Abstract
The mechanism by which 5-formylcytosine (fC) is recognised by enzymes involved in epigenetic modification and reading of DNA is not known, and recently an unusual DNA structure (F-DNA) was proposed as the basis for enzyme recognition of clusters of fC. We used NMR and X-ray crystallography to compare several modified DNA duplexes with the unmodified analogues and show that in the crystal state they all belong to the A-family, but in solution they are all members of the B-family. Contrary to the previous study, we find that 5-formylcytosine does not significantly affect the structure of DNA, though there are modest local differences at the modification sites. Hence, global conformation changes are unlikely to account for the recognition of this modified base, and our structural data favour a mechanism that operates at base-pair resolution for the recognition of 5-formylcytosine by epigenome-modifying enzymes.
Introduction
Cytosine bases at CpG sites in genomic DNA are methylated by DNA methyl transferases (DNMTs)1 to produce 5-methylcytosine (mC), resulting in epigenetic gene silencing.2 The reverse process, demethylation of mC, involves sequential oxidation to 5-hydroxymethylcytosine (hmC),3,4 5-formylcytosine (fC)5,6 and 5-carboxylcytosine (caC), by the ten-eleven translocation (TET) dioxygenase family of enzymes.3–7 The oxidized pyrimidines fC and caC may then be excised from the deoxyribose sugar by thymine DNA glycosylase (TDG)8,9 and replaced by unmodified cytosine via the base excision repair (BER) pathway.10
Remarkably, the biological roles of oxidized pyrimidines appear to extend beyond those of DNA demethylation intermediates. Recent literature suggests that oxidized derivatives of mC,8,11–14 and also thymidine,15 might function as distinct epigenetic signals. A particularly interesting case is that of fC, which, despite being a substrate of TDG, and containing an aldehyde functional group, can be a stable, possibly semi-permanent modification in the genome.12,16 Furthermore, the genomic profile of fC is distinct from mC and its other oxidized derivatives;12 it has more interacting proteins than either mC or hmC, highlighting its importance in the human genome.11,13
The mechanism by which fC is recognized by TDG and TET remains unresolved. Recently, it was reported that when fC is present in CpG repeats it causes perturbation to the DNA double helix, resulting in a unique helical conformation featuring 13 bases per turn as evidenced by an X-ray structure.17 This was the first time that epigenetically modified cytosine bases were shown to significantly alter DNA structure; other studies had not indicated major deviations from canonical B-DNA.18–21 This new conformation, named ‘F-DNA’, has been proposed as the basis for epigenetic recognition of fCpG clusters in DNA. Clearly, the validity of such a theory relies on the assumption that the F-DNA structure is unique to DNA containing fC, and is not formed by native DNA. Here we show that in the crystalline state the equivalent unmodified duplex and three sequence variants have an almost identical structure to DNA containing multiple 5-formylcytosine bases (fC-DNA), and that the structure is a member of the A-family. We also compare fC-DNA and the native analogue by NMR and show that they are both B-family helices in solution, despite CD spectra of the fC-containing duplexes displaying unusual characteristics under equivalent conditions. We suggest that this is a consequence of local electronic transition dipole moment changes caused by fC rather than gross structural perturbation to the DNA helix. This leads us to propose an alternative basis for recognition of fC-DNA that is valid for both isolated and clustered fC bases.
Results
Single crystal X-ray diffraction
The structures of six self-complementary DNA duplexes were determined at high resolution (1.4–2.3 Å) and deposited in the PDB under accession codes 5MVU [d(CTAfCGfCGfCGTAG)2], 5MVK [d(CTACGCGCGTAG)2], 5MVL [d(CbUACGCGCGTAG)2], 5MVP [d(CTAGGGCCCTAG)2], 5MVQ [d(CTACGGCCGTAG)2] and 5MVT [d(CTACGTACGTAG)2], where fC is 5-formylcytosine and bU is 5-bromouracil. All contain identical 5′- and 3′-terminal trimer sequences and different internal hexamer sequence motifs, except where bU substitutes for T (Supplementary Tables 1, 2). All six duplexes crystallized isomorphously in the P3221 space group, which is different to that of the previous structure of d(CTAfCGfCGfCGTAG)217 (Table 3).
Table 3.
Data collection and refinement statistics (molecular replacement)
| 5MVUa | 5MVK | 5MVP | 5MVT | 5MVQ | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P3221 | P3221 | P3221 | P3221 | P3221 |
| Cell dimensions | |||||
| a, b, c (Å) | 44.2, 44.2, 61.7 | 43.7, 43.7, 58.9 | 43.2, 43.2, 61.5 | 43.4, 43.3, 57.8 | 43.4, 43.4, 61.7 |
| α, β, γ(°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 38.2-2.3 (2.42-2.30)a | 37.8-1.5 (1.56-1.53) | 37.4-1.6 (1.63-1.61) | 38.4-1.9 (1.94-1.90) | 36.7-1.6 (1.63-1.60) |
| Rmeas | 7.6 (107) | 5.2 (113) | 3.4 (60.7) | 6.0 (102) | 3.7 (60.9) |
| I/σ(I) | 15.4 (2.1) | 19.7 (2.0) | 23.0 (2.3) | 21.0 (2.3) | 18.3 (2.4) |
| CC1/2 | 0.996 (0.566) | 0.998 (0.764) | 0.999 (0.837) | 1.000 (0.730) | 0.999 (0.744) |
| Completeness (%) | 100 (100) | 99.9 (99.8) | 99.7 (94.7) | 99.9 (99.1) | 91.3 (89.3) |
| Redundancy | 8.9 (9.2) | 9.2 (6.9) | 6.3 (5.8) | 9.2 (8.5) | 4.3 (3.8) |
| Refinement | |||||
| Resolution (Å) | 32.5-2.3 | 37.8-1.5 | 37.5-1.6 | 38.4-1.9 | 36.7-1.6 |
| No. reflections | 3,170 | 10,206 | 9,075 | 5,518 | 7,901 |
| Rwork / Rfree | 14.9 / 15.8 | 18.1 / 18.8 | 15.3 / 20.9 | 20.2 / 22.5 | 17.9 / 19.4 |
| No. atoms | |||||
| DNA | 498 | 486 | 486 | 486 | 486 |
| K+ | 0 | 0 | 1 | 0 | 0 |
| Co3+ | 0 | 0 | 0 | 1 | 0 |
| Mg2+ | 0 | 0 | 0 | 0 | 2 |
| Water | 3 | 67 | 75 | 31 | 75 |
| B factors | |||||
| DNA | 64.90 | 32.2 | 33.85 | 36.00 | 28.33 |
| Water | 57.36 | 40.61 | 44.27 | 39.80 | 36.97 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.008 | 0.014 | 0.014 | 0.011 | 0.011 |
| Bond angles (°) | 1.51 | 1.38 | 1.19 | 1.18 | 1.21 |
Each data set was collected from a single crystal.
crystals were twinned. See Supplementary Table 10 for twinning statistics.
Values in parentheses are for highest-resolution shell.
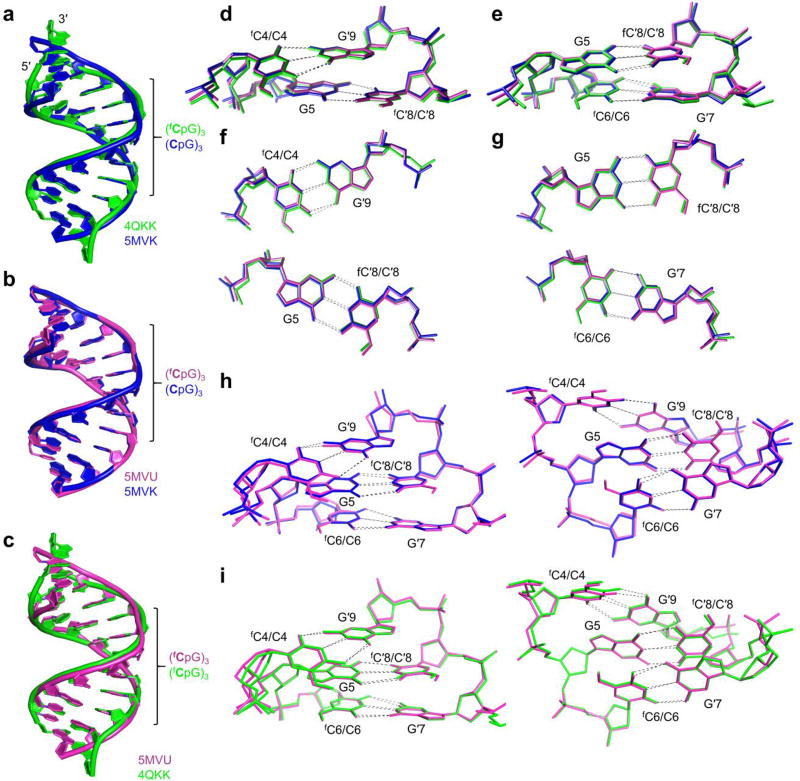
Overlays of the published crystal structure of d(CTAfCGfCGfCGTAG)2 that was found to adopt a unique helical conformation (4QKK),17 in addition to an identical duplex that we have crystallized in an alternative space group (5MVU) and its control, a non-formylated sequence analogue (5MVK) are shown in Fig. 1. All-atom rms deviations of the crystal structures from either 5MVK, ideal A-DNA or ideal B-DNA are summarized in Table 1.22 To reduce the effects of crystal packing artefacts, which are most significant at the duplex ends, rms deviations are also given for the 8-mer core which, for the structures 4QKK and 5MVU, contains the fC residues. These effects were particularly pronounced in 4QKK, whose crystallization salt concentration was an order of magnitude higher than in this study (Figure 2a). The comparison shows that deviation of the fC-containing structures from the unmodified analogue 5MVK is small; for the 8-mer core d(AfCGfCGfCGT)2 (322 atoms) the rms deviation of 5MVU and 4QKK from 5MVK was just 0.51 Å and 1.45 Å, respectively. Moreover, the rms deviation between the two identical fC-containing duplexes (1.21 Å) was more than double that between the unmodified and fC-modified duplexes that crystallized in the same space group, and is comparable to the rmsd between 4QKK and 5MVK. Thus, the crystallization conditions of 4QKK exerted a greater effect on the conformation of the duplex core than did the presence of fC. Importantly, deviation of all structures from ideal A-DNA is also small, with the unmodified duplex exhibiting the highest value (1.44 Å) and 4QKK the lowest (1.08 Å) (Supplementary Fig. 1a–d, Table 1). Consistent with the rmsd comparison, analysis of structural parameters of 4QKK, 5MVU and 5MVK (Table 2, Supplementary Fig. 2, Supplementary Tables 4,5) shows that, apart from anomalous features arising from crystal packing, all structures adopt an A-type conformation. The base pairs and dinucleotide steps in the central (CpG)3 region of d(CTACGCGCGTAG)2 (5MVK) and d(CTAfCGfCGfCGTAG)2 (4QKK & 5MVU) overlap very well (Fig. 1a–i), further emphasizing the similarity between DNA containing fCpG and CpG tracts. Thus, we found no indication that fC significantly alters base-stacking geometry in the crystal state.
Figure 1. Similarities between the crystal structures of a DNA duplex containing 5-formylcytosine and its unmodified counterpart.
A crystallographic comparison of two structures of the fC-duplex d(CTAfCGfCGfCGTAG)2 in different space groups (5MVU and 4QKK), and its non-formylated analogue (5MVK), shows that fC does not significantly affect the structure of the DNA double helix in the crystalline state. (a) Overlaid crystal structures 4QKK d(CTAfCGfCGfCGTAG)217 (green, space group P43212) and its unmodified analogue 5MVK (blue, space group P3221) showing strong overlap between the central regions of the duplexes which contain the respective formylated and non-formylated CpG repeats (rmsd of 1.33 Å, 240 atoms). Significant differences between structures occur at the duplex ends, rather than near the formylated sites, and can be attributed to differences in crystal packing interactions. (b) Overlay of crystal structures 5MVK (blue) and 5MVU (magenta), whose sequence is identical to that of 4QKK. Conformational differences between the fC-containing structure 5MVU and its unmodified counterpart 5MVK, which both crystallized in the space group P3221, are negligible (rmsd of 0.65 Å, 486 atoms). (c) Overlay of structures 5MVU and 4QKK, of an identical fC-containing duplex crystallized under different conditions; the salt content of the crystallization buffer of 4QKK was an order of magnitude higher than that of 5MVU and the other structures determined in this study. (d,e) Stacking of base pairs X4-G′9 with G5-X′8, and G5-X′8 with X6-G′7 where X = fC for 5MVU and 4QKK and C for 5MVK. The respective base pairs are shown individually in (f) and (g); differences in base-stacking geometry between all three structures are small. Overlays of the aforementioned base pairs are shown collectively in (h) for 5MVU and 5MVK and (i) for 4QKK and 5MVU, showing that differences in conformation between the CpG and fCpG steps are small and do not propagate significantly.
Table 1.
Comparison of all-atom rms deviations of the crystal structures 4QKK, 5MVK and 5MVU, and ideal A- and B-DNA of the same sequence.22 To minimize the effect of crystal packing artefacts, which is greatest at the duplex ends, an additional set of rmsd’s was determined in which the two unmodified base pairs at either end of each duplex were excluded. The comparison shows that the all-atom rmsd between the entire formylated duplex and its unmodified analogue, which crystallized in the same space group, was just 0.65 Å, and that for all core structures, deviation from ideal A-DNA is small (<1.44 Å). Notably, the 8 base-pair core of structure 4QKK, containing the entire formylated region flanked at each end by an unmodified base pair, exhibits the lowest deviation from ideal A-DNA (1.08 Å).
| d(CTAXGXGXGTAG)2 Rmsd (Å, 486 atoms) | d(AXGXGXGT)2 Rmsd (Å, 322 atoms) | |||||
|---|---|---|---|---|---|---|
| 4QKK (X=fC) | 5MVU (X=fC) | 5MVK (X=C) | 4QKK (X=fC) | 5MVU (X=fC) | 5MVK (X=C) | |
| Ideal A-DNA | 2.21 | 1.27 | 1.52 | 1.08 | 1.24 | 1.44 |
| Ideal B-DNA | 5.53 | 6.54 | 6.87 | 4.19 | 4.39 | 4.56 |
| 5MVK | 2.65 | 0.65 | -- | 1.45 | 0.51 | -- |
| 5MVU | 2.30 | -- | 0.65 | 1.21 | -- | 0.51 |
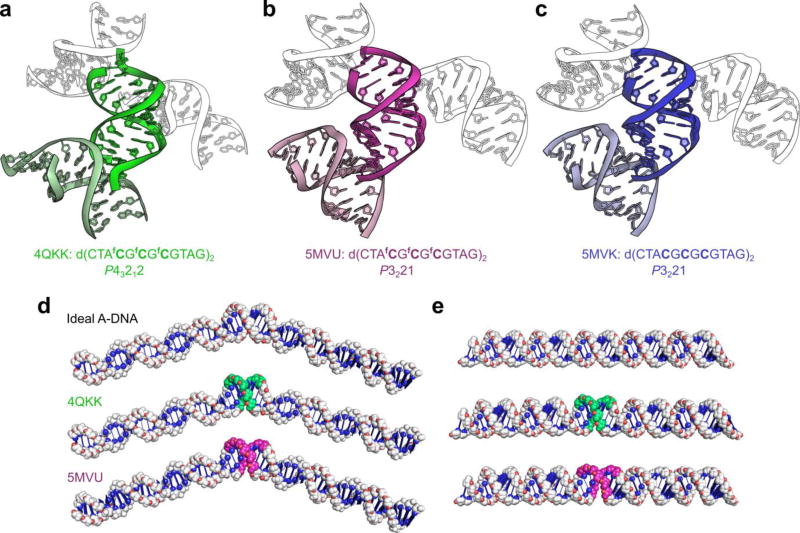
Figure 2. Crystal packing interactions exert a greater effect on DNA conformation than 5-formylcytosine in the crystalline state.
(a–c) Cartoons illustrating the crystal packing of an identical 5-formylcytosine-containing sequence crystallized in different space groups (4QKK & 5MVU), and that of its unmodified analogue 5MVK. (a) The duplex d(CTAfCGfCGfCGTAG)2, when crystallized in the P43212 space group from a buffer containing 1.8 M lithium sulfate,17 interacts strongly with neighboring duplexes within the crystal, resulting in pronounced kinking of the backbone at the duplex ends and consequently deviation from A-type geometry at the terminal nucleotides; however, the duplex’s core, which contains all of the fCpG repeats, adopts the A-form conformation. (b) The same fC-containing duplex, when crystallized in the P3221 space group under milder conditions (<100 mM salt), does not exhibit significant deviation from A-type geometry. (c) The duplex d(CTACGCGCGTAG)2, which also crystallizes in the P3221 space group under milder conditions, is very similar in conformation to its fC-containing counterpart when crystallized isomorphously (rmsd of 0.65 Å, 486 atoms) and forms similar crystal contacts. Note that some symmetry mates have been omitted for clarity. (d) Modelling of B-DNA 60-bp duplexes containing a central 8-bp region of either ideal A-DNA or the region d(AfCGfCGfCGT)2 from the crystal structures 4QKK and 5MVU (fC∙G base pairs shown in green and magenta, respectively). Alterations in the helical trajectory of B-DNA caused by the fC-containing structures closely resemble those of an ideal A-B-DNA junction. (e) Modelling of A-DNA 60-mers containing the 8-bp regions of the aforementioned crystal structures. The fC-containing structures, when flanked by ideal A-DNA, are almost indistinguishable from a continuous stretch of ideal A-DNA. See Supplementary Fig. 4 for an extended comparison.
Table 2.
Comparison of averaged structural parameters commonly used to differentiate between A- and B-DNA. To minimize crystal packing artefacts, values shown are for the 8 base-pair core of each duplex d(AXGXGXGT)2, where X = 5-formylcytosine for 5MVU and 4QKK, and cytosine for 5MVK and the ideal duplexes generated from standard parameters.22 The comparison shows that the structural parameters of all crystal structures correspond to the A-family.
| Helical twist (°) |
Base pairs per turn |
Glycosyl torsion angles (°) |
Sugar puckering | X-displacement (Å) |
Inclination (°) |
|
|---|---|---|---|---|---|---|
| 5MVU | 32.6 | 11.0 | −162 | C3′-endo | −4.74 | 17.1 |
| 4QKK | 33.9 | 10.6 | −161 | C3′-endo | −4.32 | 15.8 |
| 5MVK | 31.9 | 11.3 | −156 | C3′-endo | −5.10 | 19.7 |
| Ideal A-DNA | 32.7 | 11.0 | −157 | C3′-endo | −4.46 | 22.7 |
| Ideal B-DNA | 35.9 | 10.0 | −98 | C2′-endo | 0.50 | 2.8 |
To our knowledge, 5MVK is the only crystal structure of an unmodified, right-handed DNA duplex containing three consecutive CpG steps. To investigate whether the repeating CpG steps, which are commonly found in gene promoters,2 might give rise to significant conformational differences, we crystallized three analogues under similar conditions, in which the internal (CpG)3 motif was changed (5MVP, 5MVT and 5MVQ). It is noteworthy that certain structural parameters, such as roll angle, show an alternation of values, corresponding to the alternating pyrimidine purine sequence (Supplementary Fig. 2).23 However, the overlays and rms deviations of these structures, and the other three determined in this study (Supplementary Fig. 3, Supplementary Table 6), show that conformational differences between the duplexes are small in all cases. The largest is between 5MVQ [d(CTACGGCCGTAG)2] and 5MVT [d(CTACGTACGTAG)2], exhibiting an rmsd of just 1.26 Å (448 atoms). Significantly, the differences between the formylated duplex 5MVU and its unmodified analogue 5MVK were smaller than between two unmodified duplexes.
In summary, all duplexes are typical of the A-family, and the fC-containing structures are very similar to the unmodified control. Where significant differences occur, typically at the duplex ends, they can be attributed to crystal packing, as demonstrated by the differences observed between the two structures of an identical fC-containing duplex crystallized under very different conditions.
Effect of fC on helical coiling and trajectory
Modelling studies on the F-DNA structure and its junction with B-DNA were interpreted as showing that fC affects DNA helical coiling and trajectory and gives rise to pronounced changes in groove geometry.17 To investigate this further we modelled junctions formed between ideal B-DNA and the fC-containing structures 4QKK and 5MVU and compared them to an ideal A-DNA - B-DNA junction.22 The two base pairs at each end of the duplexes (which do not contain fC) were removed to minimise the incorporation of crystal-packing artefacts (Fig. 2d,e). Models containing the full 12-mers are shown in Supplementary Fig. 4. Although the fC-containing structures affect the helical trajectory and groove geometry at the junctions with B-DNA, equivalent effects are exhibited in the model of the A-DNA - B-DNA junctions. Therefore the reported differences in DNA curvature and groove geometry are not attributable to 5-formylcytosine, but are a consequence of the junctions formed between A- and B-DNA. Next, we built analogous models in which the structures are flanked by A-DNA. The fC-containing models closely resemble ideal A-DNA, illustrating that the fCpG repeats in 4QKK adopt A-form geometry, rather than a unique conformation. Such observations are consistent with the all-atom rms deviations between 4QKK and an A-DNA model of analogous sequence generated using standard parameters (Table 1), which show that when the outermost base pairs at each end of the 4QKK duplex are excluded from the calculation, the conformation of 4QKK resembles the ideal A-DNA model even more closely than the unmodified control 5MVK, with rmsd values of 1.08 and 1.44 Å, respectively.
Solution NMR analysis
Our X-ray diffraction results show that fC has only small effects on the A-DNA conformation in the crystal state. However, B-DNA is the dominant form in solution. To determine the influence of fC in solution, we recorded 2D NMR spectra of three DNA duplexes, the native sequence d(CTACGCGCGTAG)2 and two fC modified duplexes, d(CTAfCGfCGfCGTAG)2, and d(CTACGfCGCGTAG)2. All protons except the H5′/H5″ were assigned uniquely in each duplex using a combination of DQF-COSY (Fig. 3a and Supplementary Fig. 5) and NOESY (Supplementary Fig. 6), and the chemical shifts are given (Supplementary Table 7). As expected, the largest changes are for the modified cytosines, where the H6 resonance moves approximately 0.9 ppm downfield, reflecting the altered electronic structure of the nucleobase. In contrast, resonances of the nearest neighbour bases change only slightly (<0.05 ppm, Supplementary Fig. 7a). Similarly, the sugar protons also show only small perturbations (Supplementary Fig. 7b), suggesting, at most, small conformational changes in the vicinity of the modified bases.
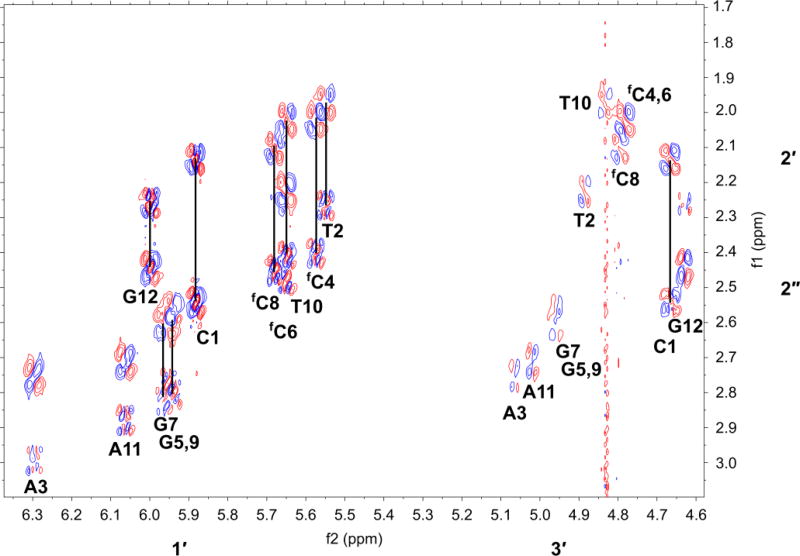
Figure 3. NMR analysis of the 5-formylcytosine-containing DNA duplex which features in the crystal structures 5MVU and 4QKK.
H1′ and H3′- H2′,H2″ region of a DQF-COSY spectrum of the duplex d(CTAfCGfCGfCGTAG)2 recorded with acquisition times of 1 s in t2 and 0.05 s in t1 with spectral widths of 6 kHz in both dimensions. The data were apodized using an unshifted Gaussian function, with zero filling to 16 k by 2 k complex data points prior to Fourier transformation. Resolution in F2=0.73 Hz/pt. Vertical black lines connect cross peaks between H1′ and H2′,H2″ of the same residue. For all non-terminal residues, 3J1′2′>3J1′2″; Σ1′>14 Hz implying the sugar pucker is ‘S’ characteristic of B- DNA. NMR spectra were recorded at 14.1 T and 293 K as described in the methods. See Supplementary Fig. 5–7 for further NMR analyses of this duplex and analogues.
Supplementary Fig. 6a shows the imino proton region of a NOESY spectrum of d(CTAfCGfCGfCGTAG)2 recorded in 93% H2O at 288 K. As expected, there are 4 GN1H and 2 TN3H. The terminal G12N1H is exchange broadened owing to fraying. The inter-imino proton and TN3H-AdeC2H NOEs indicate that all bases are involved in Watson-Crick base pairing, and except for G12:C1, are dynamically stable. We also noted cross peaks between the TN3H protons, but not the GN1H (except G12) and water, consistent with some exchange of the TN3H with water on this timescale (250 ms), typical of AT versus G:C base pairs.24 This further suggests that the fC:G base pairs are not unstable. Very similar results were obtained for the native and singly-modified duplexes.
A and B forms are readily distinguished by NMR,25 and to assess the conformational properties of the duplexes, we recorded high-resolution DQF-COSY and short (50 ms) mixing time NOESY spectra in D2O at 293 K. Fig. 3a shows the H1′-H2′/H2″ region of the DQF-COSY spectra of d(CTAfCGfCGfCGTAG)2. The number of resonances is as expected for the duplex in which the strands are equivalent (i.e. symmetric, a self-complementary sequence). The appearance of both cross peaks and their fine structure is consistent with a sugar conformation primarily in the ‘S’ domain, with the exception of the terminal C1, which shows more extensive dynamic averaging (Supplementary Table 8). We have determined the sums of coupling constants Σ1′ and where possible Σ2′ and Σ2″ from 1D spectra, the DQF-COSY and NOESY spectra (with a mixing time of 300 ms). For all non-terminal residues J1′2′>J1′2″, as expected for C2′-endo conformations that are characteristic of B-DNA, compared with C3′-endo where J1′2′<J1′2″, characteristic of the A-form, and which would be characterized by a weak or absent H1′-H2′ cross peak. The values of the couplings indicate that the dominant conformation is in the ‘S’ domain, with a moderate admixture of the ‘N’ domain.26 Furthermore, the distance r1′4′ estimated from the short mixing time NOESY was >3 Å for all non fC residues, which is inconsistent with an O4′ conformation. Similar results were obtained for the native and doubly modified duplexes (Supplementary Table 8). However this distance was noticeably shorter for the fC residues than the unmodified C in the native duplex, which also parallels the lower fraction of the ‘S’ state for these residues (Supplementary Table 8)
Supplementary Fig. 6c shows a portion of the base region of the NOESY spectrum of d(CTAfCGfCGfCGTAG)2. It is possible to trace the connectivity along the base H8/H6 protons as well as the fC formyl protons through the strand, and this is typical of a right-handed helical structure. The NOE intensities at a short mixing time (50 ms) involving the nucleobase protons (Supplementary Fig. 6b) showed very weak intensity for the H8/6-H1′ (both intranucleotide and internucleotide), strong intensity for the H6/8(i)-H2′(i) and weak intensity for the H6/8(i)-H2″(i). Furthermore, the intranucleotide H8/6-H3′ NOE intensity was also weak. These observations are consistent with a high anti glycosyl torsion angle and ‘S’-type sugar pucker for each nucleotide, which was confirmed by estimating the internucleotide distances from the NOE peak volumes at 50 ms. The critical distance is the H8/H6(i)-H2′(i) which we determined as 2.2–2.3 Å on average in the unmodified sequence and 2.2–2.5 Å in the fC modified duplexes, compared with 2.2 Å and 3.8 Å for canonical B-DNA and A-DNA respectively. The glycosyl torsion angles were in the range −90° to −110° for all sequences, as typically found in B-DNA, compared with the A form (ca. −160°). The scalar coupling and intranucleotide NOE data therefore show that the nucleotide conformations are B family in solution. The sequential internucleotide NOEs, H2′(i) and H2″(i)–H8/6(i+1) are also consistent with a B-like helical geometry. These estimated distances were >3 Å and 2.3–2.5 Å respectively, compared with the values found in standard B DNA (3.6 and 2.5 Å) versus A-DNA (2.0 and 3.5 Å).
As NOEs are very sensitive to small differences in conformation we compared the NOE volumes for the various sequences as the sums of squared differences of the NOE values for each nucleotide (Supplementary Fig. 7c,d), and the global rmsd (Supplementary Table 9). Comparing the intranucleotide NOE differences between the unmodified duplex and those containing 2 or 6 fC additions, and between the duplexes containing 2 or 6 fC additions, the differences are largest for fC compared with C, and are small for other residues. This agrees with the chemical shift perturbations, as well as the observations from the scalar couplings and the apparent r1′4′ distances. Thus the incorporation of fC influences the local nucleotide conformation, characterized by a somewhat larger glycosyl torsion angle, and a less pure C2′-endo like sugar pucker, but modest nucleotide-level conformational perturbation is not propagated throughout the helix, and thus does not have a global influence on the structure, in agreement with Szulik et al.19 The 5-formyl group has a large electronic effect on the nucleobase, as seen in the chemical shift of the fCH6 (ca. 0.9 ppm downfield of C), and in the nearest neighbour nucleotides.
In the crystal structure, some nucleotides have unusual phosphodiester torsions (Supplementary Table 4) that likely arise from packing interactions. We have recorded 31P NMR spectra of the duplexes, and find no evidence of unusual torsion angles reflected in the chemical shifts (Supplementary Fig. 7e).27
CD spectroscopy
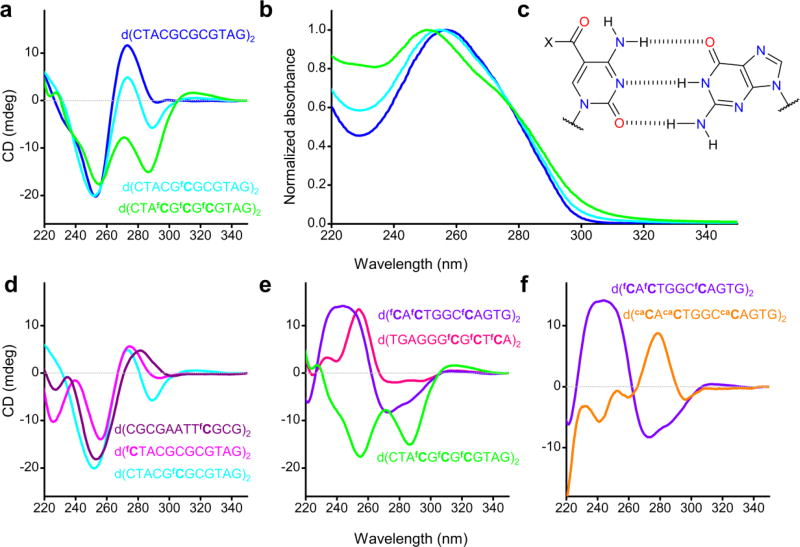
CD spectroscopy provides characteristic signatures for different conformations of nucleic acids.28 We have recorded CD spectra of various duplexes at 20 °C in PBS (Fig. 4), the buffer used for the NMR experiments, and also in the salt conditions that mimic crystallization conditions (Supplementary Fig. 8c–e). The unmodified duplex shows a CD spectrum typical of the B-form, consistent with the NMR data, whereas one or three fC residues on each strand caused a substantial change in the spectra (Fig. 4a), with the appearance of new transitions, whether or not fC was located in a CpG context (Fig. 4d, e). Furthermore, the effect of fC on the CD spectra is site-specific and dependent on the sequence context of fC. It is not consistent with a change in chirality or a global change in conformation, as shown by the small conformation perturbation observed by solution NMR. Major effects on the CD spectrum were also observed for DNA containing caC, which, like fC, has a carbonyl group conjugated to its nucleobase, and therefore can also form a hydrogen bond to the 4-amino group of cytosine (Fig. 4f). UV absorption spectra of duplexes containing fC and caC (Fig. 4b and Supplementary Fig. 8a) were also markedly different to that of unmodified DNA, indicating that the electronic environment of cytosine is strongly affected by the presence of a 5-carbonyl group.
Figure 4. Site-dependent changes to the CD spectra of oligonucleotides containing 5-formylcytosine and 5-carboxylcytosine.
(a) CD analysis of the 6× fC-containing duplex featuring in the crystal structures 4QKK and 5MVU (green), an analogous duplex containing a single, central diformylated fCpG step (cyan) and the unmodified duplex of the same sequence (blue); (b) UV absorption spectra of the duplexes featured in (a), showing that the presence of fC results in significant differences, whose magnitude increases with increasing levels of formylation, as is also the case for their CD spectra. (c) Skeletal depiction of the Watson-Crick base pairing between guanine and 5-formylcytosine (X = H) or 5-carboxylcytosine (X = OH). (d) CD analysis of duplexes containing two additions of fC, showing that differences in CD spectra are observed regardless of whether fC appears in a hemi- or diformylated step or a CpG step. (e) CD analysis of duplexes containing 6 additions of fC in various sequence contexts, demonstrating site-dependent differences in CD spectra. (f) CD analysis of a dodecamer containing either fC (violet) or caC (orange) in hemi-modified, non-CpG steps, showing that caC also strongly affects CD spectra of DNA but in a markedly different way to fC.
Discussion
The X-ray structures of the two fC-modified duplexes and their unmodified counterpart show that the formyl group has no significant influence on conformation in the crystal state. The differences between four unmodified sequence analogues, crystallized in the same space group, serve as a convenient benchmark (Supplementary Fig. 2,3). The small rms deviations between them (0.56–1.27 Å, Supplementary Table 6) reflect only small conformational changes that are associated with their different sequences. Notably, the overall rms deviation between the unmodified duplex and its fC-containing counterpart that crystallized isomorphously falls at the lower end of this range (0.65 Å). Likewise, at the local level, overlays of trinucleotide steps (Fig. 1) and rms deviation of the duplex cores (0.51 Å) show that the effect of fC on conformation is minor. The rmsd between the duplex cores of 5MVK and the fC-containing structure 4QKK is also modest (1.45 Å), though somewhat higher than 5MVK and 5MVU, and is similar that of the identical fC-containing duplexes crystallized under different conditions (1.21 Å). Conformational differences of a similar magnitude between identical dodecamer duplexes in different crystal environments have been reported, even within the same crystal lattice (rmsd of 1.93 Å),29 illustrating the significance of crystal packing forces. To summarise, the conformational differences between fCpG tracts of identical duplexes crystallized in different space groups are greater than the differences arising from the presence of fC itself. Nevertheless, the small structural differences between unmodified and fC-containing DNA do not preclude a more subtle means of recognition by proteins. Indeed, small structural differences can give rise to major biological effects30 as demonstrated by A-tract DNA; the rmsd between the 8-mer cores of the CGCAAAAAAGCG31 and d[CGCATATATGCG]2 duplexes32 is 1.16 Å.
Analysis of local structural parameters of non-terminal residues shows that all duplexes are in the A family of DNA, regardless of the space group, with minor local sequence-dependent variations that reflect the alternating CGCGCG sequence of the core. There are some specific deviations from typical A-form geometry at sites involved in crystal contacts, particularly between terminal base pairs (which do not contain fC) and the minor groove of symmetry mates. This kind of end-groove packing is common in crystals of A-DNA,33 while the distortion of terminal nucleotides resulting from crystal packing interactions is a common feature of DNA crystal structures in general.34–36 Our structures overlay well with the fC-containing structure determined by Raiber et al.17 If such structures are joined to B-DNA on both ends, a perturbation similar to that reported for F-DNA is obtained, whereas none occurs when canonical A-DNA is added to the ends (Fig. 2e).17 Our X-ray data show conclusively that any changes to the duplex conformation are not contingent on fC modification, as all the duplexes have very similar A-type structures. This similarity is also the case for fC in RNA which is also A-form,37 and in a modified Dickerson-Drew dodecamer containing fC which is B-form in the crystal state.19,21 Furthermore, our NMR data show that in solution these duplexes all adopt the B conformation, and that the differences induced by the fC modification are rather small and local, and unlikely to act as a gross structural recognition feature for TET,38 TDG39 or other enzymes. We conclude that there is no compelling evidence for a unique fC-dependent F-DNA structure, nor are the specific hydration patterns in the X-ray structure likely to be relevant for B-form duplexes.17,33 Furthermore, the hydration pattern of 5MVK was markedly different to that of 4QKK, despite exhibiting similar base-stacking geometries in the CpG and fCpG tracts (Supplementary Fig. 8f), thereby suggesting that hydration is not the dominant factor in determining the observed stacking geometries. This family of oligonucleotide duplexes represents an example of the structural differences of DNA duplexes in solution (predominantly B) and the crystal state (often A), which often forms as a result of the dehydrating conditions needed for crystallization.
The overall structural similarity between fC-DNA and unmodified DNA raises a crucial question; what features of fC-DNA do enzymes such as TDG recognize? The present results argue against a substantial shift in global conformation from B-DNA as the primary recognition feature. In the complex with the TDG the DNA is severely distorted to form a structure in which the modified base is flipped out, as observed in other DNA glycosylases,40,41 though the flanking regions remain more B- than A-like. Conceivably, there could be an equilibrium between the B-structure, and one with a base flipped out. However, Zhang et al.39 have estimated the binding of TDG to duplexes with different modified C:G base pairs compared with G:T and the apyrimidinic (Ap) product G:Ap using a catalytically inactive TDG enzyme. The highest affinity was for the product duplex (G:Ap) and the lowest was for unmodified DNA (G:C) or G:mC, which are also not cleaved by TDG. The overall reaction involves the enzyme binding to DNA, followed by bending and inserting an amino acid residue via the minor groove to help flip out the desired base, exposing the glycosyl bond for cleavage within the TDG active site. Such a structure in the absence of enzyme would be at a much higher energy than the relaxed B-DNA, and our data and those of Szulik et al19 indicate that the population of flipped out bases is very low in free fC-DNA. To achieve a productive complex, the flipped-out base must make a large number of interactions with the enzyme to compensate for the local energy of distortion of the duplex. Specific interactions of this nature between the active site residues with 5-formyl or 5-carboxyl groups are sufficient for recognition, compared with the 5H of unmodified cytosine, and have recently been observed.39,42 Once a productive complex has been reached, the nucleobase cleavage rate would depend inversely on the strength of the glycosyl bond, which is lower in fC and caC than for cytosine itself.43 We note that fC has a conformationally restrained formyl group44 and a large dipole moment with different directionality from that of C.45 It has the potential to form strong polar interactions at the TDG enzyme recognition site. It also has high N1 acidity and a weaker N1-C1′ bond, so under appropriate conditions it is a good leaving group. Thus it satisfies both the binding and excision requirements and consequently has the highest excision rate of the epigenetic cytosine derivatives. 5-Carboxylcytosine may also form strong binding interactions with TDG, but is a poorer leaving group due to the negative charge of the carboxylate anion. Therefore, its overall rate of excision is lower than that of fC, and occurs via a different mechanism.43 Interestingly a recent crystal structure of RNA polymerase II (Pol II) complexed with caC-containing DNA indicates that Pol II hydrogen-bonds directly with the 5-carboxyl group, thereby reducing transcription efficiency.46 Similar transcriptional effects were also observed for fC, likely through interactions with the 5-formyl group, though the structure of the complex has not yet been reported. We anticipate that further reports of epigenetic readers of fC will involve sensing based on direct localised interactions with the formyl group.
Although the control duplex and its fC-containing counterparts have very similar structures in solution (B-form), they exhibit drastically different CD spectra, which can be attributed to the altered electronic transitions in the modified bases. The effect is not exclusive to fCpG steps; in all sequence contexts that we examined, the CD spectra of DNA duplexes containing multiple fC bases are strongly perturbed, giving different spectra depending on the local environment of the fC bases (Fig. 4d,e). This is also true for 5-carboxylcytosine (Fig. 4f). It is highly unlikely that the various CD spectra of fC and caC DNA in Fig. 4 represent multiple different helical conformations, so the profound effects on CD must result from local factors. This is consistent with the abnormalities observed in the absorption spectra of fC and caC DNA in the same UV region (Fig. 4b and Supplementary Fig. 8a). It is also notable that the CD spectrum of a 12-mer duplex containing only a single fC base is clearly different from its unmodified counterpart (Fig. 4a, Supplementary Fig. 8e), yet is B-DNA (by NMR analysis under identical conditions). Thus assigning conformational classes of nucleic acids based on CD spectra where there are modified bases is likely to be ambiguous.
In conclusion, our studies indicate that 5-formylcytosine does not change the global conformation of DNA, and our data point to a mechanism that operates at base pair resolution for its recognition. Despite the small differences between the fC-containing duplexes and their unmodified analogue, it remains possible that fC may alter the mechanical properties of DNA. Indeed, a recent report indicates that fC increases the flexibility of DNA.47 Such an effect could lower the energetic penalty in forming certain DNA-protein complexes which involve bending of DNA, such as TDG. However, a more complete picture of the dynamic behaviour of fC-modified DNA, and any biological implications of such effects, remain to be established.
Online Methods
Oligonucleotide synthesis, purification and analysis
Standard nucleoside phosphoramidites and additional reagents were purchased from Link Technologies and Applied Biosystems Ltd. Phosphoramidite monomers of 5-formyldeoxycytidine and 5-carboxyldeoxycytidine were purchased from Glen Research. 5-Formyl-dC CE phosphoramidite (Glen catalogue number 10–1514) was used in the synthesis of all fC-containing oligonucleotides except ODN8, for which formyl dC III CE phosphoramidite (Glen catalogue number 10–1564) was used. Oligonucleotides were synthesized using an Applied Biosystems 394 automated DNA/RNA synthesizer or AKTA OligoPilot 10 using a standard 1 µmol or 15 µmol scale phosphoramidite cycle of acid-catalyzed detritylation, coupling, capping and iodine oxidation. All -cyanoethyl phosphoramidite monomers were dissolved in anhydrous acetonitrile to a concentration of 0.1 M immediately prior to use. For phosphoramidites of modified nucleosides, the coupling time was extended to 10 minutes on the ABI394 synthesizer and 20 minutes on the AKTA synthesizer. For unmodified oligonucleotides, simultaneous deprotection and cleavage from the solid support was achieved by immersion in concentrated, aqueous NH3 for 1 hour at room temperature, followed by 5 hours at 55 °C. Deprotection of oligonucleotides containing 5-formyl dC CE phosphoramidite was achieved by treating the resin-containing column with 2 mL diethylamine in acetonitrile (10%, v/v) for 3 minutes, washing with acetonitrile, drying, then immersing the resin in a 1 mL solution of 0.4 M sodium hydroxide in 4:1 methanol/water (v/v) at room temperature (RT) for 17 hours, after which the sample was neutralized with 28 µL of glacial acetic acid. The vial was briefly sonicated to break up the resin, the supernatant pipetted off, and the resin rinsed with 2 × 250 µL water, which was then combined with the methanolic solution. This solution was added to an equal volume of concentrated aqueous ammonia and heated for 2 hours at 55 °C to achieve full guanine deprotection, which typically did not proceed to completion during the sodium hydroxide treatment. The sample was then concentrated to dryness under reduced pressure, re-suspended in chilled aqueous sodium periodate (25 mM, 500 µL), vortexed and refrigerated. After 30 minutes, the solution was desalted (Nap-10 column, GE Healthcare) and freeze-dried in preparation for HPLC purification. The oxidation reaction consistently proceeded quantitatively and cleanly, as evidenced by mass spectrometry. In addition to isobutyryl-protected dG, the use of acetyl-protected dC phosphoramidites was also required. Oligonucleotides containing 5-formyl dC III CE phosphoramidite were deprotected in aqueous ammonia for 17 hours at room temperature. Due to incomplete deprotection of guanine, the samples were heated in aqueous ammonia for 2 hours at 55 °C, achieving full deprotection with no deleterious effects. After being concentrated to dryness under reduced pressure, the samples were treated with acetic acid/water (80% v/v) for 6 hours at 20 °C before being cooled over ice and neutralized by the dropwise addition of ice-cold aqueous ammonia. The oligonucleotides were desalted (Nap-25 column, GE Healthcare) and lyophilized prior to HPLC purification. For the deprotection and oxidation of 5-carboxyl-2’-deoxycytidine-containing oligonucleotides, the protocol of 5-formyl dC CE phosphoramidite was followed, except that the diethylamine wash was omitted.
Following deprotection, oligonucleotides were purified on a RP-HPLC system (Gilson, Inc.) using a Luna 10 µm C8 100 Å pore Phenomenex 10 × 250 mm column with a gradient of acetonitrile in triethylammonium bicarbonate (TEAB) buffer; Buffer A was composed of either a 0.1 M aqueous solution of triethylammonium bicarbonate or triethylammonium acetate (TEAA), prepared by dilution of a 1 M stock solution. The pH of the TEAA/TEAB solutions was adjusted to 7–8. Buffer B was composed of an equal volume of Buffer A and acetonitrile.
UPLC-MS characterization of oligonucleotides was performed on an Acquity UPLC system with a BEH C18 1.7 µm column (Waters) in conjunction with a Bruker micrOTOF mass spectrometer (ESI- mode). A gradient of methanol in TEA and hexafluoroisopropanol (HFIP) was used (Buffer A: 8.6 mM TEA, 200 mM HFIP in 5% methanol/water (v/v); Buffer B: 20% Buffer A in methanol); Buffer B was increased from 0–70% over 8 minutes, at a flow rate of 0.2 mL min-1. Raw data were processed and deconvoluted using the Bruker Daltonics Compass 1.3 software package.
Circular Dichroism (CD) and UV spectra
CD spectra were collected on a Chirascan Plus spectropolarimeter (Applied Photophysics) using a quartz cuvette with a 10-mm path length. Unless otherwise stated, DNA solutions were prepared at a final concentration of 3.5 µM (duplex) in PBS buffer (Oxoid; 137 mM sodium chloride, 3 mM potassium chloride, 8 mM disodium hydrogen phosphate, 1.5 mM potassium dihydrogen phosphate, pH 7.3). Data were collected over the range of 220–350 nm at 20 °C. Each trace was the result of the average of four scans taken with a step size of 0.5 nm, a time-per-point of 1.5 s and a bandwidth of 2 nm. The averaged trace was Savitzky-Golay-smoothed (Origin) using a polynomial order of 3 and a smoothing window of 20 points. A blank sample, consisting only of buffer, was treated in an identical manner and subtracted from the collected data. Finally, spectra were baseline-corrected using the offset at 350 nm. UV spectra were acquired on a Varian 50 Bio UV-vis spectrophotometer using a quartz cuvette with a 10-mm path length. DNA solutions were prepared at a final concentration of 1.8 µM (duplex) in PBS buffer. Data were collected at 20 °C over the range 220–350 nm and a scan rate of 60 nm per minute. A blank sample of PBS was treated in an identical manner and subtracted from the data. All oligonucleotides were annealed in PBS prior to CD and UV spectroscopic analysis by heating to 95 °C for five minutes before being cooled to room temperature at a rate of 0.1 °C per minute.
X-ray crystallography
Crystallization screens Natrix HT (Hampton Research) and Nucleix (Qiagen) were used to identify suitable crystallization conditions, which were optimised where necessary. Crystallization conditions for each structure are described in the supporting information (Supplementary Table 3).
Diffraction data collection
Crystals were flash-cooled in liquid nitrogen; additional cryoprotection was not required, due to the use of MPD or 2-propanol as a precipitant in all cases. All diffraction data were collected at 100 K at the Diamond Light Source synchrotron science facility, Harwell, on beamlines I02-I04 using Pilatus 6M hybrid pixel array detectors. For each crystal, 1800 images were typically collected (0.1° oscillation, 100 ms exposure). For data collection, X-ray wavelengths of 0.976 Å (5MVK, 5MVP, 5MVQ, 5MVU), 1.060 Å (5MVT), 0.920 and 1.240 Å (5MVL) were used.
Data processing, phase determination, model building and refinement
Data were indexed and scaled using XDS48 and AIMLESS.49 For 5MVL (containing 5-bromouracil), experimental phases were determined from the by multi-wavelength anomalous dispersion from the bromine atoms, using the SHELX50 software suite. For all other structures, initial phase estimates were generated by molecular replacement with Phaser,51 using the pdb file of 5MVL as a search model. Prior to molecular replacement, all atoms differing between the search model and the structure in question were removed from the search model to prevent model bias. After an initial stage of refinement using either REFMAC552 or PHENIX,59 missing atoms were rebuilt manually using COOT,53 followed by successive stages of further refinement.
Quantitative analysis of X-ray crystal structures
Structural parameters were calculated using the software packages 3DNA54 and Curves+.55
DNA model building
Standard A- and B-DNA models were generated and analysed using w3DNA.22 Input files for the construction of DNA models in w3DNA were created manually by combining the parameters derived from the standard models and crystal structures. Initial coordinate files of the models were then generated in w3DNA. To restore the non-idealised backbone geometry in the section of the model derived from the crystal structure, atomic coordinates were replaced manually with those of the structure following alignment in PyMOL.
NMR Spectroscopy
NMR Buffer
80 mM KCl, 22 mM NaPi in D2O pD=7.3 at 293 K. The buffer was lyophilized and redissolved in 100% D2O containing DSS as reference. DNA powders were dissolved in 0.5 mL buffer (to 2 mM), and 200 µL loaded into 3 mm NMR tubes.
1H NMR spectra were recorded at 14.1 T on an Agilent DD2 NMR spectrometer equipped with a 3 mm inverse triple resonance HCN cold probe. Spectra in D2O were recorded at 293 K, and in H2O at 288 K. 1D, DQF-COSY spectra and NOESY spectra with mixing times of 50 ms and 300 ms were recorded on each sample.
1D
The presat experiment was recorded with a 2 sec acquisition time, 3 sec delay with low power transmitter presaturation of residual HOD.
NOESY spectra were recorded with an acquisition time of 1 s in t2, 50 ms in t1, relaxation delay 1 sec, with presaturation during the 1 s relaxation delay, and with mixing times of 300, 50 ms. DQF-COSY spectra were recorded with an acquisition time of 1 s in t2, 50 ms in t1, relaxation delay 1 sec, presaturation during the delay. 2D data were transformed with 32k points in t2 (1 zero filling), with 1 linear prediction and 1 zero filling in t1. The Fids were apodized using an unshifted Gaussian function and a 1 Hz line broadening exponential.
DNA was then lyophilized and redissolved in 92.5% H2O with 7.5% D2O. NMR spectra in water were recorded using excitation sculpting for water suppression with an acquisition time of 1.5 s, and a relaxation delay of 2.5 sec at 15 °C. ES-NOESY spectra were recorded with an acquisition time of 0.386 s in t2 and 0.036 s in t1, and a mixing time of 250 ms. The Fids were Fourier transformed as 16k×2k points with 1 linear prediction in t1 and apodized using with a line broadening exponential of 1 Hz plus an unshifted Gaussian function in both dimensions.
Resonances were assigned using the NOESY and DQF-COSY spectra.56 The DQF-COSY spectra and 1D NMR spectra were used to measure coupling constants in the deoxyribose sugars, and these were analyzed to estimate the sugar conformations according to published procedures.57 The peak volumes in the 50 ms NOESY spectra were determined, and used to estimate interproton distances using either the Cyt C5H-C6H (C6H-CHO in the fC) as references. For the H1′-H4′ distance, the NOE volume of the H1′-H4′ cross peak was normalized to the H2′-H2′′ cross peak of the same residues, as this reduces effects of dynamics using the shared H1′. The NOESY data were compared with the values expected for A- and B-DNA. Furthermore, the normalized NOESY volumes were compared between the unmodified duplex and the two modified duplexes, calculated as the rmsd values to assess conformational differences.
1D 31P NMR spectra were recorded at 16.45 T on a Bruker Avance III spectrometer equipped with a 5 mm inverse HCNP cryoprobe, with an acquisition time of 1.5 s, a relaxation delay of 1.5 s and 1024 transients with proton decoupling during the acquisition period.
Data availability
The structure factors and coordinates of all structures have been deposited in the Protein Data Bank under accession codes 5MVK [d(CTACGCGCGTAG)2], 5MVU [d(CTAfCGfCGfCGTAG)2], 5MVL [d(CbUACGCGCGTAG)2], 5MVP [d(CTAGGGCCCTAG)2], 5MVT [d(CTACGTACGTAG)2] and 5MVQ [d(CTACGGCCGTAG)2]. Other data are available upon request.
Supplementary Material
Table 4.
Data collection, phasing and refinement statistics of 5MVL (MAD)
| 5MVL | ||
|---|---|---|
| Data collection | ||
| Space group | P3221 | |
| Cell dimensions | ||
| a, b, c (Å) | 43.5, 43.5, 60.8 | |
| α, β, γ(°) | 90.0, 90.0, 120.0 | |
| Peak | Remote | |
|
|
||
| Wavelength | 0.920 | 1.240 |
| Resolution (Å)a | 30.4-1.6 (1.63-1.6)a | 30.4-1.4 (1.43-1.40) |
| Rmeas | 5.5 (52.9) | 4.6 (117) |
| I/σ(I) | 18.8 (4.1) | 23.4 (2.0) |
| CC1/2 | 0.999 (0.898) | 0.999 (0.984) |
| Completeness (%) | 99.1 (100) | 99.9 (100) |
| Redundancy | 7.9 (8.0) | 9.1 (8.3) |
| Refinement | ||
| Resolution (Å) | 23.7-1.4 | |
| No. reflections | 13,446 | |
| Rwork / Rfree | 15.7 / 18.2 | |
| No. atoms | ||
| DNA | 486 | |
| Mg2+ | 1 | |
| Water | 76 | |
| B factors | ||
| DNA | 23.89 | |
| Water | 33.90 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.009 | |
| Bond angles (°) | 1.07 | |
Both data sets were collected from a single crystal.
Values in parentheses are for highest-resolution shell.
Acknowledgments
This work was supported by BBSRC sLoLa grant BB/J001694/2 (Extending the boundaries of nucleic acid chemistry), Oxford University studentship (J.S.H.) and in part by a Carmen L. Buck endowment (to A.N.L.). NMR data were recorded at Center for Environmental and Systems Biochemistry supported by the University of Kentucky, and NCI Cancer Center Support Grant (P30 CA177558).
Footnotes
Accession codes
The structure factors and coordinates of all structures have been deposited in the Protein Data Bank under accession codes 5MVK [d(CTACGCGCGTAG)2], 5MVU [d(CTAfCGfCGfCGTAG)2], 5MVL [d(CbUACGCGCGTAG)2], 5MVP [d(CTAGGGCCCTAG)2], 5MVT [d(CTACGTACGTAG)2] and 5MVQ [d(CTACGGCCGTAG)2].
Author contributions
T.B. and A.N.L. designed the project and wrote the manuscript with contributions from all authors. J.S.H. carried out oligonucleotide synthesis, performed the crystallizations and biophysical experiments. A.H.E-S. carried out oligonucleotide synthesis and biophysical experiments. A.N.L. recorded and interpreted the NMR spectra. I.T. and D.S. carried out large-scale oligonucleotide synthesis, purification and analysis. D.P. and J.S.H. acquired and analyzed X-ray crystallographic data, and solved the structures. T.B., A.N.L. and S.E.V.P. supervised the project. All authors interpreted the data and read and approved the manuscript.
Competing financial interests
T.B. is a consultant to ATDBio.
References
- 1.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annual Review of Biochemistry. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature Reviews Genetics. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Tahiliani M, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–U151. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He Y-F, et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaffeneder T, et al. The Discovery of 5-Formylcytosine in Embryonic Stem Cell DNA. Angewandte Chemie-International Edition. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 7.Ito S, et al. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raiber EA, et al. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase. Genome Biology. 2012;13:r69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiti A, Drohat AC. Thymine DNA Glycosylase Can Rapidly Excise 5-Formylcytosine and 5-Carboxylcytosine; Potential implications for active demethylation of CpG sites. Journal of Biological Chemistry. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber AR, et al. Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism. Nature Communications. 2016;7 doi: 10.1038/ncomms10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spruijt CG, et al. Dynamic Readers for 5-(Hydroxy)Methylcytosine and Its Oxidized Derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Bachman M, et al. 5-Formylcytosine can be a stable DNA modification in mammals. Nature Chemical Biology. 2015;11:555–557. doi: 10.1038/nchembio.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iurlaro M, et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biology. 2013;14:r119. doi: 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellinger MW, et al. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nature Structural & Molecular Biology. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaffeneder T, et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nature Chemical Biology. 2014;10:574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 16.Su M, et al. 5-Formylcytosine Could Be a Semipermanent Base in Specific Genome Sites. Angewandte Chemie-International Edition. 2016;55:11797–11800. doi: 10.1002/anie.201605994. [DOI] [PubMed] [Google Scholar]
- 17.Raiber EA, et al. 5-Formylcytosine alters the structure of the DNA double helix. Nature Structural and Molecular Biology. 2015;22:44–49. doi: 10.1038/nsmb.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lercher L, et al. Structural insights into how 5-hydroxymethylation influences transcription factor binding. Chemical Communications. 2014;50:1794–1796. doi: 10.1039/c3cc48151d. [DOI] [PubMed] [Google Scholar]
- 19.Szulik MW, et al. Differential Stabilities and Sequence-Dependent Base Pair Opening Dynamics of Watson-Crick Base Pairs with 5-Hydroxymethylcytosine, 5-Formylcytosine, or 5-Carboxylcytosine. Biochemistry. 2015;54:1294–1305. doi: 10.1021/bi501534x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renciuk D, Blacque O, Vorlickova M, Spingler B. Crystal structures of B-DNA dodecamer containing the epigenetic modifications 5-hydroxymethylcytosine or 5-methylcytosine. Nucleic Acids Research. 2013;41:9891–9900. doi: 10.1093/nar/gkt738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura K, Ono A, Watanabe K, Takenaka A. X-Ray analyses of oligonucleotides containing 5-formylcytosine, suggesting a structural reason for the codon-anticodon recognition of mitochondrial tRNA-Met. PDB accession code: 1VE8. 2016 doi: 10.2210/pdb1ve8/pdb. [DOI] [Google Scholar]
- 22.Zheng G, Lu XJ, Olson WK. Web 3DNA -a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Research. 2009;37:W240–6. doi: 10.1093/nar/gkp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Šponer J, Kypr J. Different intrastrand and interstrand contributions to stacking account for roll variations at the alternating purine-pyrimidine sequences in A-DNA and A-RNA. Journal of Molecular Biology. 1991;221:761–764. doi: 10.1016/0022-2836(91)80172-q. [DOI] [PubMed] [Google Scholar]
- 24.Gueron M, Leroy JL. Studies of base pair kinetics by NMR measurement of proton exchange. Nuclear Magnetic Resonance and Nucleic Acids. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 25.Gyi JI, Lane AN, Conn GL, Brown T. Solution structures of DNA.RNA hybrids with purine- rich and pyrimidine-rich strands: Comparison with the homologous DNA and RNA duplexes. Biochemistry. 1998;37:73–80. doi: 10.1021/bi9719713. [DOI] [PubMed] [Google Scholar]
- 26.Vanwijk J, Huckriede BD, Ippel JH, Altona C. Furanose Sugar Conformations in DNA from Nmr Coupling-Constants. Methods in Enzymology. 1992;211:286–306. doi: 10.1016/0076-6879(92)11017-d. [DOI] [PubMed] [Google Scholar]
- 27.Roongta VA, Jones CR, Gorenstein DG. Effect of Distortions in the Deoxyribose Phosphate Backbone Conformation of Duplex Oligodeoxyribonucleotide Dodecamers Containing GT, GG, GA, AC, and GU Base-Pair Mismatches on P-31 NMR-Spectra. Biochemistry. 1990;29:5245–5258. doi: 10.1021/bi00474a005. [DOI] [PubMed] [Google Scholar]
- 28.Kypr J, Kejnovská I, Renčiuk D, Vorlíčková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Research. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiGabriele AD, Sanderson MR, Steitz TA. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Procedings of the National Academy of Sciences United States of America. 1989;86:1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haran TE, Mohanty U. The unique structure of A-tracts and intrinsic DNA bending. Quarterly Reviews of Biophysics. 2009;42:41–81. doi: 10.1017/S0033583509004752. [DOI] [PubMed] [Google Scholar]
- 31.Nelson HCM, Finch JT, Luisi BF, Klug A. The structure of an oligo(dA)·oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 32.Yoon C, Prive GG, Goodsell DS, Dickerson RE. Structure of an alternating-B DNA helix and its relationship to A-tract DNA. Proceedings of the National Academy of Sciences USA. 1988;85:6332–6. doi: 10.1073/pnas.85.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahl MC, Sundaralingam M. Crystal structures of A-DNA duplexes. Biopolymers. 1997;44:45–63. doi: 10.1002/(SICI)1097-0282(1997)44:1<45::AID-BIP4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 34.Spink N, Nunn CM, Vojtechovsky J, Berman HM, Neidle S. Crystal structure of a DNA decamer showing a novel pseudo four-way helix-helix junction. Proceedings of the National Academy of Sciences USA. 1995;92:10767–10771. doi: 10.1073/pnas.92.23.10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Malinina L, Huynh-Dinh T, Subirana JA. The structure of the most studied DNA fragment changes under the influence of ions: a new packing of d(CGCGAATTCGCG) Febs Letters. 1998;438:211–214. doi: 10.1016/s0014-5793(98)01295-2. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Subirana JA. Structure of d(CGCGAATTCGCG) in the presence of Ca2+ ions. Journal of Biological Chemistry. 1999;274:24749–24752. doi: 10.1074/jbc.274.35.24749. [DOI] [PubMed] [Google Scholar]
- 37.Wang R, et al. Base pairing and structural insights into the 5-formylcytosine in RNA duplex. Nucleic Acids Research. 2016;44:4968–4977. doi: 10.1093/nar/gkw235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu L, et al. Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, et al. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nature Chemical Biology. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savva R, McAuley-Hecht K, Brown T, Pearl LH. The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature. 1995;373:487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]
- 41.Barrett TE, et al. Crystal structure of a G : T/U mismatch-specific DNA glycosylase: Mismatch recognition by complementary-strand interactions. Cell. 1998;92:117–129. doi: 10.1016/s0092-8674(00)80904-6. [DOI] [PubMed] [Google Scholar]
- 42.Pidugu LS, et al. Structural Basis for Excision of 5-Formylcytosine by Thymine DNA Glycosylase. Biochemistry. 2016;55:6205–6208. doi: 10.1021/acs.biochem.6b00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maiti A, Michelson AZ, Armwood CJ, Lee JK, Drohat AC. Divergent Mechanisms for Enzymatic Excision of 5-Formylcytosine and 5-Carboxylcytosine from DNA. Journal of the American Chemical Society. 2013;135:15813–15822. doi: 10.1021/ja406444x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawai G, et al. Conformational Properties of a Novel Modified Nucleoside, 5-Formylcytidine, Found at the First Position of the Anticodon of Bovine Mitochondrial tRNAMet. Nucleosides and Nucleotides. 1994;13:1189–1199. [Google Scholar]
- 45.Xu Y, Vanommeslaeghe K, Aleksandrov A, MacKerell AD, Nilsson L. Additive CHARMM force field for naturally occurring modified ribonucleotides. Journal of Computational Chemistry. 2016;37:896–912. doi: 10.1002/jcc.24307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 2015;523:621–625. doi: 10.1038/nature14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ngo TTM, et al. Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nature Communications. 2016;7:10813. doi: 10.1038/ncomms10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kabscha W. XDS. Acta Crystallographica Section D. 2009;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallographica Section D. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheldrick G. A short history of SHELX. Acta Crystallographica Section A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 51.McCoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallographica Section D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Research. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Research. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lane AN, Jenkins TC, Brown T, Neidle S. Interaction of Berenil with the EcoR1 dodecamer d(CGCGAATTCGCG)2 in Solution Studied by NMR. Biochemistry. 1991;30:1372–1385. doi: 10.1021/bi00219a030. [DOI] [PubMed] [Google Scholar]
- 57.Conte MR, Bauer CJ, Lane AN. Determination of sugar conformations by NMR in larger DNA duplexes using both dipolar and scalar data: Application to d(CATGTGACGTCACATG)(2) Journal of Biomolecular NMR. 1996;7:190–206. doi: 10.1007/BF00202036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure factors and coordinates of all structures have been deposited in the Protein Data Bank under accession codes 5MVK [d(CTACGCGCGTAG)2], 5MVU [d(CTAfCGfCGfCGTAG)2], 5MVL [d(CbUACGCGCGTAG)2], 5MVP [d(CTAGGGCCCTAG)2], 5MVT [d(CTACGTACGTAG)2] and 5MVQ [d(CTACGGCCGTAG)2]. Other data are available upon request.