [DOI] [PubMed] [Google Scholar]
- 53.Saito Y, Kondo H, Hojo Y. Granzyme B as a novel factor involved in cardiovascular diseases. J Cardiol. 2011;57:141–147. doi: 10.1016/j.jjcc.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 54.van der Burgh R, Meeldijk J, Jongeneel L, Frenkel J, Bovenschen N, van Gijn M, Boes M. Reduced serpinB9-mediated caspase-1 inhibition can contribute to autoinflammatory disease. Oncotarget. 2016;7:19265–19271. doi: 10.18632/oncotarget.8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hata J, Kubo M, Kiyohara Y. Genome-wide association study for ischemic stroke based on the Hisayama study. Nihon Eiseigaku Zasshi. 2011;66:47–52. doi: 10.1265/jjh.66.47. In Japanese. [DOI] [PubMed] [Google Scholar]
- 56.Wu L, Xi B, Hou D, Zhao X, Liu J, Cheng H, Zhou X, Shen Y, Wang X, Mi J. The SNP (rs2230500) in PRKCH decreases the risk of carotid intima-media thickness in a Chinese young adult population. PLoS One. 2012;7:e40606. doi: 10.1371/journal.pone.0040606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu L, Shen Y, Liu X, Ma X, Xi B, Mi J, Lindpaintner K, Tan X, Wang X. The 1425G/A SNP in PRKCH is associated with ischemic stroke and cerebral hemorrhage in a Chinese population. Stroke. 2009;40:2973–2976. doi: 10.1161/STROKEAHA.109.551747. [DOI] [PubMed] [Google Scholar]