Abstract
mTOR complex 1 (mTORC1) regulates cell growth and metabolism in response to multiple environmental cues. Nutrients signal via the Rag guanosine triphosphatases (GTPases) to promote the localization of mTORC1 to the lysosomal surface, its site of activation. We identified SAMTOR, a previously uncharacterized protein, which inhibits mTORC1 signaling by interacting with GATOR1, the GTPase activating protein (GAP) for RagA/B. We found that the methyl donor S-adenosylmethionine (SAM) disrupts the SAMTOR-GATOR1 complex by binding directly to SAMTOR with a dissociation constant of approximately 7 μM. In cells, methionine starvation reduces SAM levels below this dissociation constant and promotes the association of SAMTOR with GATOR1, thereby inhibiting mTORC1 signaling in a SAMTOR-dependent fashion. Methionine-induced activation of mTORC1 requires the SAM binding capacity of SAMTOR. Thus, SAMTOR is a SAM sensor that links methionine and one-carbon metabolism to mTORC1 signaling.
The mechanistic target of rapamycin complex 1 (mTORC1) protein kinase is the central component of a pathway that regulates anabolic and catabolic processes in response to environmental signals, including growth factors and nutrients (1–3). Amino acids promote the translocation of mTORC1 to the lysosomal surface, where its activator Rheb resides. This localization depends on the heterodimeric Rag GTPases, which consist of RagA or RagB bound to RagC or RagD (4, 5).
The amino acid sensing pathway upstream of mTORC1 is complicated, with several multi-component complexes regulating the Rag heterodimer, each likely conveying a distinct amino acid input. GATOR1 and FLCN-FNIP are GAPs for RagA/B and RagC/D, respectively (6, 7), whereas Ragulator tethers the Rags to the lysosomal surface and also has nucleotide exchange activity (8, 9). The KICSTOR complex binds GATOR1 and recruits it to the lysosome, and, like GATOR1, is necessary for amino acid starvation to inhibit mTORC1 signaling (7, 10, 11). The molecular function of GATOR2 is unknown, but it is required for pathway activity and might act upstream of GATOR1 (7).
Leucine and arginine are well-established activators of mTORC1 signaling, and recent work has shed light on the molecular mechanisms involved. The lysosomal transmembrane protein SLC38A9 interacts with Ragulator (12–14) and is a lysosomal arginine sensor (15), whereas Sestrin2 and CASTOR1 are cytosolic leucine and arginine sensors, respectively, that bind to and inhibit the function of GATOR2 in the absence of their cognate amino acids (16–19). Whether, and how, other amino acids affect mTORC1 signaling is unclear.
To search for proteins that bind to GATOR1 or KICSTOR, we mined the BioPlex protein-protein interaction database generated by immunoprecipitation followed by mass spectrometry of more than 5000 proteins stably expressed in human embryonic kidney (HEK)-293T cells (20). This analysis revealed C7orf60, a previously unstudied protein, as a putative interaction partner of all known components of GATOR1 (Depdc5, Nprl3, Nprl2) and KICSTOR (Kaptin, ITFG2, C12orf66, SZT2). For reasons described below, we renamed C7orf60 as SAMTOR (S-adenosylmethionine sensor upstream of mTORC1).
Using an antibody against SAMTOR to probe anti-FLAG immunoprecipitates prepared from cells having endogenously Flag-tagged components of GATOR1 (Depdc5) or GATOR2 (WDR59) or stably expressing a KICSTOR component (Flag-Kaptin), we verified that SAMTOR coimmunoprecipitated with GATOR1 and KICSTOR, but not GATOR2 (Fig. 1A). Moreover, transiently expressed SAMTOR coimmunoprecipitated endogenous GATOR1 and KICSTOR, as detected by the presence of their Nprl3 and SZT2 components, respectively. Loss of a component of GATOR1 or KICSTOR, but not of GATOR2, severely reduced the interaction of SAMTOR with KICSTOR or GATOR1, respectively (Fig. 1B). Furthermore, overexpressed GATOR1 coimmunoprecipitated SAMTOR only when KICSTOR was coexpressed (fig. S1A). Thus, SAMTOR binds to the supercomplex of GATOR1 and KICSTOR, and both complexes are required for the interaction to occur (Fig. 1C).
Figure 1. SAMTOR interacts with GATOR1 and KICSTOR.
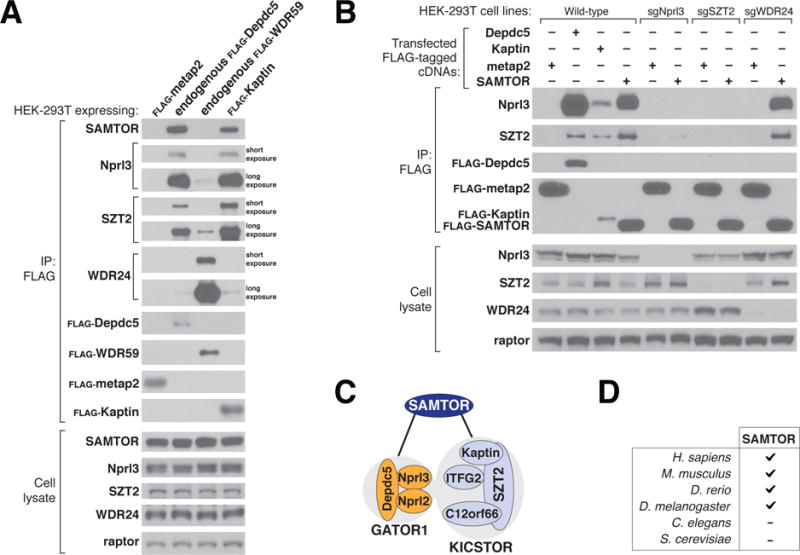
(A) GATOR1 and KICSTOR, but not GATOR2, coimmunoprecipitate SAMTOR. FLAG immunoprecipitates (IP) were prepared from HEK-293T cell lines that stably expressed FLAG-tagged metap2 or Kaptin, or had endogenously FLAG-tagged Depdc5 or WDR59. FLAG immunoprecipitates and lysates were analyzed by immunoblotting for the indicated proteins. FLAG-metap2 served as a negative control. Depdc5 and Nprl3, WDR59 and WDR24, and Kaptin and SZT2 were used as representative components of the GATOR1, GATOR2, and KICSTOR complexes, respectively; Raptor was used as a loading control. Short or long exposure indicates relative blot exposure times.
(B) SAMTOR coimmunoprecipitates GATOR1 and KICSTOR, and the interaction requires both GATOR1 and KICSTOR but not GATOR2. FLAG immunoprecipitates were prepared from wild-type, Nprl3-deficient, SZT2- deficient, or WDR24-deficient HEK-293T cells transiently expressing the indicated cDNAs. FLAG immunoprecipitates and lysates were analyzed as in (A).
(C) Model showing how SAMTOR interacts with GATOR1 and KICSTOR.
(D) Presence or absence of gene orthologs of SAMTOR in several model organisms.
Figure 2. SAMTOR is a negative regulator of mTORC1 signaling that acts upstream of the Rag GTPases, GATOR1, and KICSTOR.
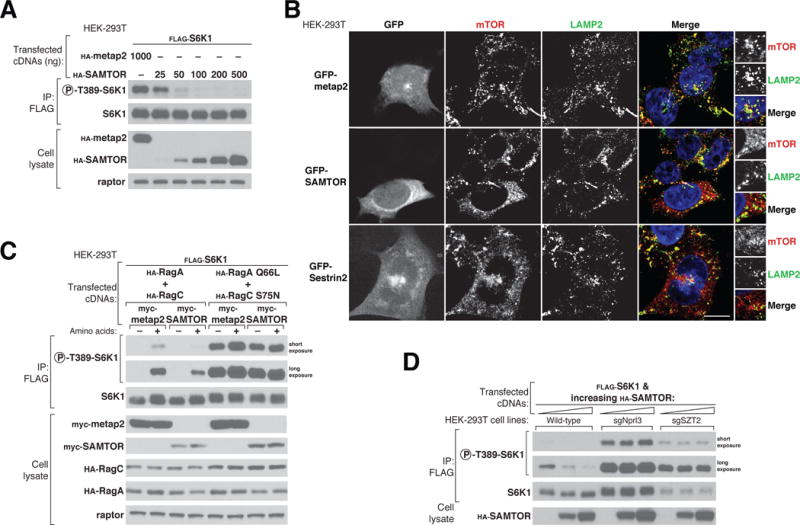
(A) Transient overexpression of SAMTOR inhibits mTORC1 signaling. FLAG immunoprecipitates were prepared from HEK-293T cells transfected with 2 ng of FLAG-S6K1 cDNA along with either hemagglutinin (HA)–tagged metap2 cDNA or increasing amounts of HA-SAMTOR cDNA. FLAG immunoprecipitates and cell lysates were analyzed by immunoblotting for the phosphorylation states and levels of the indicated proteins.
(B) Overexpression of GFP-SAMTOR displaces mTOR from lysosomes, similar to the effect of GFP-Sestrin2.Wild-type HEK-293Tcells transiently expressing GFP-metap2, GFP-SAMTOR, or GFP-Sestrin2 were processed for immunofluorescence detection of mTOR and the lysosomal marker LAMP2. In all images, insets represent selected fields magnified 5.12X as well as their overlays. Scale bar, 10 μm.
(C) SAMTOR functions upstreamof the Rag GTPases to regulate the mTORC1 pathway. HEK-293Tcells expressing the indicated cDNAs were starved of amino acids for 50 min or starved and restimulated with amino acids for 10 min. FLAG immunoprecipitates and cell lysates were analyzed as in (A).
(D) SAMTOR functions upstream of GATOR1 and KICSTOR. FLAG immunoprecipitates and cell lysates prepared from wild-type, Nprl3-deficient, or SZT2-deficient HEK-293Tcell lines expressing the indicated cDNAs were analyzed as in (A).
Orthologs of SAMTOR are encoded in the genomes of vertebrates and some invertebrates, such as Drosophila melanogaster. We could not identify SAMTOR orthologs in Caenorhabditis elegans or Saccharomyces cerevisiae (Fig. 1D).
To determine whether SAMTOR regulates mTORC1 signaling, we overexpressed SAMTOR in HEK-293T cells and monitored the phosphorylation at Thr389 of S6 Kinase 1 (S6K1), a canonical mTORC1 substrate. SAMTOR expression suppressed mTORC1 signaling in a dose-dependent fashion (Fig. 2A), establishing SAMTOR as a negative regulator of the pathway. Amino acids activate mTORC1 by promoting its localization to the lysosomal surface (4, 8). Consistent with SAMTOR inhibiting the amino acid sensing pathway upstream of mTORC1, overexpression of green fluorescent protein (GFP)-tagged SAMTOR displaced mTOR from lysosomes to an extent similar to that seen with GFP-Sestrin2, an inhibitor of GATOR2 (21, 22) (Fig. 2B).
To position the SAMTOR function within the mTORC1 pathway, we performed epistasis experiments with established mTORC1 regulators. Overexpression of SAMTOR inhibited mTORC1 signaling when coexpressed with the wild type RagA and RagC heterodimer, but not with the constitutively active mutant heterodimer (RagA Q66L and RagC S75N) that bypasses the requirement for amino acids for maintaining mTORC1 activity (Fig. 2C) (4, 5). In addition, SAMTOR did not inhibit mTORC1 signaling in cells lacking either a GATOR1 or KICSTOR component. Thus, SAMTOR acts upstream of the Rag GTPases and requires GATOR1 and KICSTOR to inhibit mTORC1 signaling (Fig. 2D). In combination with the interaction data, these results are consistent with SAMTOR promoting the function of GATOR1 and/or KICSTOR, which are both negative regulators of mTORC1 signaling.
Sequence analyses predict that SAMTOR contains a class I Rossmann fold methyltransferase domain (Fig. 3A and fig. S2) (23). These domains are known to bind S-adenosylmethionine (SAM) and exist in methyltransferases in bacteria, archaea, and eukarya (24). To determine whether SAMTOR binds SAM, we developed an equilibrium binding assay based on one we used to detect the binding of leucine to Sestrin2 (16) and determined that SAMTOR binds SAM with a dissociation constant of approximately 7 μM (Fig. 3B). A competition binding assay revealed that, as with other SAM-binding proteins, SAMTOR can also bind S-adenosylhomocysteine (SAH), the demethylated form of SAM (Fig. 3B).
Figure 3. S-adenosylmethionine binds SAMTOR to disrupt its interaction with GATOR1 and KICSTOR.
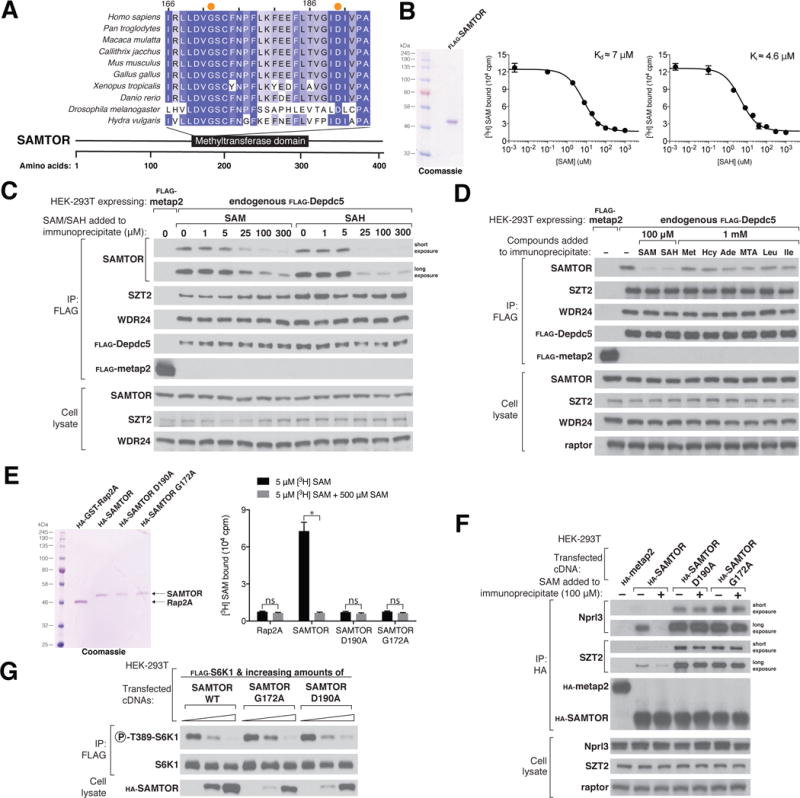
(A) Schematic of the human SAMTOR protein indicating the class I Rossmann fold methyltransferase domain. Shown is an alignment of partial sequences of this domain from SAMTOR in indicated species. Amino acid positions are colored from white to blue in order of increasing sequence similarity. Orange dots denote the Gly172 and Asp190 residues of human SAMTOR.
(B) SAMTOR binds SAM and SAH. Purified FLAG-SAMTOR protein was analyzed by SDS–polyacrylamide gel electrophoresis followed by Coomassie blue staining. Binding assays were performed with purified FLAG-SAMTOR incubated with the indicated concentrations of [3H]SAM, unlabeled SAM, or SAH.Values for each point are means ± SD of three technical replicates fromone representative experiment. The experiment was performed twice.
(C) SAM and SAH disrupt the interaction of SAMTOR with GATOR1 in vitro. FLAG immunoprecipitates were prepared from endogenously FLAG-tagged Depdc5 HEK-293Tcells. SAM and SAH were added directly to the immunoprecipitates at the indicated concentrations. FLAG immunoprecipitates and cell lysates were analyzed by immunoblotting for the levels of the indicated proteins.
(D) The interaction between SAMTOR and GATOR1 is disrupted by 100 μM SAM or SAH, but not by 1mM methionine, homocysteine, adenosine, 5-methylthioadenosine, leucine, or isoleucine. The experiment was performed and analyzed as in (C).
(E) Wild-type HA-SAMTOR, but not HA-SAMTOR G172A or D190A, binds SAM. HA-tagged wild-type and mutant SAMTOR proteins were prepared from HEK-293Tcells expressing the indicated cDNAs, and binding assays were performed as in (B). A representative experiment is shown; values are means ± SD of three technical replicates.Two-tailed t tests were used for comparisons between two groups. *P < 0.001; ns, not significant. The experiment was repeated three times.
(F) HA-SAMTOR G172A and D190A coimmunoprecipitate more endogenous GATOR1 and KICSTOR than does wild-type SAMTOR, and the interactions are insensitive to SAM added in vitro. HA immunoprecipitates and cell lysates were prepared from HEK-293Tcells transiently expressing wild-type HA-SAMTOR or its mutants G172A or D190A. SAM was added to the immunoprecipitates where indicated. HA immunoprecipitates and cell lysates were analyzed as in (C).
(G) HA-SAMTOR G172A and D190A inhibit mTORC1 activity to similar extents as wild-type SAMTOR. FLAG immunoprecipitates were prepared from HEK-293Tcells transfected with the indicated cDNAs. FLAG immunoprecipitates and cell lysates were analyzed by immunoblotting for the phosphorylation states and levels of the indicated proteins.
Given these findings, we asked whether SAM and SAH regulate the interaction of SAMTOR and GATOR1-KICSTOR. Indeed, SAM and SAH, but not methionine, homocysteine, adenosine, 5-methylthioadenosine, leucine, or isoleucine, disrupted the interaction when added directly to the immunopurified complex kept at 4°C (Fig. 3 C and D). Thus, SAM disrupts the interaction between SAMTOR and GATOR1-KICSTOR analogously to how leucine and arginine induce the release of Sestrin2 and CASTOR1 from GATOR2, respectively (16, 19). Given that SAH has the same effect, it is unlikely that a methylation event is required for SAM to dissociate SAMTOR from GATOR1-KICSTOR.
Mutagenesis of highly conserved residues in human SAMTOR yielded two mutants, Gly172->Ala (G172A) and Asp190->Ala (D190A), that no longer bound SAM (Fig. 3E and fig. S2, A and B). These mutants coimmunoprecipitated greater amounts of endogenous GATOR1 and KICSTOR than did wild-type SAMTOR, and the purified complexes were insensitive to SAM in vitro (Fig. 3F). Moreover, these mutants inhibited mTORC1 signaling comparably to wild-type SAMTOR, despite their lower expression (Fig. 3G). Thus, SAMTOR must be able to bind SAM for SAM to disrupt the interaction of SAMTOR with GATOR1-KICSTOR. In contrast, SAMTOR does not have to bind SAM to inhibit mTORC1 signaling, indicating that this function of SAMTOR does not require a methylation event.
Because SAM and SAH disrupt the interaction of SAMTOR with GATOR1-KICSTOR in vitro, we sought to determine whether this is also true in cells. The enzyme methionine adenosyltransferase (MAT) synthesizes SAM from adenosine triphosphate and methionine, an essential amino acid, so that starvation for methionine should lower SAM levels, as has been observed in other systems (25, 26). Indeed, SAM concentrations in HEK-293T cells decreased upon methionine starvation, falling from above the dissociation constant of SAMTOR for SAM to below it (Fig. 4A). In contrast, in both methionine replete and starved cells, SAH concentrations were lower than the Ki of SAMTOR for SAH (Fig. 4A), making it unlikely that SAH is a physiologically relevant modulator of the binding of SAMTOR to GATOR1-KICSTOR. Consistent with the effects of SAM on the interaction between SAMTOR and GATOR1-KICSTOR in vitro, methionine starvation strongly increased this interaction in cells. The addition to the methionine-starved cells of either methionine or SAM, which can enter cells when used at high concentrations, reduced the interaction to baseline levels (Fig. 4B).
Figure 4. SAMTOR senses SAM to signal methionine sufficiency to mTORC1.
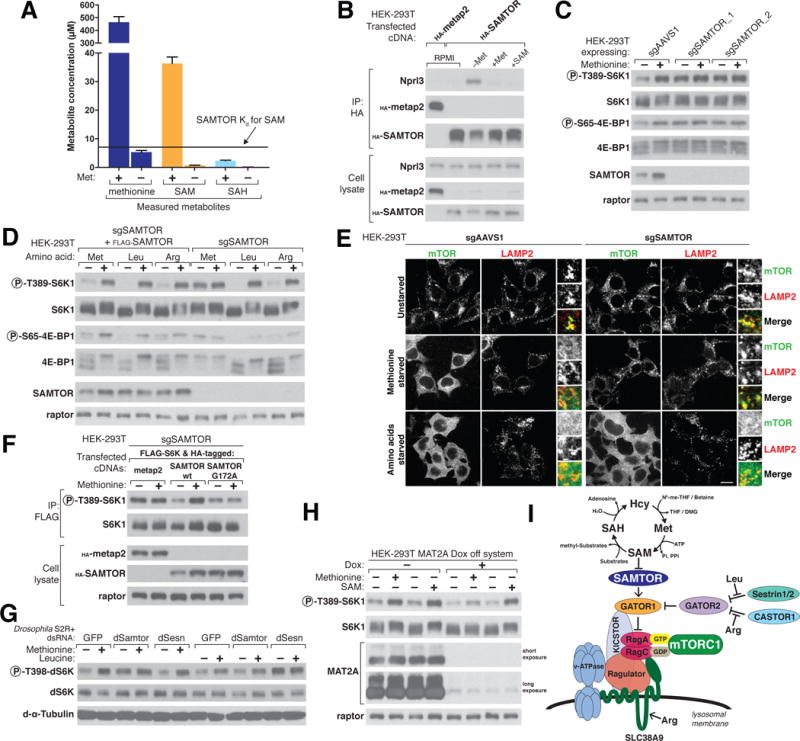
(A) HEK-293Tcells were incubated with or withoutmethionine for 2 hours before sample preparation for liquid chromatography/mass spectrometry (LC/MS)– based measurements of the absolute amounts of the indicated metabolites. The dissociation constant Kd of SAMTOR for SAM is indicated.
(B) Methionine starvation increases the interaction between SAMTOR and GATOR1. HEK-293T cells transiently expressing HA-tagged metap2 or SAMTOR were kept in growth medium (RPMI) or starved of methionine for 2 hours (–Met) and then restimulated for 20 min with 100 μM methionine (+Met) or 1 mM SAM (+SAM). HA immunoprecipitates and cell lysates were analyzed by immunoblotting for the levels of the indicated proteins.
(C) In SAMTOR-depleted cells, the mTORC1 pathway is resistant to methionine starvation. HEK-293Tcells stably coexpressing Cas9 and the indicated guides were incubated in media with or without methionine for 2 hours. Cell lysates were analyzed by immunoblotting for the phosphorylation states and the levels of the indicated proteins.
(D) The loss of SAMTOR does not affect the sensitivity of the mTORC1 pathway to leucine or arginine starvation. SAMTOR-deficient HEK-293Tcells with or without FLAG-SAMTOR expression were starved of the indicated amino acid for 2 hours. Cell lysates were analyzed as in (C).
(E) In cells without SAMTOR, mTOR colocalizes with lysosomes even upon methionine starvation. SAMTOR deficient or control HEK-293Tcells were treated as indicated for 2 hours before marker LAMP2. In all images, insets represent selected fields magnified 3.07X as well as their overlays. Scale bar, 10 μm. (F) Reexpression in SAMTOR-null cells of wild-type SAMTOR, but not the SAM-binding G172A mutant of SAMTOR, restored the capacity of the mTORC1 pathway to sense methionine sufficiency. SAMTOR-null cells were transfected with the indicated cDNAs and the cells were treated as in (C) before preparing lysates and FLAG immunoprecipitates. FLAG immunoprecipitates and cell lysates were analyzed as in (C).
(G) In Drosophila S2R+ cells depleted of dSamtor or dSesn, the dTOR pathway is resistant to methionine or leucine starvation, respectively. S2R+ cells were transfected with dsRNAs targeting the indicated mRNAs and starved of the indicated amino acids for 1 hour.
Cell lysates were analyzed as in (C).
(H) Acute loss of MAT2A using a doxycycline-suppressible (dox-off) system attenuates the capacity of mTORC1 to sense methionine but leaves SAM signaling largely intact. MAT2A dox-off HEK-293Tcells were treated with doxycycline (30 ng/ml) for 50 hours before starving them as in (C). Cell lysates were analyzed as in (C).
(I) Model depicting how SAM sensing by SAMTOR signals methionine levels to mTORC1. Substrates receiving a methyl group from SAM include DNA, RNA, proteins, and phospholipids. N5-me-THF, N5- methyl-tetrahydrofolate; DMG, dimethylglycine; Pi, inorganic phosphate; PPi, pyrophosphate.
Methionine starvation weakened the interaction between GATOR1 and GATOR2 in a SAMTOR-dependent fashion, whereas methionine addition restored the interaction to normal levels (fig. S3, A and B). Additionally, in a dose-dependent manner, SAMTOR overexpression was sufficient to disrupt the interaction between GATOR1 and GATOR2 (fig. S3, C and D).
Given that SAMTOR is an inhibitor of mTORC1 signaling and methionine starvation promotes the interaction between SAMTOR and GATOR1-KICSTOR, we hypothesized that methionine starvation would also inhibit mTORC1 signaling. Indeed, in multiple cell types, methionine starvation inhibited mTORC1 signaling in a SAMTOR-dependent fashion, as measured by the phosphorylation of the mTORC1 substrates S6K1 at Thr389 and 4E-BP1 at Ser65 (Fig. 4C and fig. S4, A to C). In contrast, loss of SAMTOR did not prevent the inhibition of mTORC1 signaling caused by withdrawal of leucine, arginine (Fig. 4D) or growth factors (fig. S4D).
Consistent with the effects of SAMTOR overexpression (Fig. 2B), methionine starvation also reduced the colocalization of mTOR with lysosomes in wild-type but not SAMTOR-null cells (Fig. 4E). Furthermore, reexpression of wild-type SAMTOR, but not a SAM-binding deficient mutant, restored the capacity of the mTORC1 pathway to sense methionine in the SAMTOR-null HEK-293T cells (Fig. 4F). Methionine starvation partially reduced SAMTOR levels in a proteasome-dependent manner (Fig. 4, B, C, and F, and fig. S4D) but this degradation was not required for mTORC1 to respond to methionine starvation (fig. S4E).
As in mammalian cells, dTOR signaling in Drosophila S2R+ cells also responds to environmental methionine and leucine levels, as detected by the phosphorylation of dS6K at residue Thr398 (Fig. 4G). Using double-stranded RNA (dsRNA)-induced RNA interference, we found that knock-down of dSamtor (encoded by the gene CG3570), but not of dSesn, prevented inhibition of dTOR signaling by methionine starvation (Fig. 4G and fig. S4F). However, the dsRNA targeting dSesn did prevent inhibition of dTOR by leucine starvation. Thus, the fly orthologs of SAMTOR and Sestrin2 have conserved roles in methionine and leucine sensing, respectively.
Our results show that SAMTOR is required for the mTORC1 pathway to detect changes in methionine levels and that this function requires its capacity to bind SAM. Moreover, the addition of SAM to methionine-starved cells reactivated mTORC1 signaling (Fig. 4H), indicating that it is the drop in SAM levels that mediates the inhibitory effects of methionine restriction on mTORC1. Given these findings, we predicted that the loss of methionine adenosyltransferase (MAT2A) would prevent mTORC1 from sensing methionine by blocking its conversion to SAM. Because MAT2A is essential in human cells (27, 28), we generated a doxycycline-repressible system in order to acutely suppress MAT2A expression (29). Consistent with SAMTOR sensing SAM rather than methionine directly, the loss of MAT2A greatly attenuated the capacity of mTORC1 to sense methionine while leaving its activation by SAM largely intact (Fig. 4H).
Several properties of SAMTOR suggest that it functions as a SAM sensor that signals methionine sufficiency to mTORC1 (Fig. 4I): (i) SAMTOR binds SAM with an affinity that is compatible with the drop in intracellular SAM concentrations caused by methionine starvation, (ii) SAMTOR is required for methionine starvation to inhibit mTORC1 signaling, and (iii) SAMTOR mutants that do not bind SAM cannot signal methionine sufficiency to mTORC1. Because SAM levels can be affected by the availability of folate, betaine, and vitamin B12, SAMTOR may also link mTORC1 signaling to the availability of these metabolites (30).
The Rag GTPase pathway senses and integrates the presence of multiple amino acids upstream of mTORC1 (4, 8). Sestrin1 and Sestrin2 detect leucine, whereas CASTOR1 and SLC38A9 sense cytosolic and lysosomal arginine, respectively (16, 19). In contrast to the Sestrins and CASTOR1, which bind to GATOR2, SAMTOR interacts with GATOR1-KICSTOR. Our genetic data suggest that SAMTOR potentiates GATOR1 function through an unknown mechanism that may involve disruption of the binding of GATOR1 to GATOR2. The interaction between SAMTOR and GATOR1 requires KICSTOR, which may reflect either a composite binding site or the requirement for KICSTOR to localize GATOR1 to the lysosomal surface. In addition, structural information will be needed if we are to understand how the binding of SAM to SAMTOR disrupts its interaction with GATOR1 and KICSTOR.
Unlike leucine and arginine, which directly bind sensors upstream of mTORC1, methionine is sensed indirectly through SAM. SAM is a central metabolite required for most methylation reactions, including that of DNA(31), histones (25, 30), and phospholipids (32), and our work highlights its additional role as a signaling molecule. Whereas Saccharomyces cerevisiae does not have a SAMTOR homolog, the yeast TOR pathway does sense methionine through the regulated methylation of the PP2A family of phosphatases (33).
In metazoans, the mTORC1 pathway senses multiple amino acids, which suggests that these nutrients were, at times, scarce during their evolution. Two inferences can be drawn from the existence of SAMTOR: (i) SAM can become limiting in certain nutritional states, and (ii) modulation of mTORC1 under these conditions is beneficial for maintaining organismal homeostasis. Indeed, diets low in methionine reduce tissue SAM levels and improve insulin sensitivity, and extend lifespan in mice and rats (34–38). It is intriguing to speculate that these benefits might be mediated in part via the SAMTOR-dependent inhibition of mTORC1, which is well appreciated for its impact on glucose metabolism and the aging process (1). Given that SAMTOR has a SAM-binding pocket, it may be possible to modulate SAMTOR function pharmacologically.
Supplementary Material
Figure S1. (A) Overexpressed GATOR1 co-immunoprecipitated transiently expressed SAMTOR only when KICSTOR was also transfected. FLAG-immunoprecipitates were prepared from wild-type HEK-293T cells transiently expressing the indicated cDNAs. FLAGimmunoprecipitates and lysates were analyzed as in Fig 1(A).
Figure S2. (A) Sequence alignment of SAMTOR homologues from various organisms. Amino acid positions are colored white and blue according to increasing sequence similarity. Two residues (G172 and D190) significant for SAM binding capacity are indicated with orange dots.
(B) Sequence alignment of human SAMTOR with three methyltransferases selected from the list of proteins predicted by HHPred as having secondary structure similarity to SAMTOR. Two residues (G172 and D190) significant for SAM binding capacity are indicated with orange dots.
Figure S3. (A) Methionine starvation increases the interaction between SAMTOR and GATOR1 but weakens that between GATOR1 and GATOR2. Stably expressed FLAG-metap2 or endogenously FLAG-tagged Depdc5 HEK-293T cells were kept in growth media (RPMI) or starved of methionine for 2 hours or starved for methionine for 2 hours and then restimulated for either 10 or 25 minutes with 100 μM methionine. FLAGimmunoprecipitates and cell lysates were analyzed by immunoblotting for the levels of the indicated proteins.
(B) Loss of SAMTOR weakens the regulation by methionine starvation of the GATOR1- GATOR2 interaction. The control and SAMTOR-deficient HEK-293T cells were treated and analyzed as in (A).
(C and D) Transiently overexpressed SAMTOR decreases the interaction between GATOR1 and GATOR2. Endogenously FLAG-tagged-Depdc5 (C) and -WDR59 (D) HEK-293T cells were transfected with a control cDNA or increasing amounts of the SAMTOR cDNA. FLAG-immunoprecipitates were analyzed as in (A).
Figure S4. (A) In HeLa cells with reduced SAMTOR expression, the mTORC1 pathway is resistant to methionine starvation. Two SAMTOR-deficient HeLa cell lines generated using CRISPR/Cas9 were treated as in Fig 4(C). Cell lysates were analyzed by immunoblotting for the phosphorylation states and levels of the indicated proteins.
(B) In MEFs with reduced SAMTOR expression, the mTORC1 pathway is resistant to methionine starvation. Cells were prepared via the stable expression of Cas9 along with the indicated guide. Cells were treated as in Fig. 4(C) and the lysates were analyzed by immunoblotting for the phosphorylation states and levels of the indicated proteins.
(C) In HEK-293T cells, restoration of SAMTOR expression in SAMTOR-deficient cells rescues methionine starvation phenotype to similar level as in wild-type cells. Wild-type, SAMTOR-deficient cell line and FLAG-SAMTOR stably expressed SAMTOR-deficient cell line were prepared and treated as in Fig. 4(C) and the lysates were analyzed by immunoblotting for the phosphorylation states and the levels of the indicated proteins.
(D) The loss of SAMTOR in HeLa cells does not impact the regulation of mTORC1 by growth factors. SAMTOR-deficient cells were incubated in the presence or absence of insulin for 1 hour. Cell lysates were analyzed by immunoblotting for the indicated proteins.
(E) Methionine starvation causes SAMTOR protein levels to drop in a proteasome dependent fashion. 10 μM of the indicated proteasome inhibitors was added to HEK-293T cells cultured in media with or without methionine for 2 hours. Cell lysates were analyzed by immunoblotting for the phosphorylation states and levels of the indicated proteins.
(F) mRNA levels of dSamtor and dSesn in Drosophila S2R+ cells after transfection of the indicated dsRNA. cDNA from transfected cells was synthesized and used for quantitative PCR. Reported values are mean ± SD of three technical replicates of ΔΔCt values, using alpha-tubulin mRNA as an internal standard.
Acknowledgments
We thank all members of the Sabatini lab for helpful insights and suggestions, in particular N. Kory for helping validate the efficacy of dsRNA knockdowns in Drosophila cells; G. Wyant for experimental discussions and advice; C. Lewis, B. Chan, and T. Kunchok for performing the LC/MS analysis; P. Jouandin and N. Perrimon for generously providing Drosophila S2R+ cells; and C. Thoreen for providing pCW57.1 vector. D.M.S., X.G., and J.M.O. are inventors on patent application submitted by Whitehead Institute for Biomedical Research that relates to SAMTOR and its role in mTORC1 signaling. Supported by NIH grants R01 CA103866 and R37 AI47389 and U.S. Department of Defense grant W81XWH-07-0448 (D.M.S.), NIH fellowships T32 GM007753 and F30 CA210373 (J.M.O.), NIH grant F31 GM121093-01A1 (R.A.S.), NSF grant 2016197106 (K.J.C.), Paul Gray UROP Fund grant 3143900 (S.M.S.), Michael J. Fox Foundation grants NS083524 and AG011085 (J.W.H.), and NIH grant U41 HG006673 (S.P.G. and J.W.H.). J.W.H. is a paid consultant for Takeda Pharmaceuticals, SV Brahma Discovery, and the American Society for Microbiology. D.M.S. is an investigator of the Howard Hughes Medical Institute and a founding member of the scientific advisory board, a paid consultant, and a shareholder of Navitor Pharmaceuticals, which is targeting for therapeutic benefit the amino acid sensing pathway upstream of mTORC1.
References and Notes
- 1.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nature reviews Molecular cell biology. 2013;14:133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsun ZY, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar-Peled L, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfson RL, et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017;543:438–442. doi: 10.1038/nature21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng M, Yin N, Li MO. SZT2 dictates GATOR control of mTORC1 signalling. Nature. 2017;543:433–437. doi: 10.1038/nature21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rebsamen M, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519:477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung J, Genau HM, Behrends C. Amino Acid-Dependent mTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9. Mol Cell Biol. 2015;35:2479–2494. doi: 10.1128/MCB.00125-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyant G, et al. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell. doi: 10.1016/j.cell.2017.09.046. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfson RL, et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxton RA, et al. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. 2016;351:53–58. doi: 10.1126/science.aad2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU, Sabatini DM. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature. 2016;536:229–233. doi: 10.1038/nature19079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chantranupong L, et al. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell. 2016;165:153–164. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huttlin EL, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chantranupong L, et al. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 2014;9:1–8. doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parmigiani A, et al. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep. 2014;9:1281–1291. doi: 10.1016/j.celrep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hildebrand A, Remmert M, Biegert A, Soding J. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009;77(Suppl 9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 24.Kozbial PZ, Mushegian AR. Natural history of S-adenosylmethionine-binding proteins. BMC Struct Biol. 2005;5(19) doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mentch SJ, et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015;22:861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan CL, et al. Targeting S-adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A. Nat Chem Biol. 2017;13:785–792. doi: 10.1038/nchembio.2384. [DOI] [PubMed] [Google Scholar]
- 27.Wang T, et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell. 2017;168:890–903 e815. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye C, Sutter BM, Wang Y, Kuang Z, Tu BP. A Metabolic Function for Phospholipid and Histone Methylation. Mol Cell. 2017;66:180–193 e188. doi: 10.1016/j.molcel.2017.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutter BM, Wu X, Laxman S, Tu BP. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell. 2013;154:403–415. doi: 10.1016/j.cell.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orentreich N, Matias JR, DeFelice A, Zimmerman JA. Low methionine ingestion by rats extends life span. J Nutr. 1993;123:269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 35.Miller RA, et al. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L, Sadighi Akha AA, Miller RA, Harper JM. Life-span extension in mice by preweaning food restriction and by methionine restriction in middle age. J Gerontol A Biol Sci Med Sci. 2009;64:711–722. doi: 10.1093/gerona/glp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown-Borg HM, et al. Growth hormone signaling is necessary for lifespan extension by dietary methionine. Aging Cell. 2014;13:1019–1027. doi: 10.1111/acel.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wanders D, et al. Role of GCN2-Independent Signaling Through a Noncanonical PERK/NRF2 Pathway in the Physiological Responses to Dietary Methionine Restriction. Diabetes. 2016;65:1499–1510. doi: 10.2337/db15-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma S, Watzinger P, Kotter P, Entian KD. Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2013;41:5428–5443. doi: 10.1093/nar/gkt195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 42.Birsoy K, et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen WW, Freinkman E, Wang T, Birsoy K, Sabatini DM. Absolute Quantification of Matrix Metabolites Reveals the Dynamics of Mitochondrial Metabolism. Cell. 2016;166:1324–1337 e1311. doi: 10.1016/j.cell.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (A) Overexpressed GATOR1 co-immunoprecipitated transiently expressed SAMTOR only when KICSTOR was also transfected. FLAG-immunoprecipitates were prepared from wild-type HEK-293T cells transiently expressing the indicated cDNAs. FLAGimmunoprecipitates and lysates were analyzed as in Fig 1(A).
Figure S2. (A) Sequence alignment of SAMTOR homologues from various organisms. Amino acid positions are colored white and blue according to increasing sequence similarity. Two residues (G172 and D190) significant for SAM binding capacity are indicated with orange dots.
(B) Sequence alignment of human SAMTOR with three methyltransferases selected from the list of proteins predicted by HHPred as having secondary structure similarity to SAMTOR. Two residues (G172 and D190) significant for SAM binding capacity are indicated with orange dots.
Figure S3. (A) Methionine starvation increases the interaction between SAMTOR and GATOR1 but weakens that between GATOR1 and GATOR2. Stably expressed FLAG-metap2 or endogenously FLAG-tagged Depdc5 HEK-293T cells were kept in growth media (RPMI) or starved of methionine for 2 hours or starved for methionine for 2 hours and then restimulated for either 10 or 25 minutes with 100 μM methionine. FLAGimmunoprecipitates and cell lysates were analyzed by immunoblotting for the levels of the indicated proteins.
(B) Loss of SAMTOR weakens the regulation by methionine starvation of the GATOR1- GATOR2 interaction. The control and SAMTOR-deficient HEK-293T cells were treated and analyzed as in (A).
(C and D) Transiently overexpressed SAMTOR decreases the interaction between GATOR1 and GATOR2. Endogenously FLAG-tagged-Depdc5 (C) and -WDR59 (D) HEK-293T cells were transfected with a control cDNA or increasing amounts of the SAMTOR cDNA. FLAG-immunoprecipitates were analyzed as in (A).
Figure S4. (A) In HeLa cells with reduced SAMTOR expression, the mTORC1 pathway is resistant to methionine starvation. Two SAMTOR-deficient HeLa cell lines generated using CRISPR/Cas9 were treated as in Fig 4(C). Cell lysates were analyzed by immunoblotting for the phosphorylation states and levels of the indicated proteins.
(B) In MEFs with reduced SAMTOR expression, the mTORC1 pathway is resistant to methionine starvation. Cells were prepared via the stable expression of Cas9 along with the indicated guide. Cells were treated as in Fig. 4(C) and the lysates were analyzed by immunoblotting for the phosphorylation states and levels of the indicated proteins.
(C) In HEK-293T cells, restoration of SAMTOR expression in SAMTOR-deficient cells rescues methionine starvation phenotype to similar level as in wild-type cells. Wild-type, SAMTOR-deficient cell line and FLAG-SAMTOR stably expressed SAMTOR-deficient cell line were prepared and treated as in Fig. 4(C) and the lysates were analyzed by immunoblotting for the phosphorylation states and the levels of the indicated proteins.
(D) The loss of SAMTOR in HeLa cells does not impact the regulation of mTORC1 by growth factors. SAMTOR-deficient cells were incubated in the presence or absence of insulin for 1 hour. Cell lysates were analyzed by immunoblotting for the indicated proteins.
(E) Methionine starvation causes SAMTOR protein levels to drop in a proteasome dependent fashion. 10 μM of the indicated proteasome inhibitors was added to HEK-293T cells cultured in media with or without methionine for 2 hours. Cell lysates were analyzed by immunoblotting for the phosphorylation states and levels of the indicated proteins.
(F) mRNA levels of dSamtor and dSesn in Drosophila S2R+ cells after transfection of the indicated dsRNA. cDNA from transfected cells was synthesized and used for quantitative PCR. Reported values are mean ± SD of three technical replicates of ΔΔCt values, using alpha-tubulin mRNA as an internal standard.


