Abstract
Small basic proteins present in most Archaea share a common ancestor with the eukaryotic core histones. We report the crystal structure of an archaeal histone-DNA complex. DNA wraps around an extended polymer, formed by archaeal histone homodimers, in a quasi-continuous superhelix, with the same geometry as DNA in the eukaryotic nucleosome. Substitutions of a conserved glycine at the interface of adjacent protein layers destabilize archaeal chromatin, reduce growth rate and impair transcription regulation, confirming the biological importance of the polymeric structure. Our data establish that the histone-based mechanism of DNA compaction predates the nucleosome, shedding light on the origin of the nucleosome.
Main text
The nucleosome consists of two (H2A-H2B) and (H3–H4) histone heterodimers assembled as an octamer that wraps 147 base pairs of DNA in 1.65 negative superhelical turns (1). Histones, the most conserved proteins known, all have a central ‘histone fold’ (HF) dimerization motif formed by three α-helices separated by two short loops (Fig. S1A). Small HF-containing proteins, present in most Archaea, likely share a common ancestor with the eukaryotic histones (2–4). Hundreds of different archaeal histone sequences are now known [Fig. S1B; (5, 6)]. Most are 70±5 amino acids long and lack HF extensions and the basic histone tails, the segments unique to each eukaryotic histone that contribute to nucleosome stability and gene regulation [Fig. S1A; (3, 7)]. Unlike the mandatory eukaryotic histone heterodimer partnerships, archaeal histones homodimerize and form heterodimers with related paralogs. Here we report the structure of archaeal histone-based chromatin, and its participation in gene expression.
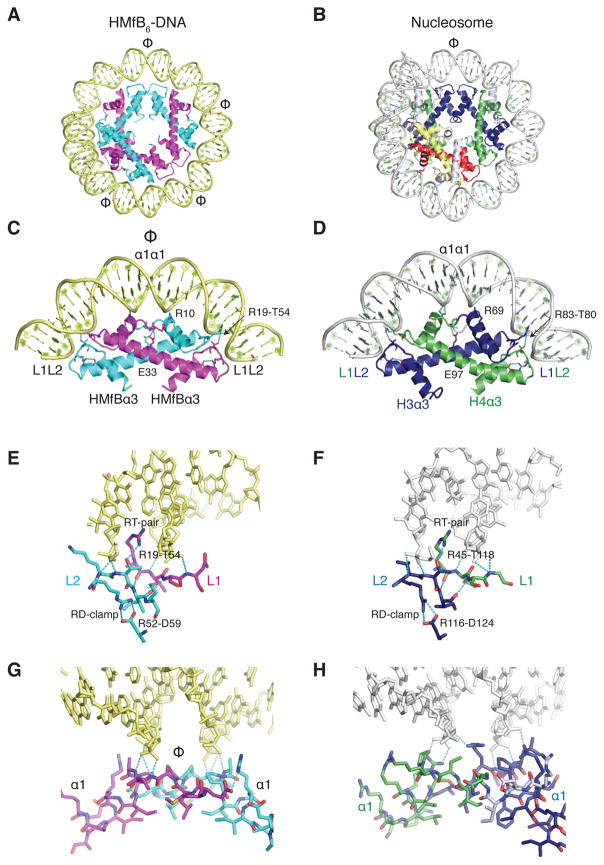
To obtain crystals, we used a DNA sequence to which homodimers of histone B from Methanothermus fervidus [(HMfB)2] bind at defined locations (8, 9). In the 4 Å crystal structure (Table S1), this 90 bp DNA wraps around three (HMfB)2 dimers (Fig. 1A) that are virtually identical when compared to each other, to (HMfB)2 dimers in the absence of DNA [(10); rmsd 0.36 Å], and to the HFs of eukaryotic (H3–H4) and (H2A–H2B) heterodimers (rmsd ~1.7 Å; Fig. 1A, 1B and Fig. S2A). Each histone fold dimer (HFD) interacts with the DNA in a very similar fashion to the eukaryotic HFDs with fully-conserved amino acid side chain interactions (RT-pair and RD-clamp, Fig. 1 and Fig. S2A, S2B) that mutagenesis has confirmed are essential for DNA binding by HMfB (11, 12). Intramolecular hydrogen bonds between the two histones in the (HMfB)2 dimer position the α1 helices and N-termini for optimal interaction with DNA and would direct an N-terminal extension appropriately through the gyres of the surrounding DNA, as seen in H2A and H3 in the nucleosome (Fig. S2C) (7).
Fig. 1. DNA binding is conserved between archaeal and eukaryotic histones.
A) The structure of three (HMfB)2 dimers bound to an 90 bp SELEX DNA is highly similar to the B) nucleosome hexasome, shown by removing one H2A–H2B heterodimer and the histone tails from the published nucleosome structure (1AOI). The axes of symmetry in both protein assemblies are indicated (Φ). C) HFs of an (HMfB) 2 dimer and (D) (H3–H4) heterodimer shown in the same orientation with associated DNA. E) The L1L2 interface of a (HMfB)2 dimer and (F) an (H3–H4) dimer is shown with conserved interactions with DNA. G) The α1α1 interface in an (HMfB)2 dimer and (H) in an (H3–H4) dimer. Further comparisons of the structures formed by HMfB and eukaryotic histones with DNA are shown in Fig. S2. In all figures, although identical, the two HMfB monomers in an (HMfB)2 dimer are colored in cyan and magenta; H3 is blue, H4 is green, H2B is red, and H2A is yellow. Regions of core histones that are not part of the histone fold are shown in white. DNA organized by HMfB is pale yellow; nucleosomal DNA is grey.
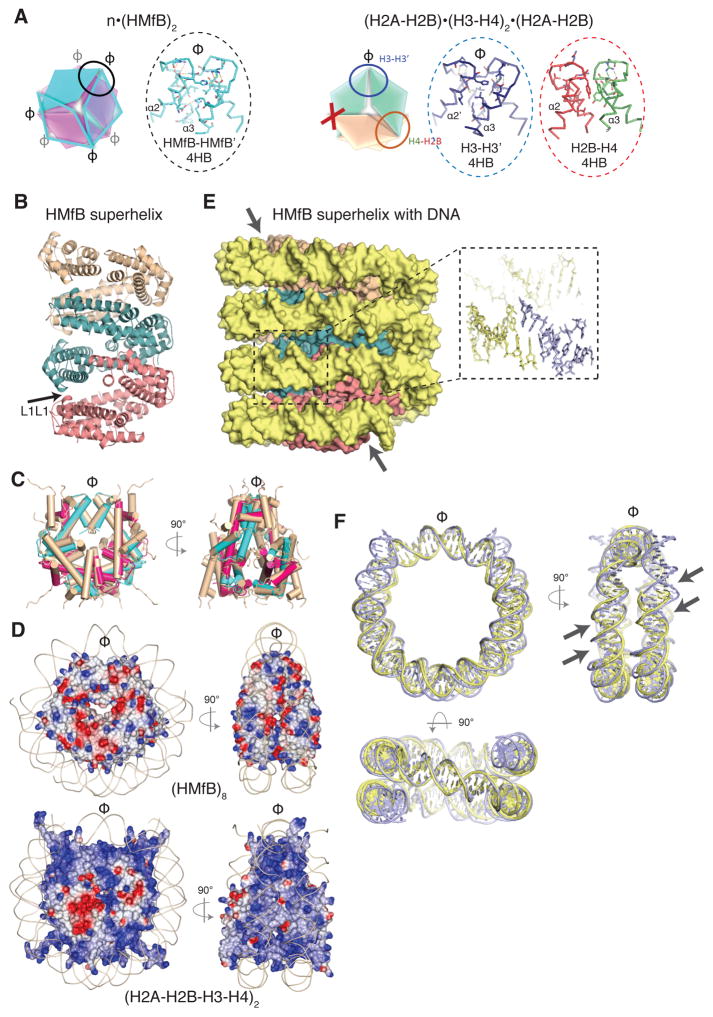
(HMfB)2 dimers are symmetric and, in the crystal lattice, polymerize through identical four α-helix bundles (4HBs; Fig. 2A) to form a continuous helical ramp (Fig. 2B). The geometry of the 4HB is conserved between HMfB-HMfB′, H3–H3′, and H4–H2B (Fig. 2A), and therefore the arrangement of any four consecutive archaeal HFDs in the crystal structure is strikingly similar to the assembly of the four HFDs in the nucleosome octamer (rmsd 2.0 Å, Fig. 2C). The surface of the complex formed by archaeal histones has however less positive charge (Fig. 2D).
Fig. 2. Archaeal histones form a continuous superhelical ramp.
A) Archaeal (HMfB)2 dimers and eukaryotic core histone heterodimers polymerize through the assembly of ‘four helix bundles’ (4HBs) involving the C-termini of α2 and α3 of the HF. While the symmetric (HMfB)2 dimers can continue to polymerize forming a protein fiber with consecutive, identical 4HB bundles (oval, and inset), the asymmetry of eukaryotic core histone dimers prevents (X) continued polymerization. B) Nine (HMfB)2 dimers are shown forming a continuous protein superhelix via 4HB interactions, with groups of three consecutive dimers shown in pink, teal and wheat. Modeling confirmed that the superhelix can also be formed by HMfA homodimers, and by HMfA+HMfB heterodimers. The arrow shows the location of the G16–G16 interaction (L1L1). C) An octamer of archaeal HFs superimposes closely with the eukaryotic histone octamer (wheat colored) in the nucleosome. Helices are shown as tubes with the archaeal histones colored magenta and cyan. D) Archaeal HMfB octamer (top panel) and eukaryotic histone octamer (bottom panel) differ in their charge distribution with a more positively charged helical ramp on the surface of the histone core (the basic histone tails are excluded for clarity). Electrostatic surfaces are calculated in the ccp4mg program and displayed from −0.5 V (red) to 0.5 V (blue). The DNA backbone is shown as a line. E) DNA (shown in space filling mode) wrapped around the HMfB superhelix shown in the same orientation as in panel B). Inset shows a close-up of the annealed 2 nt 5′ extensions. One ‘supergroove’ is indicated by two arrows. F) Superposition of 120 bp of DNA organized by four (HMfB)2 HFDs with 146 bp nucleosomal DNA, shown in three orthogonal orientations; the top two orientations are identical to the orientations shown in C). Two supergrooves (minor and major) are indicated by arrows.
In the crystal lattice, DNA wraps around the HMfB protein assembly in a quasi-continuous superhelix, through annealing of the 2 nt 5′-overhangs (Fig. 2E). The geometry, diameter, pitch and writhe of this superhelix, and the spacing between gyres, strongly resemble the nucleosomal DNA arrangement (Fig. 2F). Consequently, the alignment of DNA grooves (80 bp apart on linear DNA) across two gyres of DNA, termed nucleosomal ‘supergrooves’ (14), is also conserved (arrows in Fig. 2E). The ability of archaeal histones to form polymers was also validated in solution using (HMfB)2, and (HTkA)2 dimers from Thermococcus kodakarensis, confirming that this arrangement is not a crystallographic artefact (Fig. S3 and Table S2). Both (HMfB)2 and (HTkA)2 form complexes that protect 60, 90, 120, 150 and 180 bp fragments from MNase digestion (Fig. S3A), consistent with previous reports (8, 13, 18, 19). Ultracentrifugation further confirmed that the complexes formed on 147 bp and 207 bp DNA molecules contain the predicted number of archaeal histone dimers needed to saturate these DNAs (Fig. S3B and Table S2). In contrast, the polymerization of eukaryotic histone dimers is limited by their asymmetry to an octamer (Fig. 2A, right panel). Notably, the interactions within the archaeal superhelix do not resemble any of the nucleosome-nucleosome stacking interactions reported so far (reviewed in (15), and refs. (16, 17)).
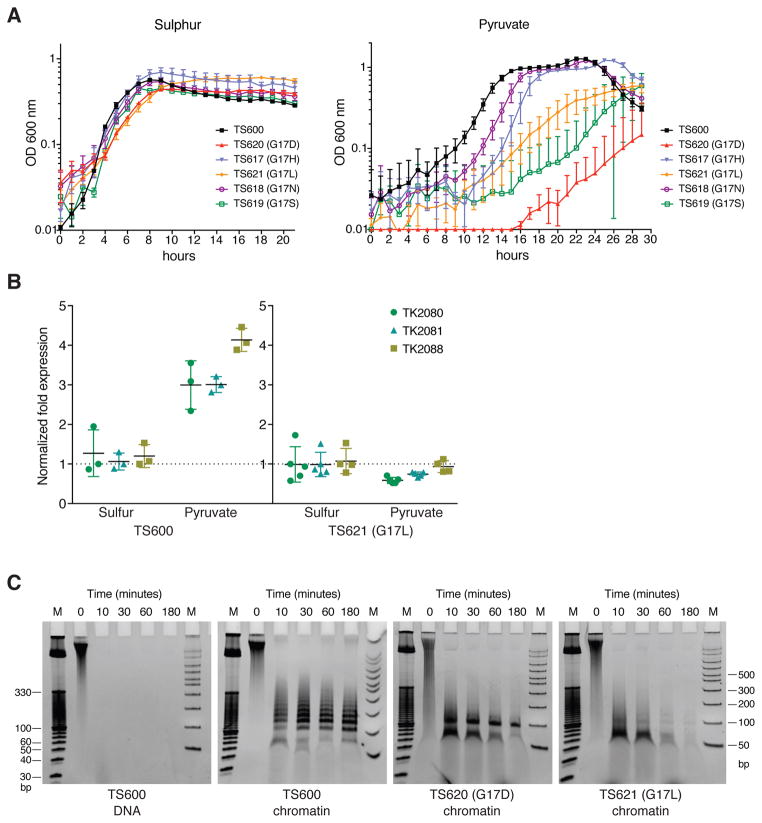
To investigate if the extended polymerization has functional significance, we sought to destabilize the superhelix in vivo without compromising the DNA-binding ability of the archaeal histone. Apart from the 4HBs, the only region of close contact between the adjacent layers of the archaeal histone polymer is where the L1 loops of dimers 1 and 4 meet (arrow in Fig. 2B and Fig. S4A), a position almost always occupied by a glycine (G16 in HMfB, G17 in HTkA; Fig. S1B). To determine if the absence of a side chain facilitates this close packing, we generated T. kodakarensis strains isogenic except for G17 substitutions in HTkA, the single histone present and essential for T. kodakarensis TS600 viability [(18,20); Fig. S5]. Cells with wild type HTkA, transferred from a S˚-containing to a S˚-free medium (pyruvate) restart growth after ~4 hr, during which time they reprogram gene expression [Fig. 3A; (21)]. The otherwise isogenic strains with HTkA G17H, G17D, G17N, G17L or G17S also grew normally in S˚, but took longer to re-start growth when transferred to medium lacking S˚, and some also grew slower (Fig. 3A and Table S3). Given the delayed response to nutrient change, we investigated transcription of the media-dependent MBH hydrogenase-encoding operon (TK2080-TK2093). As previously established (21), transcription of this operon was elevated in T. kodakarensis TS600 with wild-type HTkA when growing in the absence of S˚, but this was not the case for TS621, the strain with HTkA G17L (Fig. 3B), indicating a deregulated transcriptional program.
Fig. 3. Disturbance of layer interfaces affects chromatin structure, gene transcription and growth of T. kodakarensis.
A) Growth curves of T. kodakarensis TS600 and derivative strains with the HTkA G17 substitutions indicated, in medium containing S° and following dilution into a medium lacking S° (pyruvate). Error bars show the SD from three independent experiments, each run with triplicate cultures. B) Quantitative RT-PCR of transcripts of three genes in the hydrogenase operon (TK2080-TK2081-TK2088) present in T. kodakarensis cells containing HTkA or HTkA G17L grown in the presence or absence of S°. Transcripts of TK0895, TK1431 and TK1311 were quantified as constitutively expressed reference genes. Shown is the fold change of the hydrogenase transcripts in cells following dilution into a medium lacking S° (pyruvate). C) DNA fragments generated by MNase digestion of chromatin isolated from T. kodakarensis TS600 and derivative TS620 (G17D) and TS621 (G17L). DNA stripped from histones prior to MNase digestion is shown as a control (TS600). Size standards are in lanes M.
To determine if the negative effects of the G17 substitutions on MBH expression correlated with changes in chromatin structure, chromatin isolated from strains containing HTkA (TS600), HTkA G17L (TS621) and HTkA G17D (TS620), grown with or without S˚, was subjected to MNase digestion. As previously observed, chromatin from TS600 protected fragments ranging from 60 to ~300 bp, in increments of ~30 bp (13), with the most prominent band being 120 bp, corresponding to protection by four (HTkA)2 dimers (Fig. 3C and Fig. S4B). In contrast, digestion of chromatin from TS621 and TS620 generated only ~60 and ~90 bp protected fragments (Fig. 3C, Fig. S4B, Fig. S6A). Both SDS-PAGE and LC-MS/MS confirmed that the intracellular concentrations of HTkA, HTkA G17L and HTkA G17D were similar (Fig. S6B and Table S4). This is consistent with the differences in MNase protection resulting from the inability of the HTkA variants to form a stable extended superhelix. Apparently, substitution of leucine or aspartate for G17 prevents the close adjacent assembly of more than three (HTkA)2 dimers on DNA.
Overall, our data establish that most features of eukaryotic DNA compaction into nucleosomes are conserved in archaeal histone-based chromatin. The histone-mediated DNA geometry within these assemblies is exactly the same. However, archaeal histone-DNA complexes are not limited to one discrete structure. Unlike the defined nucleosome, archaeal histones can form complexes with variable numbers of histone dimers assembled along the DNA (18), and the resulting extended structure plays a role in gene regulation.
Why was the more flexible, variable-length archaeal chromatin structure replaced by a defined nucleosome consisting of four distinct histones very early in eukaryotic evolution? Possibly, with increasing genome size it was necessary to limit histone assembly to defined nucleosomes to allow further compaction into precisely organized but still readily accessible higher-order chromatin. With diversification into four distinct histones and numerous histone variants, plus the addition of HF extensions and tails, eukaryotes further gained the ability to selectively position nucleosomes, have a conserved chromatin architecture recognizable by regulatory proteins, and develop elaborate epigenetic regulation through post-translational modification of histone tails. Intriguingly, some recently identified archaeal histone sequences do have histone tails, hinting at the beginnings of this diversification (Fig. S1B). However, to date, there is no evidence for archaeal functional homologs and thus the ancestry of eukaryotic histone chaperones, chromatin remodelers and post-translational histone regulators remains a challenge (22).
Supplementary Material
Acknowledgments
We thank the University of Colorado BioFrontiers Institute Next-Gen Sequencing Core Facility for performing BioAnalzyer runs and the Protein Expression and Purification Facility at CSU for reagents. This work was supported by NIH grants GM 067777 (to KL), GM53185 (to JNR), GM100329 (to TJS), and GM114594 (to NGA). FM is funded by EMBO (ALTF 1267-2013) and the Dutch Cancer Society (KWF 2014-6649). KL is supported by the Howard Hughes Medical Institute.
Footnotes
FM finalized the structure, designed mutants, performed the AUC and qRT-PCR experiments, contributed to structure analysis, the MNase experiments and manuscript preparation. SB processed and phased the X-ray data, built and refined the model, and helped analyze the structure. PND and KS prepared complexes, obtained crystals and collected data. PND performed in vitro complex analysis, and AEW performed the MNase and histone extraction experiments. TJS, BWB, and KRB constructed and characterized the T. kodakarensis strains and grew biomass. TL and NGA performed mass spectrometry. TJS assisted in manuscript preparation. JNR and KL conceived and directed the project, wrote the manuscript, analyzed the structure, and prepared figures. The structure has been deposited in the protein database (PDB accession code 5T5K).
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Sandman K, Reeve JN. Archaeal histones and the origin of the histone fold. Curr Opin Microbiol. 2006;9:520–525. doi: 10.1016/j.mib.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 4.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 5.Zaremba-Niedzwiedzka K, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 6.Nishida H, Oshima T. Archaeal histone distribution is associated with archaeal genome base composition. J Gen Appl Microbiol. 2016 doi: 10.2323/jgam.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 8.Bailey KA, Pereira SL, Widom J, Reeve JN. Archaeal histone selection of nucleosome positioning sequences and the procaryotic origin of histone-dependent genome evolution. J Mol Biol. 2000;303:25–34. doi: 10.1006/jmbi.2000.4128. [DOI] [PubMed] [Google Scholar]
- 9.Sandman K, Soares D, Reeve JN. Molecular components of the archaeal nucleosome. Biochimie. 2001;83:277–281. doi: 10.1016/s0300-9084(00)01208-6. [DOI] [PubMed] [Google Scholar]
- 10.Decanniere K, Babu AM, Sandman K, Reeve JN, Heinemann U. Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus. J Mol Biol. 2000;303:35–47. doi: 10.1006/jmbi.2000.4104. [DOI] [PubMed] [Google Scholar]
- 11.Sandman K, Louvel H, Samson RY, Pereira SL, Reeve JN. Archaeal chromatin proteins histone HMtB and Alba have lost DNA-binding ability in laboratory strains of Methanothermobacter thermautotrophicus. Extremophiles. 2008;12:811–817. doi: 10.1007/s00792-008-0185-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soares DJ, Sandman K, Reeve JN. Mutational analysis of archaeal histone-DNA interactions. J Mol Biol. 2000;297:39–47. doi: 10.1006/jmbi.2000.3546. [DOI] [PubMed] [Google Scholar]
- 13.Nalabothula N, et al. Archaeal nucleosome positioning in vivo and in vitro is directed by primary sequence motifs. BMC Genomics. 2013;14:391. doi: 10.1186/1471-2164-14-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edayathumangalam RS, Weyermann P, Gottesfeld JM, Dervan PB, Luger K. Molecular recognition of the nucleosomal “supergroove”. Proc Natl Acad Sci U S A. 2004;101:6864–6869. doi: 10.1073/pnas.0401743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan S, Davey CA. Nucleosome structural studies. Curr Opin Struct Biol. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato D, et al. Crystal structure of the overlapping dinucleosome composed of hexasome and octasome. Science. 2017;356:205–208. doi: 10.1126/science.aak9867. [DOI] [PubMed] [Google Scholar]
- 17.Song F, et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 18.Maruyama H, et al. An alternative beads-on-a-string chromatin architecture in Thermococcus kodakarensis. EMBO Rep. 2013;14:711–717. doi: 10.1038/embor.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomschik M, Karymov MA, Zlatanova J, Leuba SH. The archaeal histone-fold protein HMf organizes DNA into bona fide chromatin fibers. Structure. 2001;9:1201–1211. doi: 10.1016/s0969-2126(01)00682-7. [DOI] [PubMed] [Google Scholar]
- 20.Hileman TH, Santangelo TJ. Genetics Techniques for Thermococcus kodakarensis. Front Microbiol. 2012;3:195. doi: 10.3389/fmicb.2012.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santangelo TJ, Cubonova L, Reeve JN. Deletion of alternative pathways for reductant recycling in Thermococcus kodakarensis increases hydrogen production. Mol Microbiol. 2011;81:897–911. doi: 10.1111/j.1365-2958.2011.07734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aravind L, Burroughs AM, Zhang D, Iyer LM. Protein and DNA modifications: evolutionary imprints of bacterial biochemical diversification and geochemistry on the provenance of eukaryotic epigenetics. Cold Spring Harb Perspect Biol. 2014;6:a016063. doi: 10.1101/cshperspect.a016063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.