Abstract
Cells have a remarkable ability to sense and respond to the mechanical properties of their environment. Mechanosensing is essential for many phenomena from cell movements and tissue rearrangements to cell differentiation and the immune response. Cells of the immune system get activated when membrane receptors bind to cognate antigen on the surface of antigen presenting cells. Both T and B lymphocyte signaling has been shown to be responsive to physical forces and mechanical cues. Cytoskeletal forces exerted by cells likely mediate this mechanical modulation. Here we discuss recent advances in the field of immune cell mechanobiology at the molecular and cellular scale.
Keywords: immune cells, mechanosensing, cytoskeleton, forces, T cell receptor, actin
1. Introduction
The mechanical environment of cells, such as the extracellular matrix (ECM) or other cells, plays a critical role in regulating many aspects of cell function [1, 2]. Examples of mechanical stimuli include forces due to muscle contraction, shear stresses induced by flow in blood vessels or strains induced by collective movements of tissues. Similarly, cells also encounter environments with varying physical properties, such as tissue or stiffness of the ECM, topography and fluidity. Studies have revealed that in addition to soluble chemical cues, the physical environment plays a key role in controlling cell proliferation, cell fate determination, cell migration and global organization of tissues by regulating genetic and biochemical signaling pathways [1]. Mechanical regulation of cell function appears to result from a conserved set of physical mechanisms [3]. Forces arising from the actin cytoskeleton and myosin motors generate active tension that is applied to adhesions (both cell-cell and cell-matrix), initiating biochemical signaling [4]. Given the importance of dynamic force transmission for cell and tissue physiology, the underlying mechanisms that translate forces into an appropriate cellular response need to be understood. The regulation of receptor-mediated cell signaling by cytoskeletal and external forces and the mechanical properties of the environment is important in many aspects of physiology including the immune system.
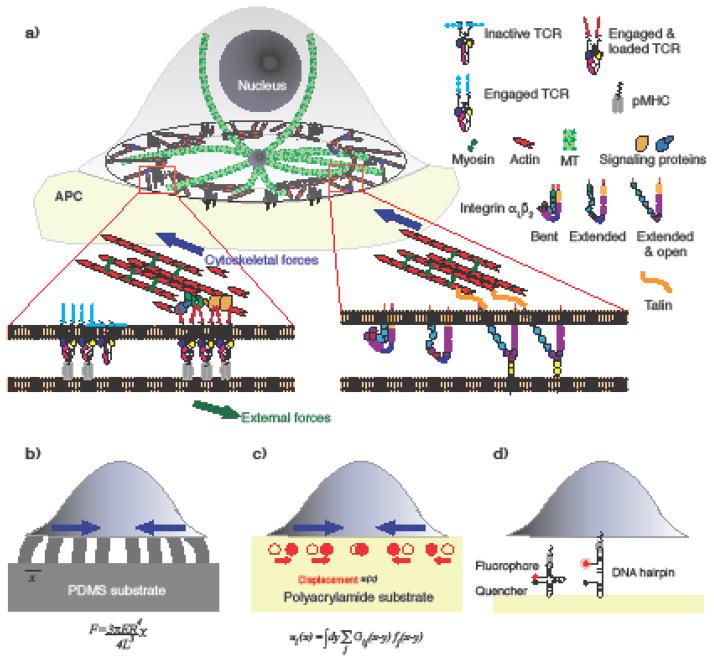
The adaptive immune system forms a strong line of defense against infections, through its ability to recognize foreign molecules (antigens), develop an appropriate response, and rapidly recall this action on subsequent re-exposure [5]. Naïve lymphocytes (T and B cells) in search for their cognate antigens encounter a variety of mechanical stimuli as they circulate through blood, lymphoid tissues, and sites of inflammation. T and B cell activation requires physical interaction with professional antigen presenting cells (APC) – dendritic cells and follicular dendritic cells respectively [6–8]. APCs display protein fragments derived from infecting pathogens on their surface. A membrane protein complex, called the T or B cell receptor (TCR/BCR), recognizes these fragments and assembles into microclusters [9–11]. Signaling proteins and adaptors accumulate at developing microclusters [11] which trigger signal transduction pathways and ultimately lead to the initiation of transcription and cell fate determination (cell activation) [12] (Fig. 1a).
Figure 1. Cytoskeletal forces during immune cell activation.
(a) Schematic diagram of a T cell spreading on the surface of an APC. The contact zone shows spatiotemporal organization of actin into Arp2/3 mediated branched networks and formin-mediated actin arcs. MT emanate from the centrosome. The insets depict conformational changes resulting from the application of actomyosin forces on TCR microclusters (left) and integrin receptors (right). One of many possible conformational changes in the TCR is shown. Cartoons of (b) a cell on PDMS microposts showing deflection of posts resulting from cellular forces, (c) a cell on a PA gel depicting movement of embedded beads as the cell exerts traction forces. The measured displacement field u(x) and the traction stress field, f(x) are related by the integral equation in the figure, with G(x–y) is the Boussinesq Green function, (d) a cell on a glass substrate with DNA-hairpin based tension sensors. The flurophore (red sphere) is near the quencher (black sphere) when the ligand attached to the hairpin is free. Ligand binding and subsequent force application moves fluorophore away from the quencher.
Several lines of evidence have converged upon the view that physical forces can trigger signaling during lymphocyte activation [13–15]. Activation also critically depends upon rearrangements of the actin cytoskeleton [16, 17]. Lymphocyte signaling is also sensitive to the mechanical properties of antigen-bearing surfaces including stiffness [18–20], mobility and topography. In this review, we explore what is currently known about mechanosensing in the immune response at different length scales ranging from the molecular scale of how mechanical forces play a role in activating immune and adhesion receptors to the cellular scale of how a cell responds functionally to mechanical forces. We then briefly review cytoskeletal dynamics in immune cells and the different ways in which cellular forces have been measured. We further discuss the potential role of cytoskeletal forces in establishing and modulating mechanotransduction. In this review, we largely focus on mechanical responses in T cells with some insights into B cells.
2. Mechanosensing at the molecular scale
2.1 Immune (antigen) receptors
The T Cell Receptor (TCR) is a multi-subunit complex expressed on the T cell membrane, which binds antigenic peptides embedded within major histocompatibility complex molecules (pMHC) on APCs during antigen recognition [21]. This heteromeric complex consists of the ligand binding TCR (α and β subunits) non-covalently associated with CD3γε, CD3δε, and CD3ζζ polypeptide chains. Structural and biophysical analyses have revealed extensive conformational changes within the TCR complex upon binding to antigenic peptides, implying the conversion of biochemical interactions into mechanical information [22].
Any proposed mechanism for antigen recognition by TCR must explain certain distinct features of TCR/pMHC interactions. A single pMHC complex can lead to TCR triggering and T cell activation (sensitivity) [23]. Further, T cells can discriminate between small numbers of agonist pMHC molecules from a large number of very similar, non-agonist pMHC molecules (specificity). How the TCR achieves this level of sensitivity and specificity is not completely understood. The TCR/pMHC bond is weak [24], suggesting that the free energy changes underlying conformational transitions likely require applied forces. Recent work has hinted that forces exerted on the TCR-pMHC bond may be critical in optimizing TCR triggering, suggesting that the TCR is a mechanosensor as it can transduce mechanical stimuli into biochemical signals through structural and conformational changes [25].
In one of the first such studies, Kim et al. [14] used optical tweezers to apply forces on beads coated with non-activating antibody or pMHC and quantified Ca2+ levels as a measure of T cell activation. They found that tangential but not normally applied forces to pMHC-coated beads induced Ca2+ signaling, indicating that TCR is mechanically triggered. A force threshold of 50 pN was required for activation. Li et al. used fibroblasts as artificial APCs to show that application of force to T cells by magnetic beads resulted in robust Ca2+ influx for ligands specific to TCR but not for integrins or non-TCR receptors [26].
In a tour-de-force study, Zhu and colleagues used a micropipette-RBC based force probe apparatus, capable of detecting <2 pN forces to examine how applied forces regulate TCR-pMHC interaction [27, 28]. If the affinity of protein-protein interaction increases with applied force up to a threshold, the interaction is called a catch-bond. Liu et al. showed that the lifetime of the bond between TCR and its cognate ligand was prolonged with application of 10 pN force, indicative of catch-bond behavior [28]. For non-specific TCR-pMHC interactions however, the affinity peaked at zero force, indicative of slip-bond behavior (weakening under applied force). At a cellular scale, high Ca2+ levels were induced with the application of force. These studies emphasize that load-induced structural transitions tune TCR–pMHC bond lifetime with high specificity. Using optical tweezers and DNA tethers, Das et al. have shown that the domains in the β-subunit of the TCR undergo force-dependent conformational transition [29]. This allosteric transition prolonged the TCR–pMHC bond lifetime, providing a possible molecular basis for the observed catch-bond behavior.
While the role of forces on the specificity and sensitivity of antigen recognition by TCR is coming to light, how information about TCR-antigen binding is transmitted into the cell is unclear. As TCRs lack large intracellular domains or intrinsic kinase activity, signal propagation across the membrane involves the CD3 intracellular domains [21] containing immunoreceptor tyrosine-based activation motifs (ITAMs) [30]. ITAM chains, buried in the hydrophobic interior of the membrane are inaccessible to Src kinases, preventing spontaneous phosphorylation [31]. Ligand binding to TCR must induce conformational transitions that propagate to CD3, extending its cytosplasmic tails for ITAM phosphorylation. Several considerations suggest that this propagation must involve forces. Soluble monomeric pMHC are poor activators of TCR triggering, while surface-bound pMHC efficiently activate TCRs, suggesting that TCR triggering requires additional steps beyond pMHC binding [25, 32]. Structural studies indicate that electrostatic interactions between the TCR-α and CD3 transmembrane regions create a pivot which can mechanically couple antigen binding and force application to conformational changes within the CD3ζζ cytosolic regions [33] (Fig. 1a, inset).
One unresolved question is the directionality of the applied force necessary for TCR triggering. While forces tangential to the cell membrane can engage the TCR-CD3 pivot driving the conformational changes leading to triggering [33], forces normal to the cell membrane along the TCR axis are required to engage the catch-bond and induce signaling [28]. Overall, these observations raise the following questions: what is the magnitude and direction of forces in situ? How do internal forces link to the TCR complex? What are the contributions of the various force-generating cytoskeletal elements to TCR activation?
Finally, we note that much less is known about whether mechanical induction is required for BCR activation and whether applied forces allow antigen discrimination and internalization. Soluble antigens are potent activators of B cell signaling but work stemming from the studies of Batista and colleagues [34, 35] have shown that surface-anchored antigens are more efficiently gathered [34, 36, 37]. Moreover, monovalent antigens are able to induce BCR clustering, potentially as a result of conformational changes induced by forces generated upon cell-substrate contact [38]. The most compelling evidence for mechanical regulation of B cell signaling comes from an elegant study combining DNA-based molecular force sensors and engineered substrates showed that B cells apply tension to the BCR-antigen bond and extract antigen from surfaces in an affinity-dependent manner [39, 40],. AFM-based force-probes showed the rupture forces of antigen/BCR bonds to be in the pN range. However, there are no direct demonstrations of force-dependent structural rearrangements in BCRs in a manner analogous to the TCR.
2.2 Integrin receptors
In addition to TCRs, integrin-dependent interaction between T cells and APC are important for signaling activation, immune synapse (IS) formation, and T cell trafficking [41, 42]. The integrin αLβ2 LFA-1, which is expressed exclusively in leukocytes and its binding to ligand, ICAM-1 is important for T cell adhesion to APC and co-stimulatory signaling enabling lymphocyte activation. In resting T cells, LFA-1 integrins are in a bent conformation, leading to low affinity binding with ICAM-1. TCR stimulation induces conformational change in LFA-1 to relieve the inhibitory conformation into an extended state, which binds ligands with intermediate affinity (inside-out signaling) [42]. Forces applied to the integrin-ligand bond induce further conformational change into an open state, which enables highest affinity binding (~100 fold increase) [43] (Fig. 1a inset). This force-induced change in affinity allows integrins to behave as mechaosensors. LFA-1 binding with ICAM-1 behaves like a catch bond [44], similar to other integrins [45]. During IS formation, ligand-engaged LFA-I undergoes spatiotemporal reorganization driven by actin flows. Different activated forms of LFA-1 partition to different regions of the IS in an actin flow dependent manner, suggesting that cytoskeletal forces are important in patterning protein distributions during early T cell activation [46]. Recent work from the Burkhardt group has revealed that actin-dependent constrained mobility of ICAM-1 on the APC surface promotes integrin (LFA-1) activation [47], further supporting the notion of force mediated mechanotransduction in T cells. T cells also express the α4β1 integrin VLA-4, which influences T cell activation. Interestingly, VLA-4 ligation to its ligand, VCAM-1, leads to a dramatic arrest of actin retrograde flow [48, 49] but its effect on cellular forces and microcluster dynamics remain to be tested.
3. Cytoskeletal dynamics during immune cell activation
Cellular mechanotransduction largely involves cell-generated forces, which result from cytoskeletal rearrangements. Below, we briefly review studies of cytoskeletal dynamics in immune cells that accompany signaling events.
3.1 Actomyosin dynamics in T cells
Among the first events after TCR engagement with antigen is the onset of actin polymerization at the contact zone between T cells and APC [50, 51] (Fig. 1a), which drives membrane deformation and cell spreading facilitating further receptor-antigen binding [10, 11, 52–54]. The cell edge is highly dynamic with extensive protrusions/retractions [55], which may allow efficient antigen sampling. Actin polymerization and myosin contraction induced by TCR engagement results in retrograde flow of actomyosin at the cell periphery which serves as a primary force-generating element at the cell-substrate interface. The advent of super-resolution imaging and novel fluorescent probes has revealed a diversity of actin structures at the T cell/substrate interface [56]. On planar substrates (and conjugates), within 2–3 minutes of stimulation, the actin network forms an annular ring at the lamellipodium and is composed of the characteristic Arp2/3 mediated branched networks as well as formin-mediated linear bundles. At the rear of the lamellipodium, the linear bundles condense into arc-like structures under the action of myosin II motors. Interestingly, unlike adherent cells, myosin activity is not required for maintenance of retrograde flow [48, 49], raising questions about the role of myosin contractility in force generation and T cell activation.
3.2 Microtubule dynamics in T cells
The initial contact of T cells and APC is also characterized by reorientation of the microtubule (MT) cytoskeleton and the centrosome towards the contact zone [57]. It is believed that the MT cytoskeleton acts as a scaffold, holding lytic granules and guiding centrosomal reorientation towards the APC for directional secretion [58] as well as reorganization of signaling microclusters [59] and other signaling pathways. However, the contribution of the MT to force generation at the synapse has been relatively unknown, despite the fact that organelle movement as well as MT deformation clearly indicates the existence of forces. We recently found that MT tip dynamics at the lamellipodial/lamellar region modulate NMII filament assembly and lamellipodial actin flow through the RhoGTPase pathway and modulates the traction forces exerted by T cells [60]. These studies suggest that centrosomal movement and microtubule deformations result from forces generated by MT motors and the actomyosin cytoskeleton [61–64], and point to the importance of interactions between the actin and MT cytoskeleton [65] in force generation and force balance at the immune synapse, but its role in TCR signaling remains to be explored.
3.3 Actin dynamics in B cells
In resting B cells, the ezrin/radixin/moesin (ERM) family proteins couple cortical actin to the plasma membrane [66], impeding BCR diffusion. Signaling cascades downstream of BCR antigen interaction cause detachment of cortical actin from the plasma membrane, increased BCR mobility and enhanced signaling [67]. Subsequently, as the B cell spreads, actin undergoes dramatic reassembly, establishing lamellipodial retrograde flow, which drives the centripetal movement of microclusters [37, 68, 69]. Additionally, de novo actin polymerization is detected at locations of BCR microcluster formation [70]. After maximal spreading, the cell undergoes contraction and BCR clusters merge into a central cluster [70, 71] leading to signaling inhibition and subsequently antigen internalization. The mechanisms underlying B cell contraction and associated actin remodeling remain elusive, but likely involve myosin contractility, signaling inhbitors [72], and actin regulatory proteins WASP and NWASP [73]. These findings implicate feedback loops coupling actin dynamics/regulation and cellular forces with signaling to drive the transition from activation to inhibition.
4. Measuring cellular force generation during immune cell activation
Recent advances in imaging and force measurements in live cells have begun to reveal the biophysics of force generation by immune cells during activation, elucidating the regulation of immune cell signaling by mechanical events. These techniques typically measure the displacement of a calibrated force sensor in response to a cellular force.
Husson et al. developed a force probe to measure forces exerted by the T cell on an antibody-coated microbead [74]. Activation resulted in a pushing response, as the T cell spread on the microbead, followed by a pulling response, accompanied by Ca2+ influx and early signaling. The typical force generated during the pushing phase was ~20–30 pN. The median loading rate during the pulling phase was ~2–3 pN/sec and increased linearly with probe stiffness, suggesting a form of mechanosensitivity. However, Ca2+ dependence on stiffness was not measured. Li and Butte used an atomic force microscope (AFM) tip coated with anti-CD3 or pMHC to stimulate primary and Jurkat T cells and measured pushing and pulling forces ~1nN [75]. Interestingly, they did not find correlations between the total Ca2+ flux and the magnitude of the applied force. but these were temporally correlated. Actin dynamics and myosin contractility were required for force generation. However, neither of these studies determined the spatiotemporal organization of forces in the plane of the T-cell/APC surrogate interface.
Two recent studies have adapted traction force microscopy (TFM) using ligand-coated elastic substrates to mimic an APC to measure forces during T cell activation. Bashour et al. [76] microfabricated polydimethylsiloxane (PDMS) pillars in a dense array (2 μm spacing) coated with stimulatory ligands (Fig. 1b). T cells spreading on the array deflected the pillars, first radially outward (similar to the pushing phase) and then radially inwards (pulling phase). Individual pillars were subject to peak forces of ~50 pN, with a total force of ~1nN. Force development required Src kinase signaling and was specific to TCR activation. Conventional TFM employs polyacrylamide (PA) substrates with embedded fluorescent marker beads and uses elasticity theory to calculate traction forces from the displacement of the beads [77–79] (Fig. 1c). We used PA gels coated with anti-CD3 antibodies to measure traction forces while simultaneously monitoring actin dynamics [19]. We found that cells exerted median average traction stresses in the range of 5–10 Pa, peak traction stresses of 10–30 Pa corresponding to ~1–2nN of total force. These forces were specific to TCR-ligand binding, and both actin dynamics (polymerization and depolymerization) and myosin activity were required for force generation. We have recently shown that microtubule tip dynamics modulate force generation in T cells [60].
While polyacrylamide substrates offer a relatively easy method to measure cell-exerted forces, they are limited in spatial resolution to about 1–2 μm2. Recently, several groups have developed DNA-based tension sensors that can measure pN forces with near-single-molecule resolution [80] (Fig. 1d). Salaita’s group has used this approach to demonstrate that individual TCR-pMHC complexes experience 10–20 pN forces exerted by cytoskeletal dynamics [81]. The magnitude of these forces modulates the sensitivity of TCRs to specific antigens, as reducing the forces reduced the response to agonist antigens. The various force measurement techniques and results are summarized in Table 1.
Table 1.
Comparison of different force measurement techniques.
| Study | Measurement technique | Force Magnitude | Signaling response | Significance |
|---|---|---|---|---|
| Husson et al. | Calibrated force probe | 20–30 pN | Ca2+ influx | Established the role of forces in T cell activation. |
| Bashour et al. | Nanopillar array | ~1 nN | Syk/ZAP70/SFK phosphorylation, IL2 secretion | First study to measure forces exerted during T cell activation. |
| Hui et al. | Polyacrylamide gel | ~1 nN | ZAP70/LAT phosphorylation | Mapped the spatiotemporal characteristics of T cell forces and showed that T cell signaling was mechanosensitive. Showed that actomyosin contractility was necessary for force generation. |
| Hu and Butte | Atomic force probe | ~ 1nN | Ca2+ influx; | An AFM based study to show that T cells are triggered by external forces and generate actin dependent forces upon stimulation. Showed temporal correlation between Ca2+ flux and force |
| Liu et al. | DNA-based tension sensors | ~ 1nN | Ca2+ influx, Lck/ZAP70 phophorylation | Performed a high-resolution (near single-molecule resolution) mapping of forces exerted during T cell activation. Showed that forces modulate sensitivity to antigen. |
Overall, these studies indicate that forces exerted by T cells are relatively weak and correspond to ~10 pN/μm2 of stress, consistent with the measured value of external force needed for TCR triggering. Based on the observed stiffness of Jurkat cells, Young’s modulus E ~ 50–100 Pa [82], the expected value of maximum force, F~E × area ~2–5 nN, suggesting that T cells which are softer than adherent cells (1–5 kPa [83]), generate weaker tractions, but these are sufficient to activate individual TCRs.
5. Mechanosensing at the cellular scale
5.1 Microcluster formation and transport in T cells
Upon receptor engagement, TCR assemble into signaling microclusters at sites of close adhesion [54, 84]. Signaling microclusters are the sites of early signaling. Activated TCR recruit a host of downstream signaling molecules into these microclusters as well as several actin regulatory proteins [52]. Signaling proteins in microclusters recruit and activate RhoGTPases, which in turn activate actin regulatory proteins. Such dense localization of actin regulators at microclusters primes them to being “hotspots” of actin dynamics. Indeed we have observed ‘actin waves’ or bursts of actin polymerization originating from signaling microclusters [49].
Initially clusters form at the periphery as the cell edge is forced outwards by actin dynamics and allows the cell to sample the antigen-presenting surface [6]. Over a time-scale of several minutes, receptor microclusters and signaling molecules organize into the ordered spatially symmetric structure of the immune synapse. Microclusters are rapidly transported centripetally towards the cell center by acto-myosin retrograde flows [10, 53, 85] and potentially MT based motors [59], forming the cSMAC (central supramolecular activation cluster) surrounded by a ring of integrins (LFA-1/ICAM bonds) in the pSMAC (peripheral supramolecular activation cluster). Importantly, TCR-MHC bonds are much smaller (10–15 nm) than LFA-ICAM bonds (45–50 nm) or other surface molecules such as phosphatases and glycoproteins. This difference in size creates energetic barriers that must be overcome for size-dependent spatial segregation to occur [86, 87]. The role of cytoskeletal forces for kinetic segregation, along with associated kinetic proofreading models for TCR signaling have been exhaustively reviewed in Comrie et al. [88]. Further evidence for active formation of microclusters comes from a recent study which used high-resolution light sheet microscopy to show that dynamic microvilli on T cells drive the formation of close contacts on the APC surface, which are colocalized with TCR microclusters [89]. This is consistent with earlier work showing the correlation between microclusters and regions of reduced membrane fluctuations [84]. These findings suggest that active forces, potentially generated by the cytoskeleton, may enhance the efficacy of TCR triggering by enabling and stabilizing contacts between the T cell membrane and APC.
5.2 Sensing substrate stiffness
The functional consequences of mechanosensitivity are best highlighted by the findings that mesenchymal stem cells differentiate into specific cell lineages depending on substrate stiffness [90]. Similar experiments in which stimulatory ligands were presented to T cells on PA substrates of varying stiffness showed that stiffer surfaces (100–200 kPa) elicited greater cytokine production in mouse CD4+ T cells than softer ones [20]. Conversely, human CD4+ cells were more strongly stimulated on 100–200 kPa PDMS substrates than stiffer ones (2 MPa), suggesting a biphasic response pattern of stiffness sensitivity [91]. In all cases, myosin inhibition led to loss of sensitivity to substrate stiffness. However, these results must be viewed against the fact that T cells are significantly softer (~1 kPa) and most models of force generation posit that cellular forces rapidly saturate at substrate stiffness values that are ~5–10x of the cell’s stiffness [92]. Using PA substrates, we showed that traction forces exerted by T cells exhibited sigmoidal behavior as a function of substrate stiffness [19], with the total exerted forces saturating on surfaces of ~5kPa stiffness. Recent studies have found that the stiffness of many immune cells varies in the range of a few kPa, suggesting that T cells need to interact with (or discriminate between) relatively soft surfaces in vivo [82]. Intriguingly, Jurkat cells showed distinct differences in the dynamics of signaling and cell morphology as a function of substrate stiffness [19]. On stiff PA substrates (~5kPa), phosphotyrosine (pY) signaling rapidly peaked at ~3 min after stimulation and declined to baseline levels, with monotonic expansion of the cell edge. On soft substrates (<1 kPa), pY signaling remained sustained even at 15 min but at lower levels, with the cell edge undergoing repeated rounds of protrusions and retractions.
The response of T cells to substrate stiffness [19] appears to obey predictions from a simple model derived from active matter theory [92]. The cell exerts an active stress (σa) resulting from actin polymerization and myosin contractility, which consumes energy by ATP hydrolysis. Force balance leads to a simple expression for the force, , where Fss is the steady state force and Fsat is the saturating force. , where A is the cell area, ksubs is the effective spring constant for the substrate (0.1 – 10 nN/μm) and kcell is the stiffness of the cell (~1 nN/μm). Thus, on compliant substrates, the steady state force exerted by the cell is predicted to linearly increase with stiffness and saturates when ksubs ≫ kcell. Assuming a linear force-velocity relationship, the loading rate can be estimated as , where η = τ(ksubs+ kcell)is the viscous dissipation in the actin gel and A is the area over which the stress is exerted. Using the estimated values for these parameters (τ~10s, ksubs~ kcell), a μm2 patch of TCRs would experience loading rates of ~2–3 pN/sec, which is similar to the force probe experiments [74].
While immune cells encounter substrates (e.g. APCs) whose stiffness is in the low kPa range [82], conventional strategies to expand clonal populations of T cells for adoptive immunotherapies use protein-coated microbeads [93], which are significantly more rigid. Alternate strategies that mimic APCs in vivo may be better in stimulating T cells. Exploiting this, the Kam group used PDMS microbeads to stimulate CD4+ and CD8+ T cells and observed a stronger proliferative response [94]. However, it is unclear whether the enhanced proliferative response was due to the nanoscale structure of the polymer or changes in substrate rigidity. Nevertheless, exploiting the mechanosensitivity of T cells may enhance strategies for T cell based immunotherapies.
B cells also appear to be mechanosensitive, exhibiting strong early BCR signaling and Ca2+ responses on stiffer antigen-coated elastic substrates, whereas cell proliferation is higher on softer substrates [95, 96]. An elegant study from the Tolar group showed that follicular dendritic cells are stiff and promote strong B cell pulling forces and stringent affinity discrimination [40]. In contrast, dendritic cells are soft and promote acquisition of low-affinity antigens through low forces. Thus, the mechanical properties of B cell synapses regulate antigen extraction, suggesting that distinct properties of presenting cells support different stages of B cell responses.
Finally, Huse and colleagues have shown that the killing responses of cytotoxic T lymphocytes are mechanosensitive and enhanced by forces [97]. Upon formation of an immune synapse like structure, CTLs secrete pore-forming proteins called perforins to lyse the target cell. Altering the membrane tension of the target cells using pharmacological means or osmotic shock and showed that perforin-induced pore formation was strongly affected. Similarly, altering the membrane tension of the target cells by changing substrate stiffness again altered cell killing, with cells on stiffer substrates being more sensitivity to perforin-induced pore formation.
5.3 Sensing ligand mobility
Lymphocyte engagement with APCs also results in extensive changes in cytoskeletal and lipid membrane composition of the APC [98–100]. Consequently, the mobility of pMHC complexes, integrin and other co-receptor ligands at the APC membrane may be modulated as they partition into lipid microdomains or associate with tetraspanin (family of small proteins with four transmembrane domains) complexes [101]. Disruption of lipid raft integrity reduces antigen presentation by class-II MHC complexes in B cells cholesterol, which modulates the diffusion of transmembrane receptors [102], affects lymphocyte signaling; tetraspanins accumulate at the immune synapse, where they form enriched membrane microdomains and cluster MHCs [101]. These studies suggest that the plasma membrane environment undergoes fluctuations in its lipid and protein composition, which may serve to modulate signaling, in part by altering ligand mobility [103].
Despite this evidence, few studies have explicitly addressed how the biophysics of ligand (or receptor) mobility influences lymphocyte signaling. Groves and colleagues used nanofabricated 10–20 nm high chromium barriers on stimulatory supported bilayer surfaces, which allow for free lipid diffusion in the bilayers but block movement of proteins with larger cytoplasmic domains. Blocking the movement of TCR microclusters from the periphery to the center prolonged pY signaling levels [104], suggesting location-dependence of TCR signaling. Intriguingly, actin dynamics in the vicinity of stalled clusters was altered compared to the regions between barriers [105]. In a complementary approach, altering the mobility of lipids and proteins by modifying the lipid composition of stimulatory bilayers showed that increased diffusivity resulted in higher Ca2+ influx, greater microcluster mobility and enhanced pY signaling as well as altered actin dynamics [106]. It is interesting to note that these two studies suggest opposite effects of mobility on signaling, however the mechanisms behind these differences remain to be explored. It is possible that the sensitivity to ligand mobility may be dependent in a non-trivial manner on spatial location in the immune synapse and this would require higher resolution studies (super resolution and single molecule imaging) to decipher.
Studies from the Batista lab have highlighted the importance of BCR diffusion in early B cell signaling [37, 67, 107], implying that ligand mobility may control signaling by modulating BCR diffusion and clustering. We examined BCR clustering, mobility and signaling when activated by mobile (on lipid bilayers) and immobile (on glass) ligands [108]. B cells interacting with mobile ligands displayed significantly greater signaling. Moreover, microcluster movement required actin polymerization, suggesting that the static actin network acts as a diffusion barrier, corralling BCRs in the initial phases of B cell activation while active polymerization is required for BCR microcluster coalescence and centralization. These studies suggest the existence of an intricate feedback loop between actin polymerization and receptor movement, which leads to global organization of membrane proteins and signaling components at the lymphocyte/APC interface.
6. Summary and future perspectives
Recent advances in the field have made it abundantly clear that the physical environment is a strong modulator of immune cell responses. The biophysical basis of mechanosensitivity remains unclear, but likely involves forces generated by actin polymerization and myosin-based contraction, interacting with the membrane, receptor-ligand bonds and microclusters. While the signaling function of microclusters is well studied, the mechanism of their formation and spatial organization is not completely understood. In adherent cells, integrin-ECM linkages lead to focal adhesions whose growth and signaling depend on locally applied cytoskeletal forces. Thus focal adhesions act as mechanosensors which couple integrin binding and actin flow with signaling. Whether TCR-associated signaling microclusters serve a similar role in T cells is not known, but it is interesting to speculate that they might do so given the similarities in the molecular players. Force transmission must involve coupling between microclusters and actin [49, 53, 104, 105, 109, 110], but what mediates this coupling is not known. Resolving this question requires measurement of forces with high spatial resolution while simultaneously monitoring actin dynamics and microcluster assembly with genetic manipulation of candidate molecules. While traction forces give a measure of the magnitude of cell-generated forces, the intracellular forces experienced by the immune receptors are expected to be stronger. It is possible to directly measure intracellular forces by measuring the displacements of small (~200 nm) beads inside cells [111] or by calibrated fluorescence resonance energy transfer (FRET) based force probes [112]. Another possibility is to measure actin flows and use computational analysis and assumptions about the viscoelasticity of actin networks to infer forces [113, 114]. Such measurements remain to be made for immune cells, but would likely yield a more accurate estimate of the forces experienced by TCRs.
In addition to stiffness and mobility, discussed here, the topography of the environment may be important in dictating immune cell function. T and B cells navigate complex topography in the lymph nodes and thymus with extensive membrane folds on APC surfaces or fibrillar structures on follicular dendritic networks. These structures present regions of high curvature, which might recruit curvature-sensing signaling proteins and actin regulators, thereby modulating cytoskeletal dynamics. The role of topography on immune cell signaling activation is an open question for future studies.
Future work must also consider the crosstalk between different receptors (e.g. TCR and integrins) and co-stimulatory or inhibitory receptors in determining the global cellular response to physical cues. Finally, while we have a better knowledge of how cellular forces, proximal signaling and certain functional responses depend on the physical environment; mechanical modulation of gene expression in immune cells is completely unstudied and is open for future research. A recent study showed that the nucleus undergoes dramatic deformation by actin-based forces upon T cell activation [115], suggesting possible changes in gene expression that may be mechanically modulated.
Acknowledgments
This work was supported by the National Science Foundation (Grants 1206060 and 1607645 to A.U.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 2.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7(4):265–75. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 3.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19(2):194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bershadsky A, Kozlov M, Geiger B. Adhesion-mediated mechanosensitivity: a time to experiment, and a time to theorize. Curr Opin Cell Biol. 2006;18(5):472–81. doi: 10.1016/j.ceb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Bonilla FA, Oettgen HC. Adaptive immunity. J Allergy Clin Immunol. 125(2 Suppl 2):S33–40. doi: 10.1016/j.jaci.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285(5425):221–7. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 7.Dustin ML. The cellular context of T cell signaling. Immunity. 2009;30(4):482–92. doi: 10.1016/j.immuni.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrasco YR, Batista FD. B cells acquire particulate antigen in a macrophage-rich area at the boundary between the follicle and the subcapsular sinus of the lymph node. Immunity. 2007;27(1):160–71. doi: 10.1016/j.immuni.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, Samelson LE. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol. 2002;158(7):1263–75. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 2006;25(1):117–27. doi: 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokosuka T, Sakata-Sogawa K, Kobayashi W, Hiroshima M, Hashimoto-Tane A, Tokunaga M, Dustin ML, Saito T. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nature immunology. 2005;6(12):1253–62. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- 12.Padhan K, Varma R. Immunological synapse: a multi-protein signalling cellular apparatus for controlling gene expression. Immunology. 129(3):322–8. doi: 10.1111/j.1365-2567.2009.03241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Z, Finkel TH. T cell receptor triggering by force. Trends Immunol. 31(1):1–6. doi: 10.1016/j.it.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim ST, Takeuchi K, Sun ZY, Touma M, Castro CE, Fahmy A, Lang MJ, Wagner G, Reinherz EL. The alphabeta T cell receptor is an anisotropic mechanosensor. The Journal of biological chemistry. 2009;284(45):31028–37. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YC, Chen BM, Wu PC, Cheng TL, Kao LS, Tao MH, Lieber A, Roffler SR. Cutting Edge: mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. Journal of immunology. 2010;184(11):5959–63. doi: 10.4049/jimmunol.0900775. [DOI] [PubMed] [Google Scholar]
- 16.Billadeau DD, Burkhardt JK. Regulation of cytoskeletal dynamics at the immune synapse: new stars join the actin troupe. Traffic. 2006;7(11):1451–60. doi: 10.1111/j.1600-0854.2006.00491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dustin ML, Cooper JA. The immunological synapse and the actin cytoskeleton: molecular hardware for T cell signaling. Nat Immunol. 2000;1(1):23–9. doi: 10.1038/76877. [DOI] [PubMed] [Google Scholar]
- 18.Bashour KT, Gondarenko A, Chen H, Shen K, Liu X, Huse M, Hone JC, Kam LC. CD28 and CD3 have complementary roles in T-cell traction forces. Proceedings of the National Academy of Sciences of the United States of America; 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hui KL, Balagopalan L, Samelson LE, Upadhyaya A. Cytoskeletal forces during signaling activation in Jurkat T-cells. Molecular biology of the cell. 2015;26(4):685–95. doi: 10.1091/mbc.E14-03-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC. Mechanosensing in T lymphocyte activation. Biophys J. 2012;102(2):L5–7. doi: 10.1016/j.bpj.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhns MS, Davis MM. TCR Signaling Emerges from the Sum of Many Parts. Frontiers in immunology. 2012;3:159. doi: 10.3389/fimmu.2012.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wucherpfennig KW, Gagnon E, Call MJ, Huseby ES, Call ME. Structural biology of the T-cell receptor: insights into receptor assembly, ligand recognition, and initiation of signaling. Cold Spring Harb Perspect Biol. 2(4):a005140. doi: 10.1101/cshperspect.a005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Brameshuber M, Zeng X, Xie J, Li QJ, Chien YH, Valitutti S, Davis MM. A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4(+) T cells. Immunity. 2013;39(5):846–57. doi: 10.1016/j.immuni.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corse E, Gottschalk RA, Allison JP. Strength of TCR-peptide/MHC interactions and in vivo T cell responses. J Immunol. 2011;186(9):5039–45. doi: 10.4049/jimmunol.1003650. [DOI] [PubMed] [Google Scholar]
- 25.Ma Z, Janmey PA, Finkel TH. The receptor deformation model of TCR triggering. Faseb J. 2008;22(4):1002–8. doi: 10.1096/fj.07-9331hyp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li YC, Chen BM, Wu PC, Cheng TL, Kao LS, Tao MH, Lieber A, Roffler SR. Cutting Edge: mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. Journal of immunology. 184(11):5959–63. doi: 10.4049/jimmunol.0900775. [DOI] [PubMed] [Google Scholar]
- 27.Hong J, Persaud SP, Horvath S, Allen PM, Evavold BD, Zhu C. Force-Regulated In Situ TCR-Peptide-Bound MHC Class II Kinetics Determine Functions of CD4+ T Cells. J Immunol. 2015;195(8):3557–64. doi: 10.4049/jimmunol.1501407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell. 2014;157(2):357–68. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, Brady SK, Wang JH, Wagner G, Reinherz EL, Lang MJ. Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci U S A. 2015;112(5):1517–22. doi: 10.1073/pnas.1424829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kane LP, Lin J, Weiss A. Signal transduction by the TCR for antigen. Curr Opin Immunol. 2000;12(3):242–9. doi: 10.1016/s0952-7915(00)00083-2. [DOI] [PubMed] [Google Scholar]
- 31.Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111(7):967–79. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Z, Sharp KA, Janmey PA, Finkel TH. Surface-anchored monomeric agonist pMHCs alone trigger TCR with high sensitivity. PLoS Biol. 2008;6(2):e43. doi: 10.1371/journal.pbio.0060043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee MS, Glassman CR, Deshpande NR, Badgandi HB, Parrish HL, Uttamapinant C, Stawski PS, Ting AY, Kuhns MS. A Mechanical Switch Couples T Cell Receptor Triggering to the Cytoplasmic Juxtamembrane Regions of CD3zetazeta. Immunity. 2015;43(2):227–39. doi: 10.1016/j.immuni.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batista FD, Iber D, Neuberger MS. B cells acquire antigen from target cells after synapse formation. Nature. 2001;411(6836):489–94. doi: 10.1038/35078099. [DOI] [PubMed] [Google Scholar]
- 35.Batista FD, Neuberger MS. Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity. 1998;8(6):751–9. doi: 10.1016/s1074-7613(00)80580-4. [DOI] [PubMed] [Google Scholar]
- 36.Fleire SJ, Goldman JP, Carrasco YR, Weber M, Bray D, Batista FD. B cell ligand discrimination through a spreading and contraction response. Science. 2006;312(5774):738–41. doi: 10.1126/science.1123940. [DOI] [PubMed] [Google Scholar]
- 37.Harwood NE, Batista FD. Early events in B cell activation. Annu Rev Immunol. 2010;28:185–210. doi: 10.1146/annurev-immunol-030409-101216. [DOI] [PubMed] [Google Scholar]
- 38.Tolar P, Sohn HW, Pierce SK. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat Immunol. 2005;6(11):1168–76. doi: 10.1038/ni1262. [DOI] [PubMed] [Google Scholar]
- 39.Natkanski E, Lee WY, Mistry B, Casal A, Molloy JE, Tolar P. B cells use mechanical energy to discriminate antigen affinities. Science. 2013;340(6140):1587–90. doi: 10.1126/science.1237572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spillane KM, Tolar P. B cell antigen extraction is regulated by physical properties of antigen-presenting cells. J Cell Biol. 2017;216(1):217–230. doi: 10.1083/jcb.201607064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pribila JT, Quale AC, Mueller KL, Shimizu Y. Integrins and T cell-mediated immunity. Annu Rev Immunol. 2004;22:157–80. doi: 10.1146/annurev.immunol.22.012703.104649. [DOI] [PubMed] [Google Scholar]
- 42.Kinashi T. Intracellular signalling controlling integrin activation in lymphocytes. Nat Rev Immunol. 2005;5(7):546–59. doi: 10.1038/nri1646. [DOI] [PubMed] [Google Scholar]
- 43.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323(5914):642–4. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 44.Chen W, Lou J, Zhu C. Forcing switch from short- to intermediate- and long-lived states of the alphaA domain generates LFA-1/ICAM-1 catch bonds. J Biol Chem. 2010;285(46):35967–78. doi: 10.1074/jbc.M110.155770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185(7):1275–84. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Comrie WA, Babich A, Burkhardt JK. F-actin flow drives affinity maturation and spatial organization of LFA-1 at the immunological synapse. J Cell Biol. 2015;208(4):475–91. doi: 10.1083/jcb.201406121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Comrie WA, Li S, Boyle S, Burkhardt JK. The dendritic cell cytoskeleton promotes T cell adhesion and activation by constraining ICAM-1 mobility. J Cell Biol. 2015;208(4):457–73. doi: 10.1083/jcb.201406120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Babich A, Li S, O’Connor RS, Milone MC, Freedman BD, Burkhardt JK. F-actin polymerization and retrograde flow drive sustained PLCgamma1 signaling during T cell activation. J Cell Biol. 2012;197(6):775–87. doi: 10.1083/jcb.201201018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam Hui K, Kwak SI, Upadhyaya A. Adhesion-dependent modulation of actin dynamics in Jurkat T cells. Cytoskeleton. 2014;71(2):119–35. doi: 10.1002/cm.21156. [DOI] [PubMed] [Google Scholar]
- 50.Burkhardt JK, Carrizosa E, Shaffer MH. The actin cytoskeleton in T cell activation. Annu Rev Immunol. 2008;26:233–59. doi: 10.1146/annurev.immunol.26.021607.090347. [DOI] [PubMed] [Google Scholar]
- 51.Gomez TS, Billadeau DD. T Cell Activation and the Cytoskeleton: You Can’t Have One Without the Other. Adv Immunol. 2008;97:1–64. doi: 10.1016/S0065-2776(08)00001-1. [DOI] [PubMed] [Google Scholar]
- 52.Barda-Saad M, Braiman A, Titerence R, Bunnell SC, Barr VA, Samelson LE. Dynamic molecular interactions linking the T cell antigen receptor to the actin cytoskeleton. Nat Immunol. 2005;6(1):80–9. doi: 10.1038/ni1143. [DOI] [PubMed] [Google Scholar]
- 53.Kaizuka Y, Douglass AD, Varma R, Dustin ML, Vale RD. Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc Natl Acad Sci U S A. 2007;104(51):20296–301. doi: 10.1073/pnas.0710258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121(6):937–50. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ritter AT, Asano Y, Stinchcombe JC, Dieckmann NM, Chen BC, Gawden-Bone C, van Engelenburg S, Legant W, Gao L, Davidson MW, Betzig E, Lippincott-Schwartz J, Griffiths GM. Actin depletion initiates events leading to granule secretion at the immunological synapse. Immunity. 2015;42(5):864–76. doi: 10.1016/j.immuni.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murugesan S, Hong J, Yi J, Li D, Beach JR, Shao L, Meinhardt J, Madison G, Wu X, Betzig E, Hammer JA. Formin-generated actomyosin arcs propel T cell receptor microcluster movement at the immune synapse. The Journal of cell biology. 2016;215(3):383–399. doi: 10.1083/jcb.201603080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin-Cofreces NB, Baixauli F, Sanchez-Madrid F. Immune synapse: conductor of orchestrated organelle movement. Trends Cell Biol. 2014;24(1):61–72. doi: 10.1016/j.tcb.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. Centrosome polarization delivers secretory granules to the immunological synapse. Nature. 2006;443(7110):462–5. doi: 10.1038/nature05071. [DOI] [PubMed] [Google Scholar]
- 59.Hashimoto-Tane A, Yokosuka T, Sakata-Sogawa K, Sakuma M, Ishihara C, Tokunaga M, Saito T. Dynein-driven transport of T cell receptor microclusters regulates immune synapse formation and T cell activation. Immunity. 2011;34(6):919–31. doi: 10.1016/j.immuni.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 60.Hui KL, Upadhyaya A. Dynamic microtubules regulate cellular contractility during T-cell activation. Proc Natl Acad Sci U S A. 2017;114(21):E4175–E4183. doi: 10.1073/pnas.1614291114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu X, Kapoor TM, Chen JK, Huse M. Diacylglycerol promotes centrosome polarization in T cells via reciprocal localization of dynein and myosin II. Proc Natl Acad Sci U S A. 2013;110(29):11976–81. doi: 10.1073/pnas.1306180110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu J, Misra G, Russell RJ, Ladd AJ, Lele TP, Dickinson RB. Effects of dynein on microtubule mechanics and centrosome positioning. Molecular biology of the cell. 2011;22(24):4834–41. doi: 10.1091/mbc.E11-07-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yi J, Wu X, Chung AH, Chen JK, Kapoor TM, Hammer JA. Centrosome repositioning in T cells is biphasic and driven by microtubule end-on capture-shrinkage. J Cell Biol. 2013;202(5):779–92. doi: 10.1083/jcb.201301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu J, Burakov A, Rodionov V, Mogilner A. Finding the cell center by a balance of dynein and myosin pulling and microtubule pushing: a computational study. Molecular biology of the cell. 2010;21(24):4418–27. doi: 10.1091/mbc.E10-07-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin-Cofreces NB, Alarcon B, Sanchez-Madrid F. Tubulin and actin interplay at the T cell and antigen-presenting cell interface. Frontiers in immunology. 2011;2:24. doi: 10.3389/fimmu.2011.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11(4):276–87. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Treanor B, Depoil D, Bruckbauer A, Batista FD. Dynamic cortical actin remodeling by ERM proteins controls BCR microcluster organization and integrity. J Exp Med. 2011;208(5):1055–68. doi: 10.1084/jem.20101125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Batista FD, Treanor B, Harwood NE. Visualizing a role for the actin cytoskeleton in the regulation of B-cell activation. Immunological reviews. 2010;237(1):191–204. doi: 10.1111/j.1600-065X.2010.00943.x. [DOI] [PubMed] [Google Scholar]
- 69.Treanor B, Harwood NE, Batista FD. Microsignalosomes: spatially resolved receptor signalling. Biochemical Society transactions. 2009;37(Pt 5):1014–8. doi: 10.1042/BST0371014. [DOI] [PubMed] [Google Scholar]
- 70.Liu C, Miller H, Orlowski G, Hang H, Upadhyaya A, Song W. Actin reorganization is required for the formation of polarized B cell receptor signalosomes in response to both soluble and membrane-associated antigens. J Immunol. 2012;188(7):3237–46. doi: 10.4049/jimmunol.1103065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu C, Miller H, Hui KL, Grooman B, Bolland S, Upadhyaya A, Song W. A balance of Bruton’s tyrosine kinase and SHIP activation regulates B cell receptor cluster formation by controlling actin remodeling. J Immunol. 2011;187(1):230–9. doi: 10.4049/jimmunol.1100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song W, Liu C, Seeley-Fallen MK, Miller H, Ketchum C, Upadhyaya A. Actin-mediated feedback loops in B-cell receptor signaling. Immunological reviews. 2013;256(1):177–89. doi: 10.1111/imr.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu C, Bai X, Wu J, Sharma S, Upadhyaya A, Dahlberg CI, Westerberg LS, Snapper SB, Zhao X, Song W. N-wasp is essential for the negative regulation of B cell receptor signaling. PLoS Biol. 2013;11(11):e1001704. doi: 10.1371/journal.pbio.1001704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Husson J, Chemin K, Bohineust A, Hivroz C, Henry N. Force generation upon T cell receptor engagement. PLoS One. 2011;6(5):e19680. doi: 10.1371/journal.pone.0019680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu KH, Butte MJ. T cell activation requires force generation. J Cell Biol. 2016;213(5):535–42. doi: 10.1083/jcb.201511053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bashour KT, Gondarenko A, Chen H, Shen K, Liu X, Huse M, Hone JC, Kam LC. CD28 and CD3 have complementary roles in T-cell traction forces. Proc Natl Acad Sci U S A. 2014;111(6):2241–6. doi: 10.1073/pnas.1315606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76(4):2307–16. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. American journal of physiology. 2002;282(3):C595–605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 79.Sabass B, Gardel ML, Waterman CM, Schwarz US. High resolution traction force microscopy based on experimental and computational advances. Biophys J. 2008;94(1):207–20. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stabley DR, Jurchenko C, Marshall SS, Salaita KS. Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nature methods. 2011;9(1):64–7. doi: 10.1038/nmeth.1747. [DOI] [PubMed] [Google Scholar]
- 81.Liu Y, Blanchfield L, Ma VP, Andargachew R, Galior K, Liu Z, Evavold B, Salaita K. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc Natl Acad Sci U S A. 2016;113(20):5610–5. doi: 10.1073/pnas.1600163113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bufi N, Saitakis M, Dogniaux S, Buschinger O, Bohineust A, Richert A, Maurin M, Hivroz C, Asnacios A. Human Primary Immune Cells Exhibit Distinct Mechanical Properties that Are Modified by Inflammation. Biophys J. 2015;108(9):2181–90. doi: 10.1016/j.bpj.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys J. 2007;93(12):4453–61. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lam Hui K, Wang C, Grooman B, Wayt J, Upadhyaya A. Membrane dynamics correlate with formation of signaling clusters during cell spreading. Biophys J. 2012;102(7):1524–33. doi: 10.1016/j.bpj.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Campi G, Varma R, Dustin ML. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J Exp Med. 2005;202(8):1031–6. doi: 10.1084/jem.20051182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Allard JF, Dushek O, Coombs D, van der Merwe PA. Mechanical modulation of receptor-ligand interactions at cell-cell interfaces. Biophys J. 2012;102(6):1265–73. doi: 10.1016/j.bpj.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee SJ, Hori Y, Groves JT, Dustin ML, Chakraborty AK. The synapse assembly model. Trends Immunol. 2002;23(10):500–2. doi: 10.1016/s1471-4906(02)02325-6. [DOI] [PubMed] [Google Scholar]
- 88.Comrie WA, Burkhardt JK. Action and Traction: Cytoskeletal Control of Receptor Triggering at the Immunological Synapse. Frontiers in immunology. 2016;7:68. doi: 10.3389/fimmu.2016.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, Weaver VM, Gerard A, Liu TL, Chen BC, Betzig E, Bartumeus F, Krummel MF. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science. 2017;356(6338) doi: 10.1126/science.aal3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 91.O’Connor RS, Hao X, Shen K, Bashour K, Akimova T, Hancock WW, Kam LC, Milone MC. Substrate rigidity regulates human T cell activation and proliferation. J Immunol. 2012;189(3):1330–9. doi: 10.4049/jimmunol.1102757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marcq P, Yoshinaga N, Prost J. Rigidity sensing explained by active matter theory. Biophys J. 2011;101(6):L33–5. doi: 10.1016/j.bpj.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hotaling NA, Tang L, Irvine DJ, Babensee JE. Strategies for Immunomodulation. Annu Rev Biomed Eng. 2015;17:317–49. doi: 10.1146/annurev-bioeng-071813-104814. Biomaterial. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lambert LH, Goebrecht GK, De Leo SE, O’Connor RS, Nunez-Cruz S, Li TD, Yuan J, Milone MC, Kam LC. Improving T Cell Expansion with a Soft Touch. Nano letters. 2017;17(2):821–826. doi: 10.1021/acs.nanolett.6b04071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wan Z, Zhang S, Fan Y, Liu K, Du F, Davey AM, Zhang H, Han W, Xiong C, Liu W. B cell activation is regulated by the stiffness properties of the substrate presenting the antigens. J Immunol. 2013;190(9):4661–75. doi: 10.4049/jimmunol.1202976. [DOI] [PubMed] [Google Scholar]
- 96.Zeng Y, Yi J, Wan Z, Liu K, Song P, Chau A, Wang F, Chang Z, Han W, Zheng W, Chen YH, Xiong C, Liu W. Substrate stiffness regulates B-cell activation, proliferation, class switch, and T-cell-independent antibody responses in vivo. Eur J Immunol. 2015;45(6):1621–34. doi: 10.1002/eji.201444777. [DOI] [PubMed] [Google Scholar]
- 97.Basu R, Whitlock BM, Husson J, Le Floc’h A, Jin W, Oyler-Yaniv A, Dotiwala F, Giannone G, Hivroz C, Biais N, Lieberman J, Kam LC, Huse M. Cytotoxic T Cells Use Mechanical Force to Potentiate Target Cell Killing. Cell. 2016;165(1):100–10. doi: 10.1016/j.cell.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kropshofer H, Spindeldreher S, Rohn TA, Platania N, Grygar C, Daniel N, Wolpl A, Langen H, Horejsi V, Vogt AB. Tetraspan microdomains distinct from lipid rafts enrich select peptide-MHC class II complexes. Nat Immunol. 2002;3(1):61–8. doi: 10.1038/ni750. [DOI] [PubMed] [Google Scholar]
- 99.Vogt AB, Spindeldreher S, Kropshofer H. Clustering of MHC-peptide complexes prior to their engagement in the immunological synapse: lipid raft and tetraspan microdomains. Immunological reviews. 2002;189:136–51. doi: 10.1034/j.1600-065x.2002.18912.x. [DOI] [PubMed] [Google Scholar]
- 100.Viola A, Schroeder S, Sakakibara Y, Lanzavecchia A. T lymphocyte costimulation mediated by reorganization of membrane microdomains. Science. 1999;283(5402):680–682. doi: 10.1126/science.283.5402.680. [DOI] [PubMed] [Google Scholar]
- 101.Rocha-Perugini V, Sánchez-Madrid F, del Hoyo GM. Function and dynamics of tetraspanins during antigen recognition and immunological synapse formation. Frontiers in immunology. 2015;6 doi: 10.3389/fimmu.2015.00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vrljic M, Nishimura SY, Moerner WE, McConnell HM. Cholesterol depletion suppresses the translational diffusion of class II major histocompatibility complex proteins in the plasma membrane. Biophys J. 2005;88(1):334–47. doi: 10.1529/biophysj.104.045989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stone MB, Shelby SA, Núñez MF, Wisser K, Veatch SL. Protein sorting by lipid phase-like domains supports emergent signaling function in B lymphocyte plasma membranes. eLife. 2017;6:e19891. doi: 10.7554/eLife.19891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310(5751):1191–3. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 105.Yu CH, Wu HJ, Kaizuka Y, Vale RD, Groves JT. Altered actin centripetal retrograde flow in physically restricted immunological synapses. PLoS ONE. 2010;5(7):e11878. doi: 10.1371/journal.pone.0011878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hsu CJ, Hsieh WT, Waldman A, Clarke F, Huseby ES, Burkhardt JK, Baumgart T. Ligand mobility modulates immunological synapse formation and T cell activation. PLoS One. 2012;7(2):e32398. doi: 10.1371/journal.pone.0032398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, Carter R, Justement LB, Bruckbauer A, Batista FD. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 2013;38(3):461–74. doi: 10.1016/j.immuni.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 108.Ketchum C, Miller H, Song W, Upadhyaya A. Ligand mobility regulates B cell receptor clustering and signaling activation. Biophys J. 2014;106(1):26–36. doi: 10.1016/j.bpj.2013.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.DeMond AL, Mossman KD, Starr T, Dustin ML, Groves JT. T cell receptor microcluster transport through molecular mazes reveals mechanism of translocation. Biophys J. 2008;94(8):3286–92. doi: 10.1529/biophysj.107.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smoligovets AA, Smith AW, Wu HJ, Petit RS, Groves JT. Characterization of dynamic actin associations with T-cell receptor microclusters in primary T cells. J Cell Sci. 2012;125(Pt 3):735–42. doi: 10.1242/jcs.092825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo M, Ehrlicher AJ, Jensen MH, Renz M, Moore JR, Goldman RD, Lippincott-Schwartz J, Mackintosh FC, Weitz DA. Probing the stochastic, motor-driven properties of the cytoplasm using force spectrum microscopy. Cell. 2014;158(4):822–32. doi: 10.1016/j.cell.2014.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466(7303):263–6. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Betz T, Koch D, Lu YB, Franze K, Kas JA. Growth cones as soft and weak force generators. Proc Natl Acad Sci U S A. 2011;108(33):13420–5. doi: 10.1073/pnas.1106145108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat Cell Biol. 2008;10(12):1393–400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fabrikant G, Gupta S, Shivashankar GV, Kozlov MM. Model of T-cell nuclear deformation by the cortical actin layer. Biophys J. 2013;105(6):1316–23. doi: 10.1016/j.bpj.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]