Abstract
Mosquitoes are vectors of pathogens that cause diseases of medical and veterinary importance. Female mosquitoes transmit these pathogens while taking a blood meal, which most species require to produce eggs. The period after a blood meal is a time of extreme physiological change that requires rapid coordination of specific tissues. Gap junctions (GJ) are intercellular channels that aid in the coordination of cells within tissues via the direct transfer of certain small molecules and ions between cells. Evolutionarily distinct groups of proteins form the gap junctions of vertebrate and invertebrate animals (connexins and innexins, respectively). Aedes aegypti mosquitoes possess six genes encoding innexins: inx1, inx2, inx3, inx4, inx7, and inx8. The goal of this study was to identify potential roles of innexins in the physiology of mosquitoes after a blood meal by using qPCR to quantify their mRNA expression in adult females at 3 h and 24 h post-blood meal (PBM) relative to non-blood-fed controls. We found that at 24 h PBM, expression levels of inx2, inx3, and inx4 mRNAs increased; inx2 was the most highly upregulated innexin in key tissues associated with blood-meal digestion and egg production (i.e., the midgut and ovaries, respectively). However, knocking down inx2 mRNA levels by over 75% via RNA interference had no significant effect on fecundity. Altogether, our results suggest that a blood meal influences the molecular expression of innexins in mosquitoes, but their specific physiological roles remain to be elucidated.
Keywords: innexin, Aedes aegypti, qPCR, RNAi, blood feeding
1. Introduction
Mosquitoes transmit many pathogens that cause deadly and debilitating diseases. The yellow fever mosquito, Aedes aegypti, is the primary vector of chikungunya, dengue, yellow fever, and Zika viruses. These pathogens are transmitted when an infected female mosquito feeds on a vertebrate host; females require vertebrate blood in order to produce eggs. Following the consumption of a blood meal, the mosquito goes through a complex series of physiological changes that require the rapid endocrine coordination of multiple tissues [1]. Intercellular channels known as gap junctions allow for endocrine signals to be rapidly shared among adjacent cells within a tissue by mediating the direct transport of ions, small molecules, and second messengers between cells [2].
Gap junctions are comprised of proteins known as connexins in vertebrates and innexins in invertebrates; they are evolutionarily distinct proteins that have convergently evolved to share similar structure and function [3,4]. Some of the broad functional roles that innexin and connexin channels contribute to include development, immune responses, and reproduction [5,6,7,8,9,10]. However, the physiological roles of gap junctions in mosquitoes have only recently begun to emerge.
In Ae. aegypti, innexins are (1) expressed in a life stage- and tissue-dependent manner; (2) implicated in the physiology of renal (Malpighian) tubules and the diuretic capacity of adult female mosquitoes; (3) critical for the survival of larval and adult female mosquitoes [11,12,13,14]. In Anopheles gambiae, inx4 (a.k.a. zero population growth) is required for proper male gonad formation, while inx1 is necessary for immune response to invading Plasmodium parasites [8,15]. As such, gap junctions appear to play important roles in mosquito biology.
2. Methods
2.1. Mosquito Rearing
Eggs of Ae. aegypti were obtained through the Malaria Research and Reference Reagent Resource Center (MR4) as part of the BEI Resources Repository (Liverpool strain; LVP-IB12 F19, deposited by M.Q. Benedict). Mosquitoes were reared to adults and the colony was maintained as described in Piermarini et al. [16]. In brief, larvae and adults were held in an environmental chamber set at 28 °C and 80% relative humidity with a 12 h:12 h light:dark cycle. Larvae were reared in plastic trays in distilled water and fed ground TetraMin tropical fish flakes (Tetra Spectrum Brands, Blacksburg, VA, USA). Adults were housed in 0.5 m3 cages with access to 10% sucrose, and fed heparinized rabbit blood (Hemostat Laboratories, Dixon, CA, USA) through a membrane feeder (Hemotek, Blackburn, UK) to produce eggs.
2.2. Blood Feeding
Mosquitoes were blood-fed three to seven days post eclosion. Twenty-four hours prior to blood feeding, sucrose was removed from the mosquito cages. Mosquitoes were then offered heparinized rabbit blood (Hemostat Laboratories) for 60 min using an artificial membrane feeding system (Hemotek). Control mosquitoes (non-blood-fed) were treated similarly and provided access to 10% sucrose instead of blood for 60 min. After blood feeding, both groups were provided with 10% sucrose. Mosquitoes for tissue level qPCR analysis were dissected at either 3 h or 24 h post-blood meal.
2.3. Dissection, RNA Extraction, and cDNA Synthesis
Dissections, RNA extraction, and cDNA synthesis were performed as described in Calkins et al. [11]. In brief, mosquitoes were anesthetized on ice before being dissected in mosquito Ringer solution (consisting of 150 mM NaCl, 3.4 mM KCl, 1.7 mM CaCl2, 1.8 mM NaHCO3, 1 mM MgCl2, 5 mM Glucose, 25 mM HEPES; pH 7.1). Tissues were isolated, transferred to 1.5-ml micro-centrifuge tubes (Thermo Fisher Scientific, Waltham, MA, USA), and preserved in TRIzol® reagent at −80 °C until utilized in RNA isolation. Total RNA was isolated using the method of Chomczynski and Sacchi [17] and quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). cDNA libraries were synthesized using 4 µg of total RNA and the GoScriptTM Reverse Transcriptase system with random primers (Promega, Madison, WI, USA), following manufacturer’s protocols. cDNA libraries were stored at −20 °C until needed for qPCR.
2.4. qPCR
qPCR was performed as described in [12]. In brief, reactions were performed in triplicate with each reaction consisting of 5 µL of GoTaq® qPCR Master Mix (Promega), 400 nM forward and reverse primers, 40 ng cDNA, and nuclease-free water (total volume = 10 µL). Primers for each innexin and our reference gene (ribosomal protein S7, RPS7) were used as in Calkins and Piermarini [12]; see Supplementary Table S1. The reactions were then subjected to the following thermocycling protocol using a C1000/CFX96 real time system (Bio-Rad Laboratories, Hercules, CA, USA): initial denaturation of 95 °C (3 min), followed by 39 cycles of 95 °C (10 s), and 58 °C (30 s), ending with a melt curve cycle. qPCR results were analyzed using the ΔCt method [18,19] and expressed as relative gene expression.
2.5. dsRNA Synthesis and Injections
dsRNA synthesis and injections were performed as described in Calkins and Piermarini [12]. In brief, primers for dsRNA template synthesis were designed to amplify 300–500 bp gene segments (Calkins and Piermarini [12]; see Supplementary Table S2) to be utilized in the T7 MEGAscript® dsRNA synthesis kit (Thermo Fisher Scientific). dsRNA was synthesized following the manufacture’s protocols and stored at −80 °C.
On the day of an injection, dsRNA was diluted to 1 µg/µL in 0.5× PBS solution (5.95 mM phosphates, 68.5 mM sodium chloride, and 1.35 mM potassium chloride; pH 7.5; Fisher Scientific, Fairlawn, NJ, USA). Eighty mosquitoes were injected with 1 µg of either inx2 dsRNA or eGFP dsRNA and returned to rearing conditions with access to 10% sucrose. After three days, three mosquitoes were removed from each treatment for knockdown assessment via qPCR and the remaining mosquitoes were utilized in fecundity assays (see Section 2.6). For tissue level knockdown analysis, 40 mosquitoes were injected with dsRNA for either inx2 or eGFP and dissected three days post injection.
2.6. Fecundity and Viability Assays
Three days after dsRNA injection, mosquitoes were offered a blood meal (see Section 2.2); those with no visible blood in their abdomens were excluded from the assays. Twenty-four hours later, the blood-fed mosquitoes were transferred to individual egg-laying tubes for the fecundity assay. The egg-laying tubes consisted of a cylindrical glass tube (21 × 70 mm; Fisher Scientific, Pittsburg, PA, USA) with a piece of coffee filter (Melitta, Clearwater, FL, USA) cut to fit the bottom of the tube. The filter was wetted with 150 µL of ddH2O (Milli-Q® filtered water, Merck KGaA, Darmstadt, Germany) and the open end was plugged with a cotton ball. The mosquitoes in their individual egg-laying tubes were returned to rearing conditions for 48 h, and the number of eggs laid by each mosquito was counted.
After counting the eggs, filter papers were consolidated according to dsRNA treatment and allowed to dry in a rearing chamber for one week. Eggs were then hatched in ddH2O under vacuum and returned to rearing conditions for 24 h. The resulting larvae were immobilized through refrigeration before counting them under a dissection stereomicroscope (World Precision Instruments, Sarasota, FL, USA/model PZMTIII-BS).
2.7. Data Analysis
All data were analyzed with GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA). Innexin mRNA expression levels between blood-fed and non-blood-fed treatments were analyzed via a two-way ANOVA with a Sidak post hoc analysis. The statistical significance of innexin mRNA knockdown after dsRNA injection was determined with t-tests comparing mosquitoes injected with eGFp and inx2 dsRNAs. Fecundity measurements (percent mosquitoes ovipositing, percent viability) were analyzed with one-way ANOVAs. Numbers of eggs laid were analyzed with a non-parametric Kruskal-Wallis ANOVA and Dunn’s post-test.
3. Results
3.1. mRNA Expression
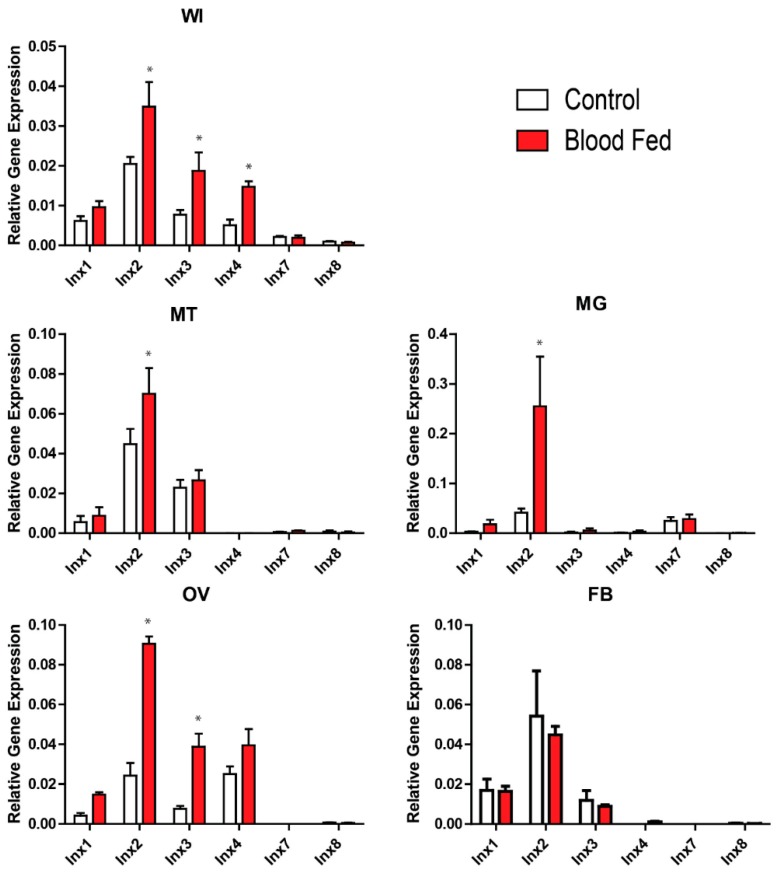
At 3 h PBM, there were no significant differences in innexin mRNA levels between blood-fed and non-blood-fed mosquitoes (Supplementary Figure S1). However, at 24 h PBM, at least one innexin was differentially expressed between blood-fed and non-blood-fed mosquitoes at the whole mosquito and tissue levels, except in the fat body (Figure 1). At the whole mosquito level, inx2, inx3, and inx4 were significantly upregulated. At the tissue level, inx2 was significantly upregulated in the Malpighian tubules, midgut, and ovaries, and inx3 was significantly upregulated in the ovaries.
Figure 1.
Effects of a blood meal on innexin mRNA expression 24 h post-blood meal. White bars indicate non-blood-fed control females and red bars indicate blood-fed females. Values are means ± SEM, n = 5. * indicate differences within a gene between blood-fed mosquitoes and non-blood-fed controls as determined by a two-way ANOVA and Newman-Keuls multiple comparison (p < 0.05). WI = whole insect; MT = Malpighian tubules; MG = midgut; OV = ovaries; FB = fat body.
3.2. RNAi
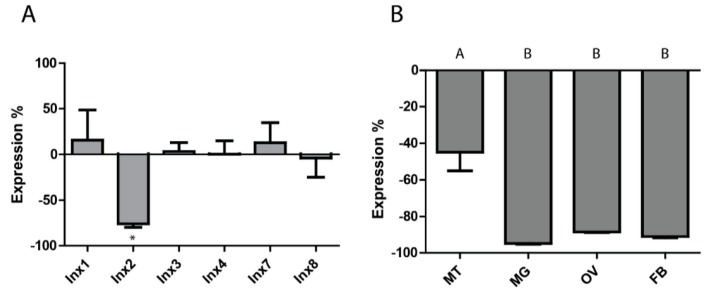
Given that inx2 was upregulated in the whole insect, Malpighian tubules, midgut, and ovaries at 24 h PBM, it was selected for RNAi experiments. Injecting 1 µg of inx2 dsRNA into adult females resulted in a 75.7 ± 3.9% reduction in mRNA expression (relative to controls injected with eGFP dsRNA) at the whole mosquito level within three days, with no significant changes in the expression levels of the other innexin mRNAs (Figure 2A). A similar degree of knockdown persisted 24 h after a blood meal (Supplementary Figure S2). At the tissue level, knockdown of inx2 was similar in the midgut (94.8 ± 0.5%), ovaries (88.5 ± 0.2%), and fat body (91.0 ± 0.7%), while knockdown of inx2 in the Malpighian tubules was significantly weaker (44.9 ± 10.1%) (Figure 2B).
Figure 2.
RNAi-induced knockdown of inx2 mRNA expression. (A) Whole insect innexin mRNA expression three days post injection of inx2 dsRNA (percent relative to eGFp dsRNA-injected controls). * indicates significant difference from eGFP dsRNA-injected mosquitoes as determined by a t-test (p < 0.05). Values are means ± SEM, n = 7; (B) Tissue-level innexin mRNA expression three days post injection of inx2 dsRNA. Abbreviations are as in Figure 1. Values are means ± SEM, n = 3. Letters represent differences as determined by one-way ANOVA and Newman-Keuls multiple comparison (p < 0.05).
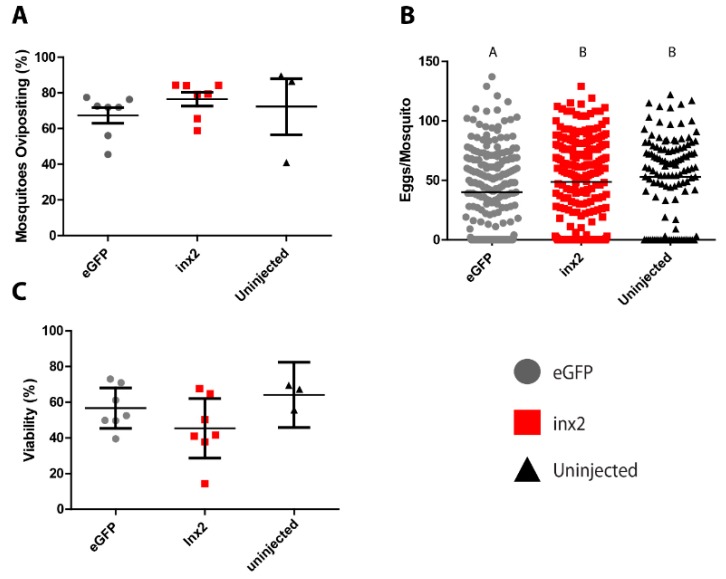
The injection of inx2 dsRNA did not significantly affect the median number of eggs laid (55 eggs per mosquito) compared to uninjected controls (60 eggs per mosquito; Figure 3A). However, mosquitoes injected with eGFP dsRNA laid significantly fewer eggs (median = 40 eggs per mosquito) than both the inx2 dsRNA-injected and uninjected mosquitoes (Figure 3A). No significant differences were found among the treatments in the viability of eggs (eGFP = 56.7 ± 4.6%; inx2 = 45.4 ± −6.8%; uninjected = 64.1 ± 4.3%; Figure 3B) or the percentage of mosquitoes that oviposited (eGFP = 67.4 ± 4.5%, inx2 = 76.5 ± 3.9%, uninjected = 72.3 ± 15.7%; Figure 3C).
Figure 3.
Effects of inx2 knockdown on fecundity of adult female mosquitoes. (A) Effect of inx2 knockdown on percentage of mosquitoes ovipositing. Values are means (horizontal black bars) ± SEM and individual data points are shown for each replicate (n = 7 for eGFP, 7 for inx2, and 3 for untreated); (B) Effects of inx2 knockdown on number of eggs laid. Horizontal black bars indicate median number of eggs laid per mosquito, and individual data points represent individual mosquitoes (n = 231 for eGFP, 235 for inx2, and 119 for untreated). Letters represent differences as determined by a non-parametric one-way ANOVA and Dunn’s multiple comparison; (C) Effect of inx2 knockdown on percentage of eggs hatching into viable larvae. Values are means (horizontal black bars) ± SEM and individual data points are shown for each replicate (n = 7 for eGFP, 7 for inx2, and 3 for untreated).
4. Discussion
Our study is the first to quantify innexin mRNA expression in adult female Ae. aegypti following a blood meal. In particular, we found that inx2 and inx3 were upregulated by 24 h PBM in the whole insect, which can in part be attributed to an upregulation of inx2 in the Malpighian tubules, midgut, and ovaries, and inx3 in the ovaries (Figure 1). Although inx4 was also upregulated by 24 h PBM in the whole insect, we did not find a corresponding upregulation in any of the tissues examined (Figure 1). Thus, the site of inx4 upregulation remains to be determined.
After a blood meal, the midgut contributes to digestion, nutrient absorption, immune responses, and xenobiotic detoxification, while the Malpighian tubules contribute to diuresis as well as xenobiotic and metabolite detoxification/excretion [20,21,22,23]. Additionally, the ovaries undergo vitellogenesis and dramatically increase in size before eggs are oviposited [24,25]. Thus, these tissues undergo profound physiological changes after the mosquito ingests a blood meal. The consistent upregulation of inx2 in all three of these tissues at 24 h PBM suggests that inx2-mediated intercellular communication may contribute to the regulation of the physiological activities in these tissues. Alternatively, inx2 could be acting as a hemichannel in the plasma membranes of these tissues to release signaling molecules that contribute to paracrine/endocrine communication after a blood meal, as suggested by Li et al. [8] for inx1 in the midgut of An. gambiae.
As such, we hypothesized that knockdown of inx2 would disrupt digestion, excretion, and/or oogenesis after a blood meal, thereby resulting in a reduction of fecundity. However, despite knocking down inx2 mRNA levels by ~75% in whole mosquitoes, ~45% in the Malpighian tubules, and ~90% in midgut and ovaries (Figure 2), we found no reduction in the percentage of mosquitoes that oviposited, the number of eggs laid per mosquito, or the viability of the eggs in inx2 dsRNA-injected females compared to uninjected mosquitoes (Figure 3). Although these results suggest that inx2 does not contribute to fecundity, we cannot rule out that inx2 protein levels were unaffected by the mRNA knockdown, thereby resulting in no detectable phenotype. If this were the case, potential contributing factors could be slow turnover rates of inx2 proteins and functional redundancy among the innexins. Innexin turnover rates are not currently known, but mRNA expression analyses of innexins in several mosquito tissues suggest that the molecular potential for functional redundancy exists [11]. Intriguingly, eGFP dsRNA-injected mosquitoes laid fewer eggs than the inx2 dsRNA-injected and uninjected control mosquitoes (Figure 3), suggesting a potential consequence of eGFP dsRNA injection on fecundity. In the honeybee Apis mellifera, injection of GFP dsRNA results in differential expression of ~10% of the insect’s transcriptome, including an upregulation of immune genes [26]. Thus, it is possible that injection of eGFP dsRNA in Ae. aegypti elicits a similar response that could potentially divert energetic resources from oogenesis.
5. Conclusions
The present study is the first to quantify the effects of a blood meal on the molecular expression of innexins in Ae. aegypti. While we found inx2 to be highly upregulated in both the midgut and ovaries following a blood meal, RNAi of inx2 yielded no reduction in fecundity. Taken together, the results from this study provide molecular evidence that innexins are potentially important in the physiology of Ae. aegypti after a blood meal, but their specific functional roles remain to be elucidated.
Acknowledgments
We thank Dr. David Denlinger (The Ohio State University, OSU) for his critical review of this manuscript and Ms. Nuris Acosta (OSU) and Ms. Edna Alfaro (OSU) for their assistance in mosquito rearing. Funding for this study was provided by grants to (1) PMP from the NIH (R03DK090186) and Mosquito Research Foundation (2014-03); (2) TLC from the OARDC SEEDS program (Grant# 2014-078; oardc.osu.edu/seeds) and Grants-in-Aid of research from both The Ohio State University and national chapters of the Sigma-Xi scientific research society. State and Federal funds appropriated to the OARDC of the Ohio State University also supported the study.
Supplementary Materials
The following are available online at www.mdpi.com/2075-4450/8/4/122/s1, Table S1: qPCR primer pairs. Each set of innexin primers was determined to be specific through melt curve analysis and DNA sequencing of PCR products, Table S2: dsRNA template synthesis primers. Each primer set consists of an innexin specific region for amplification of the target gene from plasmid, and the T7 promoter sequence (TAATACGACTCACTATAGGGAGA), Figure S1: Effects of a blood meal on innexin mRNA expression 3-h post-blood meal, Figure S2: Knockdown of inx2 in blood-fed and non-blood-fed mosquitoes.
Author Contributions
Travis L. Calkins and Peter M. Piermarini conceived and designed the experiments; Travis L. Calkins performed the experiments; Travis L. Calkins and Peter M. Piermarini analyzed the data; Travis L. Calkins and Peter M. Piermarini wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hansen I.A., Attardo G.M., Rodriguez S.D., Drake L.L. Four-way regulation of mosquito yolk protein precursor genes by juvenile hormone-, ecdysone-, nutrient-, and insulin-like peptide signaling pathways. Front. Physiol. 2014;5:103. doi: 10.3389/fphys.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phelan P., Starich T.A. Innexins get into the gap. Bioessays. 2001;23:388–396. doi: 10.1002/bies.1057. [DOI] [PubMed] [Google Scholar]
- 3.Phelan P., Stebbings L.A., Baines R.A., Bacon J.P., Davies J.A., Ford C. Shaking-B protein forms gap junctions in paired Xenopus oocytes. Nature. 1998;391:181–184. doi: 10.1038/34426. [DOI] [PubMed] [Google Scholar]
- 4.Phelan P., Nakagawa M., Wilkin M.B., Moffat K.G., O’Kane C.J., Davies J.A., Bacon J.P. Mutations in shaking-B prevent electrical synapse formation in the Drosophila giant fiber system. J. Neurosci. 1996;16:1101–1113. doi: 10.1523/JNEUROSCI.16-03-01101.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosco D., Haefliger J.-A., Meda P. Connexins: Key mediators of endocrine function. Physiol. Rev. 2011;91:1393–1445. doi: 10.1152/physrev.00027.2010. [DOI] [PubMed] [Google Scholar]
- 6.Giuliani F., Giuliani G., Bauer R., Rabouille C. Innexin 3, a new gene required for dorsal closure in Drosophila embryo. PLoS ONE. 2013;8:e69212. doi: 10.1371/journal.pone.0069212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holcroft C.E., Jackson W.D., Lin W.-H., Bassiri K., Baines R.A., Phelan P. Innexins Ogre and Inx2 are required in glial cells for normal postembryonic development of the Drosophila central nervous system. J. Cell Sci. 2013;126:3823–3834. doi: 10.1242/jcs.117994. [DOI] [PubMed] [Google Scholar]
- 8.Li M.W.M., Wang J., Zhao Y.O., Fikrig E. Innexin AGAP001476 is critical for mediating anti-Plasmodium responses in Anopheles mosquitoes. J. Biol. Chem. 2014:1–21. doi: 10.1074/jbc.M114.554519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tazuke S.I., Schulz C., Gilboa L., Fogarty M., Mahowald A.P., Guichet A., Ephrussi A., Wood C.G., Lehmann R., Fuller M.T. A germline-specific gap junction protein required for survival of differentiating early germ cells. Development. 2002;129:2529–2539. doi: 10.1242/dev.129.10.2529. [DOI] [PubMed] [Google Scholar]
- 10.Hasegawa D.K., Turnbull M.W. Recent findings in evolution and function of insect innexins. FEBS Lett. 2014;588:1403–1410. doi: 10.1016/j.febslet.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Calkins T.L., Woods-Acevedo M.A., Hildebrandt O., Piermarini P.M. The molecular and immunochemical expression of innexins in the yellow fever mosquito, Aedes aegypti: Insights into putative life stage- and tissue-specific functions of gap junctions. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2015;183:11–21. doi: 10.1016/j.cbpb.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calkins T.L., Piermarini P.M. Pharmacological and genetic evidence for gap junctions as potential new insecticide targets in the yellow fever mosquito, Aedes aegypti. PLoS ONE. 2015;10:e0137084. doi: 10.1371/journal.pone.0137084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weng X.-H., Piermarini P.M., Yamahiro A., Yu M.-J., Aneshansley D.J., Beyenbach K.W. Gap junctions in Malpighian tubules of Aedes aegypti. J. Exp. Biol. 2008;211:409–422. doi: 10.1242/jeb.011213. [DOI] [PubMed] [Google Scholar]
- 14.Piermarini P.M., Calkins T.L. Evidence for intercellular communication in mosquito renal tubules: A putative role of gap junctions in coordinating and regulating the rapid diuretic effects of neuropeptides. Gen. Comp. Endocrinol. 2014;203:43–48. doi: 10.1016/j.ygcen.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magnusson K., Mendes A.M., Windbichler N., Papathanos P.-A., Nolan T., Dottorini T., Rizzi E., Christophides G.K., Crisanti A. Transcription regulation of sex-biased genes during ontogeny in the malaria vector Anopheles gambiae. PLoS ONE. 2011;6:e21572. doi: 10.1371/journal.pone.0021572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piermarini P.M., Hine R.M., Schepel M., Miyauchi J., Beyenbach K.W. Role of an apical K, Cl cotransporter in urine formation by renal tubules of the yellow fever mosquito (Aedes aegypti) Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;301:R1318–R1337. doi: 10.1152/ajpregu.00223.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium extraction. Anal. Biochem. 1987;159:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 18.Song J., Bai Z., Han W., Zhang J., Meng H., Bi J., Ma X., Han S., Zhang Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig. Dis. Sci. 2012;57:897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 19.Silver N., Best S., Jiang J., Thein S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt R., Subramanian G., Halpern A., Sutton G., Charlab R., Nusskern D., Wincker P., Clark A., Ribeiro J., Wides R., et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 21.Sanders H.R., Evans A.M., Ross L.S., Gill S.S. Blood meal induces global changes in midgut gene expression in the disease vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2013;33:1105–1122. doi: 10.1016/S0965-1748(03)00124-3. [DOI] [PubMed] [Google Scholar]
- 22.Esquivel C.J., Cassone B.J., Piermarini P.M. Transcriptomic evidence for a dramatic functional transition of the malpighian tubules after a blood meal in the Asian tiger mosquito Aedes albopictus. PLoS Negl. Trop. Dis. 2014;8:e2929. doi: 10.1371/journal.pntd.0002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esquivel C.J., Cassone B.J., Piermarini P.M. A de novo transcriptome of the Malpighian tubules in non-blood-fed and blood-fed Asian tiger mosquitoes Aedes albopictus: Insights into diuresis, detoxification, and blood meal processing. PeerJ. 2016;4:e1784. doi: 10.7717/peerj.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koller C.N., Raikhel A.S. Initiation of vitellogenin uptake and protein-synthesis in the mosquito (Aedes aegypti) ovary in response to a blood meal. J. Insect Physiol. 1991;37:703–711. doi: 10.1016/0022-1910(91)90048-5. [DOI] [Google Scholar]
- 25.Raikhel A.S. Vitellogenesis in mosquitoes. Adv. Dis. Vector Res. 1992;9:1–39. [Google Scholar]
- 26.Nunes F.M.F., Aleixo A.C., Barchuk A.R., Bomtorin A.D., Grozinger C.M., Simões Z.L.P. Non-target effects of green fluorescent protein (GFP)-derived double-stranded RNA (dsRNA-GFP) used in honey bee RNA interference (RNAi) assays. Insects. 2013;4:90–103. doi: 10.3390/insects4010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.