SUMMARY
A mutation in the promoter of the Telomerase Reverse Transcriptase (TERT) gene is the most frequent noncoding mutation in cancer. The mutation drives unusual monoallelic expression of TERT, allowing immortalization. Here we find that DNA methylation of the TERT CpG Island (CGI) is also allele-specific in multiple cancers. The expressed allele is hypomethylated, which is opposite to cancers without TERT promoter mutations. The continued presence of Polycomb repressive complex 2 (PRC2) on the inactive allele suggests that histone marks of repressed chromatin may be causally linked to high DNA methylation. Consistent with this hypothesis, TERT promoter DNA containing 5-methyl-CpG has much increased affinity for PRC2 in vitro. Thus, CpG methylation and histone marks appear to collaborate to maintain the two TERT alleles in different epigenetic states in TERT promoter-mutant cancers. Finally, in several cancers DNA methylation levels at the TERT CGI correlate with altered patient survival.
Keywords: Telomerase, TERT promoter, Polycomb repressive complex 2, PRC2, 5-methylcytosine, allele-specific, monoallelic, CpG Island, cancer
INTRODUCTION
TERT encodes the catalytic subunit of telomerase, the ribonucleoprotein complex that maintains telomere length in stem cells and most cancer cells (Counter et al., 1992). Multiple cancers show unusual monoallelic activation of TERT by the de novo acquisition of a C>T transition on one TERT promoter (Horn et al., 2013; Huang et al., 2013, 2015; Killela et al., 2013; Stern et al., 2015). These mutations occur at −124 (occasionally at −146) base pairs (bp) from the translational start site and provide a new binding site for the GABPA/B1 transcription factor; the transcriptionally inactive TERT promoter in the same cell bears the H3K27me3 repressive mark (Bell et al., 2015; Stern et al., 2015). In many other cancers, TERT is expressed biallelically or monoallelically by molecular mechanisms that remain poorly understood (Huang et al., 2015).
In addition to H3K27me3, 5-methyl-cytosine (5mC) at CpG dinucleotides is a canonical epigenetic mark of transcriptional silencing (Baylin et al., 1998; Herman, 1999; Herman et al., 1998; Laird and Jaenisch, 1996; Merlo et al., 1995). Here, however, the TERT gene has been an outlier. TERT expression in most previously studied cancers is associated with increased 5mC in the TERT promoter CGI (Barthel et al., 2017; Dessain et al., 2000; Devereux et al., 1999) Thus, in these cancers, TERT transcription occurs despite this gain of 5mC. To explore this non-canonical relationship with 5mC in more detail, we chose to study cancers with heterozygous −124 mutations, capitalizing on the fact that these cells contain TERT alleles maintained in different transcriptional states. We reasoned that if TERT CGI methylation were a positive regulator of TERT mRNA expression in these cells as suggested by previous studies (e.g., Barthel et al., 2017), we should observe higher levels of 5mC on the active promoter-mutant allele.
Contrary to this expectation, our results indicate that 5mC levels at the TERT promoter in cancers with heterozygous −124 mutations are maintained at lower levels on the active allele than on the transcriptionally silent allele. This finding is consistent with the canonical influence of 5mC on transcription. Thus, TERT promoters with −124 mutations exhibit divergent regulatory dynamics compared to those with wild-type (wt) promoters. We find that the EZH2 subunit of PRC2, the enzyme responsible for deposition of H3K27me3, resides at the inactive TERT allele. Testing for a causal relationship between DNA methylation and histone methylation, we show that PRC2 displays a strong binding preference for methylated TERT promoter DNA in vitro. This suggests a regulatory circuit, wherein low 5mC discourages PRC2 binding on active TERT alleles in these −124 cancers.
RESULTS
Distinct 5mC Levels at the TERT CGI in −124 Cancers
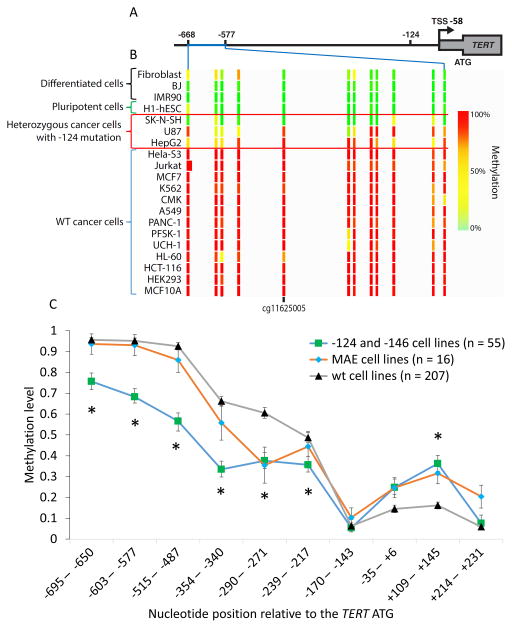
The TERT promoter contains a CGI that is methylated in cancer, and the entire feature extends from near chr5:1296000 (−838 relative to the TERT ATG) to a position near the end of exon 2 (chr5:1293450). We first studied 5mC levels in the promoter region of the TERT CGI using ENCODE reduced representation bisulfite sequencing (RRBS) data generated on a wide variety of primary cells and tumor-derived cell lines (Fig. 1A, B, Table S1A). We observed a nearly uniform lack of methylation in this region of the TERT CGI in primary cells (e.g., fibroblasts, BJ cells, IMR90) (Fig. 1B). In contrast, a nearly uniform gain of 5mC characterized the tumor-derived lines.
Figure 1. 5mC Levels at the TERT Promoter CGI Are Reduced in Cell Lines with −124 Mutations.
(A) The position of the TERT CGI relative to the −124 mutation, transcription start site (TSS) and ATG. The region of the TERT CGI displayed extends from −668 to −577 (Chr5:1,295,678 1,295,774 in HG19).
(B) ENCODE RRBS data indicate reduced 5mC at the CGI in −124 heterozygous cells vs. cancer cells with wt promoters. The position of cg11625005 is indicated (−633 from the TERT ATG). Direct bisulfite sequencing analyses are shown in Fig. S1. See Table S1 for cell types.
(C) Relative DNA methylation levels across the TERT CGI in CCLE cell lines (n=278). Cell lines are grouped by TERT promoter mutation status and monoallelic TERT expression status (MAE, wt promoter with monoallelic expression). Each value derives from RRBS data from one or more CpG; where CpG were pooled, pooling was based on nucleotide proximity. Data are median ± SEM. Significance test (see Methods) compared wt and −124/−146 cell lines; * p < 0.05.
Upon closer examination, it appeared that the three cell lines with −124 mutations (HepG2, U87 MG, and SK-N-SH) exhibited intermediate or low levels of methylation at cg11625005 (Fig. 1B). We then examined unpublished RRBS data for 55 tumor-derived cell lines with −124 or −146 mutations in the Cancer Cell Line Encyclopedia (CCLE) and compared them with 207 cell lines lacking TERT promoter mutations (Fig. 1C, Fig. S1). These cell lines were derived from a very broad range of cancer types (Table S1B). The data indicate that the average level of 5mC is significantly lower in the lines with mutated TERT promoters starting from around −220 bp upstream of the TERT ATG through −700 bp, and significantly higher within exon 1 (+109 – +145, Fig. 1C, Table S2).
Some cancer cell lines show monoallelic expression (MAE) of TERT even in the absence of promoter mutations (Huang et al., 2015). This phenotype suggested that the epigenetic conditions that facilitate TERT expression in these cells might be distinct from those with wt promoters that express TERT biallelically or cancers with promoter mutations. We therefore examined DNA methylation in 16 cell lines with MAE and found methylation levels resembling the wt cell lines at many nucleotide positions (Fig. 1C, Fig. S1). At two positions (−290 – −271 and +109 – +145), however, the MAE cells converged with the epigenotype of TERT promoter mutants, suggesting that these positions may be important for monoallelic expression.
To assess whether patient tumor samples in The Cancer Genome Atlas (TCGA) recapitulated the observed reduction in methylation in the TERT CGI seen in these promoter-mutant cell lines, we combined −124 genomic profiling with TERT promoter 5mC from Illumina Infinium HumanMethylation450 BeadChip array at cg11625005 (a specific CpG dinucleotide at −634.). We compared patient tumor samples with −124 mutations to samples without mutations in cutaneous melanoma, liver cancer and bladder cancer. Despite the Infinium array data comprising both active and inactive alleles, we detected significantly reduced levels of methylation at cg11625005 in −124 tumors (Fig. S2). Thyroid cancer and lower grade glioma did not display the same relationship between 5mC and the −124 mutation, although both of these tumor types exhibited conspicuously low TERT mRNA, and thyroid cancer also displayed overall anomalously low levels of DNA methylation (Fig. S2B).
Higher TERT Transcription in Bladder Cancer Cell Lines Associates with Reduced 5mC in the TERT CGI
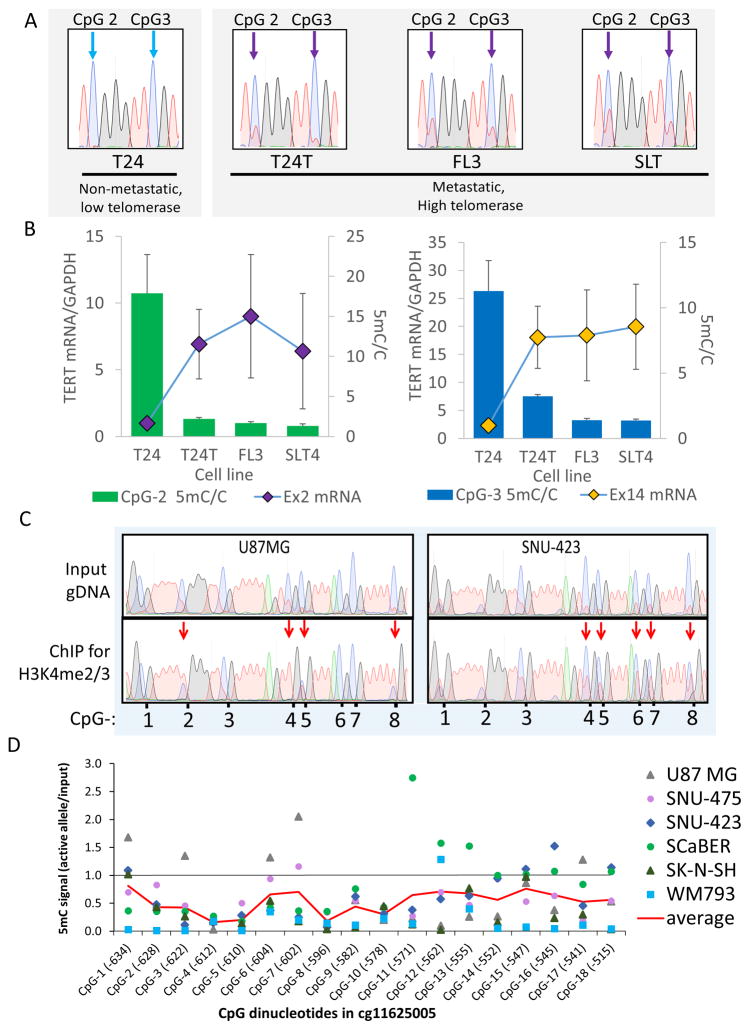
Data above indicate that both −124 mutant tumors and −124 cell lines exhibit distinct methylation patterns at the TERT CGI compared to tumors and cell lines with wt promoters. Given the importance of promoter DNA methylation in regulating transcription, we measured TERT CGI methylation and transcription in a panel of related −124 mutant bladder cancer cell lines. T24 bladder carcinoma cells are a non-metastatic tumor-derived cell line; T24T is a metastatic relative of T24 and was subsequently passaged in mice to obtain the metastatic lines FL3 and SLT4 (Gildea et al., 2000, 2002). Bisulfite sequencing analysis indicated that T24 exhibited the highest level of 5mC at the TERT CGI, while the other three lines exhibited decreased levels of 5mC at specific CpG dinucleotides (Fig. 2A, Fig. S3A). T24 expresses low levels of the telomerase enzyme, while FL3, SLT4, and T24T express much higher levels (Borah et al., 2015). We assessed the level of TERT mRNA in these cells and found that the levels were highest in the metastatic derivative lines (Fig. 2B). Thus, levels of 5mC at this locus are negatively correlated with TERT mRNA expression in this series of related bladder cancer lines.
Figure 2. Hypomethylation of the TERT CGI at −450 – −750 is Specific to Active Alleles and Associates with Higher TERT mRNA in a Panel of Related −124 Mutant Bladder Cancer Cell Lines.
(A) Sanger sequencing traces of bisulfite-converted genomic DNA from the low-telomerase, non-metastatic −124 heterozygous bladder cancer line T24 vs. its metastatic, high-telomerase relatives (FL3, SLT4, T24T). After bisulfite conversion, the ratio of the red peak (T) to the blue peak (C) at CpG sites indicates 5mC/C ratio (see Fig. S3B). Thus, T24 cells are mostly methylated at CpG2 and CpG3 (blue arrows), and the other lines are partially hypomethylated (purple arrows). CpG numbers refer to positions in Fig. 2D.
(B) T24 lines display an inverse relationship between 5mC at CpG-2 and CpG-3 within the TERT CGI vs. TERT mRNA expression measured at exon 2 (Ex2) and exon 14 (Ex14). Additional CpGs displayed a similar pattern (Fig. S3A). 5mC data are mean + SEM (n=4), TERT expression data are mean ± SEM (n=3–7).
(C) Representative Sanger sequencing traces of bisulfite-converted DNA from input genomic DNA or H3K4me2/3 ChIP-isolated DNA from two cell lines quantified in (D). Red arrows indicate reduced 5mC in ChIP DNA. Numbers below traces give relative positions of the CpG in the TERT CGI as annotated in (D).
(D) Mean 5mC levels in H3K4me2/3 ChIP-isolated DNA vs. input DNA (see Fig. S3C,D for individual cell lines and measures of variance). Each cell line bears heterozygous mutations at −124. A ratio of 1 indicates no difference in 5mC levels between input and ChIP DNA. Methylation on the actively transcribed allele across all cell lines was significantly lower (one-sample t-test, p <0.001) compared to the expected value of 1. One CpG dinucleotide in particular (CpG-4) was consistently under-methylated (p < 0.0001). The six heterozygous lines analyzed were U87 MG (glioblastoma), SK-N-SH (neuroblastoma), the HCC lines SNU-475 and SNU-423, melanoma (WM793) and the bladder carcinoma line SCaBER. CpG-1 is cg11625005. The red line indicates the average for the six lines.
Reduced 5mC Characterizes the −124 Mutant TERT Alleles
Because the levels of 5mC described in the previous datasets are a composite of both transcriptionally active and inactive alleles, we considered the possibility that tumor-derived −124 cells may have allele-specific reduction of methylation. To isolate the active alleles for bisulfite sequencing analysis, we employed chromatin immunoprecipitation (ChIP) using anti-H3K4me2/3 antibodies (Stern et al., 2015) in six −124 cell lines. DNA fragments isolated by ChIP were subjected to bisulfite sequencing. Bisulfite conversion results in the transition of cytosine to thymine only in the absence of 5mC; methylated cytosines are protected from this chemical reaction. Therefore, for each nucleotide position, the level of 5mC in ChIP samples relative to input samples can be assessed by quantifying the peak height of unconverted cytosine relative to thymine. For each sequencing sample, the bisulfite conversion was efficient as assessed by the complete conversion of neighboring non-CpG cytosines. Quantification of these data across the six cell lines revealed that the H3K4me2/3-associated active alleles commonly exhibited lower levels of CpG methylation at many positions in the TERT CGI (Fig. 2C,D, S3C). Note that these results are opposite to the expectation from the literature of increased TERT gene expression correlating with high CpG methylation in other cell types (Barthel et al., 2017; Guilleret and Benhattar, 2004), but are consistent with our data on the four related bladder cancer lines (Fig. 2A,B). To provide additional confidence in this conclusion, we cloned the PCR products from four cell lines from input and H3K4me2/3 ChIP products (Fig. S3D). The observation of higher methylation in the input samples than the ChIP samples supports our conclusion that reduced methylation characterizes the transcribed alleles in these heterozygous cell lines.
EZH2 Is Preferentially Associated with the Silent TERT Allele
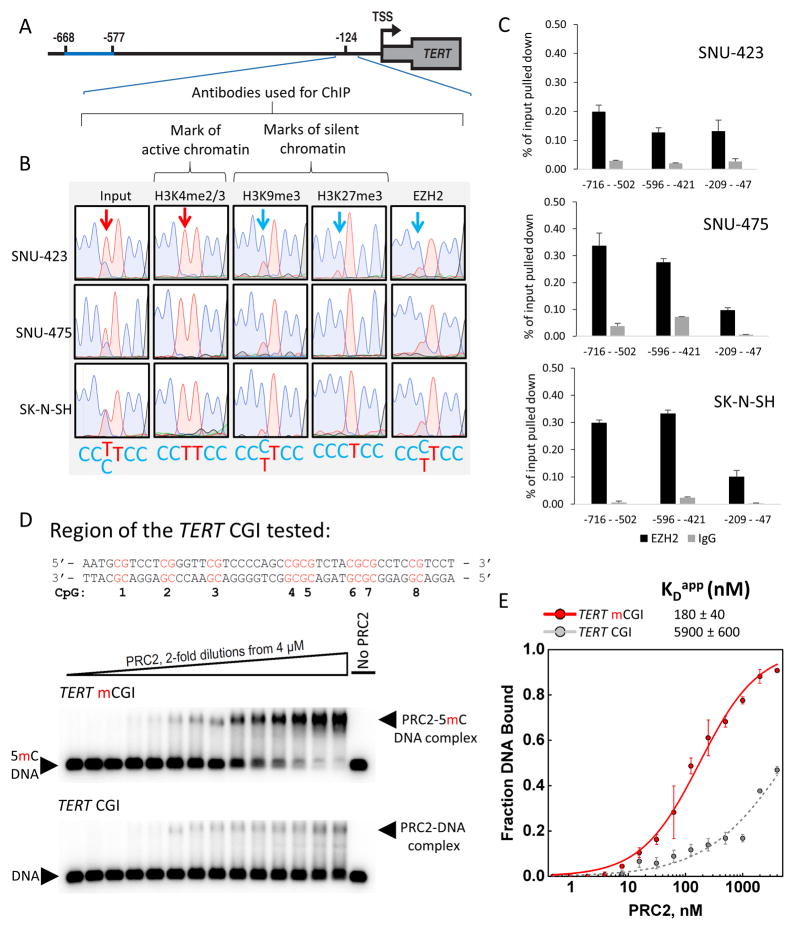
Previously we have described that transcriptionally inactive alleles in cancers with −124 mutations bear the H3K27me3 histone mark (Stern et al., 2015), which is one of the two canonical histone marks of repressive chromatin. The enzyme responsible for the deposition of H3K27me3, enhancer of zeste homolog 2 (EZH2), is the catalytic subunit of the PRC2 complex. To test if EZH2 exhibited allele-specific recruitment at TERT, we performed ChIP using antibodies directed against histone marks or EZH2, followed by DNA purification, PCR and Sanger sequencing. While DNA purified from chromatin prior to ChIP displayed both alleles in sequencing traces, ChIP for both H3K27me3 and EZH2 exhibited enrichment for the CCCTCC sequence (Fig. 3A,B) diagnostic of the silent allele. Thus, in these cells at this locus, PRC2 exhibits a preference for occupying the inactive TERT allele. The second major histone mark of repressive chromatin, tri-methylated H3K9, can co-occur with H3K27me3 (Mozzetta et al., 2015). Allele-specific ChIP using antibodies against H3K9me3 showed that it was also enriched on the inactive, wt TERT promoter in these cells (Fig. 3B). Therefore, the inactive TERT promoter is the target of both PRC2 as well as enzymes responsible for deposition of H3K9me3 such as G9a, SetDB1, and SUV39H1 and 2 (Greer and Shi, 2012).
Figure 3. PRC2 Displays a Strong Binding Preference for 5mC-Rich TERT Alleles in vivo in −124 Cells and Methylated TERT CGI DNA in vitro.
(A) Schematic illustrating the position of the allele-specific ChIP analysis (−209 – −47).
(B) Representative Sanger sequencing traces from DNA isolated by ChIP and amplified by three PCR reactions pooled prior to sequencing, showing the presence of H3K27me3, H3K9me3 and EZH2 on the inactive allele in HCC (SNU-423, SNU-475) and neuroblastoma (SK-N-SH) −124 heterozygous tumor-derived cell lines. Arrows indicate the position of the heterozygous −124 mutation.
(C) Quantitative assessment of EZH2 occupancy of TERT promoters in three −124 mutant cell lines. Data are + SEM, n=3 technical replicates.
(D) Sequence tested for PRC2 binding in vitro. CpG numbers correspond to those listed in Fig. 2D. Representative EMSA gels showing binding of purified recombinant human PRC2 to the fully CpG-methylated TERT DNA vs. the same sequence lacking 5mC modifications.
(E) Binding data fit with equilibrium binding curves, error bars are ± SD (n=3 independent experiments).
Because cell lines with wt TERT promoters typically exhibit much higher levels of 5mC at the promoter (Fig. 1B), we tested whether this might correlate with relatively higher levels of EZH2 recruitment. We therefore performed EZH2 ChIP in three lines with wt promoters: HeLa, SNU-449, and HEK-293T. In addition, since the heterozygous liver cancer cell line HepG2 displays relatively higher levels of 5mC for a TERT promoter mutant line, we also tested EZH2 occupancy in this line. Each of these lines displayed relatively high levels of EZH2 at the TERT promoter (Fig. S4B) compared to the heterozygous lines (Fig. 3C). To further test the relationship between EZH2 and 5mC, we analyzed ENCODE ChIP-seq data for HepG2 for which 5mC data are also available (Fig. S4C). These data indicate that both 5mC and EZH2 levels are higher in the 5′ region of the TERT CGI, while levels of both are reduced near the TERT TSS. Because the active allele in HepG2 cells is likely to contribute relatively little signal to these ChIP-seq data, and as we have demonstrated that in heterozygous lines the active allele is hypomethylated, we conclude that the levels of 5mC and EZH2 are correlated at this locus in this liver cancer cell line.
5mC Enhances PRC2 Binding to TERT Promoter DNA in vitro
Given that the inactive TERT allele in the −124 mutant cells accumulates both 5mC and H3K27me3, as well as EZH2, we hypothesized that a functional relationship may exist between these marks of inactive chromatin. Indeed, a number of studies have addressed the relationship between DNA methylation and repressive histone marks (Bartke et al., 2010; Lynch et al., 2012). Therefore, we tested whether 5mC-modification of DNA affects PRC2 binding in vitro. We purified a recombinant human five-protein PRC2 complex (EZH2, SUZ12, EED, RBBP4, and AEBP2; Fig. S4A) and used electrophoretic mobility shift assays (EMSA) to test its ability to bind to either a fully unmethylated or fully methylated TERT CGI DNA. PRC2 displayed >30-fold higher affinity to the 5mC-modified region of the TERT promoter over the unmethylated DNA (Fig. 3D,E). These data suggest a positive feedback relationship whereby reduced 5mC methylation at an active TERT locus may discourage PRC2 recruitment.
5mC Levels at the TERT CGI Associate with Patient Survival in Multiple Cancers
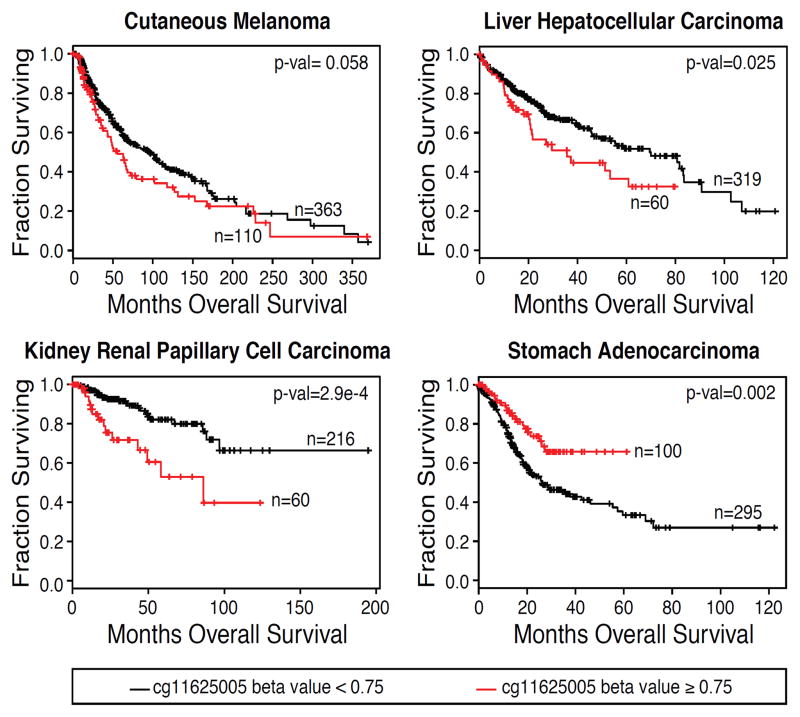
Methylation levels at cg11625005 are negatively correlated with patient survival in some cancers (Castelo-Branco et al., 2013, 2016; Gojo et al., 2017). We therefore analyzed overall survival (OS) with respect to methylation and TERT promoter mutation in patient samples for a range of cancers within the TCGA. We first analyzed the data for cutaneous melanoma (SKCM) and found that samples with cg11625005 methylation beta values ≥ 0.75 showed a trend towards poorer survival, with borderline statistical significance (p=0.058) (Fig. 4). We therefore analyzed the remaining cancer types using this threshold. These analyses revealed that in HCC (LIHC, in which −124 mutations are common) and kidney renal papillary cell carcinoma (KIRP, in which −124 mutations are uncommon), patients with methylation above 0.75 at cg11625005 had significantly poorer OS. In contrast, patients with stomach adenocarcinoma (STAD, in which −124 mutations are uncommon) exhibited significantly better survival (Fig. 4). STAD not only exhibited an opposite relationship between 5mC and survival compared to LIHC and KIRP, but STAD also had an opposite relationship between EZH2 expression levels and OS (Fig. S4D). These data suggest that the processes underlying TERT CGI hypermethylation and genome-wide activity of PRC2 may be linked and are indicative of altered patient survival in these cancers.
Figure 4. High Levels of 5mC at cg11625005 Correlate with Differences in Patient Survival in Specific Cancers.
Kaplan-Meier plots were generated for TCGA patient data with cancers that commonly harbor TERT promoter mutations (HCC, LIHC; cutaneous melanoma, SKCM) and cancers that do not typically harbor TERT promoter mutations (kidney renal papillary cell carcinoma, KIRP; stomach adenocarcinoma, STAD). Patients were stratified based on methylation (beta value) at cg11625005. Statistical comparison was done using the log-rank test (see methods for details).
DISCUSSION
A key question in understanding the indefinite proliferation of cancer cells is how they regulate the expression of their telomere maintenance machinery. Most cancers activate TERT expression, either biallelically through poorly understood mechanisms or monoallelically driven in many cases by heterozygous promoter mutations. Here we find that a major arm of epigenetic gene regulation, CpG methylation, exhibits opposing dynamics in cancers with heterozygous −124 mutations compared to cancers with wt TERT promoters. Thus, the machinery maintaining the immortal phenotype in these two classes of cancers is distinct. Reduced 5mC in the TERT promoter has been reported previously for isolated subtypes of cancers with −124 mutations (Fan et al., 2016; Lindsey et al., 2014), although the allele specificity of 5mC has not previously been reported. Our analyses of large datasets of clinical samples broaden these findings to numerous cancers across many tissue types and provide a mechanistic rationale for how DNA methylation may effect TERT gene silencing in −124 cancers.
Cancers with heterozygous −124 mutations also exhibit allele-specific deposition of H3K27me3 by EZH2/PRC2, as well as deposition of H3K9me3 (Fig. 3B). Intriguingly, there is a direct mechanistic relationship between H3K9me3 and the enzymatic machinery responsible for the 5mC modification (Cedar and Bergman, 2009; Epsztejn-Litman et al., 2008; Liu et al., 2013; McGarvey et al., 2006; Rose and Klose, 2014; Rothbart et al., 2012, 2013; Schübeler, 2015; Smith and Meissner, 2013) suggesting that the presence of H3K9me3 on the inactive allele could promote the accumulation of 5mC at the TERT promoter.
Repression facilitated by EZH2/PRC2 at the TERT locus may also be directly linked to CpG methylation (Fig. 3), with the specificity for 5mC possibly conferred by the AEBP2 subunit of PRC2 (Wang et al., 2017). Preferential PRC2 binding to methylated TERT CGI DNA is likely to be locus-dependent, because in other cases PRC2 has been found to bind unmethylated CpG-rich chromatin (Bartke et al., 2010; Lynch et al., 2012). Factors other than CpG methylation certainly affect the level of PRC2 occupancy at any given locus in vivo.
Our data reveal that cell lines with MAE expression exhibit 5mC levels similar to wt TERT promoters at many CpG positions, but at specific loci (−290 – −271 and +109 – +145) they closely resemble TERT promoter mutants. The higher level of methylation within exon 1 (+109 – +145) for cells expressing TERT monoallelically (with or without promoter mutations) may represent a mechanism to increase the fidelity of transcription initiation (Maunakea et al., 2010; Neri et al., 2017) perhaps to more efficiently utilize their single active TERT allele.
The association between 5mC levels at the TERT promoter and either poorer overall survival in liver cancer and kidney renal papillary cell carcinoma, or significantly improved overall survival in patients with stomach adenocarcinoma, suggests that these epigenotypes may be relevant to cancer progression. Association between higher methylation at cg11625005 and poorer patient survival has been previously reported (Castelo-Branco et al., 2013, 2016; Gojo et al., 2017; Seynnaeve et al., 2017); our results extend these findings to two additional cancer types, suggesting that hypermethylation of the TERT promoter may represent a broadly applicable prognostic marker. To our knowledge, our data showing a more positive outcome for stomach adenocarcinoma patients with higher methylation at cg11625005 or with high EZH2 expression and suggest that the mechanisms driving survival in these patients may be distinct.
In conclusion, we find that TERT promoters in cancers with −124 mutations exhibit allele-specific chromatin and DNA modifications that differ from those on active TERT genes in cancers with wt promoters. These findings implicate multiple mechanisms by which cancers reactivate or maintain TERT expression to achieve telomere maintenance and immortalization. Such information may be clinically relevant, because inhibitors for both 5mC deposition and EZH2 are being developed for cancer therapy (Pfister and Ashworth, 2017).
EXPERIMENTAL PROCEDURES
Further details and an outline of resources used in this work can be found in Supplemental Experimental Procedures.
Statistical Methods
For CCLE samples, comparisons were made using the Wilcoxon rank-sum test. The cg11625005 survival analysis was done using the survival R package (Therneau, 2000) (https://cran.r-project.org/web/packages/survival/citation.html). Patients were stratified based on a cg11625005 methylation beta value threshold of 0.75 and differences between the survival curves of the stratified patient groups were tested using the log-rank test. 5mC:C ratios in H3K4me2/3 ChIP of −124 samples were log transformed and compared using a single sample t-test against a hypothetical mean of 1. This analysis assumes the underlying data are distributed normally. For this analysis, the n was the number of CpGs tested (18) in the TERT CGI.
Supplementary Material
Table S1. ENCODE and CCLE lines analyzed. Related to Figure 1. (A) Cell line information for Figure 1B. (B) CCLE WT, wild-type at the TERT promoter. MUT, −124 or −146 TERT promoter mutants, mono_WT, cell lines without known TERT promoter mutations exhibiting monoallelic TERT expression (Huang et al. 2015).
Table S1A. Cell line information for Figure 1B.
Table S2. Statistical analysis of CCLE lines presented in Fig. S1. WT, wild-type at the TERT promoter. MUT, −124 or −146 TERT promoter mutants, mono_WT, cell lines without known TERT promoter mutations exhibiting monoallelic TERT expression (Huang et al. 2015).
Acknowledgments
We thank S. Borah (St. Jude’s Children’s Hospital) and members of the Cech lab for helpful discussion. We thank X. Wang (University of Colorado Boulder) for preparation of PRC2 complexes. We thank F. Lorbeer, C. Kunitoshi, and D. Hockemeyer (University of California, Berkeley) and A. Meeker and S. Yegnasubramanian (Johns Hopkins) for useful conversations relating to this work. We thank the ENCODE Consortium (Rosenbloom et al., 2013) and the ENCODE production laboratories for generating data used in this study. We thank the Novartis Institute of Biomedical Research for the use of CCLE data. J.L.S. was funded by an American Cancer Society postdoctoral fellowship; F.W.H. was funded by DOD (W81XWH-14-1-0514) and Prostate Cancer Foundation Young Investigator Award; and T.R.C. is an investigator of the Howard Hughes Medical Institute. This work was funded by National Institutes of Health grant R01 GM099705 to T.R.C.
Footnotes
AUTHOR CONTRIBUTIONS
J.L.S. designed and performed allele-specific ChIP experiments. J.L.S. and R.D.P. designed and R.D.P. performed PRC2 binding experiments. J.L.S. designed and R.N. performed experiments related to T24 lines. J.L.S. and J.C.C. designed and J.C.C. performed TCGA analyses. F.W.H. and M.G. designed and analyzed CCLE data. J.L.S. and T.R.C. wrote the manuscript.
DECLARATION OF INTERESTS
Conflict of Interest – T.R.C. is on the board of directors of Merck, Inc.
References
- Barthel FP, Wei W, Tang M, Martinez-Ledesma E, Hu X, Amin SB, Akdemir KC, Seth S, Song X, Wang Q, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49:349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- Bell RJA, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, Choi S, Hong C, He D, Pekmezci M, et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348:1036–1039. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah S, Xi L, Zaug AJ, Powell NM, Dancik GM, Cohen SB, Costello JC, Theodorescu D, Cech TR. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science. 2015;347:1006–1010. doi: 10.1126/science.1260200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco P, Choufani S, Mack S, Gallagher D, Zhang C, Lipman T, Zhukova N, Walker EJ, Martin D, Merino D, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: An integrative genomic and molecular study. Lancet Oncol. 2013;14:534–542. doi: 10.1016/S1470-2045(13)70110-4. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco P, Leão R, Lipman T, Campbell B, Lee D, Price A, Zhang C, Heidari A, Stephens D, Boerno S, et al. A cancer specific hypermethylation signature of the TERT promoter predicts biochemical relapse in prostate cancer: A retrospective cohort study. Oncotarget. 2016;7:57726–57736. doi: 10.18632/oncotarget.10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessain SK, Yu H, Reddel RR, Beijersbergen RL, Weinberg Ra. Methylation of the Human Telomerase Gene CpG Island 1. Cancer Res. 2000;60:537–541. [PubMed] [Google Scholar]
- Devereux TR, Horikawa I, Anna CH, Transcriptase R, Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA Methylation Analysis of the Promoter Region of the Human Telomerase Reverse Transcriptase (hTERT) Gene. Cancer Res. 1999:6087–6090. [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Lee S, Wu G, Easton J, Yergeau D, Dummer R, Vogel P, Kirkwood JM, Barnhill RL, Pappo A, et al. Telomerase Expression by Aberrant Methylation of the TERT Promoter in Melanoma Arising in Giant Congenital Nevi. J Invest Dermatol. 2016;136:339–342. doi: 10.1038/JID.2015.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildea JJ, Golden WL, Harding MA, Theodorescu D. Genetic and Phenotypic Changes Associated With the Acquisition of Tumorigenicity in Human Bladder Cancer. Genes Chromosom Cancer. 2000;263:252–263. doi: 10.1002/(sici)1098-2264(200003)27:3<252::aid-gcc5>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Gildea JJ, Seraj MJ, Oxford G, Harding MA, Hampton GM, Moskaluk CA, Frierson HF, Conaway MR, Theodorescu D. RhoGDI2 is an invasion and metastasis suppressor gene in human cancer. Cancer Res. 2002;62:6418–6423. [PubMed] [Google Scholar]
- Gojo J, Lötsch D, Spiegl-Kreinecker S, Pajtler KW, Neumayer K, Korbel P, Araki A, Brandstetter A, Mohr T, Hovestadt V, et al. Telomerase activation in posterior fossa group A ependymomas is associated with dismal prognosis and chromosome 1q gain. Neuro Oncol. 2017:1–12. doi: 10.1093/neuonc/nox027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilleret I, Benhattar J. Unusual distribution of DNA methylation within the hTERT CpG island in tissues and cell lines. Biochem Biophys Res Commun. 2004;325:1037–1043. doi: 10.1016/j.bbrc.2004.10.137. [DOI] [PubMed] [Google Scholar]
- Herman JG. Hypermethylation of tumor suppressor genes in cancer. Semin Cancer Biol. 1999;9:359–367. doi: 10.1006/scbi.1999.0138. [DOI] [PubMed] [Google Scholar]
- Herman JG, Umar a, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FW, Bielski CM, Rinne ML, Hahn WC, Sellers WR, Stegmeier F, Garraway LA, Kryukov GV. TERT promoter mutations and monoallelic activation of TERT in cancer. Oncogenesis. 2015;4:e176. doi: 10.1038/oncsis.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW, Jaenisch R. The Role of Dna Methylation in Cancer Genetics and Epigenetics. Annu Rev Genet. 1996;30:441–464. doi: 10.1146/annurev.genet.30.1.441. [DOI] [PubMed] [Google Scholar]
- Lindsey JC, Schwalbe EC, Potluri S, Bailey S, Williamson D, Clifford SC. TERT promoter mutation and aberrant hypermethylation are associated with elevated expression in medulloblastoma and characterise the majority of non-infant SHH subgroup tumours. Acta Neuropathol. 2014;127:307–309. doi: 10.1007/s00401-013-1225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun. 2013;4:1563. doi: 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- Lynch MD, Smith AJH, De Gobbi M, Flenley M, Hughes JR, Vernimmen D, Ayyub H, Sharpe JA, Sloane-Stanley JA, Sutherland L, et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012;31:317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- Mozzetta C, Boyarchuk E, Pontis J, Ait-Si-Ali S. Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat Rev Mol Cell Biol. 2015;16:499–513. doi: 10.1038/nrm4029. [DOI] [PubMed] [Google Scholar]
- Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, Maldotti M, Anselmi F, Oliviero S. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543:72–77. doi: 10.1038/nature21373. [DOI] [PubMed] [Google Scholar]
- Pfister S, Ashworth A. Marked for death: targeting epigenetic changes in cancer. Nat Rev Drug Discov. 2017;16:241–263. doi: 10.1038/nrd.2016.256. [DOI] [PubMed] [Google Scholar]
- Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta - Gene Regul Mech. 2014;1839:1362–1372. doi: 10.1016/j.bbagrm.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. ENCODE Data in the UCSC Genome Browser: Year 5 update. Nucleic Acids Res. 2013;41:56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD. Multivalent histone engagement by the linked tandem tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 2013;27:1288–1298. doi: 10.1101/gad.220467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- Seynnaeve B, Lee S, Borah S, Park Y, Pappo A, Kirkwood JM, Bahrami A. Genetic and Epigenetic Alterations of TERT Are Associated with Inferior Outcome in Adolescent and Young Adult Patients with Melanoma. Sci Rep. 2017;7:45704. doi: 10.1038/srep45704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Stern J, Theodorescu D, Vogelstein B, Papadopoulos N, Cech TR. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 2015;29:2219–2224. doi: 10.1101/gad.269498.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Paucek RC, Gooding AR, Brown ZZ, Ge EJ, Muir TW, Cech TR. PRC2 recruitment to DNA in chromatin and its inhibition by RNA reveal molecular mechanisms of epigenetic control. Nat Struct Mol Biol. 2017 doi: 10.1038/nsmb.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. ENCODE and CCLE lines analyzed. Related to Figure 1. (A) Cell line information for Figure 1B. (B) CCLE WT, wild-type at the TERT promoter. MUT, −124 or −146 TERT promoter mutants, mono_WT, cell lines without known TERT promoter mutations exhibiting monoallelic TERT expression (Huang et al. 2015).
Table S1A. Cell line information for Figure 1B.
Table S2. Statistical analysis of CCLE lines presented in Fig. S1. WT, wild-type at the TERT promoter. MUT, −124 or −146 TERT promoter mutants, mono_WT, cell lines without known TERT promoter mutations exhibiting monoallelic TERT expression (Huang et al. 2015).