Abstract
Small heat shock protein beta-1 (HSPB-1) plays an essential role in the protection of cells against environmental stress.Elucidation of its molecular, structural, and biological characteristics in a naturally wild-type model is essential. Although the sequence information of the HSPB-1 gene is available for many mammalian species, the HSPB-1 gene of Arabian camel (Arabian camel HSPB-1) has not yet been structurally characterized. We cloned and functionally characterized a full-length of Arabian camel HSPB-1 cDNA. It is 791 bp long, with a 5′-untranslated region (UTR) of 34 bp, a 3′-UTR of 151 bp with a poly(A) tail, and an open reading frame (ORF) of 606 bp encoding a protein of 201 amino acids (accession number: MF278354). The tissue-specific expression analysis of Arabian camel HSPB-1 mRNA was examined using quantitative real-time PCR (qRT-PCR); which suggested that Arabian camel HSPB-1 mRNA was constitutionally expressed in all examined tissues of Arabian camel, with the predominately level in the esophagus tissue. Peptide mass fingerprint-mass spectrometry (PMF-MS) analysis of the purified Arabian camel HSPB-1 protein confirmed the identity of this protein. Phylogenetic analysis showed that the HSPB-1 protein of Arabian camel is grouped together with those of Bactrian camel and Alpaca. Comparing the modelled 3D structure of Arabian camel HSPB-1 protein with the available protein 3D structure of HSPB-1 from human confirmed the presence of α-crystallin domain, and high similarities were noted between the two structures by using super secondary structure prediction.
Introduction
The one-humped camel, Camelus dromedarius (also known as Arabian camel), is one of the most important member of the Camelidae family. Arabian camel has played a major role in the culture and way of life in the Arabian Peninsula over the past couple thousand of years [1]. This animal has acclimatized itself to live in the desert, and to survive under extreme environmental conditions by promoting the expression of several genes such as small heat shock genes, which encode a family of proteins known as small heat shock proteins sHSPs [2–6]. They play a crucial role in Arabian camel defense from adverse environmental conditions by protecting other proteins from irreversible aggregation [7].
Small heat shock protein beta-1 (HSPB-1), a typical member of the sHSPs family, is a ubiquitously conserved ATP-independent protein, which is immensely preserved in a wide spectrum of organisms, ranging from bacteria to eukarya [8, 9]. Although the diversification in structure and function is a characteristic of most members of the HSP families, including HSP90 [10, 11], HSP70 [12], and HSP40 [13], remarkable diversity is noted in sHSP family ranging from a single homologue of sHSP in Saccharomyces cerevisiae to over 20 homologues of sHSPs in plants [14, 15]. However, ten well-known members of the HSPB family (HSPB-1 to HSPB-10) have been well-studied in human and mammals [16, 17]. These proteins are molecular chaperones, which commonly have a low molecular weight ranging from 12 to 30 kDa, and are generally distinguished by the presence of a typically conserved α-crystallin domain (ACD) that is flanked by a less conserved C-terminal extension (CTE) and an N-terminal domain (NTD) [18–20]. The formation of a stable dimer interface between two contiguous monomers of small heat shock proteins’ ACD facilitate the assembly of a large oligomers’ subunits [21, 22]. These molecular oligomers act as chaperones by binding to the unfolded proteins. Generally, the cellular concentration of many sHSPs is considerably increased in response to various of stresses, but they can also function fundamentally in many organisms and tissues [23].
Although HSPB-1 protein is highly conserved across species from bacteria to mammals, HSPB-1 protein from Arabian camel has not yet been characterized. This study aimed to clone and sequence a full-length of Arabian camel HSPB-1 cDNA and determine amino acid sequence as well as elucidate its protein structure. In addition, we investigated the Arabian camel HSPB-1 mRNA expression profile in ten different tissues. We believe that the study of biochemical and biophysical aspects of Arabian camel HSPB-1 gene is likely to provide molecular insights into Arabian camel physiology as well as providing annotation of Arabian camel HSPB-1 protein on which to advance further studies of Arabian camel proteins.
Materials and methods
Sample collection
Ten different Arabian camel tissue samples, including brain, lung, liver, kidney, testis, spleen, heart, stomach, skin, and esophagus, were obtained from male Arabian camel slaughtered at the main slaughter-house located in Saudi Arabia, Riyadh. This slaughter house is officially supervised by Veterinaries. Tissue samples to be used for RNA analysis were instantly immersed in RNAlater® RNA Stabilization reagent (Qiagen, Ambion, Inc, USA) to prevent RNA degradation. The samples were then stored at -80°C until further use. While those other sample tissues to be used for protein analysis were transported on ice to the laboratory.
Cell culture
Arabian camel skin fibroblast cell line (SACAS) was kindly provided by A. Alawad and routinely maintained as previously described [24]. Cells were used after they reached ≃ 70% confluency. Control cells were incubated at 37°C and experimental cell culture plates were incubated at 42°C for heat stress studies in 5% CO2 incubator at different time points 2, 4, 6, and 8 h. At each time point, cells were washed twice with cold PBS and lysed for RNA extraction using TRIZOL® Reagent [25].
Total RNA isolation and cDNA synthesis from tissues
Samples of 50 mg of each preserved tissues were subjected for RNA isolation. The tissues were homogenized in RTL lysis based buffer (Qiagen) containing 1% 2-mercaptoethanol by using steel beads (Sigma) and Tissue Lyser ⨿ (Qiagen). Nanodrop spectrophotometer (NanoDrop, ThermoScientific) was used to quantify samples at 260nm and the quality of RNA samples was evaluated using denaturing SYBR safe agarose gel 1% electrophoresis. Next, ≃ 2μg of total RNA were transcribed to first-stranded cDNA by using an ImProm-⨿ Reverse Transcription System (Promega, USA).
Examining gene expression by using PCR and qRT-PCR
Gene-specific primers (Table 1) were designed based on the data from the Arabian camel genome project (http://camel.genomics.org.cn/page/camel/index.jsp). The PCR reaction mixture was carried out in a final volume of 25 μl, containing 12.5 μl 2X GoTaq® Green Master Mix(Promega, USA), 1 μl of 5 pmol of each primer, 2 μl of cDNA. The PCR condition was 1 cycle at 94°C for 5 min, followed by 30 cycles at 94°C for 5 sec, 60°C for 30 sce, and 72°C for 45 sec. The final extension step was performed at 72°C for 10 min. The PCR products were then examined on 1.2% agarose gel stained with SYBR safe. In addition, the level of relative expression of Arabian camel HSPB-1 mRNA was evaluated by examining the ten different Arabian camel tissues by using fluorescent quantitative real-time PCR (qRT-PCR) detector ViiA 7 Real-Time PCR System. The β-actin mRNA was used as a house keeping gene control. In this experiment, Fast SYBR® Green Master Mix kit was used, and gene-specific primer pairs were designed to amplify 83 bp length of Arabian camel HSPB-1. The qRT-PCR reaction mixture were 10 μl of Fast SYBR® Green Master Mix (Cat. No., 4385612, Applied Biosystems), 1 μl of the forward primer, 1 μl of the reverse primer, 3 μl of nuclease-free water and 5 μl of cDNA target, in a total volume of 20 μl. Thermal cycling parameters were initial denaturation at 95°C for 3 min, amplification of 40 cycles at 95°C for 3 s, and 60°C for 40 s.
Table 1. Gene-specific primers: The underlined bases represent the restriction sites used for cloning.
| Usage | Primer name | Primer sequence 5′ → 3′ | Product length (bp) |
|---|---|---|---|
| ORF-PCR | Camel HSPB1-F | GAGCCACCATGGCCGAG | 759 |
| Camel HSPB1-R | GCCGGCAGGAACTTAGAACT | ||
| Camel HSPB1-pF | CACGGATCCGAGCCACCATGGCCGAG | ||
| Camel HSPB1-pR | CACGCGGCCGCTCAGCCGGCAGGAACTTAGAACT | ||
| β-Actin-F | CCCATTGAGCATGGCATCGT | 291 | |
| β-Actin-R | GTAGATGGGCACAGTGTGAG | ||
| cDNA-RACE | cDNA-RACE-5’ | CTTGGTCTTGACCGTCAGCTCCTC | 283 |
| cDNA-RACE-3’ | AGATCACCATCCCTGTCACCTTCGA | 382 | |
| qRT-PCR | Camel HSPB1-qF | GTGTCGGAGATCCAGCAGAC | 83 |
| Camel HSPB1-qR | TTCGTGCTTGCCAGTGATCT | ||
| β-Actin-qF | CCCATTGAGCATGGCATCGT | 190 | |
| β-Actin-qR | GTAGATGGGCACAGTGTGAG |
Cloning and sequencing of Arabian camel HSPB-1 cDNA
Rapid amplification of cDNA ends (RACE) was used to identify and isolate the 5′- and 3′-end of Arabian camel HSPB-1 by using a RACE kits (Invitrogen, Carlsbad, CA, USA). Total RNA was annealed with 5′- and 3′-end primers (Table 1), and reversely transcribed respectively to the respective 5′- and 3′-cDNA. The resulting first-stranded 5′- and 3′-cDNA were then utilized as templates in PCR. The cycling program was set for five cycles of 95°C for 4 min; 5 cycles of 95°C for 15 s, 70°C for 15 s, 72°C for 3 min; 5 cycles of 95°C for 15 s, 68°C for 15 s, 72°C for 3 min; 5 cycles of 95°C for 15 s, 65°C for 15 s, 72°C for 3 min; 25 cycles of 95°C for 15 s, 60°C for 15 s, 72°C for 3 min; 1 cycle of 72°C for 5 min. The purified nested PCR product was ligated into pcDNA5/FRT/TO GFP-tagged vector (a gift from Harm Kampinga; Addgene plasmid No., 19487) [26] by using BamHI (NEB R3136S) and NotI (NEB R3189S) restriction sites. Subsequently, 5 ul of the ligation mixture was used as a template to transform chemically modified DH5α competent cells (ThermoFisher Scientific). The cloned Arabian camel HSPB-1 was sequenced using Applied Biosystems 3730xl DNA Analyzer platform (Applied Biosystems, Foster City, USA). The conditions of the chain termination PCR were as follows: one cycle at 94°C for 35 s, followed by 25 cycles at 94°C for 40 s, 50°C for 35 s, and 60°C for 1 min.
Protein extraction and quantification
Proteins from brain, testis, kidney, liver, lung, and the spleen were extracted using RIPA lysis buffer. Next, 4 mg sample from each tissue was homogenized in 4 mL of RIPA lysis buffer containing 5 M NaCl, 0.5 M EDTA, 1 M Tris-HCl, NP-40, 10% soudium deoxycholate, and 10% SDS by using steel beads (Sigma) and a Tissue Lyser ⨿ (Qiagen). Lysates were then centrifuged at 14,000 rpm for 1 hour at 4°C. The supernatant fractions were then collected, and total protein quantity for each tissue was determined using the bicinchoninic acid assay (BCA).
Arabian camel HSPB-1 protein identification by using LC-MS
For this, 25 μg of Arabian camel protein lysates were subjected to one-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis (1-D SDS-PAGE) by using 4% staking and 15% resolving polyacrylamide gels (1 mm thickness gel) by running for 120 min. The 1-D SDS-PAGE was then stained overnight in a solution containing the mixture of Coomassie R-240, 40% methanol, and 10% acetic acid. The gel was subsequently destined in a solution containing 30% methanol and 10% acetic acid.
The excised band gel piece holding proteins with molecular weight of approximately 20-25 kDa was cut into cubes and incubated for 45 min in 300 μl of 1:1 mixture of 100 mM ammonium bicarbonate buffer containing 50% acetonitrile and was vortexed for 10 min; the supernatant was then discarded. The procedure was repeated until the stain was completely removed. Next, 10 mM dithiothreitol (DTT) in 100 mM ammonium bicarbonate buffer was added to the gel cubes in order to reduce the disulfide bonds; the cubes were incubated for 30 min at 56°C in an air thermostat. After they were rinsed in 100 μl of acetonitrile, 200 μl of 50 mM iodoacetamide solution was added, and the mixture was incubated for 20 min at room temperature in the dark.
The gel cubes were then dehydrated twice with 100% acetonitrile for 10 min each and then dried in a speed-vac for 10 min in order to process them ready for tryptic digestion. Trypsin (10ng/ul) solution was added to the dried gel cubes just enough to cover the gel cubes and incubated for 10 min at room temperature. Subsequently, 100 mM ammonium bicarbonate buffer was added until the gel cubes were immersed which was then incubated at 37°C for overnight. The digestion was then stopped by adding 20 μl of 5% formic acids. The digested solution (extracted peptides) was then transferred to a clean autosampler vial.
Millipore®Ziptips C18 pipette (Tip size:P10, Merck KGaA, Darmstadt, Germany) was used to prepare sample for Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry. The Ziptip pipette was washed with 100% methanol, followed by 0.1% trifluroacetic acid (TFA) solution. The tryptic-cleaved peptides mixture were then loaded onto the Ziptip pipette and then were desalted using 0.1% TFA. The loaded peptides were then eluted in 10 μl of β-cyano-4-hydroxycinnamic acid, which was used as a matrix. 1 μl of aliquots were generally sampled directly from the digest supernatant for MS fingerprint analysis by using Axima Performance ® MALDI TOF/TOF Mass Spectrometer (Shimadzu Corporation, UK). The data were searched using the MASCOT search engine (http://www.matrixscience.com).
Phylogenetic tree
The Arabian camel HSPB-1 protein sequence was used as a query to retrieve 40 sHSP sequences from the NCBI Protein Database. The α-crystalline domain was verified in all the retrieved protein sequences by using InterProScan [27] at (https://www.ebi.ac.uk/interpro/) (S3 Table). To verify whether the Arabian camel HSPB-1 protein is distinctly related to the HSPB-1 proteins family, we retrieved 40 HSP orthologues, which are conspicuously related to ten well-known sHSP families known as HSPB-1 to HSPB-10. To ensure the consistency of sampling, we retrieved all sHSPs proteins orthologues from the same species. We used Arabian camel HSPB-1 protein sequence as a query to search the NCBI Protein Database to identifying HSPB-1 proteins across diverse vertebrate species. Another set of sHSP members were sampled from the same mammalian species to ensure the consistency of sampling. The accession numbers of protein members investigated are listed in (S3 Table). Consequently, the full length amino acid sequences, including Arabian camel HSPB-1 protein, were selected for multiple alignment by using CLUSTALX 2.1 program [28]. A bootstrap re-sampling technique was used to ensure the robustness of the generated topological tree. Neighbor Joining (NJ) phylogenetic analysis was conducted in MEGA 7.0 [29]. The constructed topological trees were depicted and edited using FigTree v1.4.3. (http://tree.bio.ed.ac.uk/software/figtree/).
Structure modeling
The secondary structure of Arabian camel HSPB-1 protein sequence was generated using Geneious software v10.0.3 [30]. Consequently, a three-dimensional (3D) structure of Arabian camel HSPB-1 protein containing 201 residues was predicted after submitting the protein sequence to Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). The similarities between modeled Arabian camel HSPB-1 and human HSPB-1 structure (PDB:2YGD) were superimposed by using Pymol software. The quality of the superimposed 3D structures was assessed using PDBe on (https://swissmodel.expasy.org/interactive). The antigenicity, hydrophobicity, and flexibility of Arabian camel HSPB-1 protein were predicted according to the methods of Kolaskar, Parker, and Karplus, respectively [31, 32].
Results
Tissue-specific expression profile of Arabian camel HSPB-1 mRNA
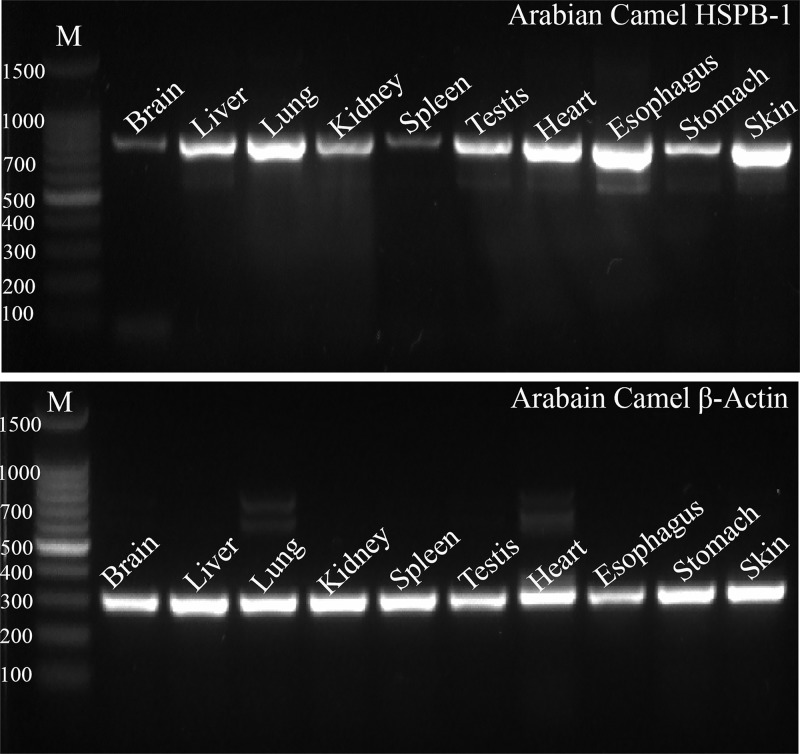
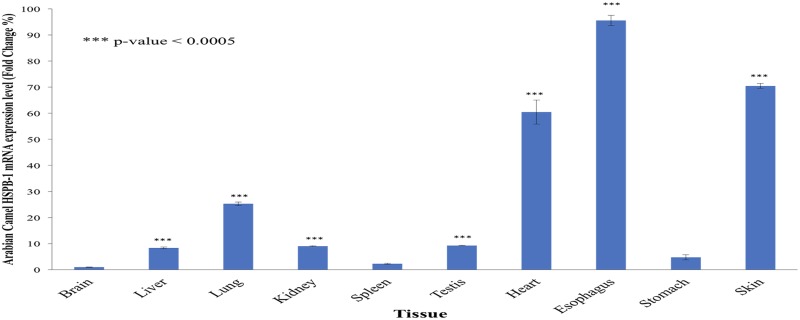
The expression of Arabian camel HSPB-1 mRNA was found in all examined tissues of Arabian camel (Fig 1), indicating its important role in cellular proteostasis. Specific primers (Table 1) were designed to amplify a single 759 bp for Arabian camel HSPB-1 and 291 bp for Arabian camel β-actin genes (as endogenous control). In addition, the level of expression of Arabian camel HSPB-1 mRNA in the ten different tissues was studied using qRT-PCR. The qRT-PCR primers were designed to amplify 83 and 190 bp for Arabian camel HSPB-1 and β-actin, respectively. Under no heat stress condition, the maximum expression of Arabian camel HSPB-1 mRNA was noted in the Arabian camel esophagus, skin, and heart, followed by nearly equally expression in the liver, kidney, testis, and lung, whereas the lowest expression was noted in the brain, spleen, and stomach tissues (Fig 2). This result is in agreement with that of a previous study investigating tissue-specific expression of HSPs in buffalo tissues [33]. These observations might be significant in understanding the differential sensitivities of Arabian camel tissues to environmental conditions.
Fig 1. Agarose gel (1.2%) electrophoresis of PCR products for HSPB-1 and β-actin Arabian camel mRNAs, 1500 bp DNA molecular weight marker was used.
Fig 2. Arabian camel HSPB-1 mRNA expression levels in different tissues.
The results are expressed relative to that of β-actin as an endogenous control.
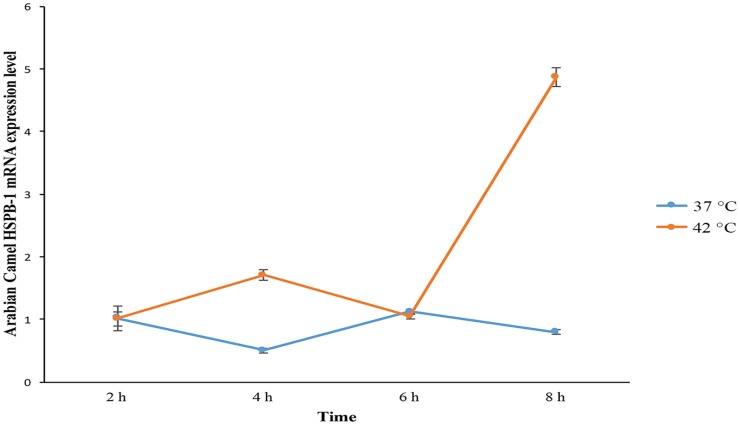
In order to investigate the effect of heat stress(42°C) on the level of expression of Arabian camel HSPB-1 mRNA, we used SACAS cells as a model system by using qRT-PCR. The camel skin fibroblasts were exposed to elevated ambient temperature (42°C) at different time points. The expression of HSPB-1 mRNA, as shown in (Fig 3), was remarkably upregulated in response to the 42°C heat stress after 6h incubation compared with that in the control at 37°C. This result showed that the induction of Arabian camel HSPB-1 mRNA expression depended on the duration and temperature of heat stress.
Fig 3. Arabian camel HSPB-1 mRNA expression levels in SACAS cells at control(37°C) and heat-stressed condition (42°C) for different time points.
The results are expressed relative to that of β-actin as an endogenous control.
Characterization and sequence of the full-length of HSPB-1 cDNA from Arabian camel
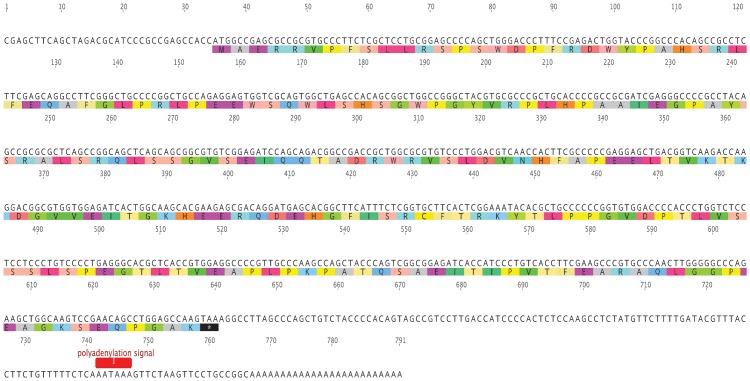
The full-length of Arabian camel HSPB-1 cDNA contained a 5′-untranslated region (UTR) of 34 bp, a 3′-UTR of 151 bp with typical polyadenylation signal (AATAAA), and with a poly(A) tail were obtained and deposited as GenBank accession No. MF278354. The open reading frame (ORF) includes 606 bp and encodes a protein of 201 residues (Fig 4). The sequence indicated a length of 791 bp, and revealed high statically significant similarity scores to many HSPB-1 nucleotide sequences from other species. The Bactrian camel (Camelus bactrianus) showed the highest homology score of 99%, followed by that of alpacas (Vicugna pacos); suggesting a close evolutionary relationship. The other mammals shared a high identity score ranging from 82% to 92%, as shown in (Table 2).
Fig 4. Nucleotide and amino acid sequences of Arabian camel HSPB-1 cDNA (GenBank accession no., MF278354).
The numbers above the nucleotide sequence show the nucleotide positions. The stop codon is represented with an asterisk(*). The putative polyadenylation signal is shown in red.
Table 2. Nucleotide homology of Arabian camel HSPB-1 with that from other species.
| Species | Common name | Accession no. | length (bp) | Identity (%) |
|---|---|---|---|---|
| Homo sapiens | Human | BC000510.2 | 867 | 85 |
| Pan troglodytes | Chimpanzee | XM_519162.5 | 947 | 85 |
| Mus musculus | Mouse | NM_013560.2 | 913 | 82 |
| Sus scrofa | Pig | NM_001007518.1 | 624 | 91 |
| Equus caballus | Horse | XM_001504478.3 | 923 | 88 |
| Bos taurus | Cattle | AB605262.1 | 672 | 92 |
| Camelus dromedarius | Arabian Camel | MF278354 | 791 | 100 |
| Capra hircus | Goat | XM_018040903.1 | 891 | 91 |
| Camelus bactrianus | Bactrian Camel | XM_010972325.1 | 849 | 99 |
| Vicugna pacos | Alpaca | XM_015236804.1 | 820 | 99 |
| Macaca mulatta | Rhesus monkey | NM_001260949.2 | 893 | 85 |
| Ailuropoda melanoleuca | Giant panda | NM_001304892.1 | 633 | 90 |
| Canis lupus familiaris | Dog | NM_001003295.2 | 864 | 87 |
| Macaca fascicularis | Crab-eating macaque | NM_001283885.1 | 845 | 85 |
Identification of Arabian camel HSPB-1 protein by using mass spectrometry
For peptide mass fingerprint mass spectrometry (PMF-MS), the targeted protein band (spot) was manually excised from the gel (S1 Fig) and was subjected to MS analysis. Of the total trypsin-digested peptide mass of Arabian camel HSPB-1 protein, seven peptides, which covered 49% of the entire protein sequence, were hit in NCBIprot database (containing 4114420 sequences) by using the Mascot peptide fingerprint search engine with Arabian camel HSPB-1 protein (accession no. ATJ03466) with a score of 125 and p < 0.05 (Fig 5).
Fig 5. MLDI-TOF MS-derived peptides (red) matched to the sequence of Arabian camel HSPB-1 protein(accession no. ATJ03466).
The mass spectrum revealed several protonated ions [M+H]+ in the peptide fragments. As listed in (Table 3), the ions at 1031.89, 1178.04, 2314.72, 1479.23, 1619.27, 1798.42, and 1831.55 were the seven trypsin digested peptides corresponding to amino acids 21-28, 29-38, 39-57, 58-71, 76-90, 93-108, and 168-184, respectively. As interpreted in (Table 3), the peptide mass profiles were obtained from NCBIprot database search engine, and amino acid sequence of individual peptides were identified from the sequence of Arabian camel HSPB-1 protein from the desired spot of this protein on the SDS-PAGE. The PMF-MS results were also homologous with that in some other animals; the second best matching protein received a score of 102 for Alpaca (accession no. XP_015092290) HSPB-1 protein. The third and fourth best matching proteins were scored with 100 and 80 for Bactrianus camel (accession no. XP_010970627) and pig (accession no. NP_001007519.1) HSPB-1 proteins, respectively.
Table 3. Calculated and observed ions of peptide masses of Arabian camel HSPB-1 protein.
| Amino Acid positions | [M+H]+ | ||
|---|---|---|---|
| Start-End | Observed(m/z) | Calculated(m/z) | Peptide Sequence |
| 21-28 | 1031.89 | 1030.46 | DWYPAHSR |
| 29-38 | 1178.04 | 1176.63 | LFEQAFGLPR |
| 39-57 | 2314.72 | 2313.1 | LPEEWSQWLSHSGWPGYVR |
| 58-71 | 1479.23 | 1477.77 | PLHPAAIEGPAYSR |
| 76-90 | 1619.27 | 1617.8 | QLSSGVSEIQQTADR |
| 93-108 | 1798.42 | 1796.93 | VSLDVNHFAPEELTVK |
| 168-184 | 1831.55 | 1829.95 | PATQSAEITIPVTFEAR |
Characterization of HSPB-1 protein from Arabian camel
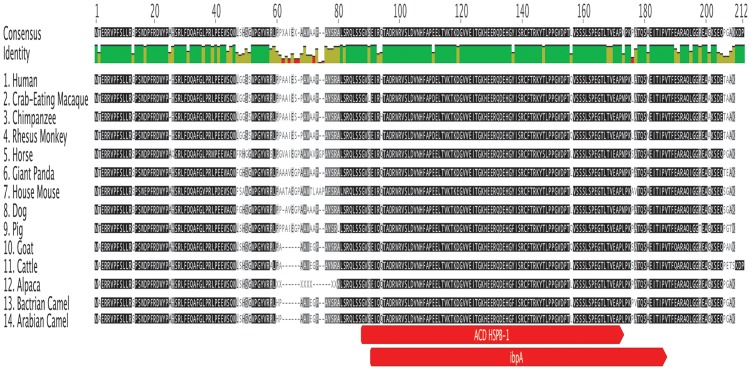
The protein sequence was compared with those of other mammalian HSPB-1 protein sequences by using ClustalW alignment [34], as shown in (Fig 6). Results of multiple sequence alignment of Arabian camel HSPB-1 protein showed two highly conserved domains of about 85 residues (from 88 to 173): ACD and IbpA domains, which were flanked by less conserved NTD and CTE across the species. The comparative analysis of Arabian camel HSPB-1 protein sequence showed high similarity with that of other vertebrates (Table 4). As expected, the highest homology was observed between Arabian camel HSPB-1 protein and the one from Bactrian camel (99%). The other vertebrates showed a high homology ranging from 86% to 95%, as shown in (Table 4). The complete amino acid sequence of Arabian camel HSPB-1 is shown in (Fig 5).
Fig 6. Multiple alignment of amino acid sequence of Arabian camel HSPB-1 protein with that in other 13 mammalian species.
Identical amino acids are marked in green color, and typical ACD and ipbA domains are showed in red.
Table 4. Amino acids homology of Arabian camel HSPB-1 with that in other species.
| Species | Common name | Protein (Accession no.) | Protein length | Identity (%) |
|---|---|---|---|---|
| Homo sapiens | Human | NP_001531 | 205 | 86 |
| Pan troglodytes | Chimpanzee | XP_519162.3 | 205 | 86 |
| Mus musculus | Mouse | NP_038588 | 209 | 86 |
| Sus scrofa | Pig | NP_001007519 | 207 | 92 |
| Equus caballus | Horse | XP_001504528 | 209 | 87 |
| Bos taurus | Cattle | NP_001020740 | 204 | 95 |
| Camelus dromedarius | Arabian Camel | ATJ03466 | 201 | 100 |
| Capra hircus | Goat | XP_017896392 | 201 | 95 |
| Camelus bactrianus | Bactrian Camel | XP_010970627 | 201 | 99 |
| Vicugna pacos | Alpaca | XP_015092290 | 197 | 94 |
| Macaca mulatta | Rhesus monkey | NP_001247878.1 | 205 | 86 |
| Ailuropoda melanoleuca | Giant panda | NP_001291821.1 | 207 | 90 |
| Canis lupus familiaris | Dog | NP_001003295.2 | 206 | 89 |
| Macaca fascicularis | Crab-eating macaque | NP_001270814.1 | 205 | 86 |
Considering the amino acid composition, the average isoelectric point (pI) for Arabian camel HSPB-1 protein calculated using a computer algorithm [35] was found to be 6.162 (S2 Fig), and its estimated molecular weight was 22.382 kDa. The basic, acidic, charged, polar, and hydrophobic amino acids were 22 (10.95%), 25 (12.44%), 58 (28.86%), 47 (23.38%) and 65 (32.34%), respectively. The hydrophobic and aromatic amino acids are overrepresented in the NTD, whereas polar and charged ones are underrepresented [36]. The instability of Arabian camel HSPB-1 protein was calculated to be 64.99, and hence this protein was classified as unstable. The molar extinction coefficient was found to be 39085±5% cm−1 M−1. The amino acids composition of Arabian camel HSPB-1 protein is shown in (S1 Table).
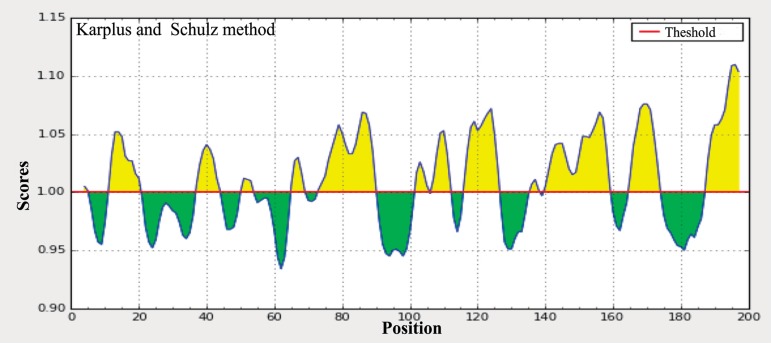
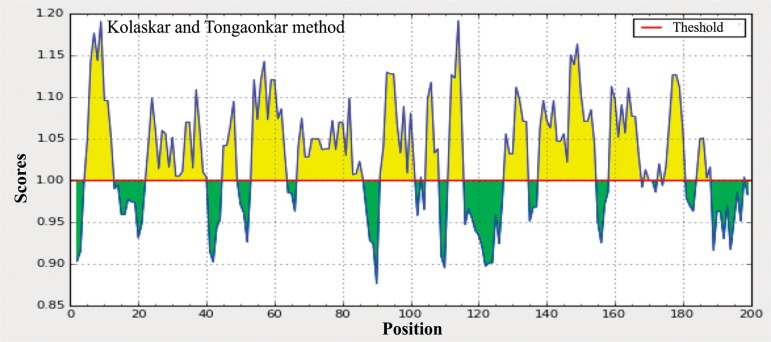
The protein structural flexibility was predicted from the amino acid sequence of Arabian camel HSPB-1 protein by using the Karplus and Schulz method [32], in which the size of window was optimized to 7 residues (Fig 7). The flexcibility analysis showed that Arabian camel HSPB-1 protein was more flexible at its C-terminal than at the N-terminal regions, and thus possibly also the surface amino acids in this protein might be considered as epitopes. In addition, Arabian camel HSPB-1 protein sequence was used as a query to identify B cell epitopes by using the Kolaskar and Tongaonkar antigenicity method [31] (Fig 8). The results showed that the average antigenic tendency value was 1.027 for the protein, with the minimum value of 0.876 and maximum of 1.192. This protein harbors nine antigenic peptides, the lengths of which range from 6 to 20 amino acids (S2 Table). The results also revealed that the two regions from 8 to 11 and 113 to 116 amino acids were the most preferred B cell epitope characteristics.
Fig 7. Karplus and Schulz flexibility prediction of Arabian camel HSPB-1 protein.
The x-axis and y-axis represent the position and score, respectively. The threshold is 1.0. The flexible regions of the protein are shown in yellow color, above the threshold value.
Fig 8. Kolashkar and Tongaonkar antigenicity prediction of the most antigenic regions of Arabian camel HSPB-1 protein.
The threshold value is 1.0. The regions above the threshold are antigenic, shown in yellow.
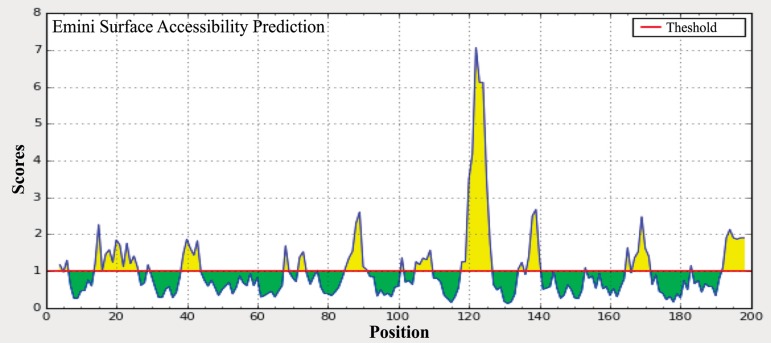
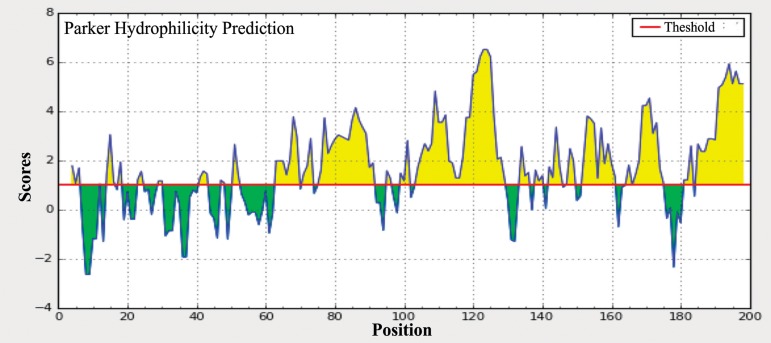
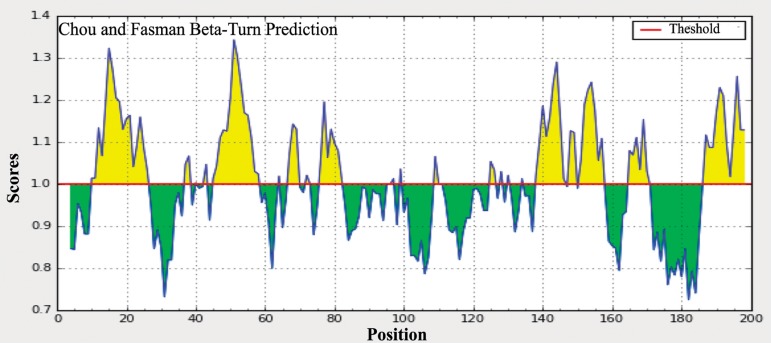
The amino acid sites that are located on the surface of Arabian camel HSPB-1 protein were predicted using the Parker hydrophilicity tool [37] and the Emini surface accessibility prediction [38]. Those sites might increase the probability of predicting the antigenic regions since they are more accessible and hydrophilic than the interior regions of the protein. The maximum surface probability value was found to be 5.823 from amino acid position 121 to 126 for Arabian camel HSPB-1 protein (Figs 9 and 10). In addition, β-turns structure in a protein are mostly hydrophilic and surface accessible in nature. The β-turns were also predicted in Arabian camel HSPB-1 protein by using Chou and Fasman Beta turn prediction [39]. The results suggested that this protein is rich in β-turns in the region between 80 to 175 residues, which is the region where β-strands are oriented in anti-parallel to form β-sheets (Fig 11).
Fig 9. Emini surface accessibility prediction of Arabian camel HSPB-1 protein.
The threshold value is 1.000. The regions above the threshold are antigenic and are shown in yellow.
Fig 10. Parker hydrophilicity prediction of Arabian camel HSPB-1 protein.
The threshold is 1.0. The regions having β-turns in the protein are shown in yellow color, above the threshold value.
Fig 11. Chou and Fasman β-turns prediction of Arabian camel HSPB-1 protein.
The threshold is 1.00. The regions having β-turns in the protein are shown in yellow color.
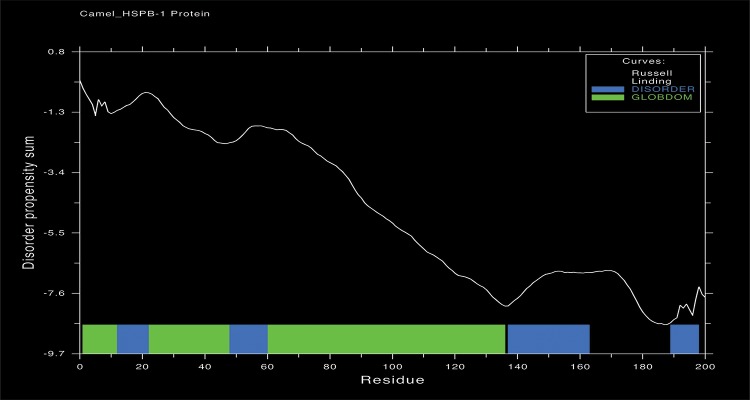
GlobPlot server (http://globplot.embl.deis) was used in order to predict the disordered and ordered (globular) regions within Arabian camel HSPB-1 protein. In this program, ordered regions are described as those have regular secondary structure (α-helices and β-strands), whereas disordered ones are that lack such structures. The Russell/Linding [40] set was selected, in which α-helices and β-strands structures are assigned as globular regions (GlobDoms), whereas random coils and β-turns structures as disordered regions. Residue ranges for the disordered regions (blue) and globular regions (green) are shown at the bottom of the graph (Fig 12).
Fig 12. Glob Plot analysis.
Blue boxes are disordered regions, and green boxes are ordered regions in the Arabian camel HSPB-1 protein.
Phylogeny and classification of Arabian camel HSPB-1 protein
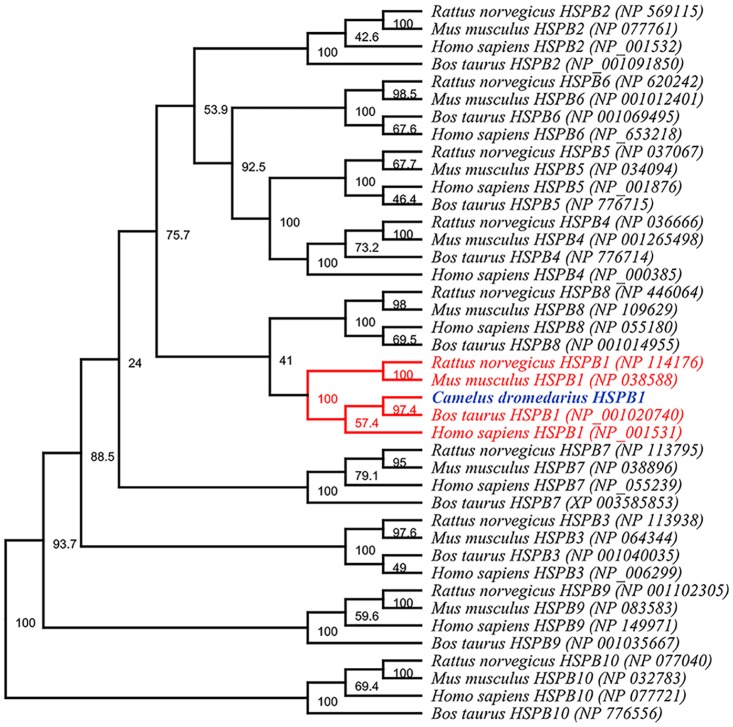
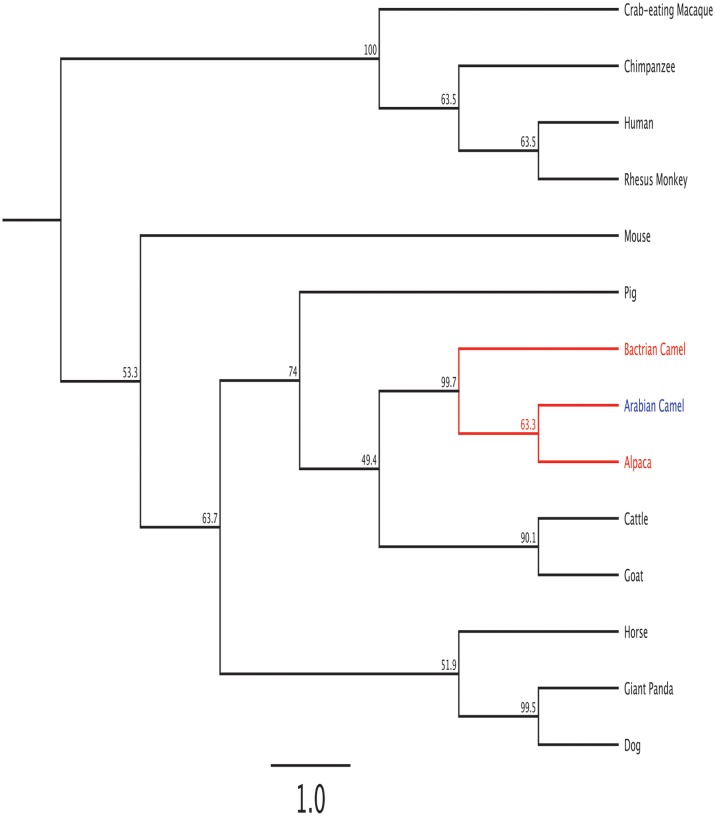
After confirming the relationship of Arabian camel HSPB-1 protein to the HSPB-1 family, we constructed phylogenetic trees by using the Arabian camel HSPB-1 protein sequence as a query to retrieve 40 orthologues sequences derived from different vertebrate species (S3 Table). the NJ phylogenetic trees were constructed based on the multiple alignment of the HSPB-1 protein sequences (Fig 13). The depicted topology showed that the Arabian camel HSPB-1 clustered closely with even-toed ungulates’ HSPB-1 into two distinct clades. In addition, the evolutionary position of Arabian camel was shown in a phylogenetic tree (Fig 14). The Arabian camel HSPB-1 was grouped more closely with the Bactrian camel, alpaca from cattle,goat and further related with pig.
Fig 13. Phylogenetic tree shows the classification of Arabian camel HSPB-1 within the sHSPB family.
Fig 14. The phylogenetic tree shows the relationship of camel HSPB-1 protein and protein sequences from other species.
Maximum likelihood tree based on complete coding sequences. Values at nodes are bootstrapping ≥ 49% obtained from 1000 resampling of the data.
Secondary and 3D structures of Arabian camel HSPB-1 protein
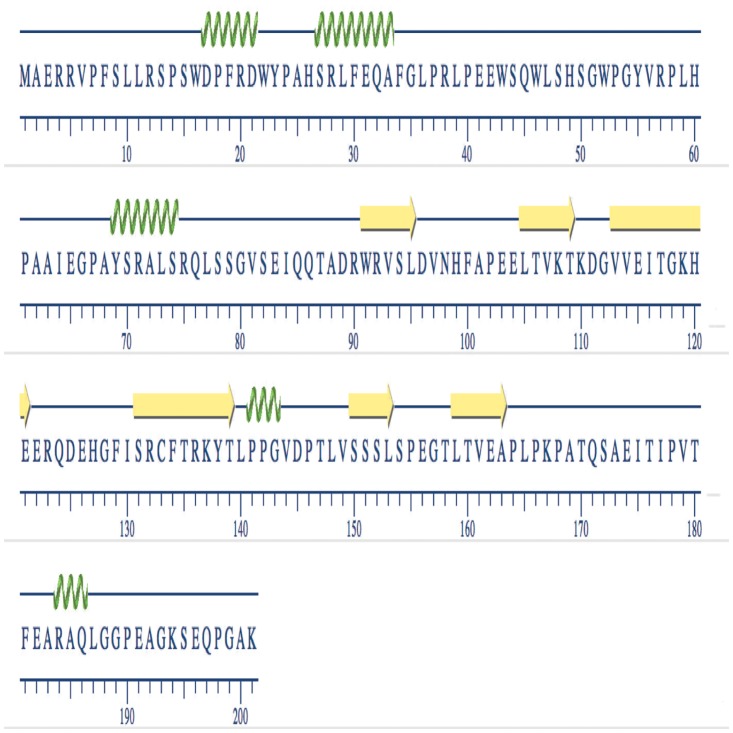
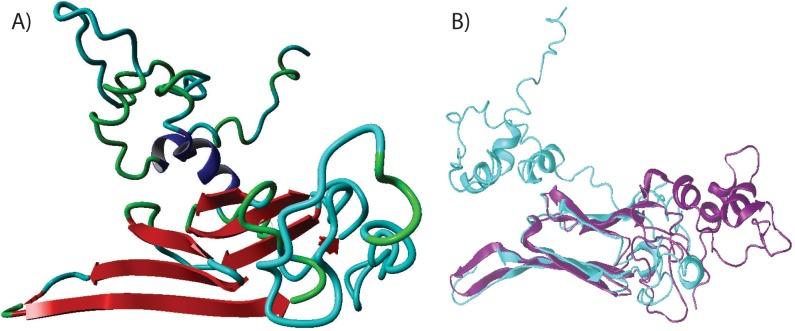
The primary structure of Arabian camel HSPB-1 protein was used to predict its secondary structure, which shows the first level of protein folding. The predicted structure suggested that Arabian camel HSPB-1 protein composed of 5 α-helices and 6 β-strands (Fig 15) in which the 6 β-strands forms a highly conserved ACD of approximately 85 residues (from 88 to 173), which is flanked by less conserved NTD and CTE. The 3D structure of this domain forms an immunoglobulin-like β-sandwich fold in the C-terminal half of the Arabian camel HSPB-1 protein (Fig 16A). The ACD domain mediates the formation Arabian camel HSPB-1 dimers via the anti-parallel pairing of the same β-strand from two monomers.
Fig 15. The secondary structure of Arabian camel HSPB-1 protein.
Fig 16. Modeled 3D structures.
(A) The 3D structure of Arabian Camel HSPB-1 protein. (B) Stereo ribbon representation of the predicted 3D structure model of Arabian camel HSPB-1 (cyan) and the superimposition with Homo sapiens α-B-crystallin chain V (purple).
To construct the 3D structural model of Arabian camel HSPB-1 protein, we generated its homology model by using Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) (Fig 16A). In this study, we used Homo sapiens α-B-crystallin chain V (PDB ID: 2YGD) [41] as a template in which 86% of amino acid residues were modeled at > 90% confidence. The 3D structural model consisted of 5 α-helices and 6 β-strands. The ACD region is folded into a compact of 6-anti-parallel-strands forming two β-sheets. It has a very similar fold and topology as those from human. The structural similarity of Arabian camel HSPB-1 with human HSPB-1 was examined by superimposing their structures by using the Pymol program (https://www.pymol.org/) (Fig 16B). The root mean square deviation between Arabian camel HSPB-1 and human α-B-crystallin chain V structures was 3.134. The Q-score is another crucial parameter to assess the similarity of the homologous structures; it represented that the quality of recognition and superimposition was 0.8435, indicating high structural identity. The Z- and P-scores of the 3D structure of Arabian camel HSPB-1 and human HSPB-1 were 44.6 and 65.9, respectively.
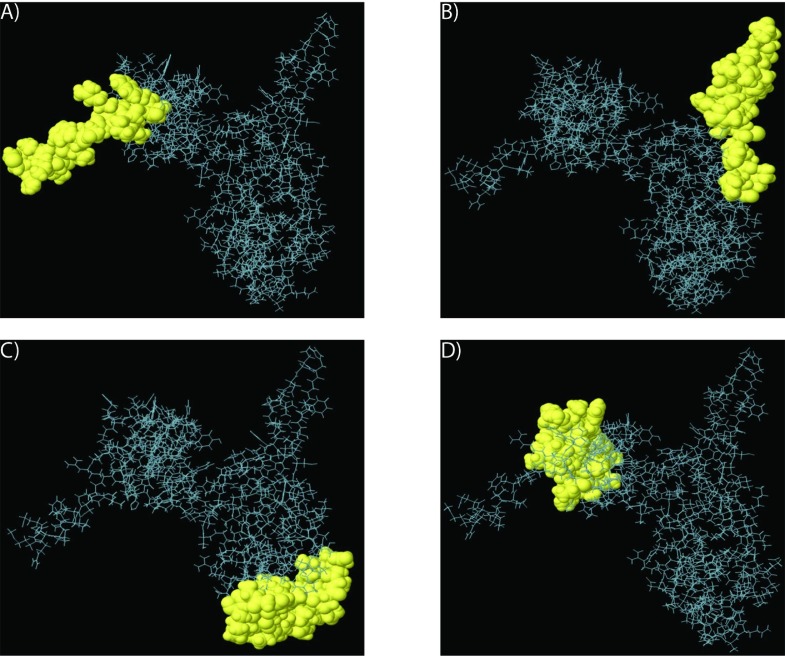
The epitope regions of Arabian camel HSPB-1 protein based on its 3D structure were predicted using Ellipro server (http://tools.iedb.org/ellipro/). Four discontinuous peptides were identified having score value of > 0.7. The highest probability of a discontinuous epitope was computed as 78.5%. Amino acids involved in discontinuous epitopes, their sequence location, number of amino acids, and scores are shown in (Table 5), whereas their positions on 3D structure of Arabian camel HSPB-1 protein are shown in (Fig 17).
Table 5. Predicted discontinous antigenic epitopes of Arabian camel HSPB-1 protein.
| Start | End | Peptide | Peptide Length | Score | 3D Structure |
|---|---|---|---|---|---|
| 1 | 17 | MAERRVPFSLLRSPSWD | 17 | 0.785 | A |
| 119 | 137 | KHEERQDEHGFISRCFTRK | 19 | 0.753 | B |
| 166 | 201 | PKPATQSAEITIPVTFEARAQLGGPEAGKSEQPGAK | 36 | 0.72 | C |
| 33 | 58 | AFGLPRLPEEWSQWLSHSGWPGYVRP | 26 | 0.706 | D |
Fig 17. 3D representation of discontinues epitopes (A to D) of Arabian camel HSPB-1.
The epitopes are shown in yellow surface, and bulk of Arabian camel HSPB-1 protein is shown in grey sticks.
Discussion
The sHSPs help maintain protein homeostasis by interplaying with unfolded substrates to prevent cellular damage [16]. The ATP-independent chaperone HSPB-1 protein is a typical example. The HSPB-1 protein is expressed in several tissue types under stress-induced conditions, where it serves as a chaperone for partly folded cellular proteins. Thus, a full structural description of Arabian camel HSPB-1 gene is an important step toward understanding its mode of action.
The molecular characterization of Arabian camel HSPB-1 gene is crucial for realizing the effect of exposure to different environmental factors on the health position of this animal. The study focused on the molecular characterization of sHSP family, mainly the HSPB-1 protein from C. dromedarius. We cloned Arabian camel HSPB-1 cDNA(791 bp) by using specific primers spanning the entire ORF, encoding 201 amino acids for the protein with size of 22.382 kDa; highly matches with several HSPB-1 protein sequences from other species were found (Table 2). Arabian camel HSPB-1 cDNA sequence was matched with other 13 other mammalian HSPB-1 sequences in GenBank and submitted in the NCBI database with the accession number MF278354.
Our findings suggest that Arabian camel HSPB-1 mRNA is highly expressed in esophagus, skin, and heart, followed by nearly equally expressed in liver, kidney, testis, and lung, whereas the brain, spleen and stomach tissues showed the lowest levels of Arabian camel HSPB-1 mRNAs, as indicated by qRT-PCR analysis (Fig 2). Heat-shock response can be determined by transcriptional activation of HSP genes and accumulation of their proteins [42, 43]. We utilized Arabian camel skin cell line as a model to investigate heat-stress responses. The effect of heat stress on the expression level of Arabian camel HSPB-1 mRNA was examined by thermal stressing the cells at 42°C during an 8 h time course (Fig 3). As can be seen in (Fig 3), the expression level of Arabian camel HSPB-1 mRNA increased after 6 h time course after 42°C heat stress, indicating that Arabian camel HSPB-1 mRNA expression depends on time and temperature exposure. Further, heat-induced HSP27 expression was shown to mainly dependent on the time and temperature of exposure in skin and lung tissues of rats [44].
Two highly conserved domains are present in Arabian camel HSPB-1 protein: ACD and IbpA domains that are localized near the C-terminal end of the protein. Multiple sequence alignment of Arabian camel HSPB-1 protein (Fig 6) showed a highly conserved ACD of approximately 85 residues (from 88 to 173), which was flanked by less conserved NTD and CTE across the species. The NTD is very diverse in protein sequence and is therefore largely responsible for the sequence variation of HSPB-1 protein between organisms. The amino acids such as Trp(W), Phe(F), and Pro(P) are overrepresented in the NTD. However, the CTE contains a highly conserved IXI motif, which is thought to be important for inter-dimer contacts [45].
In general, mammalian HSPB-1 proteins exist as polydispersed oligomeric population, and their full crystal structures have not yet been determined. Nevertheless, the crystal structure of ACD of HSPB-1 protein indicates a β-sheet rich immunoglobulin-like fold. The 3D structure of a protein provides useful information regarding its function. Comparative modeling is possible to predict the 3D structure of a protein based only on its primary structure. Therefore, we predicted the 3D structure of Arabian camel HSPB-1 protein, of which the amino acid sequence is known. The 3D structure of Arabian camel ACD domain forms an immunoglobulin-like β-sandwich fold in the C-terminal half of the protein (Fig 16A). The ACD mediates the formation Arabian camel HSPB-1 dimers via the anti-parallel pairing of the same β-strand from two monomers.
Supporting information
(EPS)
(EPS)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Mohammed Aljohi for his help in completing this project. We would also like to thank Faisal Alagrafi for his assistance in tissue sample collection and Amer Alharthi for his technical support. All authors extend their appreciation to the anonymous referees and editors for their useful suggestions.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work is supported by KACST grant 20-0078 to Manee from the Center of Excellence for Genomics (CEG), King Abdulaziz City for Science and Technology (KACST). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fowler ME. Medicine and surgery of camelids. John Wiley & Sons; 2011. [Google Scholar]
- 2. Monari M, Foschi J, Rosmini R, Marin MG, Serrazanetti GP. Heat shock protein 70 response to physical and chemical stress in Chamelea gallina. Journal of Experimental Marine Biology and Ecology. 2011;397(2):71–78. Available from: http://www.sciencedirect.com/science/article/pii/S0022098110004648. [Google Scholar]
- 3. Wu H, Guang X, Al-Fageeh M, Cao J, Pan S, Zhou H, et al. Camelid genomes reveal evolution and adaptation to desert environments. Nature Communications. 2014;5 doi: 10.1038/ncomms6188 [DOI] [PubMed] [Google Scholar]
- 4. Saeed H, Shalaby M, Embaby A, Ismael M, Pathan A, Ataya F, et al. The Arabian camel Camelus dromedarius heat shock protein 90α: cDNA cloning, characterization and expression. International Journal of Biological Macromolecules. 2015;81:195–204. doi: 10.1016/j.ijbiomac.2015.07.058 [DOI] [PubMed] [Google Scholar]
- 5. Elrobh MS, Alanazi MS, Khan W, Abduljaleel Z, Al-Amri A, Bazzi MD. Molecular Cloning and Characterization of cDNA Encoding a Putative Stress-Induced Heat-Shock Protein from Camelus dromedarius. International Journal of Molecular Sciences. 2011;12(12):4214–4236. doi: 10.3390/ijms12074214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu Q, Schett G, Li C, Hu Y, Wick G. Mechanical stress–induced heat shock protein 70 expression in vascular smooth muscle cells is regulated by Rac and Ras small G proteins but not mitogen-activated protein kinases. Circulation Research. 2000;86(11):1122–1128. doi: 10.1161/01.RES.86.11.1122 [DOI] [PubMed] [Google Scholar]
- 7. Bakthisaran R, Tangirala R, Rao CM. Small heat shock proteins: Role in cellular functions and pathology. Biochimica et Biophysica Acta (BBA)—Proteins and Proteomics. 2015;1854(4):291–319. Available from: http://www.sciencedirect.com/science/article/pii/S1570963914003355. [DOI] [PubMed] [Google Scholar]
- 8. Waters ER, Lee GJ, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. Journal of Experimental Botany. 1996;47(3):325–338. doi: 10.1093/jxb/47.3.325 [Google Scholar]
- 9. de Jong WW, Leunissen JA, Voorter CE. Evolution of the alpha-crystallin/small heat-shock protein family. Molecular biology and evolution. 1993;10(1):103–126. [DOI] [PubMed] [Google Scholar]
- 10. Sangster TA, Bahrami A, Wilczek A, Watanabe E, Schellenberg K, McLellan C, et al. Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PloS one. 2007;2(7):e648 doi: 10.1371/journal.pone.0000648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carretero-Paulet L, Albert VA, Fares MA. Molecular evolutionary mechanisms driving functional diversification of the HSP90A family of heat shock proteins in eukaryotes. Molecular biology and evolution. 2013;30(9):2035–2043. doi: 10.1093/molbev/mst113 [DOI] [PubMed] [Google Scholar]
- 12. Baringou S, Rouault JD, Koken M, Hardivillier Y, Hurtado L, Leignel V. Diversity of cytosolic HSP70 Heat Shock Protein from decapods and their phylogenetic placement within Arthropoda. Gene. 2016;591(1):97–107. doi: 10.1016/j.gene.2016.06.061 [DOI] [PubMed] [Google Scholar]
- 13. Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cellular and Molecular Life Sciences CMLS. 2006;63(22):2560–2570. doi: 10.1007/s00018-006-6192-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Waters ER. The molecular evolution of the small heat-shock proteins in plants. Genetics. 1995;141(2):785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kappé G, Leunissen JA, de Jong WW. Evolution and diversity of prokaryotic small heat shock proteins In: Small stress proteins. Springer; 2002. p. 1–17. [DOI] [PubMed] [Google Scholar]
- 16. Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nature structural & molecular biology. 2005;12(10):842–846. doi: 10.1038/nsmb993 [DOI] [PubMed] [Google Scholar]
- 17. Kappé G, Franck E, Verschuure P, Boelens WC, Leunissen JAM, de Jong WW. The human genome encodes 10 α-crystallin–related small heat shock proteins: HspB1–10. Cell Stress & Chaperones. 2003;8(1):53 doi: 10.1379/1466-1268(2003)8%3C53:THGECS%3E2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caspers GJ, Leunissen JA, de Jong WW. The expanding small heat-shock protein family, and structure predictions of the conserved “α-crystallin domain”. Journal of molecular evolution. 1995;40(3):238–248. doi: 10.1007/BF00163229 [DOI] [PubMed] [Google Scholar]
- 19. de Jong WW, Caspers GJ, Leunissen JA. Genealogy of the α-crystallin—small heat-shock protein superfamily. International journal of biological macromolecules. 1998;22(3):151–162. doi: 10.1016/S0141-8130(98)00013-0 [DOI] [PubMed] [Google Scholar]
- 20. Fu X, Jiao W, Chang Z. Phylogenetic and biochemical studies reveal a potential evolutionary origin of small heat shock proteins of animals from bacterial class A. Journal of molecular evolution. 2006;62(3):257–266. doi: 10.1007/s00239-005-0076-5 [DOI] [PubMed] [Google Scholar]
- 21. Sudnitsyna MV, Mymrikov EV, Seit-Nebi AS, Gusev NB. The role of intrinsically disordered regions in the structure and functioning of small heat shock proteins. Current protein & peptide science. 2012;13(1):76–85. doi: 10.2174/138920312799277875 [DOI] [PubMed] [Google Scholar]
- 22. Zhang K, Ezemaduka AN, Wang Z, Hu H, Shi X, Liu C, et al. A Novel Mechanism for Small Heat Shock Proteins to Function as Molecular Chaperones. Scientific Reports. 2015;5:8811 doi: 10.1038/srep08811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bepperling A, Alte F, Kriehuber T, Braun N, Weinkauf S, Groll M, et al. Alternative bacterial two-component small heat shock protein systems. Proceedings of the National Academy of Sciences. 2012;109(50):20407–20412. doi: 10.1073/pnas.1209565109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alawad A, Alhazzaa O, Alkhrayef M, Alagrafi F, Alhamdan Z, Alenazi A, et al. Sacasg and sacalg: new GFP-labelled camel skin and lung fibroblast cell lines. Journal of Camel Practice and Research. 2016;23(1):73–80. [Google Scholar]
- 25. Ma H, Young J, Shen C. RNA, DNA and protein isolation using Trizol reagent. Nature and Science. 2008;6:66–75. [Google Scholar]
- 26. Hageman J, Kampinga HH. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress & Chaperones. 2009;14(1):1–21. doi: 10.1007/s12192-008-0060-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thompson J. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular biology and evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kolaskar A, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS letters. 1990;276(1-2):172–174. doi: 10.1016/0014-5793(90)80535-Q [DOI] [PubMed] [Google Scholar]
- 32. Karplus P, Schulz G. Prediction of chain flexibility in proteins. Naturwissenschaften. 1985;72(4):212–213. doi: 10.1007/BF01195768 [Google Scholar]
- 33. Sodhi M, Kishore A, Sharma A, Shandilya U, Kumari P, Mukesh M. Differential expression of heat shock proteins in tissues of riverine buffaloes. The Indian Journal of Animal Sciences. 2015;85(4). [Google Scholar]
- 34. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947 Available from: http://dx.doi.org/10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35. Kozlowski LP. IPC–Isoelectric Point Calculator. Biology direct. 2016;11(1):55 doi: 10.1186/s13062-016-0159-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kriehuber T, Rattei T, Weinmaier T, Bepperling A, Haslbeck M, Buchner J. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. The FASEB Journal. 2010;24(10):3633–3642. doi: 10.1096/fj.10-156992 [DOI] [PubMed] [Google Scholar]
- 37. Parker J, Guo D, Hodges R. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25(19):5425–5432. doi: 10.1021/bi00367a013 [DOI] [PubMed] [Google Scholar]
- 38. Emini EA, Hughes JV, Perlow D, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. Journal of virology. 1985;55(3):836–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chow P, Fasman G. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol. 1978;47:45–148. [DOI] [PubMed] [Google Scholar]
- 40. Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11(11):1453–1459. doi: 10.1016/j.str.2003.10.002 [DOI] [PubMed] [Google Scholar]
- 41. Braun N, Zacharias M, Peschek J, Kastenmüller A, Zou J, Hanzlik M, et al. Multiple molecular architectures of the eye lens chaperone αB-crystallin elucidated by a triple hybrid approach. Proceedings of the National Academy of Sciences. 2011;108(51):20491–20496. Available from: http://www.pnas.org/content/108/51/20491.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lindquist S. The heat-shock response. Annual review of biochemistry. 1986;55(1):1151–1191. doi: 10.1146/annurev.bi.55.070186.005443 [DOI] [PubMed] [Google Scholar]
- 43. Thayyullathil F, Chathoth S, Hago A, Wernery U, Patel M, Galadari S. Investigation of heat stress response in the camel fibroblast cell line Dubca. Annals of the New York Academy of Sciences. 2008;1138(1):376–384. doi: 10.1196/annals.1414.039 [DOI] [PubMed] [Google Scholar]
- 44. Blake M, Gershon D, Fargnoli J, Holbrook N. Discordant expression of heat shock protein mRNAs in tissues of heat-stressed rats. Journal of Biological Chemistry. 1990;265(25):15275–15279. [PubMed] [Google Scholar]
- 45. Hilton GR, Lioe H, Stengel F, Baldwin AJ, Benesch JL. Small heat-shock proteins: paramedics of the cell In: Molecular Chaperones. Springer; 2012. p. 69–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(EPS)
(EPS)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.