Abstract
The ventral telencephalon is the developmental origin of the basal ganglia and the source of neuronal and glial cells that integrate into developing circuits in other areas of the brain. Radial glia in the embryonic subpallium give rise to an enormous diversity of mature cell types, either directly or through other transit-amplifying progenitors. Here, we review current knowledge about these subpallial neural stem cells and their progeny, focusing on the period of neurogenesis. We describe their cell biological features and the extrinsic and intrinsic molecular codes that guide their fate specification in defined temporal and spatial sequences. We also discuss the role of clonal lineage in the organization and specification of mature neurons.
Keywords: neural progenitors, cell fate specification, ganglionic eminences
Introduction
The subpallial structures of the cerebrum (basal ganglia, centromedial extended amygdala, septum, preoptic region) are originated from the ventral half of the telencephalon during embryonic development [1]. The embryonic subpallium is also the source of all cortical interneurons [2], which must undergo a long tangential migration before settling into and refining developing cortical circuits [2, 3].
The ventral telencephalon is subdivided into four major anatomical regions, which are the source of specific mature structures [Fig. 1]. Around the 12th embryonic day (E12) in the developing mouse brain, two ganglionic eminences, swellings of the neuroepithelium protruding into the lateral ventricles, can be distinguished in rostromedial positions along the rostrocaudal axis of the telencephalon [Fig. 1A,B]. The dorsal-most one, located adjacent to the developing neocortex, is known as the lateral ganglionic eminence (LGE), and is the origin of the striatum. The ventral-most one is located medially with respect to the LGE, and is thus known as the medial ganglionic eminence (MGE); pallidal structures (globus pallidus, ventral pallidus) are mostly derived from this region. Immediately ventral to the MGE lies the anterior entopeduncular area (AEP), composed of the septal anlage in rostromedial locations, and the embryonic preoptic area (PoA), positioned more caudally; both of these structures give rise to their mature counterparts. The LGE and MGE fuse into a third proliferative region, the caudal ganglionic eminence (CGE), towards the caudal end of the telencephalon [Fig. 1A,B] [1]. The MGE, CGE and PoA are the source of most GABAergic neurons of the cerebrum [4, 5] which, like most cells derived from the ventral telencephalon, need to undergo substantial migration before reaching their final destination and integrating into developing circuits [3] [Fig. 1A,B].
Figure 1.
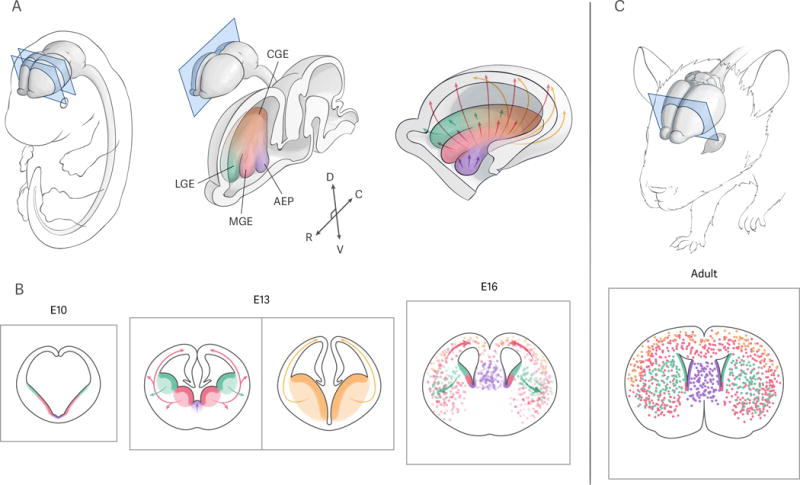
Subpallial germinal zones and their cellular output. A: left, view of a mid-neurogenesis mouse embryo, with the central nervous system shaded in gray; middle, sagittal section through the embryonic brain, highlighting the subpallial proliferative regions: lateral ganglionic eminence (LGE, green), medial ganglionic eminence (MGE, red), caudal ganglionic eminence (CGE, yellow), and anterior entopeduncular area (AEP, purple), with rostro-caudal (R-C) and dorso-ventral (D-V) axes as represented; right, medial view of an “open” right forebrain hemisphere, with the regions color-coded as indicated before. Arrows depict the main migratory routes taken by neurons derived from each germinal zone. B: coronal sections through the forebrain of mouse embryos at early (left, approximately E10), mid- (middle, approximately E13), and late (right, approximately E16) stages of subpallial neurogenesis; the middle panel shows two sections at rostromedial and caudal levels, representing the positions indicated by the coronal planes in the left panel of A. Color code as in A, with proliferative regions in solid colors and cells derived from them in dotted pattern; arrows represent the main directions of migration from the corresponding germinal zones. C: Allocation of subpallium-derived neurons in the adult brain (top), as viewed on a coronal section at the level indicated (bottom); adult neural stem cells line the ventricle (solid colors), and mature neurons derived from the embryonic subpallial germinal zones (dots) occupy different structures throughout the forebrain. Color code as in A and B.
As in other parts of the developing central nervous system, the initial population of neuroepithelial cells in the subpallium differentiates into radial glia (RG); this transition occurs around embryonic day 10 in the telencephalon[6]. RG are the origin of all neurons and glia generated in the ventral telencephalon, either directly or through other neural progenitor cell types subsequently derived from them [7] [Fig. 2C]. The cell bodies of radial glia are located in the ventricular zone (VZ), a germinal region lining the ventricle; along development, new types of progenitors are generated and establish a new proliferative area immediately basal to the VZ, the subventricular zone (SVZ) [Fig. 2]. The SVZ soon overtakes the VZ as the main site of cell proliferation within the ganglionic eminences [8].
Figure 2.
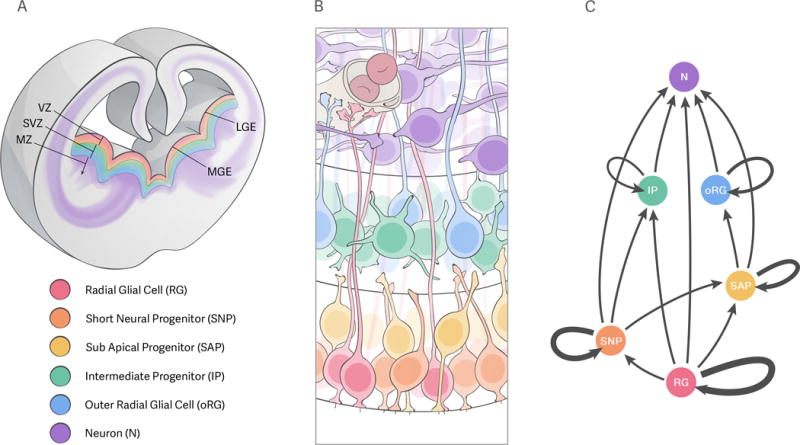
Proliferative regions and progenitor types in the ganglionic eminences. A: coronal section through rostromedial-medial levels of a mid-neurogenic mouse forebrain, depicting the main proliferative regions in the lateral and medial ganglionic eminences (LGE and MGE, respectively): the ventricular zone (VZ, warm colors) and subventricular zone (SVZ, cold colors) are the source of neurons (purple) that accumulate in the marginal zone (MZ) or migrate away from it, as represented by the purple traces. B: cartoon representing the different progenitor types potentially present in the ganglionic eminences. The apical (ventricular) surface of the tissue is down, and the basal side is up – notice that this is inverted from the depiction in A, as is standard in the field. The proliferative regions are not drawn to scale. The cell bodies of radial glia (RG, red), short neural precursors (SNP, orange) and subapical progenitors (SAP, yellow) reside in the ventricular zone; RG extend their basal processes beyond this region, to contact either blood vessels (gray structure on top left) or the pial surface (not represented). Outer radial glia (oRG, blue) and intermediate progenitors (IP, green) reside in the subventricular zone. Neurons (N, purple) born from these progenitors move into the marginal zone, where they will undergo radial and/or tangential migration to reach their final positions within the brain. C: Lineage of progenitors in the ganglionic eminences. Each progenitor type (color and letter code as in A and B) is represented correlative with its position within the proliferative zones as depicted in B. Arrows represent potential outcomes of the division of each progenitor type; the thickness of the looped arrows represents the self-renewal capacity of each progenitor type.
Neurogenesis is defined as the period of development during which neurons are generated from progenitor cells located within germinal zones. In the ventral telencephalon of the mouse, neurogenesis covers most of the second half of embryonic development (approximately from E10 to E16/17), largely overlapping with cortical neurogenesis [9]. This period overlaps partially with the subsequent process of gliogenesis, or generation of glial cells [10], which is still largely understudied in the ventral telencephalon and lies beyond the scope of this review [11].
An enormous diversity of cell types is generated from subpallial progenitors. This diversity is determined along spatial and temporal axes. The spatial axis refers to the different germinal zones and smaller subdomains within each structure, associated with different cellular outputs [12]. The temporal axis refers to the production of different cell types from subpallial progenitors in defined sequences along development, particular to each germinal area; this is analogous to the inside-out pattern of neuronal generation in the cortex [13]. For example, the LGE produces striatal medium spiny neurons following distinct spatiotemporal gradients [14], with neurons in the patch compartments being born earlier (E12–13) than those in the matrix (E13–16) [15–17]. Likewise, cortical interneurons derived from the MGE are destined for distinct cortical layers according to their time of generation, largely following the inside-out pattern characteristic of cortical pyramidal cells [18–20]. Neurons in other regions of the basal ganglia also acquire different positional and functional identities depending on their birth date, such as those in the accumbens (shell neurons are generated first, followed by core ones [21]); globus pallidus (the MGE-derived subpopulations of neurons are born earlier, followed by the LGE-derived ones [22]), and septum (formed following an “onion-skin-like” early- to late-born inside-out pattern [23]).
Much research has focused on the developing neocortex; as a result, the diversity and cell biology of cortical progenitors, the relationships between them and the molecular control and spatial organization of their cellular output are beginning to be understood [24, 25]. However, comparatively little is known about these aspects of ventral telencephalon development, with the majority of studies published so far focusing on the MGE and the production of GABAergic forebrain interneurons [7, 26]. In this review, we will discuss the cell biology of subpallial radial glia and the different progenitor types derived from them. We will focus on studies performed in vivo, most of which have used mice as experimental models. We will summarize the extrinsic and intrinsic mechanisms of molecular specification guiding the production of different mature neuron types from those progenitors, paying special attention to the transcriptional circuits underlying cell fate decisions. Finally, we will describe recent advances towards understanding the role of clonal lineage in the organization of neural circuits, i.e., how cortical interneurons derived from the same subpallial radial glial cell are distributed within the mature brain.
Cell biology of radial glia in the ventral telencephalon
Radial glia in the ventral telencephalon share the main cell biological characteristics and marker expression profiles of those in other parts of the developing central nervous system [6]. They have a bipolar morphology, with an apical process that maintains contact with the ventricular surface, and an elongated basal process reaching beyond the ventricular zone [Fig. 2B, red]. In ventral radial glia, the apical junctional complexes and their surroundings contain classic polarity proteins such as ß-catenin, N-cadherin, Numb or ZO-1 [27–29]. However, the loss of polarity protein Lgl1 causes a dramatic loss of polarity specifically in radial glia located in the ganglionic eminences [28], suggesting that there might be differences in polarity determinants between dorsal and ventral RG. These processes are kept throughout the cell cycle, during which RG perform an evolutionarily conserved movement known as interkinetic nuclear migration (INM), whereby their nuclei undergo S-phase at the basal end of the VZ and mitosis at the ventricular surface, moving between these locations through G1 and G2 [8, 30–32]. The duration of the different phases of the cell cycle of subpallial progenitors could presumably play an important role in their expansion and the diversity of their neuronal output, as it does in the developing cortex [33, 34]. As early as E10.5, progenitors in the MGE have a slightly longer cell cycle than those in the LGE, possibly reflecting an earlier onset of neurogenesis [35]. The cell cycle of slightly later (E11–E12) LGE progenitors in the VZ was studied in greater detail, and found to be longer than that of cortical progenitors at the same stages, with S-phase taking up approximately half of the total cell cycle length and a trend towards overall cell cycle lengthening along development [36]. Later studies confirmed that the SVZ of the LGE contained a separate progenitor population with similar cell cycle parameters [37]. Recently, live imaging experiments demonstrated that the progeny of RG in the LGE have progressively shorter cell cycles, presumably as an additional mechanism to amplify the output of single progenitors [31]. Changes in cell cycle parameters have significant effects on the cellular output from subpallial progenitors: in the MGE, the loss of G1/S phase-specific cyclin D2 in the MGE decreases SVZ proliferation and increases cell cycle exit, causing a reduction in the number of a subgroup of late-born cortical interneurons (parvalbumin-positive: see below) [38]. Likewise, the length of S-phase of LGE progenitors is decreased as they undergo a major developmental transition late in striatal neurogenesis, switching from producing striatonigral to striatopallidal medium spiny neurons [39]. These findings suggest that cell cycle regulation is important for the proliferation and differentiation of subpallial progenitors [40], and highlight the need for further studies taking into account the diversity of germinal zones and progenitor types in the subpallium. Throughout neurogenesis, cortical radial glia maintain contact with the basement membrane of the pia (the innermost layer of the meninges), through their endfeet, small ramifications branching out from the tip of their basal processes. In contrast, most basal processes of RG in the ganglionic eminences do not reach the pia [14, 41], but rather contact blood vessels in the SVZ or MZ [29, 42] [Fig. 2B]. The basal processes of RG in the MGE progressively lose contact with the pial basement membrane as the tissue expands during development. Except for early-born RG, which appear to preserve the pial contact, the basal endfeet of RG in the GE display very dynamic behavior, actively searching for and wrapping around periventricular or marginal zone vessels; vascular anchorage seems to be critical to maintain normal numbers of progenitor divisions and to preserve their neuronal output [29]. A similar process is observed in the dorsal telencephalon of the developing human brain, where radial glia switch basal contact from the pia to blood vessels as the greatly enlarged SVZ of the human cortex expands [43]. Future research will be necessary to clarify whether the switch in endfeet attachment is a consequence of SVZ expansion, a reflection of changes in RG identity, or both.
RG give rise to other progenitor types
During early stages of neurogenesis in the ganglionic eminences, most progenitor divisions (presumably from RG) occur at the ventricular surface. As development proceeds, some cells start dividing at basal locations, marking the appearance of the subventricular zone (around E11 in the MGE, and E12 in the LGE [8], which progressively increases in size and number of mitotic cells, until it eventually overtakes the VZ as the main site of progenitor proliferation around mid-neurogenic stages (E13–E14: [8, 14, 31]) [Fig. 2A]. The expansion of the SVZ is associated with an increase in the variety of progenitor types present in the ganglionic eminences, which in turn might be related to the production of different neuron subclasses [40].
Radial glia are the source of a variety of additional progenitor types in ventral telencephalic structures, and thus give rise, either directly or indirectly, to all subpallium-derived neural cells [Fig. 2]. Radial glia divisions in the ganglionic eminences display a cleavage plane largely perpendicular to the ventricular surface throughout neurogenesis [8, 14, 27]. Direct observation of RG behavior through time-lapse imaging confirmed that they mostly undergo asymmetric, self-renewing divisions at mid-neurogenesis, usually giving rise to a new RG and another progenitor, both in the LGE [27, 31] and the MGE [30, 44]. Four additional types of progenitors, with diverse proliferative potentials, have been described so far in the ganglionic eminences: short neural progenitors (SNPs), subapical progenitors (SAPs), intermediate progenitors (IPs), and outer radial glia (oRG) [Fig. 2B]. So far, all of these progenitors appear to have the capacity to self-renew and/or generate neurons directly, but they can be broadly categorized into more “stem-like”, such as SNPs and oRG, and more “transit-amplifying”, in the case of SAPs and IPs. The former normally undergo several rounds of cell division, usually self-renewing or generating other progenitors, while the latter tend to give rise to neurons by undergoing terminal divisions, either directly or after one or few rounds of amplification [Fig. 2C]. However, this distinction does not appear to be too sharp; the hierarchy of these progenitors is likely to be only loosely directional, as in the cortex of gyrencephalic mammals [45] [Fig. 2C]. Progenitor diversity and behavior in other ventral germinal zones, such as the CGE, the preoptic area or the septal anlage, have not been studied in detail so far.
Other VZ progenitors
Short neural precursors [Fig. 2B, orange] display an apical process contacting the ventricular surface and a short basal process that does not reach the pial surface and is retracted before mitosis. These cells were initially described in the developing neocortex [46], where they typically undergo a single round of division at the ventricular surface, giving rise to two neurons [47]. Recent studies have found evidence for the presence of SNPs in the LGE [31] and MGE [48]. In the MGE, SNPs seem to exhibit a slight bias towards neurogenic divisions, bypassing the production of IPs [48]; however, live imaging experiments performed in the LGE found that they normally undergo proliferative divisions, self-renewing and/or giving rise to further transit-amplifying progenitors (but not to RG), and rarely terminal neurogenic divisions [27, 31]. While most RG in the VZ display division planes nearly perpendicular to the ventricular surface, SNP divisions normally display a tilted spindle orientation, resulting in an oblique cleavage plane [8, 14, 27]. Randomization (both constitutive and acute) of spindle orientation in the mid-neurogenic LGE reduces the number of asymmetric self-renewing RG divisions, increasing the production of SNPs at the expense of RG without affecting the apical vs. non-apical progenitor ratio or the polarity of VZ progenitors [27]. This suggests that naturally occurring changes in the spindle orientation of RG at specific stages of development could lead to their transition to other progenitor types.
Progenitor cells in the LGE undergoing mitosis at basal locations within the VZ, rather than at the ventricular surface, were first described forty years ago as “subsurface ependymal mitoses” [8]. These cells were recently described as VZ progenitors that maintain an apical process in contact with the ventricular surface throughout their cell cycle, with or without a basal process; they divide at non-apical locations, and were thus named subapical progenitors [27, 31] [Fig. 2B, yellow]. SAPs do not seem to have the ability to generate RG or SNPs (while both of these progenitor types can generate SAPs), but rather to self-renew or give rise to SVZ progenitors such as IPs and oRG (see below), suggesting that they occupy a middle ground in the transit-amplifying progenitor lineage within the LGE [31].
SVZ progenitors
Ventral intermediate progenitors appear similar to those in the cortex: they display a multipolar/non-polar morphology, without any distinct apical or basal processes, and generally undergo a single round of terminal division in the SVZ, generating two cells with neuronal morphology and behavior [30, 31, 44] [Fig. 2B, green]. This, together with the fact that they can be generated from any of the progenitors described so far, suggests that they could be the last step within the progenitor lineage of the ganglionic eminences. However, there have also been reports of self-renewing IPs in rodent ganglionic eminences [31, 44], although the overall frequency of this type of division has yet to be determined.
Outer radial glia (oRG, also known as basal radial glia), monopolar progenitors that maintain a basal process, but no apical contact, throughout their cell cycle, have recently been described in the LGE [31] [Fig. 2B, blue]. oRG have the capacity to self-renew, and are usually derived from the basal daughters resulting from asymmetric divisions of bipolar SAPs [31]. The presence of abundant oRG and related progenitors seems to be highly correlated with SVZ enlargement in the cortex of gyrencephalic species [49]. SAPs, which give rise to oRG in the LGE, are present in the cortical VZ of gyrencephalic animals, but virtually absent in lissencephalic species, drawing interesting parallels with the ganglionic eminences of the mouse (see below).
Progenitor types in the ganglionic eminences, expansion of the proliferative zones and daughter cell fate
As discussed above, radial glia are the start of the progenitor “lineage tree” in the ganglionic eminences [Fig. 2C]. Along the period of subpallial neurogenesis, they generate a diversity of additional progenitors, presumably through increasingly asymmetric cell divisions. In general terms, RG give rise to SNPs, which in turn generate SAPs, which are a source of oRG and IPs; this sequence, however, seems to be quite flexible, in that more committed progenitors (such as IPs) can be derived from any of the more stem-like (and more RG-like) progenitor types [31] [Fig. 2C]. The current knowledge of the field is quite limited: most of the progenitor types described here, as well as their behavior and morphology, have been described very recently, and almost exclusively during mid-neurogenic stages [27, 30, 31, 44]. A recent study reported that within the MGE, SNPs and IPs are biased to produce two different subtypes of cortical interneurons (expressing the mutually exclusive markers somatostatin and parvalbumin, respectively) [48], in line with a previously suggested cell-cycle related mechanism that predicted such a VZ/SVZ bias [40]. While this study focuses only on one developmental stage, and does not account for the possible generation of neurons from progenitor types other than SNPs and IPs [48], it raises the interesting possibility that the diversification of MGE progenitors might be correlated with the extremely diverse cellular output from this region.
An increase in progenitor diversity has been correlated with the evolutionary expansion of the SVZ in the dorsolateral telencephalon of primates, ultimately leading to the enhanced complexity and computational power of their mature neocortex [45]. This suggests that the expansion (evolutionary and/or ontogenic) of the SVZ within any area of the developing brain might require an increase in the numbers and/or diversity of transit-amplifying progenitors. This is consistent with the recently discovered presence of a great variety of progenitors (including SNPs, SAPs, oRG, and IPs; Fig. 2B,C) in the ganglionic eminences of the mouse, where the SVZ largely outsizes the VZ throughout most of the neurogenic period [8, 14]. More progenitor types might be described in the rodent subpallium in the near future, such as the bipolar and monopolar RG recently described in the developing neocortex of gyrencephalic species [31] including primates [50], or proliferative IPs similar to those found in the MGE of primates [51]. Studying the different transitional forms and lineage hierarchy among these progenitors is likely to be fundamental to understand the extremely diverse cellular output derived from the developing subpallium [45]. The rodent ganglionic eminences could thus provide an unexpected model to study the evolution of SVZ expansion that might underlie the increase in size of specific brain areas in many vertebrate species [52]. In further support of this idea, the greatly expanded OSVZ in the MGE of primates contains at least one additional subtype of nonpolar progenitors, which could be similar to the proliferative IPs described in the mouse [51, 53].
Future research should address the full extent of this progenitor diversity across developmental stages (for example, the effect of spindle randomization on VZ progenitors in the LGE seems to be highly stage-dependent [27] and ventral telencephalic areas (remarkably, very little is known about the types of progenitors present in the AEP and their proliferative behavior, other than the fact that the PoA seems to lack a SVZ [5]). It would be particularly informative to study the molecular events guiding the progression between different progenitor types (for example, the production of SAPs and IPs seems to depend on the expression of the transcription factor Ascl1 [31]).
Extracellular signals and cell fate determination
As in other parts of the developing embryo, extracellular cues play fundamental roles in the specification of progenitors in the ventral telencephalon. Morphogens are diffusible molecules secreted from defined locations, or organizing centers, within a developing tissue. They act as fate determinants in a concentration-dependent manner, such that progenitor cells will respond to a morphogen according to their specific location along its diffusion gradient. In the mouse embryo, the telencephalon is specified into dorsal and ventral domains before the onset of neurogenesis, and further patterned into distinct progenitor regions through the action of multiple morphogens secreted from different organizing centers [54–56]. The environment or niche in which neural progenitors are located plays a fundamental role in their cell fate specification, providing additional signals from diverse non-neural cell types [57]; while this has been widely researched in other parts of the developing CNS, little is known about the ventral telencephalic niche. We will briefly describe the main extrinsic factors that have been shown to have key functions in the context of ventral subpallial development, focusing on their effects on progenitor proliferation and fate specification.
Sonic hedgehog (Shh), a secreted protein from the hedgehog family, is critical for the formation of ventral telencephalic structures [58, 59]. Shh specifies ventral telencephalic progenitors in a temporally defined manner [60]: early ablation of Smoothened, the canonical downstream effector of Shh, in the telencephalon, completely abrogates the formation of ventral structures and cells derived from them [61], while its deletion in the entire CNS after the onset of neurogenesis has a milder effect, causing a reduction in MGE size but largely preserving ventral patterning [62]. Shh is initially produced by cells in the ventral midline of the developing telencephalon [59]. As development proceeds, postmitotic neurons in the mantle zone of the dorsal part of the MGE (dMGE) start secreting this protein as well [63]. Progenitors in the dMGE respond to the presence of Shh, activating downstream targets [64, 65] that ensure correct cell fate specification. In the MGE, Shh is required to maintain a balanced output of distinct interneuron subgroups, through both cell-autonomous [66] and non-cell-autonomous [65] mechanisms, mostly involving the transcription factor Nkx2.1 (see below).
Retinoic acid (RA), a metabolite of vitamin A, is a morphogen known to play a critical role in CNS development [67], especially in the cortex [68]. On the ventral side of the telencephalon, components of the retinoic acid pathway are highly expressed in the developing striatum [69]. In the LGE, RA and its receptors are expressed by radial glia [70] and SVZ/MZ cells [71] from E11 and throughout the neurogenic period. Local synthesis of RA in the ventral telencephalon does not seem to play a role in patterning the ventral telencephalon [72], but it is critical for the specification of LGE-derived GABAergic neurons [73], including the DARPP32-positive medium spiny neurons of the striatum, in a process regulated by the transcription factor Gsx2 [70, 74]. The detection of RA by different receptors might underlie the differentiation of LGE progenitors into specific subtypes of striatal neurons [75, 76].
Wnt is another family of morphogen proteins critical for early telencephalon patterning, secreted from the dorsomedial-most aspect of the developing forebrain, or cortical hem [77]. The canonical Wnt signaling pathway acts through the transcriptional coactivator ß-catenin. Loss of ß-catenin before the onset of neurogenesis leads to severe dorsoventral patterning defects, causing ventralization of the telencephalon; if the loss occurs after E11.5, no major patterning defects are observed [78]. This pathway is involved in maintaining the MGE/PoA progenitors in the cell cycle, since ß-catenin deletion in this region greatly decreases their proliferation, without seemingly altering the fate of differentiated cells derived from it [79]. Furthermore, the Wnt pathway is at least partially responsible for cell fate switches in progenitors in the MGE, where the Wnt3a protein acts through the Ryk receptors expressed in nestin-positive radial glia to promote neuronal over oligodendroglial differentiation [80].
Fibroblast growth factors (FGFs) are another family of growth factors critically involved in telencephalon determination (mice without FGF receptors fail to develop a telencephalon [81] and patterning [82]. Two main FGF ligands, Fgf8 and Fgf15, act downstream of SHH via FoxG1 to promote regional MGE and CGE identity, respectively [83, 84]. Loss of Fgf8 reduces proliferation in ventral structures as early as E9, causing a great decrease in the expression of Shh and other ventral markers by E12.5, with the consequent dorsalization of the telencephalon [85]. Similar dorsalizing effects were observed when FGF receptors 1 and 3 were deleted at the same time, in a process downstream but independent of SHH signaling [86]. Beyond patterning, the FGF pathway acts to maintain the radial glia-like identity and proliferation of progenitors in the ganglionic eminences, either through Notch activity [87] or through specific adaptor proteins called FRS, which are dispensable for the formation of the MGE but necessary for the proliferation of cells in its VZ and especially SVZ, which might reflect different effects of FGF signaling on different progenitor types [88]. This pathway seems to be critical for the formation of the septum: cells derived from the partially overlapping Fgf8- and Fgf17-expressing lineages contribute to form the mature septum [89], which is completely absent in mutants lacking the former gene [85].
It is highly likely that other types of signaling events take place in concert with these pathways to convey extrinsic information to subpallial progenitors, contributing to establish the fate and behavior of their progeny [57, 90, 91]. However, at present there are still few published studies addressing either short-range interactions, such as those mediated by Notch signaling [16, 92], attractive/repulsive signals like Slit/Robo [93] or direct contact of the basal process of radial glia with blood vessels [29], or long-range signaling by other diffusible factors such as TGF-ß [94], EGF [95], BMPs [96], or other cytokines [97] in the context of the ventral telencephalon. More research will be necessary to understand the effect of these and other extrinsic cues on different progenitor types within distinct progenitor zones, their potentially dynamic contributions to cell fate specification across developmental stages, and especially the interactions between different pathways [55].
Intrinsic determination of subpallial cell fate
Throughout the developing nervous system diversity arises from spatial and/or temporal differences in the specification of neural progenitors. This is classically illustrated in the developing spinal cord, where combinatorial codes of homeodomain transcription factors define spatially segregated progenitor domains, which generate different neural cell types along development [98, 99]. Transcription factor codes appear to correspond to both anatomical boundaries and the fate potential of spatially restricted ventral telencephalic radial glia and progenitors. Several transcription factors are broadly expressed in all subpallial progenitor zones responsible for the production a wide diversity of neural cell types [Fig. 3A]. This is the case of homedomain Dlx genes [100, 101], the proneural bHLH transcription factor Ascl1 (the expression of which appears to be restricted to more differentiated progenitors rather than radial glia [102] or the bHLH transcription factor Olig2, which is often co-expressed with Dlx2 and Ascl1 [103].
Figure 3.

Transcriptional codes guiding cell fate specification in the ganglionic eminences. A: coronal section through rostromedial-medial levels of the left cerebral hemisphere of a mid-neurogenesis mouse embryo. Different transcription factors are expressed in distinct spatial domains throughout the lateral and medial ganglionic eminences (LGE and MGE, respectively) and septal anlage (Sep), as represented by the color patterns (see legend at the bottom of the panel). The complex interplay of different fate determinants controls the production of an enormous diversity of neural cell types from these regions. B: Proposed “transcriptional logic” of lineage progression in the LGE (top) and the MGE (bottom), whereby the levels of expression of different transcriptional regulators (represented by the curves; same colors as in A) guide the transition of the progenitor lineage through its different cell types, represented by silhouettes (legend at bottom; see Fig. 2) and located in the areas represented below the lineage axis (VZ: ventricular zone; SVZ: subventricular zone; MZ: marginal zone) within each proliferative region. The exact molecular mechanisms informing the transition between different progenitor types or the progeny they can produce are still largely unknown.
While there has been significant progress in identifying region-specific patterns of gene expression, translating these patterns into a combinatorial transcription factor “code” for cell fate specification has been challenging [104]. One issue is that many of the subpallium-specific transcription factors studied so far are expressed in gradients; thus, the effects of these factors on cell fate specification are likely to depend not only on their expression in a particular location, but the level at which they are expressed across a given spatial axis [56, 105]. Another issue is that besides the spatial bias guiding the fate of radial glia and other progenitors, there is a precise temporal sequence dictating when distinct subgroups of neurons and glial cells are produced. Here, we will summarize the current knowledge about the fate specification of progenitors in the different subpallial germinal zones [Fig. 3], and try to describe the “molecular logic” that is beginning to arise from recent studies [12, 106, 107].
LGE
The lateral ganglionic eminence is the primary source of medium spiny GABAergic projection neurons of the striatum and adult neural stem cells that will produce the majority of postnatal olfactory bulb interneurons in rodents. Isl1 and Sp9 appear to be differentially expressed in the early-born striatonigral and late-born striatopallidal lineages respectively [108, 109]. The zinc-finger protein Helios controls the transition of LGE progenitors through the production of these two lineages [39]. The dorsal portion of the LGE (dLGE) lies immediately adjacent to the cortex, ventral to the pallial-subpallial boundary, and is characterized by expression of Gsx2, Er81 and low levels of Pax6. Pax6 and Gsx2 appear to work cooperatively during patterning to set the pallial-subpallial boundary [110]. The ventral domain of the LGE is distinguished by the expression of Gsx2 and Nkx6.2 [Fig. 3A]; whether there are functional differences between neurons derived from dLGE and vLGE is still unclear [111]. Recent clonal lineage fate mapping experiments suggest that striatal projection neurons and olfactory interneurons are derived from common radial glial cells prior to E13 in the mouse [112].
MGE
A wide variety of GABAergic neurons are born from progenitors located in the MGE. This region is the main source of cortical interneurons, which can be categorized into two broad but non-overlapping subgroups distinguished by their expression of parvalbumin (PV) or somatostatin (SST), which encompass a collection of more distinct interneuron subtypes (for example, the late-born Chandelier cells, a subset of which are PV+ [113]). The expression of these histological markers also corresponds with distinctive physiological properties: PV+ interneurons are fast spiking, while SST+ neurons are primarily regular spiking and low threshold spiking cells [114]. Nkx2.1 is a homeodomain transcription factor expressed in progenitors and subsets of postmitotic neurons in the MGE, PoA and part of the septal anlage [101]. Loss of Nkx2.1 in the MGE causes cells to adopt more dorsal (LGE- and CGE-like) fates [115, 116]. Nearly all of the cholinergic pallidal projection neurons derived from the ventral MGE and POA are also lost in Nkx2.1 mutants [116, 117]. These findings have led to the idea that Nkx2.1 functions as a master regulator of MGE progenitor cell identity; however, this transcription factor alone is not sufficient to account for the specification of the wide diversity of GABAergic neurons derived from the MGE. There is evidence that the MGE can be divided along its dorsoventral axis into at least two distinct spatial domains that show clear biases in the production of SST and PV interneuron subgroups, with progenitors in the dorsal MGE (dMGE) biased toward the production of SST+ interneurons, and those in the ventral MGE (vMGE) towards PV+ subtypes [65]. Nkx6.2 is expressed in the dMGE progenitor zone and extends dorsally across the interganglionic sulcus, into the ventral LGE [Fig. 3A], and caudally to the ventral CGE [111, 118, 119]. While a wide diversity of interneuron subgroups is derived from the Nkx6.2 progenitor domain, there is a bias towards the production of SST+ interneurons in this region [111]. This is further substantiated with transplantation studies, where fluorescent labeled dMGE precursors transplanted into recipient animals primarily develop into SST+ interneurons [120]. Conversely, transplantation of ventral MGE (vMGE) precursors yields mostly PV+ subtypes [120].
CGE
The caudal ganglionic eminence is the second largest source of cortical interneurons, producing subtypes that express serotonin receptor 3c (5-HTR3c), together with reelin (RLN), calretinin (CR) and/or vasointestinal peptide (VIP) [121]. Fate mapping and transplantation studies have shown that most neurogliaform and bipolar GABAergic cortical interneurons are derived from the CGE [122]. Despite the different fates of neurons originating from the CGE, a distinct transcription factor code defining this region has thus far been elusive: molecularly, the CGE is defined by its expression of pan-subpallial transcription factors and the lack of Nkx2.1 [11]. Many of the genes that have been shown to be involved in the specification of CGE interneurons have overlapping patterns of expression in other GE domains; these include Gsx2, which is highly enriched in the LGE, and Prox1, which is also expressed in MGE and LGE progenitor zones [123, 124]. Prox1 expression, however, is maintained in CGE-derived interneurons throughout their maturation, while it shuts down in the MGE-derived ones shortly after they leave the cell cycle [124].
PoA
The preoptic area (PoA) produces approximately 10% of the total population of cortical interneurons, in subtypes largely overlapping with those derived from other regions, including PV+, SST+ and RLN+ interneurons, as well as the majority of the cholinergic neurons in the globus pallidus and striatum [117, 125, 126]. Like the neighboring MGE, the PoA also expresses Nkx2.1, but only low levels of Olig2 [5]. The anatomical structure of the PoA is unique from the MGE and LGE in that it lacks a discernable subventricular zone, suggesting there may be key differences in the composition of progenitor types and the patterns of cell division in these regions [5]. Nkx5.1 and Dbx1 appear to demarcate two distinct progenitor domains within the PoA [125]. However, PoA-derived PV+ and SST+ neurons are restricted to deep cortical layers, suggesting they may have important functional differences with their MGE-derived counterparts [125].
Septum
The embryonic subpallial septum is the source of the GABAergic and cholinergic projection neurons that will make up the medial and lateral divisions of the mature septum and the diagonal band nuclei [23, 127]. The progenitors of the septal anlage express pan-subpallial transcription factors (Dlx, Ascl1, Olig2, Vax1) [104, 128]; interestingly, the loss of Vax1 results in complete ablation of the septum while only partially depleting cells derived from other ventral proliferative regions, suggesting that this factor plays a key role in septal development [129]. The septal anlage is characterized by the expression of transcription factors from the Zic family that are required for its proper development [130, 131] [Fig. 3A]. Part of the subpallial septum also expresses Nkx2.1 [131], which is required for the production of cholinergic projection neurons in the septum [132]. However, the fate potential of progenitors derived from these discrete molecular domains has only begun to be explored. A small subset of cells in the dorsal medial septum is pallial in origin, as evidenced by their expression of the transcription factors Emx1 and Tbr1; these cells are derived from the cortical hem, a dorsomedial pallial domain that also expresses Zic family transcription factors [1, 101, 130]. Septal progenitors do not produce cortical interneurons, but recent findings suggest that Zic-positive postnatal neural stem cells may descend from embryonic septal progenitors [131, 133].
Differential interpretation of transcription factor codes
The ability of subpallial progenitors to acquire distinct cell fates changes across development, as they transition through a sequence of competence states to produce their diverse neural progeny [12]. This implies that within each spatially distinct subpallial proliferative zone all cell types generated throughout development very likely arise from common radial glial progenitors. As discussed above, the sequential production of different cell populations is a constant theme throughout subpallial development, and is reflected by the different combinations of transcription factors expressed at each stage. For example, striatal patch neurons are produced during early neurogenesis, when Ascl1 is expressed in the proliferative zones of the LGE, while functionally distinct matrix neurons are produced later from Ascl1+, Dlx1/2+ progenitors [134]. However, the generation of the enormous diversity of neuronal and glial output from subpallial progenitor domains is likely to require additional levels of cell fate control, beyond the expression of a relatively reduced set of spatially confined transcriptional regulators. Epigenetic changes in radial glia and their progeny are likely to influence their fate potential, since modifications of chromatin structure can significantly alter the availability of enhancer/promoter regions of genes regulated by a particular transcription factor. Recent work examining the molecular function of NKX2.1 during specification of MGE interneurons beautifully illustrates how the synergistic activity of co-expressed transcription factors along with dynamic changes in chromatin can produced coordinated cell type and stage specific patterns of gene expression [107]. Nkx2.1 in VZ progenitors binds to distal regulatory elements and functions to promote repressive chromatin state. As progenitors differentiate, Nkx2.1 coordinates with the postmitotic transcription factor Lhx6 to promote permissive chromatin states and transcriptional activation of cell type specific gene programs. Future research is necessary to understand the mechanisms that underlie the changes in temporal competence guiding the generation of different progenitor types within a given lineage [Fig. 3B]. This should comprise efforts to understand not only the complex transcriptional codes present in each germinal region [Fig. 3A], but also how they relate to the dynamic changes in chromatin that coordinate cell type and stage specific gene expression programs. Understanding the changes in progenitor types and competence should be an important step towards understanding how the enormous diversity of subpallium-derived neural cell types is generated.
Integration of signals for lineage progression
Despite the growing body of knowledge about the extracellular cues and the intrinsic molecular programs guiding progenitor cell fate specification [55, 135], little is known about how these signals influence each other to ensure that subpallial radial glia produce other progenitors and/or postmitotic cells in the right numbers and following appropriate developmental sequences [90]. Each progenitor subtype within the developing telencephalon has unique cell biological features that can influence its ability to receive extracellular signals. As an example, RG maintain basal and apical processes throughout their cell cycle, which allow them to contact blood vessels and the ventricular surface, respectively [Fig. 2B]. Given the extensive self-renewing potential of these progenitors, it could be speculated that extrinsic cues conveyed through either of those contact sites have an effect on the ability of any progenitor type to proliferate. In line with this idea, other progenitor types that keep contact with either the ventricle (SNPs, SAPs) or blood vessels, either in the SVZ/MZ or in the meninges (oRG), seem to have greater proliferative potential than those that have no contacts with either surface (IPs) [Fig. 2B,C]. This raises the possibility that signals produced in different parts of the developing embryo might guide telencephalic progenitors through their temporally defined competence states. However, recent in vitro experiments have shown that it is possible to produce exclusively GABAergic neurons of different subtypes from pluripotent stem cells by manipulating their cell expression programs, thus proving that cell fate can be directly determined solely by intrinsic mechanisms [136].
Clonal lineage and fate specification
There are numerous examples in the developing nervous system whereby a cell’s lineage is indicative of its ultimate fate and organization within a circuit [137–139]. Tracking the clonal lineage of ventral telencephalic progenitors has been more technically challenging than in cortical progenitors, due to the tremendous migratory capacity of a large proportion of the cells derived from ventral regions and the protracted period during which they acquire their subtype-specific identities. The wide dispersion of cells derived from the same progenitor lineage presents significant problems for the assignment of clonal boundaries and relationships [140]. In the early 1990s there was a major technical advance with the development of a retroviral lineage tracing approach that utilizes a library of retroviruses carrying DNA ‘barcodes’ [141]. These retroviruses infect dividing progenitors and tag all of their subsequent progeny with an unambiguous barcode label, allowing for the assignment of lineage relationships between widely dispersed cells. The first iterations of these retroviral libraries labeled pallial and subpallial progenitors indistinctly, and included a transgene for either beta galactosidase or alkaline phosphatase to identify the location and morphology of the progeny of infected cells [140–143]. In these early studies it was observed that radially distributed clones consisted of cells with pyramidal morphology, while widely dispersed cells had non-pyramidal morphologies [144]. We now appreciate that those radial clones were most likely derived from cortical radial glia, while the widely dispersed clones came from ventral telencephalic radial glia [2, 145]. AP histochemical labeling made it possible to differentiate pyramidal and non-pyramidal cell types by morphology, but precluded further assessment of neuron subtype-specific identities within these two broad categories [144]. The existence of distinct lineages for pyramidal and non-pyramidal neurons was further confirmed with the characterization of lineage-specific transcription factors such as Nkx2.1, Gsx2, Olig2 or Ascl1, which are confined to ventral telencephalic progenitor zones and produce a vast array of non-pyramidal neuron subtypes [105, 119, 146–148]. However, the question of whether diverse GABAergic neuron subtypes are derived from the same progenitor lineage remained unaddressed until recently [18, 30, 44, 149]. Recent technical advances allowed the specific targeting of genetically defined subgroups of ventral progenitors, such as RG in the MGE/PoA, which were labeled either with pseudotyped retroviruses encoding fluorescent protein markers [18, 30, 44, 149] or by sparse labeling through the use of inducible genetic reporters [18, 149]. The progeny of MGE/PoA radial glia could then be studied across the numerous forebrain structures that it populates, although most efforts were focused on the cerebral cortex. The Shi laboratory used low titers of GFP-encoding pseudotyped retroviruses to target MGE/PoA radial glia, studying both the behavior of these progenitors and the fate of their progeny; based on the presence and distribution of GFP-expressing cells in the adult, the authors concluded that interneurons formed horizontal and vertical clusters according to their clonal lineage [30]. This was followed by an elegant study from the Marín lab, where they confirmed and refined the concept of the inside-out layering of MGE/PoA-derived interneurons, suggesting that cells sharing a birth date tend to cluster in the same or adjacent cortical layers [18]. These two landmark publications were followed by concurrent studies by the Fishell group and our own; using newly developed pseudotyped barcoded viruses, these papers showed that MGE/PoA-derived interneurons unequivocally derived from the same RG were distributed widely across the forebrain, rather than forming local clusters within a single structure, suggesting that cells derived from the same clonal lineage are unlikely to have chemical or electrical synaptic circuit relationships [44, 149]. However, the precise role of lineage in the organization of inhibitory circuits is far from settled, and there are many questions that have yet to be addressed, such as whether interneurons derived from the same clonal lineage but located in different forebrain regions share similar functional properties. [150–152]. There was however agreement that most ventral progenitors are not restricted to producing one neuronal subtype exclusively, since PV+ and SST+ interneurons could be derived from the same radial glial cell in the MGE/PoA [18, 30, 44, 149].
While there is consensus about the diversity of fates that can derive from individual clonal lineages, it is still unknown whether there is a strict temporal structure to the production of distinct GABAergic cell types within a single clone. From a cell biological point of view, it would be especially informative to investigate if two different cell types can be derived from the same progenitor undergoing a terminal division, similar to what has been shown for intermediate progenitors in the retina [153], and what mechanisms might guide such precise cell fate specification. Many of the defining characteristics of cortical interneurons are not acquired until weeks into postnatal development, long after cells have settled into their final positions [154]. This raises the interesting possibility that newborn interneurons might maintain some level of plasticity in their fate, depending upon extrinsic factors such as the cellular environment and neuronal activity in the tissue where they are allocated for the fine-tuning of their functional identity [154, 155]. It is possible that clonally related cells share a set of core functional features or a tendency to integrate preferentially into specific circuit motifs in the developing cortex, where their subtype would be further refined in a context-dependent manner. The clonal lineages of subpallial progenitors have not yet been investigated in other germinal zones such as the LGE or the AEP. Clonal relationships could play a much more relevant role in organizing neuronal cell types in regions where they do not need to undergo massive tangential migration to reach their final destination, but still have a clear temporal sequence of production [156]. Presumably, cells born at similar times and located in restricted histological areas, such as the LGE-derived matrix or patch compartments of the mature striatum, could share functionality or connectivity patterns depending on their clonal lineage.
Conclusions and perspectives
As we have discussed here, subpallial radial glia are the source of an extraordinary diversity of neural cell types. This happens through an array of progenitor subtypes derived from RG, the complexity of which is just starting to be understood. Integration of an extensive array of extrinsic signals guides the transition of subpallial progenitors through a series of spatially and temporally defined competence states that will determine the fate of their progeny. The exact mechanisms and any overarching “molecular logic” behind these progressions are not understood. Recent studies aiming to understand the molecular signature of different subpallial regions in order to gain genetic access to specific neural subtypes therein shows great promise for the field [107, 157]. Understanding the basic biology of subpallial radial glial and their progeny is necessary to fully harness their therapeutic potential for modeling and treatment of complex neurological diseases in humans [158, 159]. The many ongoing efforts to utilize subpallial progenitors in transplantation-based regenerative therapies [160] are likely to improve their already promising results [161] if researchers are able to select progenitor sources or target genetic pathways for producing pure populations of neural cell types with the greatest efficacy for the treatment of specific disease conditions.
Acknowledgments
Research in the Harwell laboratory is supported by NIH grants K01NS089720 and R01NS102228. M.T.G. is partially supported by funding from the Ellen R. and Melvin J. Gordon Center for the Cure and Treatment of Paralysis. Figures by Digizyme Inc. (Geoffrey Cheung & Gaël McGill).
References
- 1.Medina LAAA. Subpallial Structures. In: Watson GPC, Puelles L, editors. The Mouse Nervouse System. Elsevier; London; San Diego, CA: 2012. pp. 173–220. [Google Scholar]
- 2.Anderson SA, et al. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278(5337):474–6. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 3.Marin O. Tangential Migration in the Telencephalon. In: Paxinos G, editor. The Rat Nervous System. Elsevier; London; San Diego, CA: 2015. pp. 45–58. [Google Scholar]
- 4.Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7(9):687–96. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- 5.Gelman DM, et al. The embryonic preoptic area is a novel source of cortical GABAergic interneurons. J Neurosci. 2009;29(29):9380–9. doi: 10.1523/JNEUROSCI.0604-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anthony TE, et al. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41(6):881–90. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- 7.Marin O, Muller U. Lineage origins of GABAergic versus glutamatergic neurons in the neocortex. Curr Opin Neurobiol. 2014;26:132–41. doi: 10.1016/j.conb.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smart IH. A pilot study of cell production by the ganglionic eminences of the developing mouse brain. J Anat. 1976;121(Pt 1):71–84. [PMC free article] [PubMed] [Google Scholar]
- 9.Fairen A, Cobas A, Fonseca M. Times of generation of glutamic acid decarboxylase immunoreactive neurons in mouse somatosensory cortex. J Comp Neurol. 1986;251(1):67–83. doi: 10.1002/cne.902510105. [DOI] [PubMed] [Google Scholar]
- 10.Anthony TE, Heintz N. Genetic lineage tracing defines distinct neurogenic and gliogenic stages of ventral telencephalic radial glial development. Neural Dev. 2008;3:30. doi: 10.1186/1749-8104-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kessaris N, Pringle N, Richardson WD. Specification of CNS glia from neural stem cells in the embryonic neuroepithelium. Philos Trans R Soc Lond B Biol Sci. 2008;363(1489):71–85. doi: 10.1098/rstb.2006.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bandler RC, Mayer C, Fishell G. Cortical interneuron specification: the juncture of genes, time and geometry. Curr Opin Neurobiol. 2017;42:17–24. doi: 10.1016/j.conb.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berry M, Rogers AW. The migration of neuroblasts in the developing cerebral cortex. J Anat. 1965;99(Pt 4):691–709. [PMC free article] [PubMed] [Google Scholar]
- 14.Fentress JC, Stanfield BB, Cowan WM. Observation on the development of the striatum in mice and rats. Anat Embryol (Berl) 1981;163(3):275–98. doi: 10.1007/BF00315705. [DOI] [PubMed] [Google Scholar]
- 15.Graybiel AM, Hickey TL. Chemospecificity of ontogenetic units in the striatum: demonstration by combining [3H]thymidine neuronography and histochemical staining. Proc Natl Acad Sci U S A. 1982;79(1):198–202. doi: 10.1073/pnas.79.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason HA, et al. Notch signaling coordinates the patterning of striatal compartments. Development. 2005;132(19):4247–58. doi: 10.1242/dev.02008. [DOI] [PubMed] [Google Scholar]
- 17.van der Kooy D, Fishell G. Neuronal birthdate underlies the development of striatal compartments. Brain Res. 1987;401(1):155–61. doi: 10.1016/0006-8993(87)91176-0. [DOI] [PubMed] [Google Scholar]
- 18.Ciceri G, et al. Lineage-specific laminar organization of cortical GABAergic interneurons. Nat Neurosci. 2013;16(9):1199–210. doi: 10.1038/nn.3485. [DOI] [PubMed] [Google Scholar]
- 19.Flames N, Marin O. Developmental mechanisms underlying the generation of cortical interneuron diversity. Neuron. 2005;46(3):377–81. doi: 10.1016/j.neuron.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 20.Valcanis H, Tan SS. Layer specification of transplanted interneurons in developing mouse neocortex. J Neurosci. 2003;23(12):5113–22. doi: 10.1523/JNEUROSCI.23-12-05113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadikot AF, Sasseville R. Neurogenesis in the mammalian neostriatum and nucleus accumbens: parvalbumin-immunoreactive GABAergic interneurons. J Comp Neurol. 1997;389(2):193–211. [PubMed] [Google Scholar]
- 22.Nobrega-Pereira S, et al. Origin and molecular specification of globus pallidus neurons. J Neurosci. 2010;30(8):2824–34. doi: 10.1523/JNEUROSCI.4023-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei B, et al. The onion skin-like organization of the septum arises from multiple embryonic origins to form multiple adult neuronal fates. Neuroscience. 2012;222:110–23. doi: 10.1016/j.neuroscience.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Gao P, et al. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 2014;159(4):775–88. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146(1):18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sultan KT, Brown KN, Shi SH. Production and organization of neocortical interneurons. Front Cell Neurosci. 2013;7:221. doi: 10.3389/fncel.2013.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falk S, et al. Time-Specific Effects of Spindle Positioning on Embryonic Progenitor Pool Composition and Adult Neural Stem Cell Seeding. Neuron. 2017;93(4):777–791 e3. doi: 10.1016/j.neuron.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klezovitch O, et al. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18(5):559–71. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan X, et al. Vascular Influence on Ventral Telencephalic Progenitors and Neocortical Interneuron Production. Dev Cell. 2016;36(6):624–38. doi: 10.1016/j.devcel.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown KN, et al. Clonal production and organization of inhibitory interneurons in the neocortex. Science. 2011;334(6055):480–6. doi: 10.1126/science.1208884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pilz GA, et al. Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nat Commun. 2013;4:2125. doi: 10.1038/ncomms3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strzyz PJ, Matejcic M, Norden C. Heterogeneity, Cell Biology and Tissue Mechanics of Pseudostratified Epithelia: Coordination of Cell Divisions and Growth in Tightly Packed Tissues. Int Rev Cell Mol Biol. 2016;325:89–118. doi: 10.1016/bs.ircmb.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8(6):438–50. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 34.Hindley C, Philpott A. Co-ordination of cell cycle and differentiation in the developing nervous system. Biochem J. 2012;444(3):375–82. doi: 10.1042/BJ20112040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magnani D, et al. The Gli3 hypomorphic mutation Pdn causes selective impairment in the growth, patterning, and axon guidance capability of the lateral ganglionic eminence. J Neurosci. 2010;30(41):13883–94. doi: 10.1523/JNEUROSCI.3650-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhide PG. Cell cycle kinetics in the embryonic mouse corpus striatum. J Comp Neurol. 1996;374(4):506–22. doi: 10.1002/(SICI)1096-9861(19961028)374:4<506::AID-CNE3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 37.Sheth AN, Bhide PG. Concurrent cellular output from two proliferative populations in the early embryonic mouse corpus striatum. J Comp Neurol. 1997;383(2):220–30. [PubMed] [Google Scholar]
- 38.Glickstein SB, et al. Selective cortical interneuron and GABA deficits in cyclin D2-null mice. Development. 2007;134(22):4083–93. doi: 10.1242/dev.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Ibanez R, et al. Helios expression coordinates the development of a subset of striatopallidal medium spiny neurons. Development. 2017;144(8):1566–1577. doi: 10.1242/dev.138248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross ME. Cell cycle regulation and interneuron production. Dev Neurobiol. 2011;71(1):2–9. doi: 10.1002/dneu.20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamamaki N, et al. Radial glia is a progenitor of neocortical neurons in the developing cerebral cortex. Neurosci Res. 2001;41(1):51–60. doi: 10.1016/s0168-0102(01)00259-0. [DOI] [PubMed] [Google Scholar]
- 42.Misson JP, et al. Identification of radial glial cells within the developing murine central nervous system: studies based upon a new immunohistochemical marker. Brain Res Dev Brain Res. 1988;44(1):95–108. doi: 10.1016/0165-3806(88)90121-6. [DOI] [PubMed] [Google Scholar]
- 43.Nowakowski TJ, et al. Transformation of the Radial Glia Scaffold Demarcates Two Stages of Human Cerebral Cortex Development. Neuron. 2016;91(6):1219–27. doi: 10.1016/j.neuron.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harwell CC, et al. Wide Dispersion and Diversity of Clonally Related Inhibitory Interneurons. Neuron. 2015;87(5):999–1007. doi: 10.1016/j.neuron.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dehay C, Kennedy H, Kosik KS. The outer subventricular zone and primate-specific cortical complexification. Neuron. 2015;85(4):683–94. doi: 10.1016/j.neuron.2014.12.060. [DOI] [PubMed] [Google Scholar]
- 46.Gal JS, et al. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26(3):1045–56. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stancik EK, et al. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci. 2010;30(20):7028–36. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petros TJ, et al. Apical versus Basal Neurogenesis Directs Cortical Interneuron Subclass Fate. Cell Rep. 2015;13(6):1090–5. doi: 10.1016/j.celrep.2015.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borrell V, Gotz M. Role of radial glial cells in cerebral cortex folding. Curr Opin Neurobiol. 2014;27:39–46. doi: 10.1016/j.conb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 50.Betizeau M, et al. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80(2):442–57. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 51.Hansen DV, et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16(11):1576–87. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Charvet CJ, Striedter GF. Causes and consequences of expanded subventricular zones. Eur J Neurosci. 2011;34(6):988–93. doi: 10.1111/j.1460-9568.2011.07818.x. [DOI] [PubMed] [Google Scholar]
- 53.Ma T, et al. Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci. 2013;16(11):1588–97. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- 54.Azzarelli R, Hardwick LJ, Philpott A. Emergence of neuronal diversity from patterning of telencephalic progenitors. Wiley Interdiscip Rev Dev Biol. 2015;4(3):197–214. doi: 10.1002/wdev.174. [DOI] [PubMed] [Google Scholar]
- 55.Hebert JM. Only scratching the cell surface: extracellular signals in cerebrum development. Curr Opin Genet Dev. 2013;23(4):470–4. doi: 10.1016/j.gde.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hebert JM, Fishell G. The genetics of early telencephalon patterning: some assembly required. Nat Rev Neurosci. 2008;9(9):678–85. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bjornsson CS, et al. It takes a village: constructing the neurogenic niche. Dev Cell. 2015;32(4):435–46. doi: 10.1016/j.devcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiang C, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383(6599):407–13. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 59.Ericson J, et al. Sonic hedgehog induces the differentiation of ventral forebrain neurons: a common signal for ventral patterning within the neural tube. Cell. 1995;81(5):747–56. doi: 10.1016/0092-8674(95)90536-7. [DOI] [PubMed] [Google Scholar]
- 60.Sousa VH, Fishell G. Sonic hedgehog functions through dynamic changes in temporal competence in the developing forebrain. Curr Opin Genet Dev. 2010;20(4):391–9. doi: 10.1016/j.gde.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuccillo M, et al. Temporal requirement for hedgehog signaling in ventral telencephalic patterning. Development. 2004;131(20):5031–40. doi: 10.1242/dev.01349. [DOI] [PubMed] [Google Scholar]
- 62.Machold R, et al. Sonic hedgehog is required for progenitor cell maintenance in telencephalic stem cell niches. Neuron. 2003;39(6):937–50. doi: 10.1016/s0896-6273(03)00561-0. [DOI] [PubMed] [Google Scholar]
- 63.Flandin P, et al. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron. 2011;70(5):939–50. doi: 10.1016/j.neuron.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wonders CP, et al. A spatial bias for the origins of interneuron subgroups within the medial ganglionic eminence. Dev Biol. 2008;314(1):127–36. doi: 10.1016/j.ydbio.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu Q, et al. Sonic hedgehog signaling confers ventral telencephalic progenitors with distinct cortical interneuron fates. Neuron. 2010;65(3):328–40. doi: 10.1016/j.neuron.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Q, Wonders CP, Anderson SA. Sonic hedgehog maintains the identity of cortical interneuron progenitors in the ventral telencephalon. Development. 2005;132(22):4987–98. doi: 10.1242/dev.02090. [DOI] [PubMed] [Google Scholar]
- 67.Rhinn M, Dolle P. Retinoic acid signalling during development. Development. 2012;139(5):843–58. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 68.Siegenthaler JA, Pleasure SJ. We have got you ‘covered’: how the meninges control brain development. Curr Opin Genet Dev. 2011;21(3):249–55. doi: 10.1016/j.gde.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ruberte E, et al. Retinoic acid receptors and cellular retinoid binding proteins. III. Their differential transcript distribution during mouse nervous system development. Development. 1993;118(1):267–82. doi: 10.1242/dev.118.1.267. [DOI] [PubMed] [Google Scholar]
- 70.Toresson H, et al. Retinoids are produced by glia in the lateral ganglionic eminence and regulate striatal neuron differentiation. Development. 1999;126(6):1317–26. doi: 10.1242/dev.126.6.1317. [DOI] [PubMed] [Google Scholar]
- 71.Li H, et al. A retinoic acid synthesizing enzyme in ventral retina and telencephalon of the embryonic mouse. Mech Dev. 2000;95(1–2):283–9. doi: 10.1016/s0925-4773(00)00352-x. [DOI] [PubMed] [Google Scholar]
- 72.Molotkova N, Molotkov A, Duester G. Role of retinoic acid during forebrain development begins late when Raldh3 generates retinoic acid in the ventral subventricular zone. Dev Biol. 2007;303(2):601–10. doi: 10.1016/j.ydbio.2006.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chatzi C, Brade T, Duester G. Retinoic acid functions as a key GABAergic differentiation signal in the basal ganglia. PLoS Biol. 2011;9(4):e1000609. doi: 10.1371/journal.pbio.1000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waclaw RR, Wang B, Campbell K. The homeobox gene Gsh2 is required for retinoid production in the embryonic mouse telencephalon. Development. 2004;131(16):4013–20. doi: 10.1242/dev.01272. [DOI] [PubMed] [Google Scholar]
- 75.Liao WL, et al. Modular patterning of structure and function of the striatum by retinoid receptor signaling. Proc Natl Acad Sci U S A. 2008;105(18):6765–70. doi: 10.1073/pnas.0802109105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rataj-Baniowska M, et al. Retinoic Acid Receptor beta Controls Development of Striatonigral Projection Neurons through FGF-Dependent and Meis1-Dependent Mechanisms. J Neurosci. 2015;35(43):14467–75. doi: 10.1523/JNEUROSCI.1278-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bielen H, Houart C. The Wnt cries many: Wnt regulation of neurogenesis through tissue patterning, proliferation, and asymmetric cell division. Dev Neurobiol. 2014;74(8):772–80. doi: 10.1002/dneu.22168. [DOI] [PubMed] [Google Scholar]
- 78.Backman M, et al. Effects of canonical Wnt signaling on dorso-ventral specification of the mouse telencephalon. Dev Biol. 2005;279(1):155–68. doi: 10.1016/j.ydbio.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 79.Gulacsi AA, Anderson SA. Beta-catenin-mediated Wnt signaling regulates neurogenesis in the ventral telencephalon. Nat Neurosci. 2008;11(12):1383–91. doi: 10.1038/nn.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhong J, et al. The Wnt receptor Ryk controls specification of GABAergic neurons versus oligodendrocytes during telencephalon development. Development. 2011;138(3):409–19. doi: 10.1242/dev.061051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paek H, Gutin G, Hebert JM. FGF signaling is strictly required to maintain early telencephalic precursor cell survival. Development. 2009;136(14):2457–65. doi: 10.1242/dev.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hebert JM. FGFs: Neurodevelopment’s Jack-of-all-Trades - How Do They Do it? Front Neurosci. 2011;5:133. doi: 10.3389/fnins.2011.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Borello U, et al. FGF15 promotes neurogenesis and opposes FGF8 function during neocortical development. Neural Dev. 2008;3:17. doi: 10.1186/1749-8104-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martynoga B, et al. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol. 2005;283(1):113–27. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 85.Storm EE, et al. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development. 2006;133(9):1831–44. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- 86.Gutin G, et al. FGF signalling generates ventral telencephalic cells independently of SHH. Development. 2006;133(15):2937–46. doi: 10.1242/dev.02465. [DOI] [PubMed] [Google Scholar]
- 87.Yoon K, et al. Fibroblast growth factor receptor signaling promotes radial glial identity and interacts with Notch1 signaling in telencephalic progenitors. J Neurosci. 2004;24(43):9497–506. doi: 10.1523/JNEUROSCI.0993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nandi S, et al. FGF-dependent, context-driven role for FRS adapters in the early telencephalon. J Neurosci. 2017 doi: 10.1523/JNEUROSCI.2931-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoch RV, Rubenstein JL, Pleasure S. Genes and signaling events that establish regional patterning of the mammalian forebrain. Semin Cell Dev Biol. 2009;20(4):378–86. doi: 10.1016/j.semcdb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 90.Peyre E, Silva CG, Nguyen L. Crosstalk between intracellular and extracellular signals regulating interneuron production, migration and integration into the cortex. Front Cell Neurosci. 2015;9:129. doi: 10.3389/fncel.2015.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lun MP, Monuki ES, Lehtinen MK. Development and functions of the choroid plexus-cerebrospinal fluid system. Nat Rev Neurosci. 2015;16(8):445–57. doi: 10.1038/nrn3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26(2):395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 93.Andrews W, et al. The role of Slit-Robo signaling in the generation, migration and morphological differentiation of cortical interneurons. Dev Biol. 2008;313(2):648–58. doi: 10.1016/j.ydbio.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 94.Maira M, et al. Role for TGF-beta superfamily signaling in telencephalic GABAergic neuron development. J Neurodev Disord. 2010;2(1):48–60. doi: 10.1007/s11689-009-9035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burrows RC, Lillien L, Levitt P. Mechanisms of progenitor maturation are conserved in the striatum and cortex. Dev Neurosci. 2000;22(1–2):7–15. doi: 10.1159/000017422. [DOI] [PubMed] [Google Scholar]
- 96.Mukhopadhyay A, et al. Differential effects of BMP signaling on parvalbumin and somatostatin interneuron differentiation. Development. 2009;136(15):2633–42. doi: 10.1242/dev.034439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deverman BE, Patterson PH. Cytokines and CNS development. Neuron. 2009;64(1):61–78. doi: 10.1016/j.neuron.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 98.Briscoe J, et al. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101(4):435–45. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 99.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1(1):20–9. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 100.Bulfone A, et al. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993;13(7):3155–72. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Puelles L, et al. Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol. 2000;424(3):409–38. doi: 10.1002/1096-9861(20000828)424:3<409::aid-cne3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 102.Castro DS, et al. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25(9):930–45. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Petryniak MA, et al. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55(3):417–33. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Flames N, et al. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27(36):9682–95. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miyoshi G, et al. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27(29):7786–98. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McKinsey GL, et al. Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron. 2013;77(1):83–98. doi: 10.1016/j.neuron.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sandberg M, et al. Transcriptional Networks Controlled by NKX2-1 in the Development of Forebrain GABAergic Neurons. Neuron. 2016;91(6):1260–75. doi: 10.1016/j.neuron.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu KM, et al. Dual role for Islet-1 in promoting striatonigral and repressing striatopallidal genetic programs to specify striatonigral cell identity. Proc Natl Acad Sci U S A. 2014;111(1):E168–77. doi: 10.1073/pnas.1319138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang Q, et al. The Zinc Finger Transcription Factor Sp9 Is Required for the Development of Striatopallidal Projection Neurons. Cell Rep. 2016;16(5):1431–44. doi: 10.1016/j.celrep.2016.06.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128(2):193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- 111.Sousa VH, et al. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cereb Cortex. 2009;19(Suppl 1):i1–10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fuentealba LC, et al. Embryonic Origin of Postnatal Neural Stem Cells. Cell. 2015;161(7):1644–55. doi: 10.1016/j.cell.2015.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taniguchi H, Lu J, Huang ZJ. The spatial and temporal origin of chandelier cells in mouse neocortex. Science. 2013;339(6115):70–4. doi: 10.1126/science.1227622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rudy B, et al. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71(1):45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Butt SJ, et al. The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron. 2008;59(5):722–32. doi: 10.1016/j.neuron.2008.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sussel L, et al. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126(15):3359–70. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 117.Flandin P, Kimura S, Rubenstein JL. The progenitor zone of the ventral medial ganglionic eminence requires Nkx2-1 to generate most of the globus pallidus but few neocortical interneurons. J Neurosci. 2010;30(8):2812–23. doi: 10.1523/JNEUROSCI.4228-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fogarty M, et al. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 2007;27(41):10935–46. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23(1):167–74. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Inan M, Welagen J, Anderson SA. Spatial and temporal bias in the mitotic origins of somatostatin- and parvalbumin-expressing interneuron subgroups and the chandelier subtype in the medial ganglionic eminence. Cereb Cortex. 2012;22(4):820–7. doi: 10.1093/cercor/bhr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miyoshi G, et al. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J Neurosci. 2010;30(5):1582–94. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nery S, Fishell G, Corbin JG. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci. 2002;5(12):1279–87. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- 123.Miyoshi G, et al. Prox1 Regulates the Subtype-Specific Development of Caudal Ganglionic Eminence-Derived GABAergic Cortical Interneurons. J Neurosci. 2015;35(37):12869–89. doi: 10.1523/JNEUROSCI.1164-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rubin AN, Kessaris N. PROX1: a lineage tracer for cortical interneurons originating in the lateral/caudal ganglionic eminence and preoptic area. PLoS One. 2013;8(10):e77339. doi: 10.1371/journal.pone.0077339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gelman D, et al. A wide diversity of cortical GABAergic interneurons derives from the embryonic preoptic area. J Neurosci. 2011;31(46):16570–80. doi: 10.1523/JNEUROSCI.4068-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vucurovic K, et al. Serotonin 3A receptor subtype as an early and protracted marker of cortical interneuron subpopulations. Cereb Cortex. 2010;20(10):2333–47. doi: 10.1093/cercor/bhp310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Risold PY, Swanson LW. Chemoarchitecture of the rat lateral septal nucleus. Brain Res Brain Res Rev. 1997;24(2–3):91–113. doi: 10.1016/s0165-0173(97)00008-8. [DOI] [PubMed] [Google Scholar]
- 128.Furusho M, et al. Involvement of the Olig2 transcription factor in cholinergic neuron development of the basal forebrain. Dev Biol. 2006;293(2):348–57. doi: 10.1016/j.ydbio.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 129.Taglialatela P, et al. Compromised generation of GABAergic interneurons in the brains of Vax1−/− mice. Development. 2004;131(17):4239–49. doi: 10.1242/dev.01299. [DOI] [PubMed] [Google Scholar]
- 130.Inoue T, et al. Zic1 and Zic3 regulate medial forebrain development through expansion of neuronal progenitors. J Neurosci. 2007;27(20):5461–73. doi: 10.1523/JNEUROSCI.4046-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rubin AN, et al. The germinal zones of the basal ganglia but not the septum generate GABAergic interneurons for the cortex. J Neurosci. 2010;30(36):12050–62. doi: 10.1523/JNEUROSCI.6178-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Magno L, et al. NKX2-1 Is Required in the Embryonic Septum for Cholinergic System Development, Learning, and Memory. Cell Rep. 2017;20(7):1572–1584. doi: 10.1016/j.celrep.2017.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Merkle FT, et al. Adult neural stem cells in distinct microdomains generate previously unknown interneuron types. Nat Neurosci. 2014;17(2):207–14. doi: 10.1038/nn.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Long JE, et al. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 2009;512(4):556–72. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nord AS, et al. Genomic perspectives of transcriptional regulation in forebrain development. Neuron. 2015;85(1):27–47. doi: 10.1016/j.neuron.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang N, et al. Generation of pure GABAergic neurons by transcription factor programming. Nat Methods. 2017;14(6):621–628. doi: 10.1038/nmeth.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cepko C. Intrinsically different retinal progenitor cells produce specific types of progeny. Nat Rev Neurosci. 2014;15(9):615–27. doi: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- 138.Hobert O. A map of terminal regulators of neuronal identity in Caenorhabditis elegans. Wiley Interdiscip Rev Dev Biol. 2016;5(4):474–98. doi: 10.1002/wdev.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kohwi M, Doe CQ. Temporal fate specification and neural progenitor competence during development. Nat Rev Neurosci. 2013;14(12):823–38. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Golden JA, Fields-Berry SC, Cepko CL. Construction and characterization of a highly complex retroviral library for lineage analysis. Proc Natl Acad Sci U S A. 1995;92(12):5704–8. doi: 10.1073/pnas.92.12.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Walsh C, Cepko CL. Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science. 1992;255(5043):434–40. doi: 10.1126/science.1734520. [DOI] [PubMed] [Google Scholar]
- 142.Reid CB, Walsh CA. Evidence of common progenitors and patterns of dispersion in rat striatum and cerebral cortex. J Neurosci. 2002;22(10):4002–14. doi: 10.1523/JNEUROSCI.22-10-04002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ware ML, et al. Coexistence of widespread clones and large radial clones in early embryonic ferret cortex. Cereb Cortex. 1999;9(6):636–45. doi: 10.1093/cercor/9.6.636. [DOI] [PubMed] [Google Scholar]
- 144.Reid CB, Liang I, Walsh C. Systematic widespread clonal organization in cerebral cortex. Neuron. 1995;15(2):299–310. doi: 10.1016/0896-6273(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 145.Noctor SC, et al. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409(6821):714–20. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 146.Battiste J, et al. Ascl1 defines sequentially generated lineage-restricted neuronal and oligodendrocyte precursor cells in the spinal cord. Development. 2007;134(2):285–93. doi: 10.1242/dev.02727. [DOI] [PubMed] [Google Scholar]
- 147.Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126(3):525–34. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 148.Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506(1):16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- 149.Mayer C, et al. Clonally Related Forebrain Interneurons Disperse Broadly across Both Functional Areas and Structural Boundaries. Neuron. 2015;87(5):989–98. doi: 10.1016/j.neuron.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mayer C, Bandler RC, Fishell G. Lineage Is a Poor Predictor of Interneuron Positioning within the Forebrain. Neuron. 2016;92(1):45–51. doi: 10.1016/j.neuron.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sultan KT, et al. Clonally Related GABAergic Interneurons Do Not Randomly Disperse but Frequently Form Local Clusters in the Forebrain. Neuron. 2016;92(1):31–44. doi: 10.1016/j.neuron.2016.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Turrero Garcia M, Mazzola E, Harwell CC. Lineage Relationships Do Not Drive MGE/PoA-Derived Interneuron Clustering in the Brain. Neuron. 2016;92(1):52–58. doi: 10.1016/j.neuron.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hafler BP, et al. Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc Natl Acad Sci U S A. 2012;109(20):7882–7. doi: 10.1073/pnas.1203138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wamsley B, Fishell G. Genetic and activity-dependent mechanisms underlying interneuron diversity. Nat Rev Neurosci. 2017;18(5):299–309. doi: 10.1038/nrn.2017.30. [DOI] [PubMed] [Google Scholar]
- 155.De Marco Garcia NV, Karayannis T, Fishell G. Neuronal activity is required for the development of specific cortical interneuron subtypes. Nature. 2011;472(7343):351–5. doi: 10.1038/nature09865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yu YC, et al. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458(7237):501–4. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.He M, et al. Strategies and Tools for Combinatorial Targeting of GABAergic Neurons in Mouse Cerebral Cortex. Neuron. 2016;92(2):555. doi: 10.1016/j.neuron.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 158.Birey F, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545(7652):54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Parent JM, Anderson SA. Reprogramming patient-derived cells to study the epilepsies. Nat Neurosci. 2015;18(3):360–6. doi: 10.1038/nn.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tyson JA, Anderson SA. GABAergic interneuron transplants to study development and treat disease. Trends Neurosci. 2014;37(3):169–77. doi: 10.1016/j.tins.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Braz JM, et al. Forebrain GABAergic neuron precursors integrate into adult spinal cord and reduce injury-induced neuropathic pain. Neuron. 2012;74(4):663–75. doi: 10.1016/j.neuron.2012.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]


