Abstract
TRPML3 channels are mainly localized to endolysosomes and play a critical role in the endocytic pathway. Their dysfunction causes deafness and pigmentation defects in mice. TRPML3 activity is inhibited by low endolysosomal pH. Here we present cryoelectron microscopy structures of human TRPML3 in the closed, agonist-activated, and low-pH-inhibited states, with resolutions of 4.06, 3.62 and 4.65 Å, respectively. The agonist ML-SA1 lodges between S5 and S6 and opens an S6 gate. A polycystin-mucolipin domain (PMD) forms a luminal cap. S1 extends into this cap, forming a ‘gating rod’ that connects directly to a luminal pore-loop, which undergoes dramatic conformational changes in response to low luminal pH. S2 extends intracellularly and interacts with several intracellular regions to form a ‘gating knob’. These unique structural features, combined with electrophysiological studies, reveal a new mechanism thereby luminal pH and other physiological modulators such as PIP2 regulate TRPML3 by changing S1 and S2 conformations.
INTRODUCTION
The endocytic pathway is essential for cell signaling and physiology1. It is regulated by a wide array of ion channels that are present in the endocytic vesicles2. The mucolipin subfamily of transient receptor potential (TRP) channels (TRPML) are primarily localized in endosomes and lysosomes3–8. TRPML channels are tetramers. Each subunit has six transmembrane segments (S1–S6) and a pore-loop between S5 and S6, which form a voltage-sensor-like domain (VSLD) and a pore domain. A long luminal/extracellular linker exists between S1 and S2 (the S1–S2 linker). This linker is named the polycystin-mucolipin domain (PMD)9 and is a unique but shared feature of the TRPML and TRPP subfamilies of TRP channels9–12. TRPML channels are nonselective cation channels that mainly conduct Ca2+ and monovalent cation currents from the lumen to the cytoplasm4–7,13–17. They play a crucial role in membrane trafficking4,5,18–22, autophagy4,5,19,22–26, exocytosis4,5,15,22,24 and ion homeostasis27,28.
There are three TRPML subtypes, TRPML1-3. They share ~40% amino acid sequence identity (Supplementary Fig. 1) and have overlapping tissue distribution and biophysical properties4–8,17. The founding member TRPML1 was identified as a genetic determinant of mucolipidosis type IV (MLIV)29–31, a severe lysosomal storage disorder (LSD) that display cognitive, linguistic, visual and motor deficits4,32–34. No human diseases have been linked to TRPML2 and TRPML3, but two spontaneous gain-of-function mutations in TRPML3 (A419P and I362T) cause deafness and coat color dilution in mice6,7,35–39. These mutations are called varitint-waddler mutations (Va, A419P and VaJ, A419P + I362T). The severe effects of TRPML1 and TRPML3 mutations underscore the crucial functional importance of these channels in cell physiology.
The lumen of endosomes and lysosomes has a lower pH than the cytoplasm and extracellular milieu do. Late endosomes have an acidic pH of 5.5–6.0, and lysosomes have a more acidic pH of 4.5–5.02,5,40–42, a condition necessary to maintain the activity of lysosomal hydrolases17,43. In accordance with their endolysosomal localization, TRPML1 and TRPML3 have adopted common strategies to fine tune their activities to serve their physiological functions in the endocytic pathway. Thus, TRPML1 and TRPML3 are activated by PI(3, 5)P216, which is enriched in the endolysosomal membrane17,44. On the other hand, these channels are suppressed by PI(4, 5)P245 (H. Xu, personal communication), which is more abundant in the plasma membrane. Two separate clusters of positively charged amino acids in the N-terminus of TRPML1 are critical for these effects45. Another common strategy is to employ the low endolysosomal pH to regulate their activities. In the presence of Ca2+, TRPML1 currents are greatly enhanced by H+ 4,13–17,46,47. Three aspartic residues in a luminal pore-loop are crucial for this enhancement9. In contrast, TRPML3 currents are inhibited by H+ 48. A histidine residue (H283) in the PMD is crucial for this inhibition48. A unique property of TRPML3 is that it is inhibited by Na+ 37,48,49, whose concentration in lysosomes may be as high as in the extracellular solution50. Therefore, under physiological conditions, endolysosomal TRPML1 channels are active whereas TRPML3 channels are inactive. However, upon lysosomal damage or breakdown of lysosomal Na+ and H+ gradients, TRPML3 channels become activated. The ensuing Ca2+ influx triggers the exocytosis of neutralized lysosomes. This mechanism plays a role in the expulsion of pathogens from infected cells51.
A large number of synthetic small molecule agonists of TRPML channels have been identified52,53. Many of these molecules, such as ML-SA1, activate all three subtypes of TRPML channels but show specificity toward other types of ion channels53,54. These molecules are useful tools in studying the physiological functions of TRPML channels.
To better understand the molecular mechanisms of TRPML channel function and regulation, we determined the structures of full length human TRPML3 in the apo, ML-SA1-bound, and low-pH-inhibited states by using cryoelectron microscopy (cryo-EM). We previously obtained high-resolution crystal structures of an isolated TRPML1 PMD at three different pH conditions that correspond to the pH in lysosomes, endosomes and the extracellular milieu9. Our new structures show striking differences as well as expected similarities between TRPML3 and TRPML1 PMDs and reveal novel structural features and conformational changes that shed light on TRPML3 activation and regulation.
RESULTS
Structure determination and general architecture
Full-length human TRPML3 was heterologously expressed in Tni inset cells. Purified TRPML3 protein in the apo and ML-SA1-bound forms at pH 7.4 was used for single particle cryo-EM analysis (Supplementary Fig. 2–4). With C4 symmetry imposed, the structures of the two forms were determined to an overall resolution of 4.06 Å and 3.62 Å, respectively. The central regions were better resolved than the peripheries (Supplementary Fig. 2, 3 and 5a). Most of the 553 amino acids were modeled in both structures, but some amino acids were not modeled due to poor density, including 32 residues in the distal N-terminus, 27 residues in the distal C-terminus, and 18 residues in the PMD (Fig. 1, Supplementary Fig. 1, and Table 1).
Figure 1. Structures of Apo and Agonist-bound TRPML3.
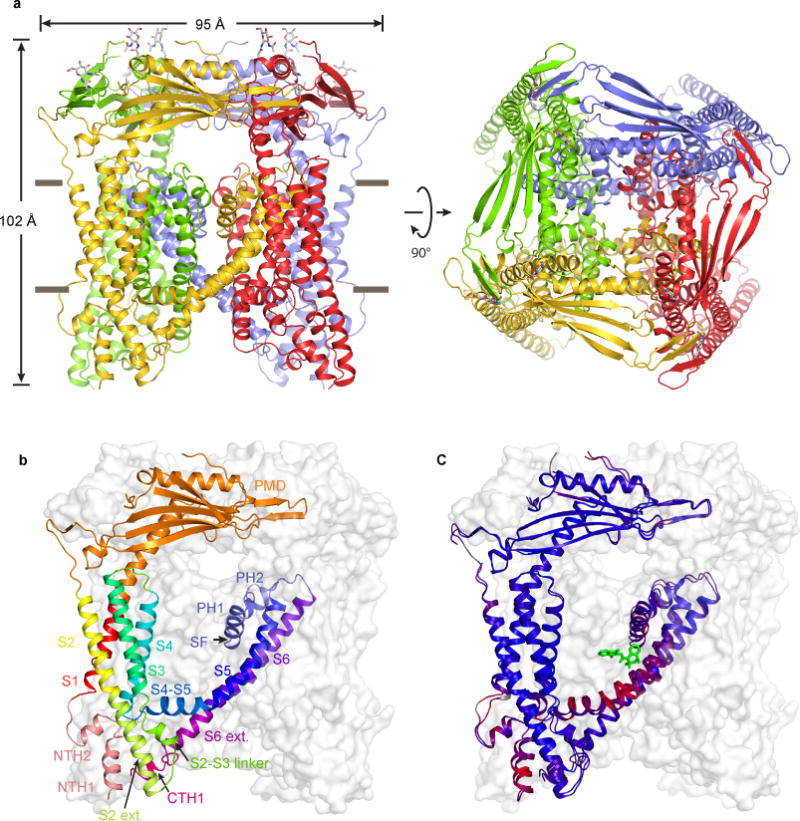
(a) ML-SA1-bound structure, viewed parallel to the membrane (left) and from the luminal/extracellular side of the membrane (right). (b) Structure of a protomer, viewed parallel to the membrane. Different regions are illustrated in different colors. The other three subunits are shown in grey surface representations. (c) Superposition of the apo and ML-SA1-bound structures. Blue and red indicate similarity and divergence, respectively.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| Apo TRPML3 at pH 7.4 (EMDB 7018, PDB 6AYE) | TRPML3 / ML-SA1 (EMDB 7019, PDB 6AYF) | Apo TRPML3 at pH 4.8 (EMDB 7020, PDB 6AYG) | |
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 22,500 | 105,000 | 105,000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 50 | 50 | 50 |
| Defocus range (μm) | −1.5 – −3.0 | −1.0 – −2.5 | −1.0 – −2.5 |
| Pixel size (Å) | 1.32 | 1.338 | 1.338 |
| Symmetry imposed | C4 | C4 | C4 |
| Initial particle images (no.) | 1,059,673 | 697,805 | 171,031 |
| Final particle images (no.) | 43,542 | 50,726 | 42,559 |
| Map resolution (Å) | 4.06 | 3.62 | 4.65 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | — | — | — |
| Refinement | |||
| Initial model used | — | — | — |
| Model resolution (Å) | 4.06 | 3.62 | 4.65 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Model resolution range (Å) | 211.20 – 4.06 | 214.08 – 3.62 | 214.08 – 4.65 |
| Map sharpening B factor (Å2) | −228 | −180 | −244 |
| Model composition | |||
| Nonhydrogen atoms | 15664 | 15780 | 10456 |
| Protein residues | 1904 | 1904 | 1756 |
| Ligands | 0 | 8 | 0 |
| B factors (Å2) | |||
| Protein | 116 | 118 | 153 |
| Ligand | — | 120 | — |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.009 | 0.005 | 0.013 |
| Bond angles (°) | 1.44 | 1.11 | 1.71 |
| Validation | |||
| MolProbity score | 2.11 | 2.18 | 2.62 |
| Clash score | 15.6 | 17.0 | 17.4 |
| Poor rotamers (%) | 0.9 | 0.7 | 3.3 |
| Ramachandran plot | |||
| Favored (%) | 93.8 | 93.0 | 91.9 |
| Allowed (%) | 5.6 | 6.8 | 7.6 |
| Disallowed (%) | 0.6 | 0.2 | 0.5 |
The channel is a four-fold symmetric homotetramer with a central ion-conducting pore and a dimension of 95 Å × 95 Å × 102 Å (Fig. 1a). The transmembrane domain (TMD) has a typical architecture, with S1–S4 and the S4–S5 linker forming a VSLD and S5, S6 and the pore-loop forming the pore domain (Fig. 1b). The pore-loop contains the ion selectivity filter (SF), which is flanked by two pore helices (PH1 and PH2). The VSLD of one subunit interacts with the pore domain of a neighboring subunit in a canonical domain-swapped configuration (Fig. 1a). The PMD of each subunit interacts with both of its neighbors and forms a luminal (i.e., extracellular) cap on top of the TMD (Fig. 1a). The N- and C-termini are located intracellularly.
The apo and ML-SA1-bound structures are overall very similar (Fig. 1c); the two structures superimpose well in the PMD (rmsd=0.63 for amino acids 81–275) and S1–S4 (rmsd=0.60 for amino acids 61–80 and 284–395). But the two structures diverge significantly in the S4–S5 linker, S5, S6 and the pore-loop.
Recently a 5.4 Å-resolution cryo-EM structure of the mouse TRPML1 has been obtained55. This structure shows a similar general architecture as our TRPML3 structures.
The ML-SA1 binding pocket
Comparison of the electron density maps of the apo and ML-SA1-bound structures shows clear differences in a crevice surrounded by S5, S6 and pore helix 1 of one subunit and S6 of a neighboring subunit (Supplementary Fig. 5b). The normalized difference map between the two structures reveals an extra density in the ML-SA1-bound structure. We posit that this density represents a bound ML-SA1 (Fig. 2a). Supporting this proposition, ML-SA1 binding produces significant conformational changes in the regions surrounding the crevice (Supplementary Video 1): S5 and S6 are pushed outward and the S4–S5 linker is pushed downward by 2 to 4 Å, the pore-loop is pulled downward by ~2 Å, and S6 undergoes a 27 degree counterclockwise rotation (top down view). Due to the limited resolution, we were unable to unambiguously define the orientation of ML-SA1 and its molecular interactions with the channel, but we identified two amino acids, Y423 in S5 and F497 in S6, as key residues participating in ML-SA1 binding (Fig. 2a).
Figure 2. The ML-SA1 Binding Site.
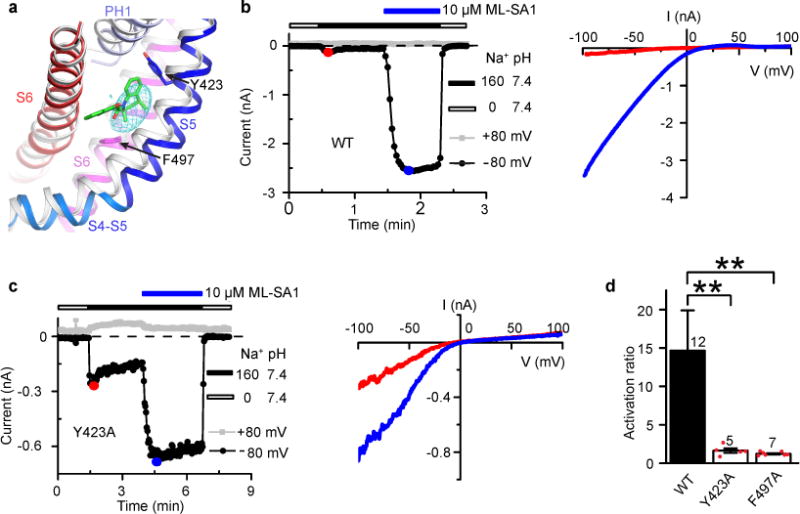
(a) Close-up view of the indicated regions in the apo (gray) and ML-SA1-bound (color) structures. The mesh shows the normalized difference density map between the apo and ML-SA1-bound structures. ML-SA1 is shown in green. (b) Time course (left) and current-voltage (I-V) relationships (right) of WT channel currents in response to ML-SA1. In this and subsequent figures, the Na+ concentration (in mM) and pH of the bath solution is indicated. The I-V curves are obtained at the color-marked time points. (c) Time course (left) and I-V relationships (right) of Y423A channel currents in response to ML-SA1. (d) Bar graph showing the ratio of the ML-SA1-activated current and the basal current for WT, Y423A and F497A channels. Number of recordings is shown above the bars. Error bars represent SEM. Red dots represent original data points for Y423A and F497A.
To verify that ML-SA1 indeed binds in this crevice, we examined the effect of ML-SA1 on WT and mutant channels carrying the Y423A or F497A mutation. WT and mutant channels were expressed in HEK 293T cells and studied by whole-cell recording, largely following the protocols of previous studies39,48. A basal current was usually detected upon changing the bath solution from a Na+-free to a 160 mM Na+ solution (Fig. 2b,c). Bath application of 10 μM ML-SA1 greatly increased the WT channel current (Fig. 2b), as has been reported53,54. Both the basal current and ML-SA1-induced current showed strong inward rectification (Fig. 2b). Inwardly rectifying current was absent in nontransfected cells. The same concentration of ML-SA1 also increased the currents of Y423A (Fig. 2c) and F497A; however, instead of a 15-fold increase for the WT channel, it was only 1.6-fold for Y423A and 1.2-fold for F497A channels (Fig. 2d), indicating that ML-SA1’s effect was greatly weakened by the mutations.
It is interesting to note that the ML-SA1 binding site is close to the A419P Va mutation site and that both ML-SA1 binding and A419P mutation make the channel stay open. It is also interesting to note that in the ML-SA1 binding site, there are electron densities in the apo structure that cannot be accounted for by the surrounding amino acid side chains (Supplementary Fig. 5b). Whether these densities represent endogenous molecules such as lipids or small ligands awaits further investigation. Cheminformatics analysis categorizes TRPML3 agonists into 9 chemical scaffolds and 20 singletons6,52. It will be interesting to determine where different classes of molecules bind in TRPML3. This knowledge may aid the development of selective TRPML activators that may ultimate yield clinical benefits for ML-IV and other LSDs.
Unique structures of S1 and S2
The TRPML3 structures show several striking and important features (Fig. 3). First, S1 extends extracellularly into the PMD by forming a continuous α helix with the first 19 amino acids (designated as α1) of the PMD (Supplementary Fig. 3a). This observation confirms our previous speculation of such a rigid connection based on the crystal structures of an isolated TRPML1 PMD9. The S1 extension connects directly to the luminal pore-loop, which has a divergent amino acid sequence among the three TRPML subunits (Supplementary Fig. 1) and contains three negatively charged amino acids (D108, D111 and D112 in TRPML3) (Supplementary Fig. 3a). With this rigid physical connection, conformational changes occurring in the luminal pore-loop likely propagate to S1 and, through it, to other transmembrane regions. We will revisit this mechanism later. Considering its shape and role in gating, we name the continuous S1+α1 helix the “gating rod”. TRPML1 also has such a gating rod55.
Figure 3. Unique Structural Features of TRPML3.
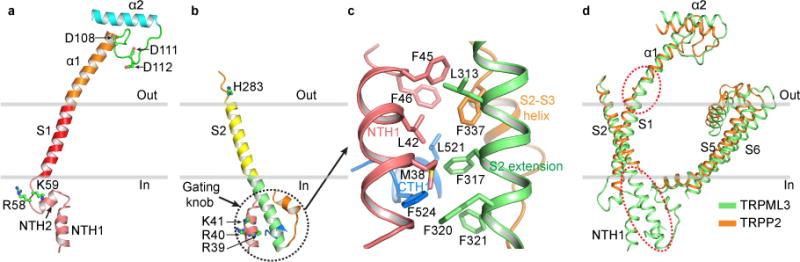
(a) Structure of the S1 gating rod and its end connections, viewed parallel to the membrane. (b) Structure of the S2 gating knob, viewed parallel to the membrane. (c) Close-up view of the gating knob, illustrating the regions and hydrophobic interactions involved in forming the knob. (d) Comparison of selected regions of TRPML3 and TRPP2 (PDB accession number: 5T4D), highlighting the lack of gating rod and gating knob in TRPP2.
Second, S2 extends intracellularly beyond the inner membrane boundary, forming a continuous α helix with the first 21 amino acids of the S2–S3 linker (Supplementary Fig. 3b). This S2 extension interacts with an N-terminal helix (NTH1), a C-terminal helix (CTH1) and another helix in the S2–S3 linker to form an irregular four-helix bundle (Fig. 3c). Given its shape (Fig. 3b) and possible role in regulating channel gating, we name this helix bundle the “gating knob”. Conformational changes occurring in any part of the gating knob are likely transmitted to S2 and hence to other parts of the VSLD. The gating knob is formed through extensive hydrophobic interactions (Fig. 3c), and the involved hydrophobic amino acids are well conserved in the TRPML subfamily (Supplementary Fig. 1), suggesting a conserved structural configuration and functional mechanism. H283 is situated at the luminal end of S2 (Fig. 3b). Thus, conformational changes triggered by the protonation of H283 are likely transmitted to the VSLD and gating knob through S2.
Third, the amino acids important for PI(3, 5)P2 activation of TRPML1 are R61 and K6245; the corresponding residues in TRPML3 are R58 and K59 (Supplementary Fig. 1). They are located at the intracellular end of S1 and near the inner membrane surface (Fig. 3a). The amino acids important for PI(4, 5)P2 inhibition of TRPML1 are R42, R43 and R4445; the analogous residues in TRPML3 are R39, R40 and K41 (Supplementary Fig. 1). They are located on NTH1, which is a part of the gating knob (Fig. 3b). We speculate that PI(3, 5)P2 activates TRPML3 by pulling on S1 and PI(4, 5)P2 inhibits TRPML3 by pulling on the gating knob and consequently on S2. Precisely how PI(3, 5)P2 and PI(4, 5)P2 interact with TRPML3 to alter its activity remains to be elucidated structurally.
The direct connections of the VSLD to both the luminal (extracellular) and intracellular domains through continuous rigid α helices revealed in TRPML3 are a unique feature in the voltage-gated ion channel superfamily. Through such connections, luminal and intracellular modulators can change the conformation of the VSLD and thus change channel activity. These connections are not observed even in the closely related TRPP2. In TRPP2 structures10–12, S1 and α1 are interrupted by a loop, and the distal C-terminal end of S2 and the S2–S3 linker are not well resolved (Fig. 3d), suggesting that these regions are flexible.
The ion conduction pathway and activation gate
The ion conduction pathway in the TMD consists of the ion selectivity filter (SF), the central cavity and the inner pore (Fig. 4a,b). The SF is formed by four conserved amino acids N456, G457, D458 and D459 (Fig. 4c and Supplementary Fig. 1). Starting from the luminal entrance to the central cavity, it is lined by the side chains of D459 and D458 and the backbone carbonyls of G457 and N456 (Fig. 4c). The SF external entrance is thus highly electronegative and hence attracts cations. The SF lumen is 5.4–7.3 Å wide in the apo state and 6.1–8.3 Å wide in the ML-SA1-bound state. These features are consistent with TRPML3 being a Ca2+-permeable, nonselective cation channel36–39. They also explain why the D458K and D458K/D459K mutations abolish channel activity39,56.
Figure 4. The Pore and Gate.
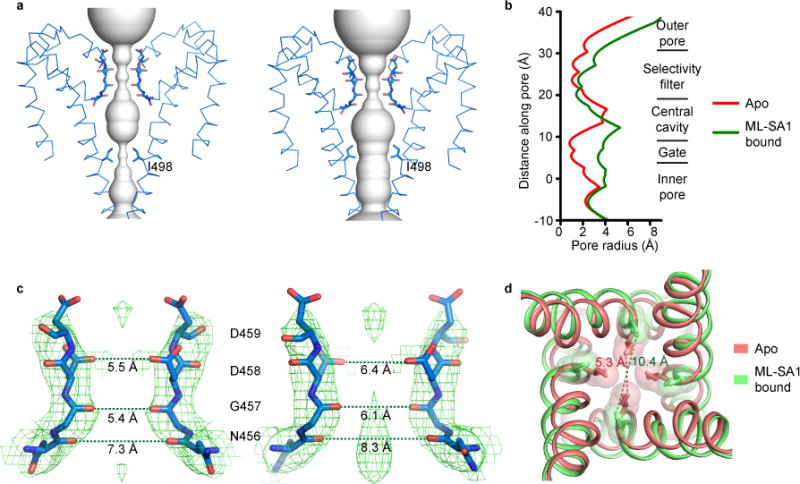
(a) Solvent-accessible pathway in the TMD in the apo (left) and ML-SA1-bound (right) structures. Only two diagonally opposed subunits are shown. (b) Pore-size profile generated with the HOLE program of different sections of the solvent-accessible pathway. The origin of the pore axis is set at the cytoplasmic end of S6. (c) Close-up view of the selectivity filter in the apo (left) and ML-SA1-bound (right) structures. Only two diagonally opposed subunits are shown. The cryo-EM density map and modeled amino acids are shown in mesh and sticks, respectively. (d) Superposition of S6 near I498 in the apo and ML-SA1-bound structures. The I498 side chains are shown in both stick and space-filling representations. In (c) and (d), the distances between the atoms are measured as the center-to-center distance of two diagonally opposed atoms.
A strong electron density is present in the center of the SF just above the central cavity in the ML-SA1-bound structure, but not in the apo structure (Fig. 4c). As Na+ was the only cation in the cryo-EM protein sample, this density may represent a Na+ ion binding site.
The luminal cap atop the TMD has multiple passageways for cations to flow to the SF, including a 11.4 Å-wide luminal pore in the center and four 10 Å-wide portals on the side (Supplementary Fig. 6a). These entryways are electronegative and sufficiently wide for hydrated monovalent and divalent cations to pass through.
The pH7.4 apo and ML-SA1-bound structures most likely represent closed and open states, respectively. In the apo structure, the narrowest constriction of the ion conduction pathway is formed by the side chains of I498 of S6 and is 5.3 Å wide (Fig. 4d). These side chains point to the center of the inner pore and form a hydrophobic seal that blocks monovalent and divalent cation conduction. In the ML-SA1-bound structure, S6 moves outward and rotates 27 degrees counterclockwise, thereby placing the I498 side chains sideways. These movements dilate much of the inner pore (Fig. 4b and Supplementary Video 1) and expand the I498 hydrophobic seal to 10.4 Å (Fig. 4d and Supplementary Video 1), which is wide enough to allow conduction of hydrated monovalent and divalent cations.
It was recently proposed that the closely related TRPP2 channel has two gates, one in the SF and one in S6; the latter is formed by L677 and N68110–12. We do not consider the SF to be a gate in TRPML3 since the SF has similar structures in the closed and open states and its physicochemical properties should allow it to pass partially hydrated monovalent and divalent cations in both states (Fig. 4b,c). Our work suggests that TRPML3 has only one activation gate, i.e., the S6 gate located at I498.
The polycystin-mucolipin domain
Each PMD protomer has a structural fold that consists of four α-helices (α1-α4) and eight β-strands (Fig. 1b and Supplementary Fig. 1 and 6b). The α1 and α2 helices are packed tightly against a 5-stranded β sheet (β1, β4, β5, β6 and β7) (Supplementary Fig. 6b). Between α1 and α2 is the luminal pore-loop (Fig. 3a and Supplementary Fig. 6b). Although the overall structure of the TRPML3 PMD is highly similar to that of the TRPML1 PMD (Supplementary Fig. 6b), their luminal pore-loops have different conformations (Supplementary Fig. 6c–e). Structural divergence in this 16-amino acid loop is largest from position 5 to position 10, corresponding to MDRMDD in TRPML3 and SDGADD in TRPML1. As we will show and discuss below, different luminal pore-loop conformations correspond with vastly different functional consequences. Two glycosylation sites in the PMD (N138 and N172) were clearly glycosylated in the ML-SA1-bound structure (Fig. 1a).
Figure 6. Low-pH-induced Conformational Changes in the PMD and TMD.
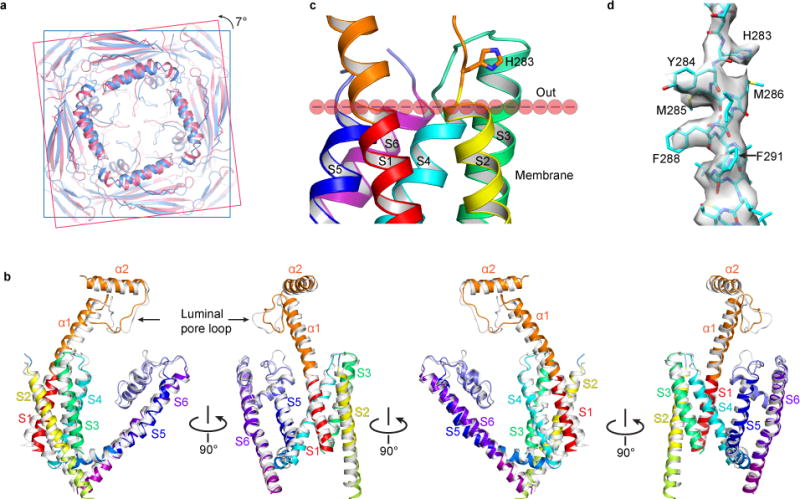
(a) Superposition of the PMD in the pH 4.8 (creamy) and 7.4 (sky blue) apo structures, viewed from the luminal/extracellular side of the membrane. The former shows a 7-degree counterclockwise rotation relative to the latter. (b) Superposition of the TMD and selected key regions in the PMD in the pH 4.8 (gray) and 7.4 (color) apo structures, viewed parallel to the membrane in four different orientations. (c) Location of H283 relative to the membrane. The boundary of the outer leaflet of the membrane is defined by the ends of S2, S4, S5 and S6. (d) Cryo-EM density map and atomic model of the regions near H283.
Two modes of inhibition by low luminal pH
Previous studies show that TRPML3 channels are inhibited by low luminal pH and that a histidine (H283) in the PMD is critical for this inhibition48. We reproduced these results but also found that low luminal pH actually produced two different modes of inhibition. In whole-cell recordings of WT TRPML3 channels, changing the bath solution from a 160 mM Na+ pH 7.4 solution to a 160 mM Na+ pH 4.6 solution fully inhibited the currents (Fig. 5a). This inhibition was largely irreversible as long as the channels were continuously bathed in the high Na+ neutral pH solution. However, the currents could recover partially (Fig. 5a) or fully (data not shown) after the channels were bathed in a Na+-free pH 7.4 solution. Surprisingly, the currents mediated by the H283A mutant channel were still inhibited by low pH (Fig. 5b). However, this inhibition was largely reversible even though the channels were continuously bathed in the high Na+ neutral pH solution (Fig. 5b). Thus, in the presence of a high luminal concentration of Na+, as is the case in lysosomes50, low luminal pH inhibits TRPML3 channels in a reversible mode (mode 1) and a long-lasting mode (mode 2). Only the long-lasting mode was abolished by the H283A mutation, indicating that additional amino acids in TRPML3 can sense luminal pH.
Figure 5. Low-pH-induced Current Inhibition and Conformational Change in the Luminal Pore-loop.
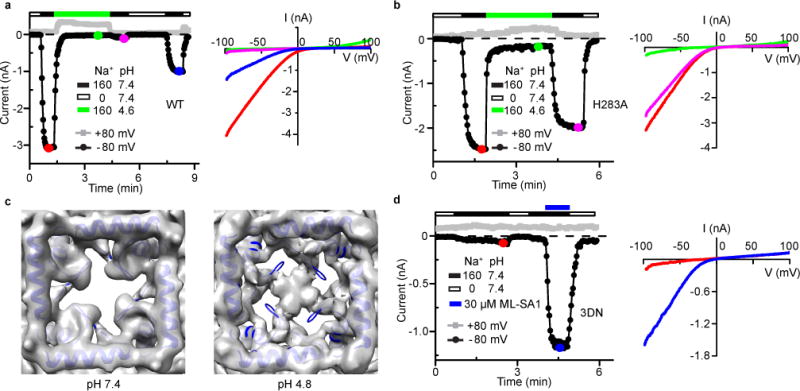
(a) Time course (left) and I-V relationships (right) of WT channel currents in response to low pH. Similar results were obtained in 6 recordings. (b) Time course (left) and I-V relationships (right) of H283A channel currents in response to low pH. Similar results were obtained in 10 recordings. (c) Comparison of the cryo-EM density map of the luminal pore-loop in the pH 7.4 (left) and 4.8 (right) apo structures, viewed from the luminal/extracellular side of the membrane. The modeled pH 7.4 apo structure is superimposed. (d) Time course (left) and I-V relationships (right) of 3DN channel currents in response to ML-SA1. Similar results were obtained in 4 recordings.
Low pH structure reveals a dynamic luminal pore-loop
To elucidate the structural basis of low pH inhibition, we obtained a cryo-EM structure of apo TRPML3 at pH 4.8 at 4.65 Å resolution. The low-pH apo structure is markedly different from the pH 7.4 apo structure in both the PMD and TMD (Fig. 5c and 6a,b). Strikingly, the luminal pore-loop reorients and forms intersubunit bridging interactions that completely occlude the central luminal pore; meanwhile, four new side luminal pores are formed, which are large enough to allow hydrated cations to go through (Fig. 5c).
To investigate whether luminal pore-loop bridging plays a role in low pH inhibition of TRPML3, we simultaneously mutated to asparagine three aspartic residues (D108, D111 and D112) located in the luminal pore-loop. This mutant channel, named TRPML3-3DN, did not produce significant basal currents (Fig. 5d). Thus, we were unable to examine whether the 3DN mutation affects low pH inhibition. However, TRPML3-3DN was strongly activated by ML-SA1, indicating that it folded properly and was expressed robustly on the plasma membrane (Fig. 5d). These results indicate that a proper conformation of the luminal pore-loop is critical for TRPML3 activity. We speculate that the 3DN mutation causes a conformational change in the luminal pore-loop and this conformational change transmits through α1 to S1 and stabilizes the closed state.
Low pH induces global conformational changes
In addition to the dramatic conformational change in the luminal pore-loop, comparison of the pH 4.8 and pH 7.4 apo structures shows that the PMD undergoes a 7 degree counterclockwise rotation while the VSLD undergoes a 3 to 6 degree clockwise rotation, when viewed top down from the luminal side (Fig. 6a and Supplementary Video 2). Further comparison of selected key regions of the PMD and TMD (including S1–S6, α1, α2 and the luminal pore-loop) reveals significant conformational changes in many regions (Fig. 6b and Supplementary Video 2): (1) The luminal pore-loop extends into the center, as shown in Figure 5c. (2) α1 bends downward and towards the center. (3) S1, especially its intracellular end, bends outward. (4) S2 rotates upward and bends outward. (5) S3 also bends outward. (6) The S4–S5 linker moves upward. (7) S5 and S6 move inward toward the pore. These concerted movements presumably make an energetically more stable closed state.
It should be noted while the pH 7.4 apo structure was obtained from TRPML3 in amphipol, the pH 4.8 structure was obtained from TRPML3 in the detergent LMNG. It is possible that some of the differences in the two structures are due to this different condition. It is also possible that some of the conformational changes caused by the low pH are not triggered by the protonation of H283 and the luminal pore-loop aspartates since the entire TRPML3 channel was bathed in the pH 4.8 solution during protein sample preparation for structure determination.
In contrast to the large conformational change of the luminal pore-loop, the structural change of the luminal end of S2 is relative small (Fig. 6b). H283 is located immediately above the luminal end of S2 (Fig. 6c) and does not interact with other amino acids in the pH 7.4 apo structure (Fig. 6c). It does not appear to interact with any other amino acids in the pH 4.8 structure either. These observations contract functional data showing the importance of H283 in low pH inhibition of TRPML348 (Fig. 5a,b). The reason for this discrepancy probably lies in the condition in which the low pH structure was obtained –the cryo-EM protein sample was not in a lipid bilayer. A possibility is that in native cells protonation of H283 enables it to interact with certain lipids in the outer leaflet of the membrane and these interactions change the conformation of S2. This then causes conformational changes in the VSLD and pore domain and produces a long-lasting inhibited state. Validation of this hypothesis requires further functional studies and determination of TRPML3 structures in the presence of lipids in nanodisccs57.
DISCUSSION
Our study elucidates the structural basis of TRPML3 assembly, ion permeation, agonist activation and low-pH inhibition. It is interesting that TRPML3 and TRPML1 channels are both regulated by luminal pH but with the opposite effects. Low pH inhibits TRPML3 and this inhibition appears to involve large, global conformational changes in the PMD and TMD (Fig. 6). On the other hand, low pH potentiates TRPML1. Our previous work on an isolated TRPML1 PMD shows that changing pH does not change the structure of the PMD and that the conformation of the luminal pore-loop is identical at pH 7.4, 6.0 and 4.69. We proposed that H+ potentiates TRPML1 currents by protonating the luminal pore-loop aspartates and attenuating block by Ca2+. According to this proposal, the effect of low pH in TRPML1 is local and does not involve conformational changes in the PMD or TMD. Verification of this view awaits determination of full length TRPML1 structures at different pH values.
Our structural and electrophysiological data suggest that low luminal pH inhibits TRPML3 through two different and independent mechanisms. In mode 1 (Fig. 7a), H+ ions protonate the three aspartic residues in the luminal pore-loop, thereby causing a conformational change that propagates through α1 to S1 and subsequently to S2, S3, S4–S5 linker, S5 and S6 (Fig. 5c and 6a,b, and Supplementary Video 2). These conformational changes produce an inhibited state that is readily reversible when the luminal pH becomes neutral. Although we have not functionally confirmed that it is the protonation of the luminal pore-loop aspartates that produce mode 1 inhibition, the reversible inhibition by low pH of the H283A channel (Fig. 5b) indicates that there is indeed another pH sensor in addition to H283. Two other histidines (H252 and H273) were reported to play a role in low pH inhibition of TRPML348, but functional studies suggest that their role was mainly to affect the protonation of H28348. Moreover, both the open and closed state structures show that they are either buried in the protein interior (H252) or exposed to the surface (H273), suggesting that they may sense pH changes in the lumen.
Figure 7. Model of Two Modes of Low-pH Inhibition of TRPML3.
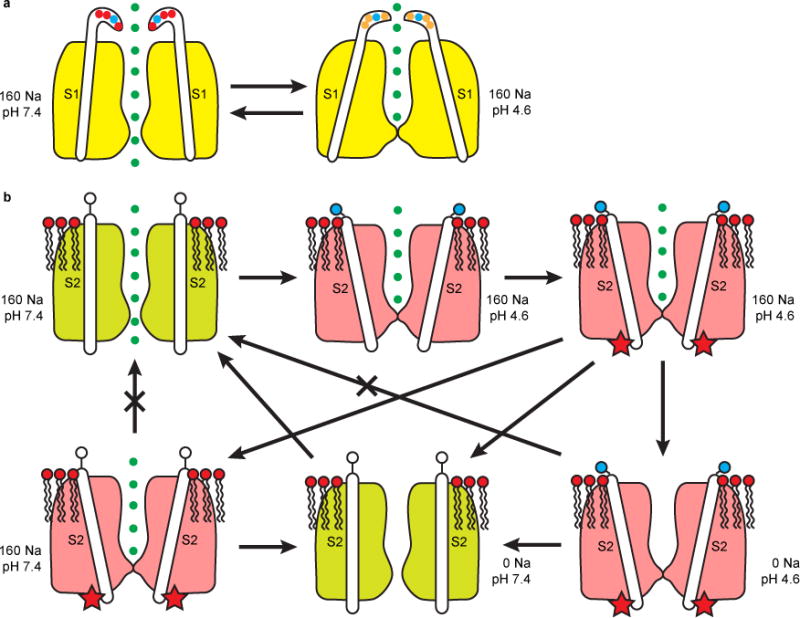
(a) Mode 1, reversible inhibition induced by protonation of the luminal pore-loop aspartates and transmitted through S1. (b) Mode 2, long-lasting inhibition induced by protonation of H283 and transmitted through S2. Protonated H283 (filled blue circle) interacts with negatively charged head groups of lipids in the outer leaflet of the membrane, causing a conformational change of S2 and a modification of the channel (indicated by the red star). This inhibition can be reversed only by bathing the channel in a Na+-free, neutral pH solution. The existence and properties of some of the proposed transition states need experimental verification.
In mode 2 (Fig. 7b), H+ ions protonate H283. The protonated, positively charged H283 side chain then interacts with the negatively charged head groups of certain outer leaflet lipids, thereby causing a conformational change in S2 and consequently in the VSLD and pore domain. Importantly, S2 movement also induces a conformational change in the gating knob. These conformational changes cause the channel to undergo an undefined modification and be in a long-lasting inhibited state that can only recover after the channels are pre-bathed in a Na+-free neutral pH luminal solution. The modification might be phosphorylation/dephosphorylation, sumoylation, or binding to a protein partner. How incubation in a Na+-free neutral pH luminal solution enables the channel to recover from the long-lasting inhibited state remains to be elucidated.
Structural studies on TRPML3 may aid the development of new treatment strategies for ML-IV. All ML-IV-causing mutations are loss-of-function mutations, and most of them are splicing, deletion or frame-shift mutations that result in a total loss of functional TRPML1 proteins in cells4,29–34. One potential strategy for ML-IV treatment is gene therapy, as is for several other LSDs58. However, application of gene therapy in ML-IV patients will likely be an arduous process. Another potential strategy is to develop means to functionally substitute TRPML1 with TRPML3 by taking advantage of their endolysosomal colocalization and shared ion permeation properties4–6,52. The principal hypothesis here is that if one is able to selectively and judiciously activate the normally dormant TRPML3 in endosomes and lysosomes, one might be able to restore, at least partially, the functions of the endocytic pathway in ML-IV patients. Structural information of TRPML3 in different states and bound with different activators may be useful in this equally arduous endeavor.
ONLINE METHODS
Molecular biology
The DNA fragment encoding full-length human TRPML3 channel was cloned into a modified pFastBac1 vector. A maltose binding protein (MBP) tag was added before the N terminus of TRPML3, and a linker sequence containing the tobacco etch virus (TEV) protease recognition sequence was inserted between the MBP tag and TRPML3.
For electrophysiology experiments, the full-length TRPML3 gene was cloned into a pIRES2-EGFP vector. All site-specific mutants were subsequently generated in this TRPML3_pIRES2-EGFP construct by PCR-based overlapping extension.
Protein purification
The baculovirus of TRPML3 was generated with Sf9 cells using the standard Bac-to-Bac method (Invitrogen). Tni insect cells grown in suspension in ESF921 medium (Expression Systems) were infected with the TRPML3 virus. Forty-eight hours after infection, cells were harvested by centrifugation at 4 °C, and suspended in a buffer containing 50 mM NaH2PO4-Na2HPO4 pH 8.0, 500 mM NaCl and 5% glycerol (buffer A) in the presence of cOmplete protease inhibitor cocktail (Roche). After cell disruption by sonication, cell debris was removed by centrifugation at 3,200g for 10 min at 4 °C and cell membrane was pelleted from the supernatant by ultracentrifugation at 150,000g for 1 h at 4 °C. Membrane was suspended in buffer A containing protease inhibitor cocktail and homogenized with a glass dounce homogenizer. TRPML3 protein was extracted with 1% Lauryl Maltose Neopentyl Glycol (LMNG, Anatrace) for 1 h at 4 °C. The solubilized membrane was clarified by ultracentrifugation at 150,000g for 30 min and incubated with amylose resin (NEB) for 2 h at 4 °C with gentle agitation. Subsequently, the resin was collected by low speed spin at 800g, transferred into a gravity column, and washed with buffer A containing 0.5 mM LMNG and 0.1 mg/ml soybean lipids (Avanti polar lipids). MBP-tagged TRPML3 protein was eluted from amylose resin with 20 mM maltose in the above wash buffer.
For samples at pH 7.4, the eluted protein was mixed with amphipol A8–35 (Anatrace) at 1:6 (w/w) ratio and incubated overnight at 4 °C with gentle agitation. Detergent was removed by incubation with Bio-Beads SM-2 (Bio-Rad) for 8 h. After removal of Bio-Beads, the protein sample was mixed with TEV protease (100 μg/mg TRPML3) and incubated at 4 °C overnight to cleave the MBP tag. TRPML3 protein was concentrated and further purified on a Superose 6 (GE Healthcare) column in a buffer containing 20 mM HEPES-NaOH (pH7.4) and 150 mM NaCl. Fractions containing the TRPML3 protein were examined by negative staining EM, and then pooled and concentrated. ML-SA1 was added to for the complex before cryo sample was prepared (see below).
For sample at pH 4.8 sample, the protein eluted from amylose resin in detergent solution was mixed with TEV protease (100 μg/mg TRPML3) and incubated overnight at 4 °C. Then the protein was concentrated and further purified on a superpose 6 column in a buffer containing 20 mM HEPES-NaOH (pH7.4), 150 mM NaCl, and 0.5 mM LMNG. Fractions containing the TRPML3 protein were pooled and concentrated, and the pH was changed to 4.8 before cryo sample preparation (see below).
Proteins were concentrated by ultrafiltration using the Amicon Centrifugal Filter Units (EMD Millipore), and their concentrations were measured by using a Bradford Protein Assay kit (Bio-Rad).
Cryo-EM sample preparation and data acquisition
Cryo-EM samples of TRPML3 in three different states were prepared.
For the apo TRPML3 sample in pH 7.4 solution, a drop of 4 μl sample at a concentration of 1.9 mg/ml was loaded onto glow-discharged Quantifoil holey carbon grid (R1.2/1.3, 300 mesh, ELECTRON MICROSCOPY SCIENCES). After waiting for 3s, the grid was blotted for 4s under 100% humidity at 8℃ using FEI Vitrobot (double-sided, blot force 1) and immediately plunged into liquid ethane cooled by liquid-nitrogen. Cryo sample of the TRPML3/ML-SA1 complex was prepared the same way as the apo sample, except that 100 mM ML-SA1 in DMSO was added to protein sample to a final concentration of 100 μM before freezing. For TRPML3 sample at pH 4.8, protein in 20 mM HEPES-NaOH (pH7.4), 150 mM NaCl, and 0.5 mM LMNG was concentrated to 2 mg/ml. 1 M Na acetate pH 4.6 buffer was added to the protein solution to a final concentration of 100 mM. The pH of the mixed HEPES and acetate buffer was verified to be 4.8.
The samples of TRPML3/MLSA1 complex and TRPML3 at pH 4.8 were observed using a Titan Krios microscope (FEI) operated at 300 kV, equipped with a K2 Summit direct electron detector (Gatan) working in super-resolution counting mode with a pixel size of 0.669 Å. The detector was placed at the end of a GIF Quantum energy filter (Gatan), operated in zero-energy-loss mode with a slit width of 20 eV. The Etas (developed by Bo Shen at Xueming Li lab) was used for data collection under a defocus ranged from −1.0 μm to −2.5 μm. Each micrograph was dose-fractionated to 32 frames with 0.25s exposure time in each frame. The dose rate was 8.2 counts per physical pixel per second, resulting in a total dose of ~ 50 e−/Å2. The data acquisition condition for the TRPML3 protein at pH 7.4 was mostly the same as described above except that the Titan Krios used was not equipped with GIF Quantum energy filter, the pixel size was 0.66 Å and the defocus ranged from −1.5 μm to −3.0 μm.
Image processing
The cryo-EM super-resolution micrographs were 2×2 binned. Motion correction was performed using the MotionCorr2 program59 to generate integrated micrographs for further processing. CTFFIND3 program was used to determine defocus values60. As for the data of apo TRPML3 at pH 7.4, the full data set of 2,721 micrographs was divided into two subsets with 726 and 1,995 micrographs, respectively, each subset was subjected to two rounds of two-dimensional classification and then the selected particles were classified into four classes in the three-dimensional classification procedure. The map of CNG channel61 low-pass filtered to 60 Å was used as the initial model. Particle heterogeneity was observed among the resulting 3D classes, which presented two typical conformations: a near-perfect tetramer in the first subset and a defective tetramer with two missing/flexible domain in the second subset. So two rounds of two-references 3D classification was performed as described previously61. Finally, 43,542 particles corresponding to the near-perfect tetramer were gathered and subjected to final 3D refinement with C4 symmetry imposed, which resulted in a final map at 4.06 Å resolution. As for TRPML3 at pH 4.8 buffer and TRPML3/MLSA1 complex, the image processing procedure was almost the same, after two rounds of 2D classification and one round 3D classification, the best one or two 3D classes were selected and gathered for final refinement. All the 2D classification, 3D classification and refinement procedure were performed by RELION 1.462. The final refinement of TRPML3/ML-SA1 complex was done by using THUNDER, a software developed by Mingxu Hu, Hongkun Yu and colleagues in Xueming Li Lab and is in preparation for publication. The 3D reconstruction obtained by THUNDER had a 3.62 Å resolution, higher than the 3.82 Å resolution obtained by RELION 1.4. As for the sample at pH 4.8, a 4.65 Å map was reconstructed. All resolutions were estimated by the gold-standard Fourier shell correlation (FSC) = 0.143 criterion. ResMap63 was used to calculate the local resolution map. The normalized difference map between the ML-SA1-bound pH 7.4 and apo pH 7.4 structures was calculated by using THUNDER and was filtered to 6.0 Å.
Code Availability
The THUNDER software is in preparation for publication and is available from X.L. upon request.
Model building and refinement and validation
Atomic models were built in Coot64. The 3.62 Å density map of the TRPML3/ML-SA1 complex was used for de novo manual model building of the transmembrane. Densities of the side chains for bulky residues (Trp, Tyr and Phe) and predicted transmembrane ranges were used to guide amino acid assignment. The homologous crystal structure of the I-II linker of human TRPML1 (PDB ID: 5TJA) was docked into the density map using USCF chimera65, mutated into human TRPML3 sequence and rebuilt in Coot. Atomic model for membrane-proximal N- and C- termini and cytoplasmic II-III linker was built, although the densities of the side chains of some amino acids were missing. Atomic model was built for one subunit, and then the tetramer model of the channel was generated by 4-fold rotational symmetry operation. This model was then used as an initial model and was rebuilt into the density map of apo TRPML3. Extra densities in the map of the TRPML3/ML-SA1 complex was found where an atomic model of the ligand ML-SA1 (coordinates downloaded from https://pubchem.ncbi.nlm.nih.gov) was tentatively put in. For TRPML3 at pH 4.8, individual transmembrane fragments and the PMD domain from the apo model were fitted into the density in UCSF chimera. The model was then manually rebuilt in COOT. Residues missing side chain densities were mutated to alanine. All three models were refined in reciprocal-space using Phenix66. Coordinates and individual B-factors were refined using maximum likelihood refinement, with secondary structure restraints and strict NCS constraints.
Overfitting of the atomic model was checked by using methods described previously67. The coordinates were randomly displaced up to 0.2 A, and the new model was then refined against one of the two half maps generated. FSC values were calculated between the resulting model and the two half maps, as well as the averaged map of two half maps.
The quality of the models was evaluated by MolProbity68. PyMOL and UCSF Chimera were used to make figures. The pore radius was calculated with HOLE software69.
Electrophysiology
Human embryonic kidney (HEK) 293T cells (American Type Culture Collection (ATCC), not tested for mycoplasma) were grown in DMEM (Gibco) plus 10% Fetal Bovine serum (VWR) and penicillin (100 U/ml)/streptomycin (0.1 mg/ml) (SIGMA). HEK 293T cells were transiently transfected with WT or mutant TRPML3 using LipoD293 (SignaGen Laboratories) and used in 48 h.
All experiments were performed at room temperature (~22 °C). Pipettes were fabricated from borosilicate glass (Corning PYREX) using a micropipette puller (PC-10, NARISHIGE), and were fire-polished to resistances of 2~3 MΩ for whole-cell recording. Whole-cell currents were elicited by 400-ms voltage ramps from −100 to +100 mV at a frequency of 0.5 Hz with a holding potential of 0 mV. Currents were amplified by Axopatch 200B and digitized by Digidata 1322A (Molecular Devices). Currents were low-pass filtered at 1 kHz and sampled at 1 kHz. pCLAMP 8.2 software (Molecular Devices) was used for data acquisition and analysis. The standard pipette solution contained (in mM) 120 Cesium methanesulfonate, 10 BAPTA, 2 MgCl2, 20 HEPES, pH 7.2 adjusted with CsOH. The pH 7.4 standard bath solution contained (in mM) 160 NaCl, 5 KCl, 10 D-glucose, 20 HEPES, pH adjusted with NaOH. The pH 4.6 standard bath solution contained (in mM) 160 NaCl, 5 KCl, 10 D-glucose, 10 HEPES, 10 MES, pH adjusted with NaOH. The pH7.4 Sodium-free solution contained (in mM) 165 NMDG, 10 D-glucose, 10 HEPES, 10 MES, pH adjusted with HCl.
Data are presented as mean±s.e.m. Statistical significance was evaluated by using two-tailed Student’s t-test, with **P < 0.01.
Supplementary Material
Acknowledgments
This work was supported by grants to J.Y. from the National Basic Research Program of China (2014CB910301), the National Institutes of Health (R01GM085234), the National Natural Science Foundation of China (31370821), the Top Talents Program of Yunnan Province (2011HA012), and the High-level Overseas Talents of Yunnan Province; to X.L. from the China Youth 1000-Talent Program of the State Council of China, Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, and the National Natural Science Foundation of China (31570730).
Footnotes
AUTHOR CONTRIBUTIONS
M.L and J.Y. conceived and initiated the project. X.Z., M.L., D.S., Q.J., H.L., X.L. and J. Y. designed the experiments, analyzed the results, and wrote the manuscript. M.L. performed all molecular biology and biochemical experiments and built the atomic models. X.Z. and X.L. performed all cryo-EM experiments, including data acquisition and processing. D.S., Q.J. and H. L. performed electrophysiology experiments. All authors contributed to manuscript discussion, preparation and editing.
COMPETINGFINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession codes. 3D cryo-EM density maps of the apo TRPML3 at pH 7.4, the ML-SA1-bound TRPML3 at pH 7.4, and the apo TRPML3 at pH 4.8 have been deposited in the Electron Microscopy Data Bank under the accession number EMD-7018, EMD-7019 and EMD-7020, respectively. The coordinates of the atomic model of the apo TRPML3 at pH 7.4, the ML-SA1-bound TRPML3 at pH 7.4, and the apo TRPML3 at pH 4.8, have been deposited in the Protein Data Bank under the accession number 6AYE, 6AYF and 6AYG, respectively.
References
- 1.Di Fiore PP, von Zastrow M. Endocytosis, signaling, and beyond. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu H, Martinoia E, Szabo I. Organellar channels and transporters. Cell Calcium. 2015;58:1–10. doi: 10.1016/j.ceca.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimm C, Butz E, Chen CC, Wahl-Schott C, Biel M. From mucolipidosis type IV to Ebola: TRPML and two-pore channels at the crossroads of endo-lysosomal trafficking and disease. Cell Calcium. 2017 doi: 10.1016/j.ceca.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Venkatachalam K, Wong CO, Zhu MX. The role of TRPMLs in endolysosomal trafficking and function. Cell Calcium. 2015;58:48–56. doi: 10.1016/j.ceca.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu H, Ren D. Lysosomal physiology. Annu Rev Physiol. 2015;77:57–80. doi: 10.1146/annurev-physiol-021014-071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimm C, Barthmes M, Wahl-Schott C. Trpml3. Handb Exp Pharmacol. 2014;222:659–674. doi: 10.1007/978-3-642-54215-2_26. [DOI] [PubMed] [Google Scholar]
- 7.Noben-Trauth K. The TRPML3 channel: from gene to function. Adv Exp Med Biol. 2011;704:229–237. doi: 10.1007/978-94-007-0265-3_13. [DOI] [PubMed] [Google Scholar]
- 8.Grimm C, Hassan S, Wahl-Schott C, Biel M. Role of TRPML and two-pore channels in endolysosomal cation homeostasis. J Pharmacol Exp Ther. 2012;342:236–244. doi: 10.1124/jpet.112.192880. [DOI] [PubMed] [Google Scholar]
- 9.Li M, et al. Structural basis of dual Ca2+/pH regulation of the endolysosomal TRPML1 channel. Nat Struct Mol Biol. 2017;24:205–213. doi: 10.1038/nsmb.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grieben M, et al. Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2) Nat Struct Mol Biol. 2017;24:114–122. doi: 10.1038/nsmb.3343. [DOI] [PubMed] [Google Scholar]
- 11.Shen PS, et al. The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs. Cell. 2016;167:763–773e711. doi: 10.1016/j.cell.2016.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkes M, et al. Molecular insights into lipid-assisted Ca2+ regulation of the TRP channel Polycystin-2. Nat Struct Mol Biol. 2017;24:123–130. doi: 10.1038/nsmb.3357. [DOI] [PubMed] [Google Scholar]
- 13.Kiselyov K, et al. TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J Biol Chem. 2005;280:43218–43223. doi: 10.1074/jbc.M508210200. [DOI] [PubMed] [Google Scholar]
- 14.Dong XP, et al. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455:992–996. doi: 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong XP, et al. Activating mutations of the TRPML1 channel revealed by proline-scanning mutagenesis. J Biol Chem. 2009;284:32040–32052. doi: 10.1074/jbc.M109.037184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong XP, et al. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat Commun. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong XP, Wang X, Xu H. TRP channels of intracellular membranes. Journal of neurochemistry. 2010;113:313–328. doi: 10.1111/j.1471-4159.2010.06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat Cell Biol. 2016;18:404–417. doi: 10.1038/ncb3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, et al. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc Natl Acad Sci U S A. 2015;112:E1373–1381. doi: 10.1073/pnas.1419669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng X, et al. The intracellular Ca(2)(+) channel MCOLN1 is required for sarcolemma repair to prevent muscular dystrophy. Nat Med. 2014;20:1187–1192. doi: 10.1038/nm.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miedel MT, et al. Membrane traffic and turnover in TRP-ML1-deficient cells: a revised model for mucolipidosis type IV pathogenesis. The Journal of experimental medicine. 2008;205:1477–1490. doi: 10.1084/jem.20072194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samie M, et al. A TRP Channel in the Lysosome Regulates Large Particle Phagocytosis via Focal Exocytosis. Dev Cell. 2013;26:511–524. doi: 10.1016/j.devcel.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, et al. MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat Commun. 2016;7:12109. doi: 10.1038/ncomms12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaPlante JM, et al. Lysosomal exocytosis is impaired in mucolipidosis type IV. Molecular genetics and metabolism. 2006;89:339–348. doi: 10.1016/j.ymgme.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Venkatachalam K, et al. Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell. 2008;135:838–851. doi: 10.1016/j.cell.2008.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vergarajauregui S, Connelly PS, Daniels MP, Puertollano R. Autophagic dysfunction in mucolipidosis type IV patients. Human molecular genetics. 2008;17:2723–2737. doi: 10.1093/hmg/ddn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrity AG, et al. The endoplasmic reticulum, not the pH gradient, drives calcium refilling of lysosomes. Elife. 2016;5 doi: 10.7554/eLife.15887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahoo N, et al. Gastric Acid Secretion from Parietal Cells Is Mediated by a Ca2+ Efflux Channel in the Tubulovesicle. Dev Cell. 2017;41:262–273e266. doi: 10.1016/j.devcel.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun M, et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Human molecular genetics. 2000;9:2471–2478. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- 30.Bargal R, et al. Identification of the gene causing mucolipidosis type IV. Nature genetics. 2000;26:118–123. doi: 10.1038/79095. [DOI] [PubMed] [Google Scholar]
- 31.Bassi MT, et al. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. American journal of human genetics. 2000;67:1110–1120. doi: 10.1016/s0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weitz R, Kohn G. Clinical spectrum of mucolipidosis type IV. Pediatrics. 1988;81:602–603. [PubMed] [Google Scholar]
- 33.Bach G. Mucolipidosis type IV. Molecular genetics and metabolism. 2001;73:197–203. doi: 10.1006/mgme.2001.3195. [DOI] [PubMed] [Google Scholar]
- 34.Wakabayashi K, Gustafson AM, Sidransky E, Goldin E. Mucolipidosis type IV: an update. Molecular genetics and metabolism. 2011;104:206–213. doi: 10.1016/j.ymgme.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Palma F, et al. Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc Natl Acad Sci U S A. 2002;99:14994–14999. doi: 10.1073/pnas.222425399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grimm C, et al. A helix-breaking mutation in TRPML3 leads to constitutive activity underlying deafness in the varitint-waddler mouse. Proc Natl Acad Sci U S A. 2007;104:19583–19588. doi: 10.1073/pnas.0709846104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HJ, et al. Gain-of-function mutation in TRPML3 causes the mouse Varitint-Waddler phenotype. J Biol Chem. 2007;282:36138–36142. doi: 10.1074/jbc.C700190200. [DOI] [PubMed] [Google Scholar]
- 38.Nagata K, et al. The varitint-waddler (Va) deafness mutation in TRPML3 generates constitutive, inward rectifying currents and causes cell degeneration. Proc Natl Acad Sci U S A. 2008;105:353–358. doi: 10.1073/pnas.0707963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu H, Delling M, Li L, Dong X, Clapham DE. Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proc Natl Acad Sci U S A. 2007;104:18321–18326. doi: 10.1073/pnas.0709096104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stauber T, Jentsch TJ. Chloride in vesicular trafficking and function. Annu Rev Physiol. 2013;75:453–477. doi: 10.1146/annurev-physiol-030212-183702. [DOI] [PubMed] [Google Scholar]
- 41.Kiselyov KK, Ahuja M, Rybalchenko V, Patel S, Muallem S. The intracellular Ca(2)(+) channels of membrane traffic. Channels (Austin) 2012;6:344–351. doi: 10.4161/chan.21723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel S, Cai X. Evolution of acidic Ca(2)(+) stores and their resident Ca(2)(+)-permeable channels. Cell Calcium. 2015;57:222–230. doi: 10.1016/j.ceca.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 43.Appelqvist H, Waster P, Kagedal K, Ollinger K. The lysosome: from waste bag to potential therapeutic target. J Mol Cell Biol. 2013;5:214–226. doi: 10.1093/jmcb/mjt022. [DOI] [PubMed] [Google Scholar]
- 44.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Li X, Xu H. Phosphoinositide isoforms determine compartment-specific ion channel activity. Proc Natl Acad Sci U S A. 2012;109:11384–11389. doi: 10.1073/pnas.1202194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raychowdhury MK, et al. Molecular pathophysiology of mucolipidosis type IV: pH dysregulation of the mucolipin-1 cation channel. Human molecular genetics. 2004;13:617–627. doi: 10.1093/hmg/ddh067. [DOI] [PubMed] [Google Scholar]
- 47.Cantiello HF, et al. Cation channel activity of mucolipin-1: the effect of calcium. Pflugers Arch. 2005;451:304–312. doi: 10.1007/s00424-005-1448-9. [DOI] [PubMed] [Google Scholar]
- 48.Kim HJ, et al. A novel mode of TRPML3 regulation by extracytosolic pH absent in the varitint-waddler phenotype. EMBO J. 2008;27:1197–1205. doi: 10.1038/emboj.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimm C, Jors S, Guo Z, Obukhov AG, Heller S. Constitutive activity of TRPML2 and TRPML3 channels versus activation by low extracellular sodium and small molecules. J Biol Chem. 2012;287:22701–22708. doi: 10.1074/jbc.M112.368876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, et al. TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell. 2012;151:372–383. doi: 10.1016/j.cell.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miao Y, Li G, Zhang X, Xu H, Abraham SN. A TRP Channel Senses Lysosome Neutralization by Pathogens to Trigger Their Expulsion. Cell. 2015;161:1306–1319. doi: 10.1016/j.cell.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimm C, et al. Small molecule activators of TRPML3. Chem Biol. 2010;17:135–148. doi: 10.1016/j.chembiol.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen D, et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat Commun. 2012;3:731. doi: 10.1038/ncomms1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng X, Xiong J, Lu Y, Xia X, Zhu MX. Differential mechanisms of action of the mucolipin synthetic agonist, ML-SA1, on insect TRPML and mammalian TRPML1. Cell Calcium. 2014;56:446–456. doi: 10.1016/j.ceca.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang S, Li N, Zeng W, Gao N, Yang M. Cryo-EM structures of the mammalian endo-lysosomal TRPML1 channel elucidate the combined regulation mechanism. Protein Cell. 2017 doi: 10.1007/s13238-017-0476-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim HJ, Yamaguchi S, Li Q, So I, Muallem S. Properties of the TRPML3 channel pore and its stable expansion by the Varitint-Waddler-causing mutation. J Biol Chem. 2010;285:16513–16520. doi: 10.1074/jbc.M109.078204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biffi A. Gene therapy for lysosomal storage disorders: a good start. Human molecular genetics. 2016;25:R65–75. doi: 10.1093/hmg/ddv457. [DOI] [PubMed] [Google Scholar]
- 59.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 61.Li M, et al. Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature. 2017;542:60–65. doi: 10.1038/nature20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bharat TA, Russo CJ, Lowe J, Passmore LA, Scheres SH. Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging. Structure. 2015;23:1743–1753. doi: 10.1016/j.str.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14376:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


