SUMMARY
Aneuploidy disrupts cellular homeostasis. However, the molecular mechanisms underlying the physiological responses and adaptation to aneuploidy are not well understood. Deciphering these mechanisms is important because aneuploidy is associated with diseases including intellectual disability and cancer. Although tumors and mammalian aneuploid cells including several cancer cell lines show altered levels of sphingolipids, the role of sphingolipids in aneuploidy remains unknown. Here we show that ceramides and long-chain bases, sphingolipid molecules that slow proliferation and promote survival, are increased by aneuploidy. Sphingolipid levels are tightly linked to serine synthesis, and inhibiting either serine or sphingolipid synthesis can specifically impair the fitness of aneuploid cells. Remarkably, the fitness of aneuploid cells improves or deteriorates upon genetically decreasing or increasing ceramides, respectively. Combined targeting of serine and sphingolipid synthesis could be exploited to specifically target cancer cells, the vast majority of which are aneuploid.
eTOC Blurb
Hwang et al. demonstrate that aneuploid yeast cells rely on the synthesis of the amino acid serine for their viability. Serine is used for the synthesis of sphingolipids that control the fitness of aneuploid cells. Aneuploid cells are vulnerable to combined inhibition of serine and sphingolipid biosynthesis.
INTRODUCTION
Chromosome missegregation events leading to aneuploidy cause severe developmental defects in organisms (Torres et al., 2008). In the absence of other genomic alterations, losing chromosomes is usually lethal to cells while gaining chromosomes disrupts cellular homeostasis and hampers proliferation. At both the cellular and organismal levels, the deleterious effects of gaining chromosomes correlate with an increased number of encoding genes (Torres, 2015). However, the effects of aneuploidy on cell physiology can depend on the microenvironment as aneuploidy can confer a proliferative advantage under stress conditions or resistance to a particular drug (Pavelka et al., 2010; Selmecki et al., 2006; Yona et al., 2012). Aneuploidy is a common characteristic of cancer cells, and gaining or losing chromosomes provides a mechanism by which cells gain copies of oncogenes or lose tumor suppressor genes, thereby driving tumorigenesis (Davoli et al., 2013). Importantly, the mechanisms by which cancer cells overcome the deleterious consequences associated with aneuploidy are not known.
To investigate how aneuploidy affects cellular physiology in eukaryotes, we generated and characterized a series of aneuploid yeast strains, each carrying an extra copy of a given chromosome (referred to as disomes) (Torres et al., 2007). A direct consequence of acquiring an extra chromosome is the increased expression of the duplicated genes (Torres et al., 2016). On average, duplicated transcripts are translated, leading to proportional increases in protein abundance, with the notable exception of subunits of macromolecular complexes whose stability depends on complex assembly (Dephoure et al., 2014; McShane et al., 2016). The general increase in protein synthesis can cause several phenotypes shared by all aneuploid cells independent of the identity of the extra chromosome (Oromendia et al., 2012; Torres et al., 2007). Such phenotypes include decreased proliferation rates, increased glucose utilization, and signs of proteotoxic stress, all of which are also observed in aneuploid human cells (Santaguida and Amon, 2015; Stingele et al., 2012).
We previously identified aneuploidy-tolerating spontaneous mutations that improve the fitness of aneuploid cells (Torres et al., 2010). Among these, loss of function mutation in the deubiquitinating enzyme Ubp6 was shown to improve the fitness of 4 out of 12 aneuploid strains. Global proteome quantification revealed that loss of UBP6 leads to the attenuation of the levels of overexpressed proteins, likely through a general increase in proteasome activity (Bashore et al., 2015; Dephoure et al., 2014; Hanna et al., 2006). This provides at least one mechanism by which altering a cellular process - increasing protein turnover - improves the fitness of aneuploid cells independent of karyotype.
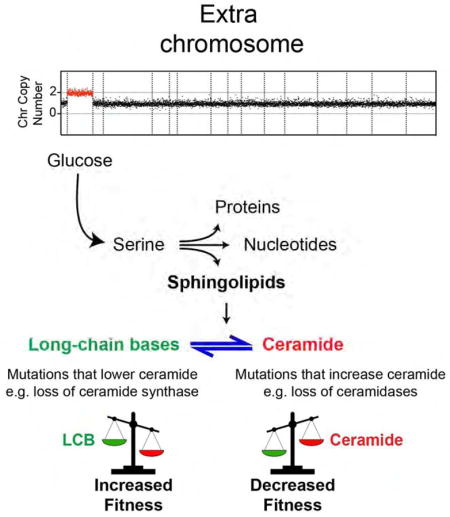
Among the other aneuploidy-tolerating mutations in yeast were three independent spontaneous mutations in a gene that regulates sphingolipid synthesis (SVF1), implicating these molecules in the physiological responses to aneuploidy (Brace et al., 2007; Torres et al., 2010). Aneuploid primary mouse embryonic fibroblasts (MEFs), cancer cell lines, and tumors show elevated levels of sphingolipids, yet a functional link between changes in sphingolipid levels and aneuploidy is not known (Erez-Roman et al., 2010; Guillermet-Guibert et al., 2009; Morad and Cabot, 2013; Tang et al., 2017). Sphingolipids are synthesized from serine and palmitoyl-CoA (Figure 1A). Both long-chain bases (LCBs) and ceramides increase rapidly upon stress and function as signaling molecules (Dickson et al., 1997; Jenkins et al., 1997). While ceramides serve to slow proliferation by delaying entry into the cell cycle (Nickels and Broach, 1996), LCBs activate transcriptional responses and signaling pathways associated with cell wall integrity and survival (Cowart et al., 2003; Dickson and Lester, 2002). Here, we find that the levels of both LCBs and ceramides are elevated in aneuploidy and that increased serine synthesis accounts for the accumulation of sphingolipids in aneuploid cells. Remarkably, the proliferation of several aneuploid strains improves significantly upon genetically decreasing the levels of ceramides and increasing those of LCBs. Transcriptome and proteome analyses of disomes harboring a mutation that improves their fitness indicate that sphingolipids regulate membrane protein composition, RNA biosynthesis, and metabolic pathways associated with energy production and biosynthesis.
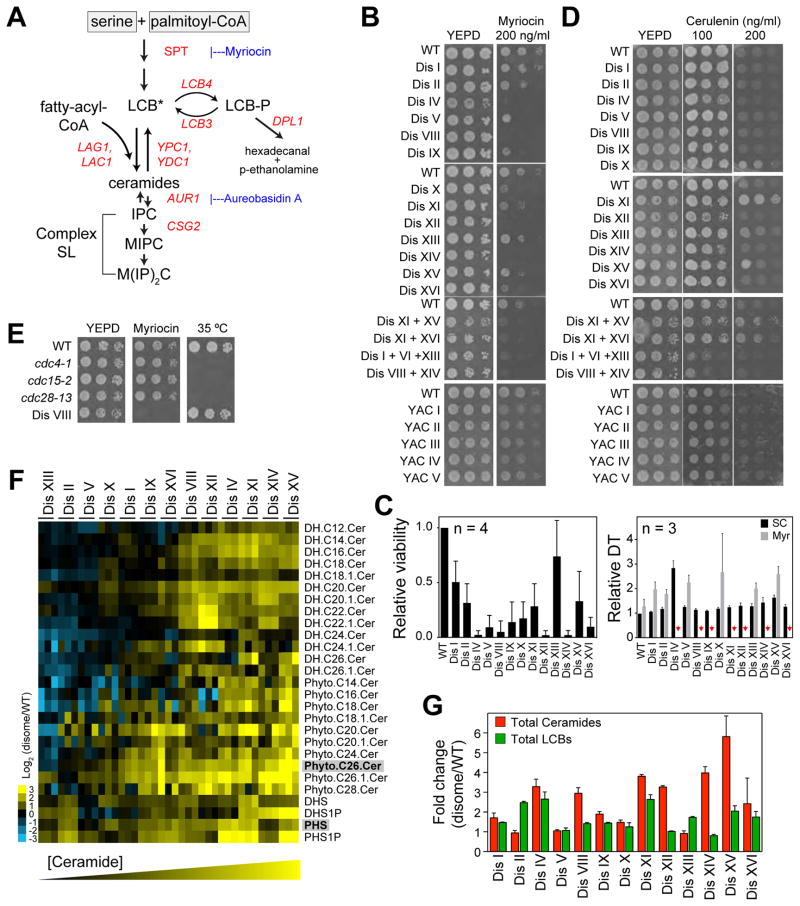
Figure 1. Aneuploidy Increases Sphingolipid Biosynthesis.
(A) Biochemical pathway of de novo synthesis of sphingolipids in yeast. Genes used in this study are shown in red. SPT, serine palmitoyltransferase; LCB, long-chain bases, asterisk (*) indicates that LCBs need to be phosphorylated/dephosphorylated to be converted to ceramide; IPC, inositol-phosphorylceramide, MIPC, mannosyl-IPC; M(IP)2C, mannosyl-diinositol-phosphorylceramide.
(B) Proliferative capability of wild type cells (WT), disomes and strains harboring YAC in the presence of myriocin.
(C) Quantification of the viability of cells treated with 200 ng/ml myriocin in the left panel. Right panel shows the doubling times of disomes in synthetic complete media alone (SC, black bars) and with 200 ng/ml myriocin (Myr, gray bars) relative to WT. Red arrows point to strains that did not grow in culture.
(D) Proliferative capability of WT, disomes and strains harboring YAC in the presence of cerulenin.
(E) Proliferative capability of WT, cell cycle mutants, and disome VIII in the presence of myriocin (200 ng/ml) and at a restrictive temperature for the cell cycle mutants.
(F) LC/LC-MS analysis of LCBs and ceramides in the disomes compared to WT. Columns represent experiments (3 biological replicates shown). Rows represent lipid species. DH, dihydro; Cer, ceramide; DHS, dihydrosphingosine; PHS, phytosphigosine. Most abundant LCB and ceramide are in gray boxes.
(G) Fold change of total LCBs and ceramides in the disomes relative to WT.
Error bars represent +/− standard deviation (SD). See Supplementary Information for details on the strains used in all figures. See also Figure S1 and Tables S1 and S2.
RESULTS
Aneuploid Cells Rely on Increased De Novo Synthesis of Sphingolipids to Proliferate
To determine how regulation of sphingolipids affects the viability of aneuploid cells, we treated cells with myriocin, a specific inhibitor of serine palmitoyltransferase (SPT), which catalyzes the first and rate-limiting step of sphingolipid synthesis (Figure 1A). We found that 10 out of 13 disomes are hypersensitive to myriocin compared to wild type cells (IV, V, VIII, IX, X, XI, XII, XIV, XV and XVI, Figures 1B and 1C). Four other aneuploid strains harboring extra copies of either 2 or 3 chromosomes were also sensitive to myriocin treatment (Figure 1B). Furthermore, we found that 33 of 35 aneuploid yeast strains derived from random meiosis and that harbor complex karyotypes are sensitive to myriocin (Figure S1A)(Pavelka et al., 2010). These results indicate that despite the phenotypic variability and non-genetic individuality of aneuploid cells (Beach et al., 2017; Pavelka et al., 2010), the majority of aneuploid strains are sensitive to inhibition of sphingolipid synthesis, independent of karyotype identity.
Similar sensitivities were not observed in strains carrying yeast artificial chromosomes (YAC) that contain mammalian DNA with no known protein-coding genes (Figure 1B), indicating that enhanced myriocin sensitivity results from increased gene expression in aneuploid cells rather than increased DNA content. To determine whether sensitivity to inhibition of sphingolipid synthesis is a reflection of a more general need for lipids, we treated cells with cerulenin, an inhibitor of fatty acid synthase. We found that cerulenin does not specifically affect the viability of aneuploid cells, suggesting that lipid synthesis, in general, is not affected by aneuploidy (Figures 1D and S1B). Notably, three hypomorphic alleles of essential cell cycle genes that exhibit reduced proliferation rates at the permissive temperature were not sensitive to myriocin (Figure 1E), indicating that impaired proliferation itself does not confer myriocin sensitivity. These results indicate that aneuploid cells rely on increased sphingolipid production for their viability.
Aneuploid Cells Have Elevated Levels of Long-chain Bases and Ceramides
To test whether aneuploidy alters the cellular composition of LCBs or ceramides, we performed quantitative mass spectrometry (MS) on lipid extracts from 13 disomes as well as wild type cells (Table S1)(Bielawski et al., 2009). We found that the levels of LCBs or ceramides are increased in the aneuploid strains compared to wild type cells (Figures 1F, 1G, and S1C). Interestingly, disomes with modest cell cycle delays, including disomes I, II, V, IX, X and XIII (Torres et al., 2007), exhibited the smallest increases in ceramide levels (Figure 1G). Indeed, increases in ceramide levels correlated with decreased proliferation rates of the disomes (Figure S1D, Pearson r = 0.65, p-value = 0.008). These results support the hypothesis that accumulation of ceramides plays a role in slowing the cell cycle in response to aneuploidy.
To investigate whether other lipids also increase in aneuploid cells, we comprehensively examined the cellular lipid composition using a MS-based global lipidomic technique in several strains (6 disomes analyzed: V, XI, XII, XIV, XV, XVI, and wild type cells) (Ejsing et al., 2009). These measurements confirmed a consistent and significant increase in ceramide levels in the disomes compared to wild type cells. This is in contrast to most other lipids, the levels of which were only mildly and inconsistently altered (Figures S1E, S1F, and Table S1). Lastly, we used a third approach based on quantitative high-performance liquid chromatography (qHPLC) to specifically quantify LCBs and their phosphorylated forms LCB-Ps (Lester and Dickson, 2001). We validated that the levels of LCBs and LCB-Ps are elevated in several disomes relative to wild type cells (5 disomes analyzed: V, XI, XII, XV and XVI, and wild type cells; Figures S1G, S1H, S1I, and Table S2). Altogether, these results demonstrate, via three independent methodologies, that the levels of both LCBs and ceramides are increased in aneuploid cells and indicate that altered sphingolipid metabolism is a general feature of aneuploid cells.
Increased Serine Synthesis Accounts for Higher Levels of Sphingolipids in Aneuploid Cells
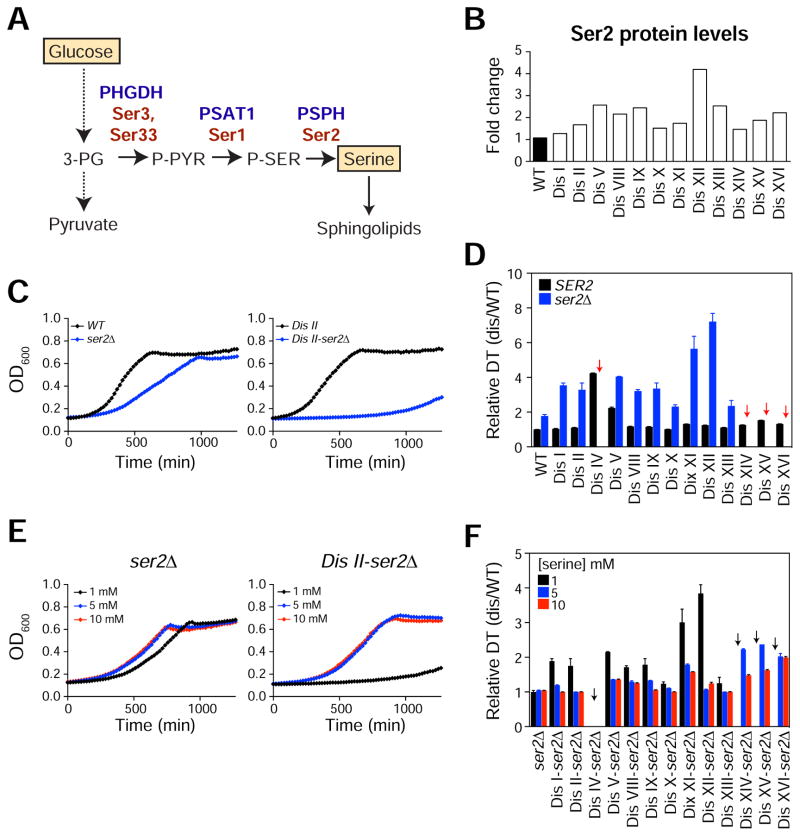
We next sought to understand the mechanisms leading to increased sphingolipid levels in aneuploid cells. Dozens of genes are involved in the regulation of sphingolipid biosynthesis (Table S3) (Cherry et al., 2012). With the exception of the genes present in the duplicated chromosomes, mRNA and protein expression of this network of genes do not show significant differences between disomes and wild type cells, indicating that transcriptional regulation does not play a significant role in the accumulation of sphingolipids in aneuploid cells (Figures S2A and S2B) (Dephoure et al., 2014). We next examined whether aneuploidy increases SPT enzymatic activity. We found that with the exception of disome IV, which contains two copies of the catalytic subunit of the SPT complex (LCB2), SPT activity is not significantly altered in the disomes compared to wild type cells (Figures S2C and S2D). This indicates that signaling pathways known to regulate SPT activity do not play a critical role in the cellular responses to aneuploidy (Roelants et al., 2011).
Because substrate availability is rate-limiting for the de novo synthesis of sphingolipids (Alvarez-Vasquez et al., 2005; Cowart and Hannun, 2007), we next examined the serine requirements for the proliferation of the disomes. We recently identified Ser2, the phosphoserine phosphatase that catalyzes the final and rate-limiting step in the synthesis of serine from glucose, to be upregulated in the disomes, indicating that serine biosynthesis from glucose may be critical for aneuploid cells (Figures 2A, 2B, S2A, and S2B)(Dephoure et al., 2014). Strikingly, deletion of SER2 significantly hampered the proliferation of all disomes tested, whilst only causing a modest effect on the proliferation of wild type cells (Figures 2C and 2D). Disomes harboring the largest chromosomes were most affected by ser2Δ, and four of these (IV, XIV, XV and XVI) did not grow in liquid medium. Standard synthetic medium contains 1 mM serine, a condition under which the disomes proliferate very poorly. Supplementation of 5 or 10 mM serine significantly improved the proliferation of the disomes while minimally affecting that of wild type cells (Figures 2E and 2F). Addition of other amino acids did not result in similar effects (Figure S2E). These data indicate that the disomes exhibit a higher metabolic demand for serine and that the lack of serine alone and not the accumulation of metabolic intermediates upon SER2 loss of function is deleterious to aneuploid cells. Similar results were obtained when the phosphoserine aminotransferase SER1 was deleted in the disomes (Figure S2F), thus corroborating our findings.
Figure 2. Aneuploid Cells Require Increase Serine Biosynthesis for Their Survival.
(A) Biochemical pathway of de novo serine synthesis from glucose. Human genes (blue) and yeast genes (red) are shown. 3PG, 3-phosphoglycerate; P-PYR, phosphohydroxypyruvate; P-SER, phosphoserine.
(B) Ser2 protein levels in the disomes relative to WT (Dephoure et al., 2014).
(C) Growth curves of cells with wild type SER2 (black line) or harboring ser2Δ (blue line). WT and disome II are shown.
(D) Doubling times of disomes (black bars) and disomes harboring ser2Δ (blue bars) relative to WT. Red arrows point to strains that did not grow in culture. Error bars = +/− SD, n = 3.
(E) Growth curves of cells harboring ser2Δ in medium containing 1 mM serine (black line), 5 mM serine (blue line) and 10 mM serine (red line).
(F) Doubling times of disomes harboring ser2Δ in medium containing 1 mM (black), 5 mM (blue) or 10 mM (red) serine. Error bars = +/− SD, n = 3. Black arrows point to strains that did not grow in culture.
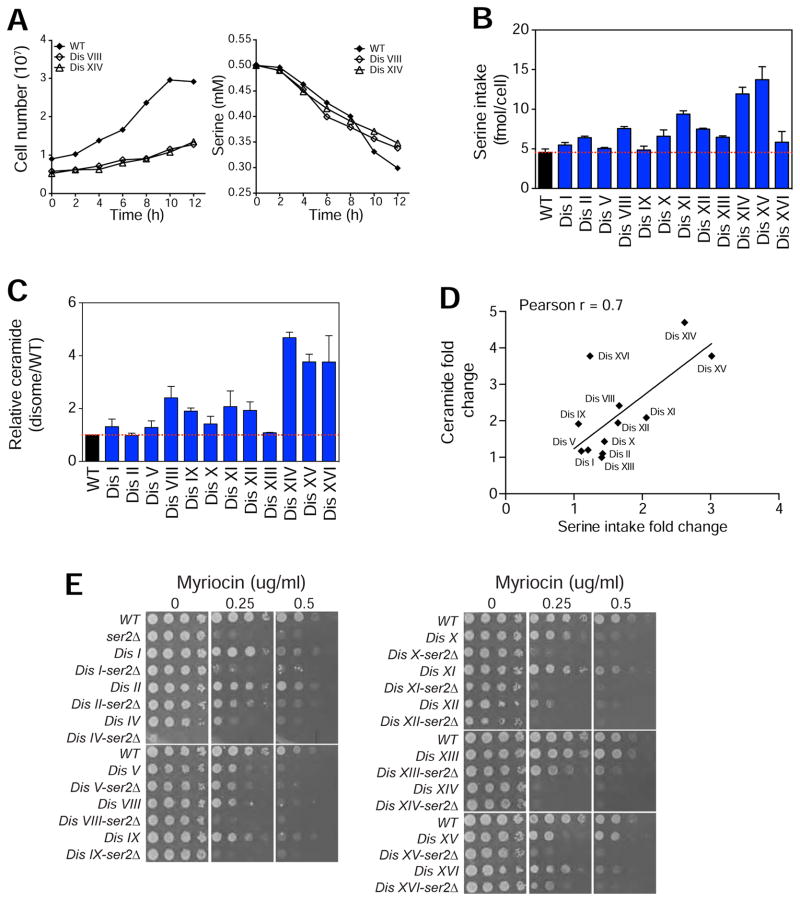
Increased levels of sphingolipids and higher dependence on serine synthesis suggest that compared to euploid cells, aneuploid cells utilize more serine for the production of sphingolipids. Indeed, we found that despite their impaired proliferation, most disomes (10 out of 12: I, II, VIII, X, XI, XII, XIII, XIV, XV, XVI) have increased serine utilization compared to wild type cells (Figures 3A and 3B). To determine whether changes in serine metabolic flux leads to increased sphingolipid synthesis, we quantified 14C-serine incorporation into ceramides in wild type cells and disomes. Using this approach, we detected increased serine incorporation into ceramides in 10 out of 12 disomes (I, V, VIII, IX, X, XI, XII, XIV, XV, XVI, Figure 3C). The differences in serine intake account for the increased ceramide synthesis, since the fold change in serine intake strongly predicts the fold changes in ceramide levels (Pearson r = 0.7, Figure 3D). To validate whether impaired serine synthesis affects the synthesis of sphingolipids, we used quantitative MS to measure the levels of LCBs in four strains harboring the ser2Δ (WT and 3 representative disomes: V, XI and XIV, Figure S3A) and confirmed that indeed LCBs are reduced when serine synthesis is impaired. These results show that aneuploid cells rely on increased serine synthesis for their survival and indicate that increased serine synthesis due to the upregulation of the phosphoserine phosphatase Ser2 is a major path through which aneuploid cells accumulate sphingolipids.
Figure 3. Aneuploid Cells Use Serine for the Synthesis of Sphingolipids.
(A) Growth curves of WT, disomes VIII and disome XIV are shown in the left panel. Amount of serine in the medium as a function of time (right panel).
(B) Serine utilization per cell during exponential growth of WT and disomes. Error bars = +/− SD, n = 3 biological replicates.
(C) Relative amount of serine incorporated into ceramides in the disomes relative to WT. Error bars = +/− SD, n = 3 biological replicates.
(D) Linear correlation between the fold change in serine intake and fold change in ceramide synthesis in the disomes.
(E) Proliferative capability of WT, ser2Δ, disomes, and the disomes harboring ser2Δ in the presence of myriocin.
See also Figure S3.
Impaired Serine Synthesis Enhances Sensitivity to Inhibition of Sphingolipid Synthesis
Our results raise the possibility that impaired de novo serine biosynthesis may render cells vulnerable to drugs that target sphingolipid synthesis. Indeed, we found that loss of either SER2 or SER1 enhanced sensitivity of all strains to myriocin (Figures 3E and S3B). Serine also serves as a precursor molecule for the synthesis of nucleotides. Therefore, it is possible that aneuploid cells show synthetic interaction with ser2Δ or ser1Δ because these cells have increased serine demand for nucleotide biosynthesis. To assess whether nucleotide biosynthesis is rate-limiting for the proliferation of the disomes, we treated cells with drugs that inhibit dihydrofolate reductase (DHFR) activity thereby blocking dTMP synthesis from serine (Methotrexate and Aminopterin, Figure S3C). Surprisingly, we found that while most disomes are sensitive to the DNA damaging agent 4-Nitroquinoline 1-oxide (4NQ), the majority of disomes do not display enhanced sensitivity to DHFR inhibitors (Figure S3D). Nonetheless, impairing serine synthesis did enhance sensitivity to DHFR inhibitors in the disomes (Figure S3E). Our results indicate that serine flux into sphingolipid synthesis plays an important role in the cellular responses to aneuploidy and that inhibiting serine biosynthesis can sensitize aneuploid cells to drugs that target nucleotide or sphingolipid synthesis.
Reducing Ceramide Synthesis Improves the Proliferation of Aneuploid Cells
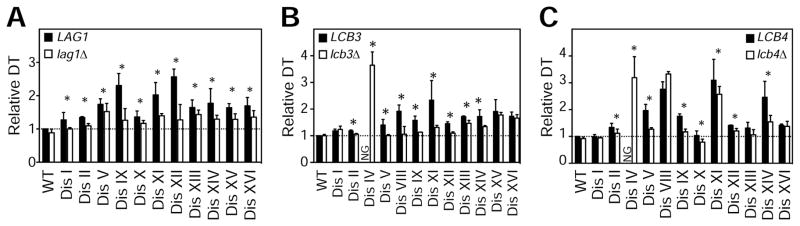
To investigate the physiological consequences of the altered levels of sphingolipids in aneuploid cells, we performed epistasis analysis between genes that regulate ceramide synthesis and aneuploidy. If increased ceramide levels serve to slow cell cycle progression, deleting a gene that lowers ceramide synthesis may enhance the proliferation of aneuploid cells. To test this hypothesis, we deleted the ceramide synthase LAG1 in 12 disomes and compared the doubling times with and without the LAG1 deletion (lag1Δ). Remarkably, we found that loss of LAG1 enhanced the proliferation of 9 disomes at 30°C (Figure S4A). Consistently, at 37°C where the difference in proliferation between disomes and wild type cells is more apparent, all disomes showed improved proliferation upon loss of LAG1 (Figures 4A and S4B).
Figure 4. Reduced Ceramide Synthesis Improves the Fitness of Aneuploid Cells.
(A) Doubling times of disomes (black bars) and disomes harboring lag1Δ (open bars) relative to WT at 37°C.
(B) Doubling times of disomes (black bars) and disomes harboring lcb3Δ (open bars) relative to WT at 37°C.
(C) Doubling times of disomes (black bars) and disomes harboring lcb4Δ (open bars) relative to WT at 37°C.
The average of 3 biological replicates is shown. Error bars represent +/− standard deviation (SD). * represent p-value < 0.05, paired Student’s t-test.
See also Figure S4.
SVF1, the gene mutated in evolved isolates of disome XIV cells acts in concert with the LCB kinase LCB4 and the LCB-P phosphatase LCB3 to control the phosphorylation of LCBs (Brace et al., 2007). The phosphorylation-dephosphorylation cycle of LCBs is required for their proper compartmentalization and acylation by ceramide synthases (Funato et al., 2003; Qie et al., 1997). Consequently, loss of LCB3, LCB4, or SVF1 lowers ceramides and may suppress cell cycle defects in aneuploid cells (Figures S4D and S4E)(Mandala et al., 1998). Remarkably, we found that deletion of any one of these three genes suppresses the proliferation defects of several aneuploid strains. Indeed, in addition to disome XVI, the proliferation of 5 other disomes improved upon deleting SVF1 at 30°C (Figure S4C). Moreover, deletion of LCB3 significantly improved the proliferation of 7 disomes at 30°C (Figure S4D) and that of 9 disomes at 37°C (Figure 4B). Furthermore, while LCB4 loss of function did not affect the fitness of wild type cells, it significantly improved the fitness of 5 disomes at 30°C (Figure S4E) and that of 8 disomes at 37°C (Figure 4C). The se results indicate that distinct gene mutations leading to lowered ceramide synthesis have beneficial effects on the proliferation of the aneuploid strains.
Increased Ceramide Levels Can Be Exploited to Target Aneuploid Cells
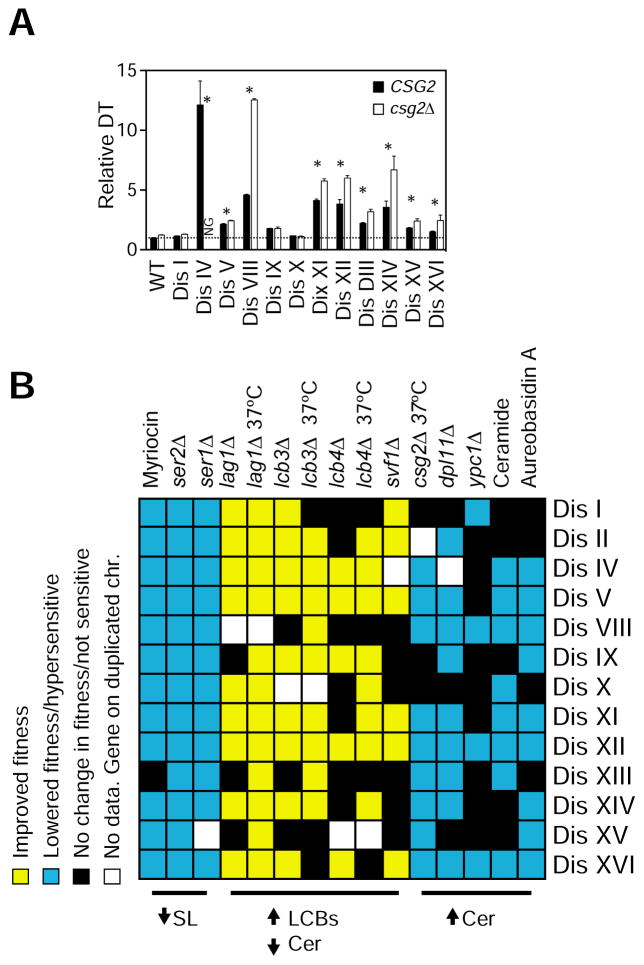
Because ceramides are elevated in the disomes and ceramides specifically arrest cells in G1 (Figure S5A), we next examined whether further increases in ceramide levels could be exploited to kill aneuploid cells. To this end, we performed three different genetic approaches that can result in elevated ceramide levels. First, we analyzed the consequences of deleting CSG2, a protein that regulates the synthesis of complex sphingolipids (Beeler et al., 1997). We found that 9 out of 12 disomes harboring csg2Δ show impaired fitness and that their proliferation is significantly affected at 37°C (Figure 5A). Second, we examined the effects o f inhibiting the hydrolysis of sphingolipids by deleting the LCB lyase DPL1 (Cowart et al., 2010; Saba et al., 1997). We found that loss of DPL1 is deleterious to the majority of the disomes (9 out of 12 disomes, Figure S5B). Lastly, we examined the effects of deleting the ceramidase YPC1 and found that this deletion reduces the fitness of four disomes (I, VIII, XII and XVI; Figure S5C). We hypothesize that expression of the other yeast ceramidase YDC1 may compensate for the loss of YPC1. We could not test the effects of deleting both ceramidases due to lethality to yeast.
Figure 5. Effects of Shifting the Balance between Ceramide and Long-chain Bases in Aneuploid Strains.
(A) Doubling times of disomes (black bars) and disomes harboring csg2Δ (open bars) relative to WT at 37°C. The average of 3 biological replicates is shown. Error bars represent +/− standard deviation (SD). * represent p-value < 0.05, paired Student’s t-test.
(B) Summary of the effects of several genes deletions and drugs that target the synthesis of sphingolipids on the fitness of thirteen disomes.
See also Figure S5.
To validate these findings, we compared the viability of cells grown in rich medium alone or in the presence of 20 μM C2-ceramide. We found that the proliferation of the majority of the disomes is impaired compared to wild type cells in the presence of C2-ceramide (IV, V, VIII, X, XI, XII, XIII, XIV, XV and XVI; Figure S5D). We next examined the effects of increasing ceramide levels in the disomes by inhibiting their conversion to complex sphingolipids by treating cells with Aureobasidin A, a specific inhibitor of AUR1, the inositol phosphorylceramide synthase (Nagiec et al., 1997). We found that most disomes are hypersensitive to this drug compared to wild type cells (IV, V, VIII, IX, XI, XII, XIV, XV, and XVI; Figure S5E). These results demonstrate that the majority of aneuploid strains are sensitive to drugs that elevate ceramide levels, with a few exceptions. Given the complexity and number of genes involved in the regulation of ceramide synthesis, expression of one or more genes on the duplicated chromosomes may confer resistance to these drugs or help cells tolerate increases in ceramide levels.
In sum, our studies reveal that loss of function of either LAG1, LCB3, LCB4 or SVF1, which leads to increased LCB and lowered ceramide levels, improves the proliferation of the majority of aneuploid strains; in contrast, increasing ceramide levels either genetically or pharmacologically, decreases their fitness (Figure 5B). The concomitant increase in the production of LCBs and ceramides in response to aneuploidy supports the hypothesis that ceramides slow down the proliferation of aneuploid cells while LCBs promote their survival.
Loss of LCB3 Remodels Membrane Protein Composition of Aneuploid Cells
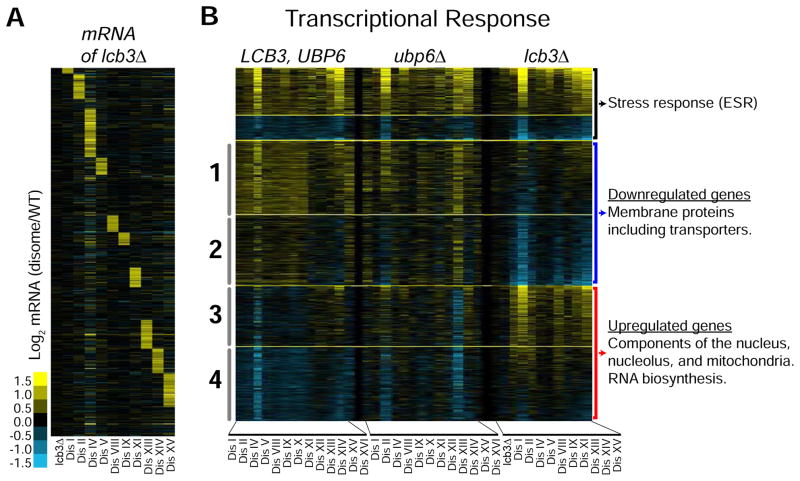
To gain mechanistic insight into how sphingolipids improve the fitness of aneuploid cells, we analyzed the transcriptome profiles of 10 disomes harboring lcb3Δ compared to wild type cells (Figures 6A and Table S4). We focused on the effects of LCB3 loss for two reasons: a- competition assays revealed that whereas the fitness of wild type cells is not affected by lcb3Δ, that of 9 disomes improved significantly at 30°C (Figures S6A); b- MS measurements revealed that loss of LCB3 restores ceramide levels close to those of wild type, while significantly increasing LCB levels (Figures S6B and S6C). This allows us to determine gene expression changes mainly driven by increases in LCBs. In addition, we included in our analysis the transcriptional profiles of 13 disomes without any deletions (LCB3, UBP6) and 13 disomes harboring ubp6Δ (Dephoure et al., 2014). We refer to the latter 26 strains as control strains.
Figure 6. Loss of LCB3 Remodels Membrane Protein Composition and Promotes RNA biosynthesis.
(A) Gene expression analysis of disomes harboring lcb3Δ grown in batch culture, ordered by chromosome position. Experiments (columns) are ordered by the number of the chromosomes present in two copies. Data were normalized to account for the extra chromosome present in disomic strains. Upregulated genes are show in yellow and downregulated ones in blue.
(B) Gene expression of disomes, disome harboring ubp6Δ, and disomes harboring lcb3Δ. Columns represent experiments. Rows are genes organized by pattern of expression of disomes-lcb3Δ compared to the control strains (13 disomes and 13 disomes-ubp6Δ). Top two clusters are up- and downregulated in all 36 strains. Clusters 1 and 2 are downregulated by lcb3Δ. Clusters 3 and 4 are upregulated by lcb3Δ. See Table S4 for GO enrichment analysis details.
We found that 62% of the transcriptome of the disomes with lcb3Δ show significant changes compared to wild type cells (3,107 of 5,016 genes). Sorting by intensities, we identified groups of genes with similar profiles and categorized them into several clusters (Figure 6B, see Supplemental Experimental Procedures). Cluster 1 includes 655 genes that are upregulated in the control strains but are significantly attenuated upon loss of LCB3. Cluster 2 includes 624 genes whose expression is not altered in the control strains but are downregulated by the lcb3Δ. Gene Ontology enrichment (GO) analysis revealed that genes encoding membrane proteins are significantly enriched in clusters 1 and 2 (n = 496, p-value = 8E-26, Table S4). Among these are several transmembrane transporters (n = 145, p-value = 5E-08, Table S4). Notable downregulated genes include ATP-binding cassette (ABC) transporters and transporters of metabolites such as amino acids, uracil, copper, and phosphate. LCBs, whose levels increase upon loss of LCB3, are known to downregulate nutrient intake by mechanisms that are not fully understood (Chung et al., 2001; Skrzypek et al., 1998). Our results indicate that transcriptional regulation of transporters is one mechanism by which LCBs regulate nutrient intake.
Loss of LCB3 Upregulates Genes Involved in Transcription and Metabolic Processes
Cluster 3 includes 533 genes that do not significantly change in the disomes but are upregulated by lcb3Δ. Cluster 4 includes 657 genes that are downregulated in the disomes and their levels increase upon loss of LCB3. GO analysis revealed a significant enrichment for genes encoding proteins that localize to the nucleus in these clusters (n = 558, p-value = 3E-17), including nucleolar proteins (n = 96, p-value = 3E-06), and proteins that regulate RNA biosynthesis (n = 361, p-value = 7E-11)(Figure 6B and Table S4). In addition, within the most upregulated genes, we found a significant enrichment for those that regulate metabolic processes including mitochondrial genes (p-value = 1E-06). Our results indicate that accumulation of LCBs upon loss of LCB3 activates signaling pathways that drive the described transcriptional changes, raising the hypothesis that changes in sphingolipids improve the fitness of aneuploid cells by altering membrane physiology, promoting RNA biosynthesis, and affecting metabolic functions. Notably, genes whose levels increase or decrease in all 36 aneuploid strains are associated with a transcriptional response termed the environmental stress response and are not affected by the loss of LCB3 in the disomes (Gasch et al., 2000).
Consequences of LCB3 Loss of Function on Cellular Protein Composition of Aneuploid Cells
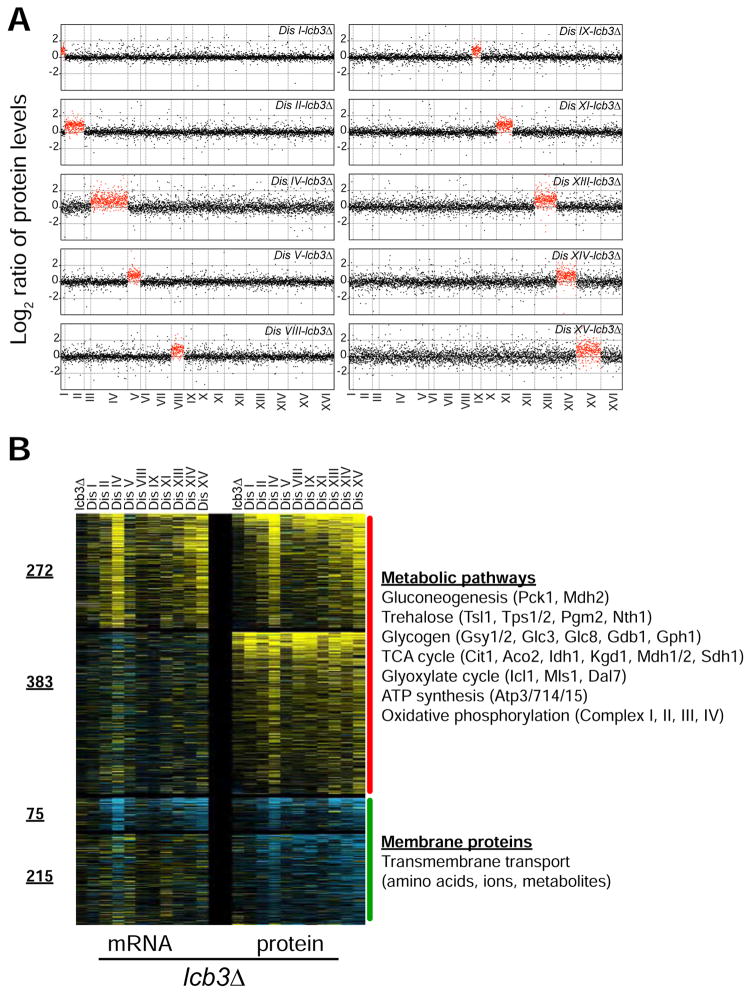
To gain further insight into how sphingolipids improve fitness of aneuploid cells, we used TMT-based quantitative mass spectrometry to determine the proteome profiles of 10 disomes harboring lcb3Δ (72% of verified ORFs, n = 3,682, Table S5). As previously shown for the control strains, we found that, on average, increases in gene copy number lead to proportional changes in protein levels in 10 disomes analyzed, all harboring lcb3Δ (Figure 7A). In addition, we found that protein levels of duplicated genes that code for subunits of macromolecular complexes are attenuated to the same extent as in the control strains (Figure S7A).
Figure 7. Loss of LCB3 Remodels Membrane Protein Composition and Promotes Anabolism and Mitochondrial Function.
(A) The plots show the log2 ratio of the relative protein abundance of disomes-lcb3Δ compared to WT. Protein levels are shown in the order of the chromosomal location of their encoding genes. Protein levels of duplicated chromosomes are shown in red.
(B) Transcript and protein abundances of genes that are specifically up- and down-regulated in the disomes upon the loss of LBC3. See Table S5 for GO enrichment analysis details.
To reveal the most relevant changes in protein levels elicited by the loss of LCB3, we sorted by the averaged log2 ratios and identified a subset of proteins that specifically change in the disomes upon loss of LCB3 (Figure 7B). We found that 655 and 290 proteins are upregulated and downregulated, respectively, in the disomes harboring lcb3Δ. Consistent with the transcriptome analysis, we found that the subset of downregulated proteins is enriched for membrane proteins while those that are upregulated are significantly enriched for metabolic processes (Figure 7B). GO term analysis of the upregulated proteins revealed more specific cellular processes than the transcriptome analysis because as much as 58% (383 proteins) of the upregulation in protein levels takes place independent of transcription. Remarkably, several metabolic pathways associated with cellular anabolism and energy production were induced by the loss of LCB3 (Figure 7B). Among the most enriched proteins were enzymes involved in gluconeogenesis, the TCA and glyoxylate cycles, trehalose and glycogen biosynthesis and breakdown, and ATP synthesis. In addition, among the most upregulated proteins was Hsp12, a membrane protein involved in maintaining membrane integrity and required for dietary restriction-induced lifespan extension (Herbert et al., 2012). These results are consistent with our conclusions from the transcriptome analysis, and indicate that LCBs improve the fitness of aneuploid cells through transcriptional and posttranscriptional mechanisms to remodel membrane protein composition and promote metabolic pathways associated with anabolism and ATP synthesis.
Importantly, loss of LCB3 did not attenuate the levels of overexpressed proteins in the disomes, indicating that the mechanisms by which changes in sphingolipids improve the fitness of aneuploid cells are distinct from those induced by the loss of UBP6 (Figure S7B). In support for this, we found that concomitant loss of UBP6 and LCB3 enhances the fitness of aneuploid cells to a greater extent than the loss of either alone, indicating a cooperation between both gene deletions (Figure S7C).
DISCUSSION
Aneuploidy Increases De Novo Synthesis of Sphingolipids
Here, we found that aneuploid cells accumulate higher levels of LCBs and ceramides and that this accumulation plays a key role in the cellular response to aneuploidy. Our studies support the hypothesis that LCBs activate survival pathways and ceramides function to slow down cell cycle progression in response to aneuploidy. Remarkably, we found that the changes in lipid composition elicited by aneuploidy are specific to sphingolipids, rather than any other lipid class. Furthermore, we provide the important finding that changes in sphingolipid levels are driven by increased serine biosynthesis, which itself is driven by enhanced glucose utilization and upregulation of the Ser2 protein levels. In addition, we found that decreased serine synthesis significantly alters sphingolipid composition indicating that sphingolipid levels are sensitive to the amount of intracellular serine available for their synthesis. These results indicate that altered serine metabolic demand can elicit dramatic physiological responses associated with sphingolipid biology. Serine serves as a precursor molecule for nucleotides, amino acids, and other lipids, and its synthesis is increased in a large proportion of human tumors (Locasale et al., 2011; Possemato et al., 2011). In cancer, it is recognized that serine fuels metabolic pathways associated with the one-carbon cycle (Locasale, 2013). Here we demonstrate that as compared to euploids, aneuploid cells are dependent on increased serine utilization, which can lead to altered sphingolipid content and improved cellular fitness. As a result, serine and sphingolipid synthesis represent an important metabolic dependency of aneuploid cells.
Lowering Ceramides Improves the Fitness of Aneuploid Cells
Inhibition of the synthesis of both LCBs and ceramides by myriocin treatment is lethal to aneuploid cells. However, lowering ceramide synthesis while increasing LCB enhances their proliferation. These results suggest that aneuploid cells strictly rely on the survival functions of LCBs. If ceramide slows down the cell cycle, why would aneuploid cells accumulate higher levels of this molecule? Endogenous levels of LCBs are coupled to those of ceramides, as these molecules are generated from each other and are in a physiological balance. As it happens in response to extrinsic stresses, both the “good” (LCBs) and the “bad” (ceramide) lipids increase in response to aneuploidy. Thereby, higher ceramide levels may increase the selective pressure to acquire mutations to lower their synthesis. Lipidomics measurements indicate that the combined cellular amounts of LCBs and ceramides are two orders of magnitude lower than their products, the complex sphingolipids (0.1% vs. 15% of total lipids, Figure S1). This suggests that a high metabolic flux exists through the pathway and that minor changes in enzyme activities or substrate concentrations could lead to significant changes in the levels of LCBs and ceramides. Four enzymatic activities control the interconversion of LCBs and ceramides, each encoded by at least 2 genes: LCB kinase (LCB4 and LCB5), LCB-P phosphatase (LCB3 and YRS3), ceramide synthase (LAG1 and LAC1) and ceramidase (YPC1 and YDC1). Given this genetic redundancy, we found that reducing ceramide levels while increasing those of LCBs (through deletion one of two genes that hamper LCB kinase activities, LCB-P phosphatase or ceramide synthase), improves the fitness of aneuploid cells. Increasing ceramide levels by lowering ceramidase activity has the opposite effect. Interestingly, most of the deletions included in our studies have little effect on the proliferation of wild type cells under non-stress conditions. This suggests that the genetic redundancy of this essential pathway that is conserved and expanded in humans -where at least 6 different genes code for ceramide synthases and 5 for ceramidases (Coant et al., 2016; Wegner et al., 2016)- allows flexibility of regulation during cellular responses to stress.
Long-chain Bases Promote Anabolism and Energy Production in Aneuploid Cells
Identifying a specific cellular process affected by changes in sphingolipid levels is experimentally challenging, because sphingolipids regulate many aspects of cellular physiology. Because the lcb3 deletion 1) improves the fitness of aneuploid cells without affecting wild type cells, and 2) increases LCB levels while restoring ceramide levels closer to those of wild type levels, we were able to identify specific cellular processes affected by LCBs in aneuploid cells. Nearly 50% of the cellular transcriptome is affected in the disomes when LCB3 is deleted. We found that nutrient transporters are transcriptionally downregulated by LCBs, indicating that transcription factors can act downstream of LCBs. Counter intuitively, restriction of nutrient intake could be beneficial for aneuploid cells. In a manner similar to how dietary restriction promotes metabolic changes contributing to increased longevity and resistance to stress (Fontana et al., 2010), remodeling the cell’s membrane protein composition in aneuploid cells may promote metabolic pathways associated with biosynthesis and energy production. These metabolic changes in turn are beneficial to aneuploid cells because gaining an extra chromosome increases the metabolic demands driven by enhanced transcription and protein synthesis, folding, and turnover. Consistently, loss of LCB3 or LAG1 (Longevity Assurance Gene) induces resistance to stress and increases lifespan in yeast (D’Mello N et al., 1994; Skrzypek et al., 1999). Our results provide important insights into the molecular mechanisms by which several mutations in the sphingolipid pathway can promote longevity and enhance resistance to stress.
The mechanisms by which loss of LCB3 improve the fitness of aneuploid cells are distinct from those elicited by the loss of UBP6. This highlights the complexity of the cellular responses to aneuploidy and indicates that parallel pathways can be acted upon to overcome the negative effects of aneuploidy. It is important to note that the fitness of the disomes harboring both deletions is lower than that of wild type cells, indicating that other mutations could be acquired to further improve fitness.
Targeting sphingolipids in human diseases
Aneuploid primary MEFs, cancer cell lines, and human tumors show elevated levels of sphingolipids. Despite numerous studies exploiting this correlation to target cancer cells, a general mechanism explaining how cells accumulate higher levels of sphingolipids is lacking. Here we show that aneuploidy increases the biosynthesis of sphingolipids and that aneuploid cells are highly dependent on serine and sphingolipid synthesis. This metabolic codependency could therefore be exploited therapeutically to specifically target cancers, the vast majority of which are aneuploid. Furthermore, individuals with Trisomy 21 suffer from several immunological disorders, increased risk for leukemias, premature aging, and neurodegenerative disorders such as Alzheimer’s. Consequently, our results also provide a novel framework to study the mechanisms affecting sphingolipid metabolism in Down Syndrome patients.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Conditions
All stains are derivatives of W303 (E187). Detailed methodologies, a list of strains and media conditions are provided in Supplemental Experimental Procedures.
Growth Rate Measurements
All cells were grown overnight to OD600 between 0.5 to 2 and then diluted to a starting OD600 of 0.2 in 100 μl of fresh medium in 96 well-plates. Cells growth was monitored using a Tecan Infinite PRO microplate reader at indicated temperatures with continuous shaking. Relative doubling times (DT) are simply the ratio of the DT of the strain divided by the DT of the WT measured in identical experimental conditions.
Mass Spectrometry of Sphingolipids
Lipodomics of wild type cells and thirteen disomes were outsourced to the Lipidomics Shares Resource at the Medical University of South Carolina. Cells were treated with 5% trichloroacetic acid for 10 minutes on ice and washed with water 3 times before shipping in dry ice. Three independent cultures were analyzed for each strain and relative levels of all lipids were normalized to total cell numbers which were determined using a Beckman Coulter Counter. Protocols for lipid extraction and analysis are described in Bielawski et al. (2009)(Bielawski et al., 2009).
Global Lipodomics of Yeast Cells
The lipid compositions of total cell extracts were determined by quantitative shotgun lipidomic analysis as previously described Ejsing et al. (2009)(Ejsing et al., 2009; Klose et al., 2012)(Supplemental Experimental Procedures).
Gene Expression and Proteome Quantification
Cells were grown overnight at 30°C in selective medium. Batch cultures were diluted to OD600 = 0.2 into YEPD medium the next day and harvested once they reached an OD600 = 1.0. Gene expression was analyzed using Agilent microarrays as previously described in Torres et al. 2007. Proteome analysis was performed described in Dephoure et al. 2014. See Supplementary Experimental Procedures for details.
Supplementary Material
Highlights.
Aneuploid cells rely on increased serine synthesis to proliferate
Increased serine synthesis leads to the accumulation of sphingolipids
Mutations that lower ceramide levels increase the fitness of aneuploid cells
Combined inhibition of serine and sphingolipid synthesis is lethal to aneuploid cells
Acknowledgments
We are thankful to Sofia Ordonez, Raquelle Torres, and Aracelli Acevedo for technical assistance. We are grateful to Jennifer Benanti and Angelika Amon for reagents. We thank Nada Kalaany for critical reading of the manuscript. This research was supported by the Richard and Susan Smith Family Foundation and the Searle Scholars Program to EMT. This work was also supported by a grant from the National Institutes of Health grant 1R01GM118481-01A1 to EMT.
Footnotes
Accession Numbers
The accession number for the microarray data is GSE93762.
Author Contributions
E.M.T. designed the study and E.M.T., S.H., H.T.G., C.O., and G.B. performed all experiments. X.H. and R.C.D. contributed to the design of lipidomics experiments and performed qHPLC. C.K. and A.S. contributed to the design of lipidomics experiments and performed LC–MS. P.C and N.D. contributed to the design and performed proteomics experiments. All authors contributed to data analysis and discussion. E.M.T. wrote the paper and all authors contributed to editing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez-Vasquez F, Sims KJ, Cowart LA, Okamoto Y, Voit EO, Hannun YA. Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature. 2005;433:425–430. doi: 10.1038/nature03232. [DOI] [PubMed] [Google Scholar]
- Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC, Martin A. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nature structural & molecular biology. 2015;22:712–719. doi: 10.1038/nsmb.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach RR, Ricci-Tam C, Brennan CM, Moomau CA, Hsu PH, Hua B, Silberman RE, Springer M, Amon A. Aneuploidy Causes Non-genetic Individuality. Cell. 2017;169:229–242. e221. doi: 10.1016/j.cell.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler TJ, Fu D, Rivera J, Monaghan E, Gable K, Dunn TM. SUR1 (CSG1/BCL21), a gene necessary for growth of Saccharomyces cerevisiae in the presence of high Ca2+ concentrations at 37 degrees C, is required for mannosylation of inositolphosphorylceramide. Mol Gen Genet. 1997;255:570–579. doi: 10.1007/s004380050530. [DOI] [PubMed] [Google Scholar]
- Bielawski J, Pierce JS, Snider J, Rembiesa B, Szulc ZM, Bielawska A. Comprehensive quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods Mol Biol. 2009;579:443–467. doi: 10.1007/978-1-60761-322-0_22. [DOI] [PubMed] [Google Scholar]
- Brace JL, Lester RL, Dickson RC, Rudin CM. SVF1 regulates cell survival by affecting sphingolipid metabolism in Saccharomyces cerevisiae. Genetics. 2007;175:65–76. doi: 10.1534/genetics.106.064527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic acids research. 2012;40:D700–705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung N, Mao C, Heitman J, Hannun YA, Obeid LM. Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. The Journal of biological chemistry. 2001;276:35614–35621. doi: 10.1074/jbc.M105653200. [DOI] [PubMed] [Google Scholar]
- Coant N, Sakamoto W, Mao C, Hannun YA. Ceramidases, roles in sphingolipid metabolism and in health and disease. Adv Biol Regul. 2016 doi: 10.1016/j.jbior.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart LA, Hannun YA. Selective substrate supply in the regulation of yeast de novo sphingolipid synthesis. The Journal of biological chemistry. 2007;282:12330–12340. doi: 10.1074/jbc.M700685200. [DOI] [PubMed] [Google Scholar]
- Cowart LA, Okamoto Y, Pinto FR, Gandy JL, Almeida JS, Hannun YA. Roles for sphingolipid biosynthesis in mediation of specific programs of the heat stress response determined through gene expression profiling. The Journal of biological chemistry. 2003;278:30328–30338. doi: 10.1074/jbc.M300656200. [DOI] [PubMed] [Google Scholar]
- Cowart LA, Shotwell M, Worley ML, Richards AJ, Montefusco DJ, Hannun YA, Lu X. Revealing a signaling role of phytosphingosine-1-phosphate in yeast. Molecular systems biology. 2010;6:349. doi: 10.1038/msb.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Mello NP, Childress AM, Franklin DS, Kale SP, Pinswasdi C, Jazwinski SM. Cloning and characterization of LAG1, a longevity-assurance gene in yeast. The Journal of biological chemistry. 1994;269:15451–15459. [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife. 2014:e03023. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC, Lester RL. Sphingolipid functions in Saccharomyces cerevisiae. Biochimica et biophysica acta. 2002;1583:13–25. doi: 10.1016/s1388-1981(02)00210-x. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Nagiec EE, Skrzypek M, Tillman P, Wells GB, Lester RL. Sphingolipids are potential heat stress signals in Saccharomyces. The Journal of biological chemistry. 1997;272:30196–30200. doi: 10.1074/jbc.272.48.30196. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez-Roman R, Pienik R, Futerman AH. Increased ceramide synthase 2 and 6 mRNA levels in breast cancer tissues and correlation with sphingosine kinase expression. Biochemical and biophysical research communications. 2010;391:219–223. doi: 10.1016/j.bbrc.2009.11.035. [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K, Lombardi R, Vallee B, Riezman H. Lcb4p is a key regulator of ceramide synthesis from exogenous long chain sphingoid base in Saccharomyces cerevisiae. The Journal of biological chemistry. 2003;278:7325–7334. doi: 10.1074/jbc.M209925200. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillermet-Guibert J, Davenne L, Pchejetski D, Saint-Laurent N, Brizuela L, Guilbeau-Frugier C, Delisle MB, Cuvillier O, Susini C, Bousquet C. Targeting the sphingolipid metabolism to defeat pancreatic cancer cell resistance to the chemotherapeutic gemcitabine drug. Molecular cancer therapeutics. 2009;8:809–820. doi: 10.1158/1535-7163.MCT-08-1096. [DOI] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Herbert AP, Riesen M, Bloxam L, Kosmidou E, Wareing BM, Johnson JR, Phelan MM, Pennington SR, Lian LY, Morgan A. NMR structure of Hsp12, a protein induced by and required for dietary restriction-induced lifespan extension in yeast. PloS one. 2012;7:e41975. doi: 10.1371/journal.pone.0041975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GM, Richards A, Wahl T, Mao C, Obeid L, Hannun Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. The Journal of biological chemistry. 1997;272:32566–32572. doi: 10.1074/jbc.272.51.32566. [DOI] [PubMed] [Google Scholar]
- Lester RL, Dickson RC. High-performance liquid chromatography analysis of molecular species of sphingolipid-related long chain bases and long chain base phosphates in Saccharomyces cerevisiae after derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate. Anal Biochem. 2001;298:283–292. doi: 10.1006/abio.2001.5368. [DOI] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nature reviews Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandala SM, Thornton R, Tu Z, Kurtz MB, Nickels J, Broach J, Menzeleev R, Spiegel S. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:150–155. doi: 10.1073/pnas.95.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane E, Sin C, Zauber H, Wells JN, Donnelly N, Wang X, Hou J, Chen W, Storchova Z, Marsh JA, et al. Kinetic Analysis of Protein Stability Reveals Age-Dependent Degradation. Cell. 2016;167:803–815. e821. doi: 10.1016/j.cell.2016.09.015. [DOI] [PubMed] [Google Scholar]
- Morad SA, Cabot MC. Ceramide-orchestrated signalling in cancer cells. Nature reviews Cancer. 2013;13:51–65. doi: 10.1038/nrc3398. [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, Lester RL, Dickson RC. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. The Journal of biological chemistry. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- Nickels JT, Broach JR. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes & development. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes & development. 2012;26:2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qie L, Nagiec MM, Baltisberger JA, Lester RL, Dickson RC. Identification of a Saccharomyces gene, LCB3, necessary for incorporation of exogenous long chain bases into sphingolipids. The Journal of biological chemistry. 1997;272:16110–16117. doi: 10.1074/jbc.272.26.16110. [DOI] [PubMed] [Google Scholar]
- Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19222–19227. doi: 10.1073/pnas.1116948108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba JD, Nara F, Bielawska A, Garrett S, Hannun YA. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. The Journal of biological chemistry. 1997;272:26087–26090. doi: 10.1074/jbc.272.42.26087. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nature reviews Molecular cell biology. 2015;16:473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Inhibition of amino acid transport by sphingoid long chain bases in Saccharomyces cerevisiae. The Journal of biological chemistry. 1998;273:2829–2834. doi: 10.1074/jbc.273.5.2829. [DOI] [PubMed] [Google Scholar]
- Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J Bacteriol. 1999;181:1134–1140. doi: 10.1128/jb.181.4.1134-1140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Molecular systems biology. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Yuwen H, Wang K, Bruno PM, Bullock K, Deik A, Santaguida S, Trakala M, Pfau SJ, Zhong N, et al. Aneuploid Cell Survival Relies upon Sphingolipid Homeostasis. Cancer research. 2017;77:5272–5286. doi: 10.1158/0008-5472.CAN-17-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E. Yeast as Models of Mitotic Fidelity. Recent Results Cancer Res. 2015;200:143–164. doi: 10.1007/978-3-319-20291-4_7. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres EM, Springer M, Amon A. No current evidence for widespread dosage compensation in S. cerevisiae. eLife. 2016;5:e10996. doi: 10.7554/eLife.10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner MS, Schiffmann S, Parnham MJ, Geisslinger G, Grosch S. The enigma of ceramide synthase regulation in mammalian cells. Prog Lipid Res. 2016;63:93–119. doi: 10.1016/j.plipres.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Yona AH, Manor YS, Herbst RH, Romano GH, Mitchell A, Kupiec M, Pilpel Y, Dahan O. Chromosomal duplication is a transient evolutionary solution to stress. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21010–21015. doi: 10.1073/pnas.1211150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.