Abstract
Microbial rhodopsins are a family of photoactive retinylidene proteins widespread throughout the microbial world. They are notable for their diversity of function, using variations of a shared seven-transmembrane helix design and similar photochemical reactions to carry out distinctly different light-driven energy and sensory transduction processes. Their study has contributed to our understanding of how evolution modifies protein scaffolds to create new protein chemistry, and their use as tools to control membrane potential with light is fundamental to the transformative technology of optogenetics. We review the currently known functions, and present more in-depth assessment of three functionally and structurally distinct types discovered over the past two years: (i) anion-conducting channelrhodopsins (ACRs) from cryptophyte algae, enabling efficient optogenetic neural suppression, (ii) cryptophyte cation-conducting channelrhodopsins (CCRs), structurally distinct from the green algae CCRs used extensively for neural activation, and (iii) enzymerhodopsins, with light-gated guanylylcyclase or kinase activity promising for optogenetic control of signal transduction.
Keywords: retinylidene proteins, ion pumps, photosensors, channelrhodopsins, optogenetics
INTRODUCTION
Microbial Rhodopsins in Nature
The microbial rhodopsin family is comprised of >7000 photochemically reactive proteins in prokaryotes and lower eukaryotes found throughout the oceans from tropical to arctic, lakes and rivers, soil, and on the leaf surfaces of plants (Figure 1). Family members share a membrane-embedded seven-helix architecture forming an internal pocket for the chromophore retinal bound in a protonated Schiff base linkage to the ε-amino group of a lysyl residue in the middle of the 7th helix. Microbial rhodopsins provide a vivid example of evolution modifying a single protein scaffold to produce diverse new chemical functions. Photochemical reactions energized by photoisomerization of the retinylidene chromophore drive distinctly different processes in different microbial rhodopsins. Their biological functions fall into two categories: (1) photoenergy transducers that convert light into electrochemical potential to energize cells, namely light-driven ion pumps catalyzing outward active transport of protons, inward chloride transport, and outward sodium transport; (2) photosensory receptors that use light to gain information about the environment to regulate cell processes (Figure 2). Known modes of microbial sensory rhodopsin signaling are protein-protein interaction with membrane-embedded transducers, interaction of their cytoplasmic domain with soluble transducer proteins, enzymatic activity encoded in their cytoplasmic domain, and signaling by light-gated passive ion channel conduction.
Figure 1.
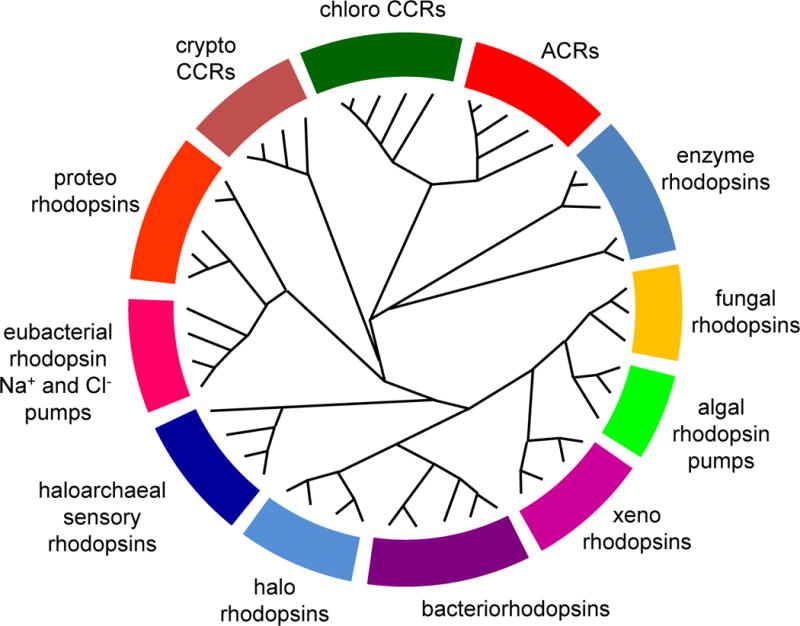
A cladogram of the microbial rhodopsin superfamily. For a list of sequences, accession numbers and source organisms see Supplementary Table 1.
Figure 2.
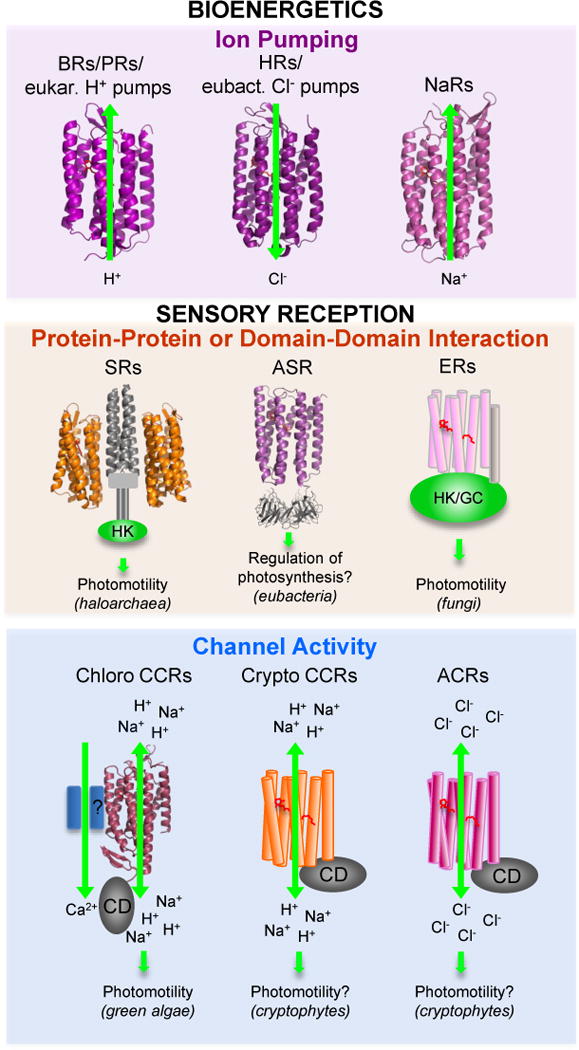
Functional types of microbial rhodopsins. For molecules shown as ribbons high-resolution crystal structures have been obtained. Abbreviations: BRs, bacteriorhodopsins; PRs, proteorhodopsins; HRs, halorhodopsins; NaRs, Na+-pumping rhodopsins; SRs, sensory rhodopsins; ASR, Anabaena sensory rhodopsin; ER, enzymerhodopsins; CCRs, cation channelrhodopsins; ACRs, anion channelrhodopsins; eukar., eukaryotic; eubact., eubacterial; HK, histidine kinase; GC, gunanylyl cyclase; CD, cytoplasmic domain.
The microbial rhodopsins are so named because of their structural similarity to animal visual pigments, such as mammalian rod rhodopsin, which also consist of seven transmembrane helices forming an interior protonated retinylidene Schiff base chromophore also linked to a lysyl residue in the 7th helix. For both the microbial and animal rhodopsins, their apoproteins are referred to as “opsins”, and when complexed with the retinal moiety, “rhodopsins”. Microbial rhodopsins and animal rhodopsins exhibit mechanistic as well as structural similarities, but no sequence homology. Based on their distinctly different phylogeny they have been designated type 1 and type 2 rhodopsins, respectively (1). The question of whether types 1 and 2 derive from convergent evolution or diverged from a common seven-helix retinylidene ancestor is still unresolved, but the latter possibility has recently received new support (2).
Use as Optogenetics Tools
Since their discovery in the 1970s and 1980s, the temporal and spatial precision available from using light as a stimulus, and the convenience of having a natural spectroscopic reporter group in the photoactive site, i.e. the retinylidene chromophore, have enabled microbial rhodopsins to contribute substantially to our understanding of membrane protein structure/function, photochemistry, bioenergetics, sensory signaling, protein evolution, and the diversity of modes of interaction of organisms with light. One of their most significant contributions is in laying the chemical foundation for the new biotechnology of optogenetics. Optogenetics, an approach that uses light to control cell membrane potential in neurons and other excitable cells, has revolutionized neuroscience research, especially studies of brain function (3–4). The chemical basis of optogenetics is genetically targeted expression of microbial rhodopsins, whose photochemical reactions enable precise spatial and temporal photocontrol of transmembrane ion currents to regulate neuronal action potentials. This new technology has transformed the study of neural circuitry in flies, worms, rodents, and other animal models and has greatly accelerated the pace of discoveries in brain functions. Phototaxis receptors from algae with light-gated channel activity (channelrhodopsins) have been the most important contributors to optogenetics. It is worthy of note that the development of optogenetics is a beautiful example of a revolutionary biotechnology growing out of purely basic research, in this case research primarily on the chemical mechanisms of phototaxis reception by microorganisms.
Scope of the Review
Comprehensive reviews have appeared on retinylidene proteins in general (both types 1 and 2) (1, 5), and on various aspects of particular type 1 rhodopsins ((6–11), and other references cited in specific sections below). This focused review will first briefly cover all of the known molecular functions of microbial rhodopsins including history of their discovery and particularly, in some cases, uniquely interesting aspects of their properties that have advanced our understanding of photobiochemistry and photobiology.
Second, we present more in-depth review of type 1 rhodopsins with new functions discovered in the past two years in this rapidly moving field, namely: (i) anion-conducting channelrhodopsins (ACRs) from cryptophyte algae, notable for their unparalleled efficiency of hyperpolarization and silencing of neural firing; (ii) CCRs from cryptophyte algae, structurally distinct from the green algae CCRs and closely related to haloarchaeal proton pumps, an example of convergent evolution of channel function via two independent paths; and (iii) enzymerhodopsins, microbial rhodopsins with a catalytic domain, specifically in the best understood case a light-gated guanylylcyclase activity used by a fungus for phototaxis, and promising for optogenetic control of cGMP (cyclic guanosine monophosphate) signaling processes.
THE KNOWN MOLECULAR FUNCTIONS OF MICROBIAL RHODOPSINS
Light Energy Capture by Light-Driven Ion Pumps
Proton Pumps
Bacteriorhodopsin (BR)
In the late 1960s Walther Stoeckenius was interested in electron microscopy of the archaeal organism Halobacterium salinarum (at that time classified as a bacterium) because of reports that its cytoplasmic membrane may have a subunit structure. It turned out that its unusual surface structure was due to 2D-crystalline arrays of a then unknown protein pigment forming purple patches in the cytoplasmic membrane. In the early 1970s Oesterhelt and Stoeckenius discovered that purple membrane contained a retinylidene proton pump that they named bacteriorhodopsin (BR) (12). Like visual pigments BR consisted of an apoprotein that formed a pigment with visible absorption upon binding retinal. Within a few years BR gained great importance as a simple single-polypeptide primary transporter obtainable in a stable concentrated form amenable to optical and molecular spectroscopic measurements and crystallography. Indeed it became the first protein in which transmembrane alpha helices were directly observed in a pioneering application of cryo-electron crystallography (13). Close relatives of BR were found in other haloarchaea, such as archaerhodopsin-3 (also known as AR-3 or Arch), which shows more promiscuous expression in heterologous systems than BR and has found use as a tool for neural photosuppression in optogenetics (14).
Proteorhodopsins (PRs)
Proteorhodopsins from proteobacteria, the largest sub-family of microbial rhodopsins, are light-driven proton pumps with the characteristic carboxylate proton acceptors and donors (Asp-85 and Asp-96 in BR) characteristic of haloarchaeal proton pumps. The first PR was discovered by environmental sequencing of Pacific coastal waters (15) followed by several more from Hawaiian surface and deep ocean samples (16). Now thousands of PR genes have been identified in essentially all of the earth’s oceans by shotgun sequencing (17). Estimated from Mediterranean samples, 13% of the bacterial cells in the photic zone contain a PR gene (18). The measured concentration of PRs in picoplankton (16) indicates that solar energy absorption by PRs on the earth’s surface waters continuously converts light energy into transmembrane proton electrochemical potential at a rate of ~1013 W, roughly equal to the energy consumption rate of fossil fuels by the human population. Remarkably this significant amount of solar energy capture was completely unknown before 2000 (15), when chlorophyll-based photosynthetic systems were the only known source of energy-transducing membranes in the ocean. A property of PRs so far unique to microbial rhodopsins is the finding of oligomeric forms with cross-protomer interactions with the photoactive site of adjacent protomers modulating transport function (19–20).
Other proton pumping rhodopsins
In addition to large numbers of PRs in proteobacteria, there are PR-related rhodopsins in actinobacteria primarily in fresh water lakes (21) and several examples of PR-like variants called xanthorhodopsins, unusual in that they contain carotenoid accessory pigments serving as light-harvesting pigments for energy transfer to the retinylidene chromophore (22). Eukaryotic microorganisms, some fungi and algae, also contain rhodopsin proton pumps (23), although the majority of eukaryotic type 1 rhodopsins so far studied have photosensory function.
Chloride pumps
In 1977 Mukohata and Matsuno-Yagi reported the existence of light-induced proton fluxes in a variant of H. salinarum cells with little or no BR (24) and in 1981 showed that the activity was caused by a distinct pigment they named halorhodopsin (25). In contrast to BR that carries out electrogenic ejection of protons from the cell, HR illumination caused a passive influx of protons indicating membrane hyperpolarization by electrogenic transport of another ion. They and other groups suggested HR was a primary Na+ pump or a BR-like pigment coupled to a H+/Na+ antiporter, but Schobert and Lanyi (26) discovered by light-scattering measurements and ion dependencies of photocurrents in cells that HR was an inwardly directed Cl− pump. A dramatic demonstration of the close relationship of the haloarchaeal HR chloride transport and BR proton-pumping mechanisms was the finding that a single mutation of BR, replacement of its protonated Schiff base proton acceptor Asp85 with threonine, which is in the homologous position in HR, converted BR into a light-driven chloride pump (27). Like the proton pump Arch, HR from Natronomonas pharaonis (NpHR) has been used in many studies as an optogenetic suppressor of neural firing (28).
An inwardly directed chloride-pumping rhodopsin CIR, the primary structure of which shows a phylogenetic lineage very distant from HR, was recently found in a marine bacterium (29–30). UV-vis absorption measurements indicate that the CIR binds Cl− near the retinal chromophore (31) as is known for HR. The common feature of CIR and HR is that the Schiff base remains protonated throughout the pumping cycle whereas Cl− uptake kinetics differs (31). A crystal structure of the CIR from Nonlabens marinus shows greater similarity to the structure of the light-driven Na+ pump KR2 (32) than to those of archaeal ion pumps, consistent with convergent evolution of Cl−-pumping within the archaeal and eubacterial type 1 rhodopsin subfamilies.
A third version of Cl—pumping rhodopsins that combines structural features of BR and HR, but also contains a number of unique residues, was discovered in cyanobacteria (33). In this protein the proton donor residue, Asp96 in BR, is conserved, in contrast to haloarchaeal HRs and CIR. Its functional characterization has just begun, but already suggested a unique mechanism of Cl− transport involving an interplay of Cl− and H+ transfers, significantly different from that in HR (Harris, A., Hughes-Visentin, A., Saita, M., Resler, T., Maia, R., Sellnau, F., Bondar, A.-N., Heberle, J., and Brown, L.S., personal communication).
Sodium pumps
An outwardly-directed sodium ion pumping rhodopsin named KR2 was discovered in 2013 in the marine flavobacterium Krokinobacter (also known as Dokdonia) eikastus (34). It was recognized by light-induced Na+-dependent passive H+ influx upon expression of the corresponding gene in E. coli, as well as by major effects of Na+ on the photochemical reaction cycle of the purified protein. KR2 contains the “NDQ motif” near the retinylidene Schiff base noted in marine eubacteria (35), so named for their contrast with the DTD and DTE motifs in haloarchaeal proton pumps and proteorhodopsins. More than 10 homologs, termed “NaRs”, have been reported in the literature (11) and functional studies in E. coli cells expressing four different NaRs have been conducted (29, 34, 36–37). KR2 was shown to outwardly pump H+ in the absence of Na+ in the medium (34) and its pumping of either ion was shown to involve a BR-like outward displacement of helix 6 during the photocycle (38), indicating a close mechanistic relationship to rhodopsin proton pumps. Nearly all measurements of Na+ and H+ transport by NaRs have been conducted by recording passive and active light-driven proton fluxes, respectively, in live cell suspensions of the native organism or heterologously transformed E. coli. One, IaNaR from Indibacter alkaliphilus (the first two italicized letters indicate the genus and species name of the source organism), has been studied in a purified in vitro unilamellar vesicle system, demonstrating that the dual light-driven H+/Na+ pumping functions are intrinsic to the single rhodopsin protein and providing a system in which ion flux measurements are not influenced by bioenergetics processes in living cells (37).
In the conserved NTQ motif of NaRs versus DTD in BR, Asn112 in KR2 corresponds to D85 in BR, which is the retinylidene Schiff base counterion and acceptor of the Schiff base proton in the BR pumping cycle. BR’s Thr89 is not directly involved in proton transfer, but the corresponding residue Asp116 in KR2 has been shown to be the Schiff base proton acceptor (11). During the photocycle of BR, the proton transfers from the Schiff base to Asp85 on the extracellular side of the protein, and Schiff base reprotonation from Asp96, the third “D” in the DTD motif, from the cytoplasmic side cause vectorial translocation of the proton across the membrane. In contrast, a mechanism of flipping of the proton acceptor has been proposed for Na+ transport based on a KR2 crystal structure (39). In the model, the ionized Asp116 serves as the Schiff base counterion, and during the pumping cycle the protonated Asp116 flips away from the Schiff base opening a space for Na+ transport. Reorientation of Asp116 toward the Schiff base was observed after soaking the crystal, obtained in acidic conditions, in alkaline conditions. In agreement, a simultaneously reported crystal structure in acidic conditions showed Asp116 oriented away from the Schiff base (40). A central role for Asp116 was proposed from the original findings that the mutation D116N red-shifts the pigment 40 nm and blocks light-induced Schiff base deprotonation and ion pumping (34).
Sensory Rhodopsins– Diverse Signaling Mechanisms
Membrane-Embedded Sensory Rhodopsin/Transducer Complexes (SR-Htrs)
Sensory Rhodopsin I (SRI)
Light-modulated swimming behavior (phototaxis in the general sense of the term) is a well-known photosensory response among motile microorganisms. The first phototaxis receptor, which was also the first light-sensing receptor identified in a microorganism, was found in studies of H. salinarum photomotility responses (41–42). Initially called “slow-cycling rhodopsin”, the pigment is now known as sensory rhodopsin I (SRI). The photochemical reactions of SRI (43) differed fundamentally from those of the other microbial rhodopsins known at the time, the ion pumps BR and HR. In the pumps linear unbranched photochemical reaction cycles have been optimized by evolution to be rapid (~10 ms half time) with short-lived intermediates. In SRI a signaling conformer of the protein accumulates as a long-lived (~800 ms) spectrally shifted intermediate in a one-photon photochemical reaction cycle. The signaling conformer is photochemically reactive and is efficiently photoconverted back to the unphotolyzed (“dark”) state in ~70 ms by a second photon excitation of the molecule. This photochromic interplay of 1-photon formation and 2-photon reversion results in color-sensitive signaling enabling color-discriminating phototaxis by the organism (43). The single SRI molecule both guides the cell towards higher intensities of long wavelength light useful for photoenergy capture by its light-driven pumps, while guiding the cell away from near-UV light, minimizing photooxidative damage. Relatively long-lived signaling states are a general property of later discovered sensory rhodopsins. For example, channelrhodopsins exhibit a similar color-discriminating mechanism with similarly slow kinetics enabling the experimenter to control the lifetime of the spectrally shifted signaling conformer (the conductive state) by 1- and 2-photon excitation. The channelrhodopsins therefore can be used in optogenetics as bistable optical switches (44), photoactivated by one color of light and rapidly reset to the dark state by light of a different wavelength.
A methylated membrane protein, HtrI (“halobacterial transducer for SRI”) was identified in the SRI signaling pathway by mutant analysis, partial sequence was obtained from the protein, and its gene cloned (45). The htrI gene was found to be immediately upstream of sopI (“sensory opsin I”), which had been cloned previously. The genes encoding SRII and HtrII are similarly arranged, and SR-Htr bicistronic operons are found also in eubacteria. HtrI’s very close homology to chemotaxis receptors (45) combined with behavioral effects of mutation of chemotaxis signaling components (46), led to the currently accepted signaling pathway from the receptors to the flagellar motor (9).
Sensory Rhodopsin II (SRII)
Takahashi and coworkers proposed the existence of a second phototaxis receptor in H. salinarum based on the action spectrum for repellent responses in highly aerobic conditions in which SRI (and BR) are produced at much lower levels (47). Spectroscopic and biochemical analyses identified the new pigment, which was simultaneously named phoborhodopsin (48) and SRII (49).
The first atomic structures of a sensory rhodopsin, that of SRII (50–51), and with a fragment of its Htr transducer (52), and later that of Anabaena sensory rhodopsin (53), revealed that these sensory rhodopsins are built on the same scaffold as the light-driven proton pump BR with photoactive sites nearly identical to that of BR. SRI is capable of efficient but slow light-driven proton transport, but in its natural state its bound transducer HtrI inhibits its pumping activity (54). Transducer-free SRII is also capable of proton pumping (55). Structural changes caused by HtrII, its cognate transducer, binding to SRII have been identified by mutagenesis, vibrational spectroscopy, and motility behavior studies, after which the elucidation of the chemical requirements for signaling by SRII was sufficiently precise to enable mutagenic conversion of BR into a robust phototaxis receptor, signaling through the SRII transducer with 35% of native SRII efficiency (56).
Functional conversion by mutagenesis of BR into HR function (27), SRI into BR function (54), BR into SRII function (56), and recent studies of interconversions of ion specificity in eubacterial pumps (33, 57) have shed light at the atomic level on how natural selection has modified their common design to create the distinctly different consequences of their photoactivation. Interconversions illustrate that even small changes are capable of modifying existing protein scaffolds to create distinctly different protein chemistry, as recently discussed (58).
Anabaena Sensory Rhodopsin – Membrane to Cytoplasm Signaling
The first sensory rhodopsin found in eubacteria (the cyanobacterium Anabaena, also known as Nostoc) was ASR, so named based on its lack of pumping sequence motifs, and its cotranscription from an operon with a soluble protein (later named ASR transducer, ASRT) that binds to it and alters its photoreactions (59). ASR exhibits photoreactions so far unique among type 1 rhodopsins in that illumination of its all-trans retinylidene chromophore form (with the position of the absorption maximum, λmax, 550 nm for detergent-purified protein) produces a stable spectrally shifted 13-cis-retinal form (λmax 537 nm), which illumination reconverts to the all-trans-retinal form (53, 60). This type of photochromism is analogous to that well known in phytochromes. The physiological function of the ASR-ASRT pair is not fully elucidated. A study in E. coli showed that the ASR-ASRT complex could regulate expression of a reporter gene controlled by an Anabaena phycocyanine promoter (61). More recently, indicating physiological relevance of the E. coli study, biochemical and genetic evidence in Anabaena point to a role of ASRT in chromatic adaptation through regulation of expression of genes encoding components of the phycobilin complex and a circadian clock gene (62). Specific reviews on ASR photochemical studies and physiological function are available (63–64).
Enzymerhodopsins– Kinases and Cyclases in Algae and Fungi
Algal histidine kinase rhodopsins
The microbial rhodopsins discussed above are single-domain proteins. In both CCRs and ACRs discussed in the next sections the rhodopsin domain is linked to a bulky C-terminal extension in which no known functional domains have been recognized. However, the genomes of some algae and fungi encode multidomain proteins, called “enzymerhodopsins”, that comprise an N-terminal rhodopsin domain followed by domains homologous to proteins of two-component signaling systems (histidine kinases and response regulators), and an adenylyl/guanylylcyclase domain (Figure 3). The first members of this class were identified in the genome of the green alga Chlamydomonas reinhardtii (65) that contains at least four such genes encoding histidine kinase rhodopsins (HKRs). HKR sequences have also been found in the genomes of several other algae (66).
Figure 3.
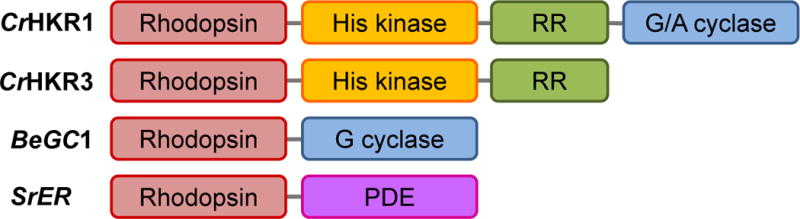
The domain structure of enzymerhodopsins. CrHKR1 and CrHKR3, histidine kinase 1 and 3, respectively, from the green alga C. reinhardtii; BeGC1, rhodopsin guanylylcyclase from the water mold B. emersonii; SrER, enzymerhodopsin from the choanoflagellate S. rosetta; RR, response regulator domain; G/A cyclase, gunanylyl/adenylyl cyclase domain.
HKR1 from C. reinhardtii is so far the only HKR, the rhodopsin domain of which has been heterologously expressed in Pichia, purified and spectroscopically studied (67). It is a photochromic pigment with two forms, Rh-UV and Rh-Bl, with absorption maxima at 380 nm and 490 nm, respectively, that are efficiently interconverted by light. In the dark, thermal equilibration of the two forms occurred with a time constant of ~3 days at room temperature (68). Resonance Raman spectroscopy showed that Rh-UV contains 13-cis, 15-anti retinal bound to the apoprotein without forming a protonated Schiff base (67). Photoconversion from Rh-UV to Rh-Bl proceeds in a branched reaction leading to two thermally interconvertible forms with protonated Schiff bases containing 13-trans, 15-anti or 13-cis, 15-syn retinal. Rh-Bl shows typical photochemistry as observed in other microbial rhodopsins. HKR1 fragments extending beyond the rhodopsin domain failed to fold properly in the heterologous systems tested (67), so its enzymatic function could not be studied. Immunofluorescent microscopy showed its localization in the eyespot of C. reinhardtii (67), but no rhodopsins besides CCRs have been found in this organelle by proteomic analysis (69). Cellular functions of HKR1 are not known.
Fungal guanylylcyclase rhodopsins
Motile zoospores and gametes of water molds such as Allomyces macrogynus and Blastocladiella emersonii exhibit phototaxis similar to that of green flagellate algae. The action spectrum of this response and its reconstitution after bleaching with exogenous retinal suggested a rhodopsin photoreceptor(s) (70–71). The genomes of these microbes and its relative Catenaria anguillulae harbor genes that encode enzymerhodopsins consisting of a rhodopsin domain and a guanylylcyclase domain (without histidine kinase or response regulator domains) (71). In contrast to all other type 1 rhodopsins, the rhodopsin domains of all five known fungal enzymerhodopsins (three from A. macrogynus and one from each B. emersonii and C. anguillulae) contain a predicted additional transmembrane helix in the N terminus (helix 0) (72). The cytoplasmic localization of the N terminal region has been confirmed by bimolecular fluorescence complementation, and its role in inhibition of the dark cyclase activity has been demonstrated by measurements from an N-terminally truncated version of the protein (72).
The results of pharmacological manipulation of the intracellular cGMP concentration and immunofluorescence microscopy in intact B. emersonii zoospores suggest the role of enzymerhodopsin (BeGC1) as the phototaxis receptor (71). It is thought that BeGC1 initiates a signaling cascade that leads to the elevation of the intracellular cGMP concentration, which regulates opening of cGMP-gated K+ channels identified in the genome of B. emersonii (73).
Unlike algal HKRs, the entire codon-optimized coding regions of fungal enzymerhodopsins have been functionally expressed and studied in animal cells. BeGC1 (under the names RhGC (74) and CyclOp (72)) produced the most robust elevation of the intracellular cGMP levels upon illumination of all tested homologous proteins, whereas its dark cyclase activity was very low, and no intrinsic ion channel or pumping activity was detected (72, 74). High specificity of its cyclase domain for cGMP over cAMP (cyclic adenosine monophosphate) was demonstrated by co-expression with two different subtypes of cyclic nucleotide-gated channel, each of which is specifically gated by one of the two cyclic nucleotides (74). Flash spectroscopy of the dark-adapted BeGC1 (λmax 525 nm) produced at least three photocycle intermediates, including a blue-shifted M-like state characteristic of a deprotonated retinylidene Schiff base (74).
A third member of the enzymerhodopsin class, a gene encoding a microbial rhodopsin domain followed by a phosphodiesterase (PDE) domain, has been found in the genome of the choanoflagellate Salpingoeca rosetta (71), but studies of its molecular characteristics have not yet been reported.
Cation-conducting Channelrhodopsins in Green Algae (Chlorophyte CCRs)
General properties and functions in native cells
The photoinduction of electrical potentials involved in phototaxis was discovered by electrophysiological recording from algal cells (75). The first report regarding the chemical nature of the photoreceptors was the demonstration in 1984 that retinal restored phototaxis to carotenoid-deficient C. reinhardtii mutants (76). The molecular identity of the phototaxis receptor proteins remained elusive until two type 1 rhodopsins cloned from partial sequences in a C. reinhardtii EST database were each shown to mediate phototaxis responses by depolarizing the algal membrane upon illumination (77). When expressed in animal cells, the algal phototaxis receptors function as light-gated cation channels, for which they were named “channelrhodopsins” (ChR1 and ChR2) (78–79). The use of CrChR2 for photoinduction of action potentials in neurons (80) brought about the era of optogenetics (3–4). Since a separate class of neuron-silencing anion channelrhodopsins (ACRs) has been recently discovered ((81), see the next section), here we will refer to cation channelrhodopsins as CCRs.
CCRs in green algae are the only group of eukaryotic microbial rhodopsins the physiological function of which in native cells is well characterized. Depolarization of the plasma membrane by CCRs triggers a signaling cascade that eventually leads to initiation of photomotility responses (82). The CCR-mediated photoreceptor current in algal cells is comprised of two components (83). The fast (early) component is attributable to the later shown direct channel activity of the CCRs. The second (late) component is carried by Ca2+ ions and makes a major contribution to the membrane depolarization extending the photosensitivity of the algae by three orders of magnitude (84). RNAi knock-down experiments in C. reinhardtii demonstrated that both CCRs play the role of photomotility receptors (77, 85), and that short wavelength-absorbing CrChR2 predominantly activates secondary Ca2+ channels by a yet unknown mechanism (84).
More than 50 different natural CCRs from different chlorophyte species are known at present, but only a few of them have been investigated in any detail (58, 86). Most mechanistic studies have been carried out using CrChR2 as a prototype CCR, and are summarized in recent excellent reviews (10, 87).
The photoactive site and proton transfer reactions
The X-ray crystal structure of a CCR chimera made of CrChR1 and CrChR2, called C1C2, shows that its photoactive site strongly resembles that of BR and NpSRII (88). It is generally accepted that in CCRs the photocycle initiated by the all-trans form leads to channel opening, but functional relevance of the photocycle of the 13-cis form is currently debated (89–90). The ultrafast processes upon CrChR2 photoexcitation have been reviewed elsewhere (10). In all so far studied CCRs all-trans to 13-cis retinal isomerization is manifested by the formation of a red-shifted K-like intermediate(s) (called P500 according to the wavelength position of its absorption maximum in CrChR2) (91–93). Its decay leads to the appearance of an M-like intermediate (P390) blue-shifted by deprotonation of the Schiff base (94–95). M formation proceeds in two kinetically distinct phases (92, 96–97) suggesting the presence of two substates probably similar to M1 and M2 intermediates in the photocycle of BR.
Electrophysiological measurements of intramolecular proton transfers in CaChR1 and VcChR1 from Chlamydomonas augustae and Volvox carteri, respectively, showed that both active site carboxylates can serve as Schiff base proton acceptors (96). A novel two-step proton relay mechanism that transfers a proton from the Asp85 homolog to the Asp212 homolog during the primary phototransition and from the Schiff base to the Asp85 homolog during M formation has been proposed for CaChR1 based on FTIR (Fourier Transform Infrared) difference spectroscopy (98). Intramolecular proton transfer currents are not detected by patch clamp recording from CrChR2 and other high-efficiency CCRs, although an outward intramolecular proton transfer is observed in weaker CCRs such as CaChR1 (96). The results from FTIR spectroscopy of CrChR2 are somewhat conflicting. One study has concluded that only the Asp212 homolog serves as the Schiff base proton acceptor in this protein (97), whereas another has reported parallel protonation of both active site carboxylates simultaneously with Schiff base deprotonation, thus suggesting that both of these residues might act as proton acceptors also in CrChR2 (99).
Conversion of the M intermediate to a red-shifted N/O species (P520) reflects reprotonation of the Schiff base (94–95). Time-resolved FTIR spectroscopy has identified Asp156 (corresponding to Asp115 in BR; Figure 4, left) as the proton donor in CrChR2 (97, 100). However, this conclusion has been challenged by the observation that the kinetics of Asp156 deprotonation does not match the Schiff base reprotonation (87). FTIR spectroscopy suggested a hydrogen-bonding interaction between Asp156 and Cys128 (Thr90 in BR), the “DC gate”; (101) disruption of which results in a dramatic reduction of the channel closing rate (102).
Figure 4.
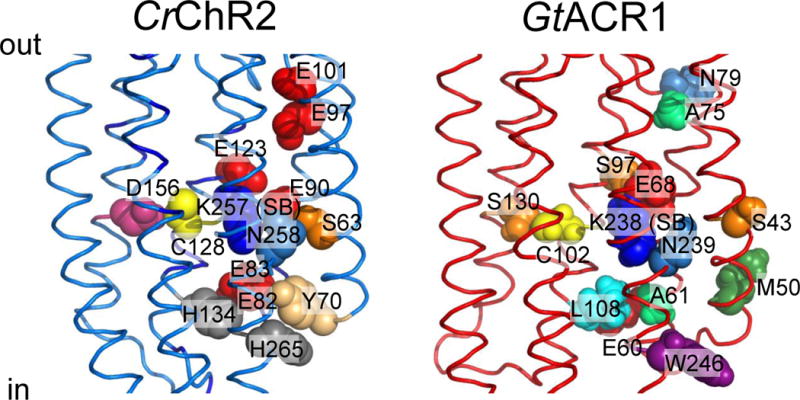
Functionally important residues in chlorophyte CCRs and cryptophyte ACRs discussed in the text. Left, C1C2 crystal structure (3ug9) with residues numbered according to CrChR2 sequence. Right, GtACR1 homology model built using 3ug9 as a template. The side chains are colored according to their identity.
Flash photolysis reveals complexity due to branching within CCR photocycles. In both CrChR2 (97) and PsChR2 (93) P520 decays in ~10 ms, but only ~75% of the molecules return to the unphotolyzed state, whereas the remaining 25–30% convert to the seconds-long-lasting P480. Furthermore, analysis of photochemical conversions in the slow CrChR2_C128T mutant has suggested the existence of two stable unphotolyzed states, one of which contains all-trans,15-anti retinal, and another, 13-cis, 15-syn retinal (103). These forms have been modeled as the parent states of two parallel photocycles, each of which contains P390, P520 and P480 intermediates, with the two photocycles linked by interconversion of the long-lived P480 states. This scheme has been also extended to the wild type, for which a two-photocycle model was earlier deduced from electrophysiological data with the difference that the two cycles are connected via unphotolyzed states, not long-lived P480 states (104). The latter scheme was also suggested by a combination of flash photolysis, nuclear magnetic resonance and resonance Raman spectroscopy data (89).
Channel gating
The C1C2 crystal structure of the closed state shows that helices 1–3 and 7 form a water-filled cavity at the extracellular side of the membrane (88). This cavity is blocked near the Schiff base by the “central gate” formed by the side chains of Ser63, Glu90 and Gln258 (CrChR2 numbering; Figure 4, left). There is also a constriction (called the “inner gate”) near the intracellular membrane surface formed by the side chains of Tyr70, Glu82, Glu83, His134 and His265 (CrChR2 numbering; Figure 4, left). Large conformational changes in the peptide backbone occur rapidly upon retinal isomerization (105–106). Double electron-electron resonance (DEER) spectroscopy showed that the intracellular end of helix 2, and of helix 6 to a lesser degree, move outward upon illumination (107–108). The results of time-resolved measurements of fluorescence anisotropy are consistent with an outward tilt of helix 2 (109). Projection maps obtained by cryo-electron microscopy suggested in addition a photoinduced movement of helix 7 (110). The outward movement of helix 6 (accompanied in BR by more subtle rearrangements of the cytoplasmic portions of helices 3, 5, and 7) is the major conformational change that occurs during the M1→M2 transition in BR (111), NaR (38), SRI (112) and SRII (112–113). However, structural rearrangement of helix 2 appears to be unique for CCRs and is thought to play a major role in formation of a conducting pore (99).
P520 is generally accepted as the main conductive state, whereas contribution of P390 (corresponding to BR’s M), which is in equilibrium with P520, has also been implicated (102). Time resolved FTIR analysis has shown that water influx upon photoactivation proceeds in two temporally separated steps with time constants of 10 and 200 μs (114).
Photocurrents of all so far studied CCRs exhibit inactivation (also called desensitization), i.e., a decrease in the photocurrent amplitude to a stationary level during prolonged light stimulation. Inactivation is explained by accumulation of a long-lived nonconductive state(s) P480 (94–95). A slow (tens of seconds) time course of the photocurrent peak recovery in the dark reflects slow relaxation of P480 to the unphotolyzed state. The central gate Glu90 deprotonates during the photocycle and, according to one view, this event initiates formation of the conductive pore (99, 115). However, other authors have proposed that deprotonation of Glu90 occurs only during the formation of the nonconductive P480 intermediate (97, 100).
Conductance and selectivity
Stationary noise analysis has yielded the value of 40 femtosiemens as an estimate of the unitary conductance of CrChR2 (116), and a ~3-fold greater value was obtained for PsChR2 from Platymononas (Tetraselmis) subcordiformis (117). All so far tested CCRs are primarily H+ channels: their relative permeability for this ion is ~6 orders of magnitude greater than that for monovalent metal cations (79). The Na+/H+ permeability ratio is, nevertheless, different for different ChRs (58). In any case under physiological conditions a large fraction of CCR current is carried by Na+ because the concentration of Na+ in physiological solutions is several orders of magnitude higher than that of H+ (118).
Utility for optogenetics
CCRs are widely used to depolarize the membrane and stimulate action potential generation in excitable cells, and, less frequently, to alter the intracellular ionic composition. Many excellent reviews cover this topic in detail (3–4, 119–120); therefore we will touch upon it only briefly. Despite the great variety of available CCRs, CrChR2 and its derivatives, such as CrChR2_H134R, remain the most frequently used activation molecules in optogenetic experiments (121). Extensive engineering efforts have yielded synthetic variants with red- (122) or blue-shifted absorbance (123), altered current kinetics (44), or increased relative permeability for individual cation species (124). Moreover, by introducing strategically placed mutations CCRs have been converted into light-gated Cl− channels (discussed in the next section). Systematic comparative analysis of the optogenetic utility of various natural and artificial CCRs have provided the guidelines for selection of optimal tools for a particular experimental purpose (125–127).
A promising direction to improve the penetration depth of optical stimulation is two-photon excitation of CCRs with near-infrared light (128). Promising strategies are being developed for specific targeting of CCRs to subcellular domains (129), for combining two spectrally separated CCRs for independent optical stimulation of distinct neuronal populations in the same study (86), and for using a CCR as actuator and an engineered fluorescent microbial rhodopsin as a reporter to achieve powerful all-optical recording of neuronal activity (130).
Anion-conducting Channelrhodopsins (ACRs) – Natural Chloride Channels in Cryptophyte Algae
Conductance and diversity
Photocurrents very similar to those in green flagellates have also been recorded from the phylogenetically distant phototactic cryptophyte Cryptomonas sp. (131). The only cryptophyte the genome of which has been completely sequenced is the marine alga Guillardia theta. Among 53 predicted microbial-type rhodopsins in this organism there is a cluster showing closer homology to chlorophyte CCRs than to other G. theta rhodopsins. Surprisingly, photocurrents generated by these rhodopsins upon expression in animal cells were carried exclusively by anions (Cl− under physiological conditions), with no conduction of protons or metal cations (81). Therefore, these proteins were named Anion Channel Rhodopsins, or ACRs.
The unitary conductance of GtACRs estimated by stationary noise analysis was ~25-fold greater than that of CrChR2 (81). The spectral sensitivities of GtACR1 and GtACR2 photocurrents peak at 515 and 470 nm, respectively. Another cryptophyte alga, Proteomonas sulcata, contained a channelrhodopsin initially named PsChR1 (86), but renamed PsuACR1 (also known as PsACR1) when shown to conduct exclusively anions (132–133). Screening sequences obtained by an ongoing transcriptome sequencing projects (134–135) expanded the list of functional ACRs to include 20 proteins derived from various marine cryptophyte species. These proteins showed large variation of the amplitude, spectral sensitivity, and kinetics of their photocurrents (Govorunova, E.G., Sineshchekov, O.A., Rodarte, E.M., Janz, R., Morelle, O., Melkonian, M., Wong, G.-K., and Spudich, J.L., manuscript under review). One variant, “ZipACR”, is particularly promising for inhibitory optogenetics because of its combination of large current amplitudes and an unprecedentedly fast conductance cycle (current half-decay time 2–4 ms depending on voltage). ZipACR expressed in cultured rat hippocampal neurons enabled precise photoinhibition of individual spikes in trains of up to 50 Hz frequency (Govorunova, E.G., Sineshchekov, O.A., Rodarte, E.M., Janz, R., Morelle, O., Melkonian, M., Wong, G.-K., and Spudich, J.L., manuscript under review). Neither subcellular localization, nor functions of ACRs in algal cells have yet been tested.
Residue determinants of anion selectivity
A conspicuous feature of ACRs is a non-carboxylic residue in the position of the primary proton acceptor from the retinylidene Schiff base of BR (Asp85; Figure 4, right), as is also observed in haloarchaeal HRs and chloride pumps from eubacteria. In GtACR1 replacement of the corresponding Ser with Glu (found at this site in most CCRs) led to a dramatic reduction of the current amplitude in response to the first excitation flash suggesting a critical importance of a non-carboxylate residue at this position for ACR channel function (136). However, the lack of a carboxylate residue in this position itself does not confer anion selectivity: e.g. ChR1 from Dunaliella salina (DsChR1) has an Ala, but is a proton channel (137).
Glu90 and Asn258 of the central gate in CCRs are also conserved in all so far confirmed ACRs, and the position of Ser63 is occupied by Ser or Cys (Figure 4, right). Glu90 is a major determinant of cation selectivity in CCRs (115, 138). However the presence of Glu in the corresponding position in ACRs (Figure 5) is obviously not a barrier to anion permeation, and its replacement with Gln or Arg did not change anion permeability of GtACR1 (139). Therefore, the Ser, Glu, Asn triad does not appear to function as an ion selective gate in ACRs.
Figure 5.
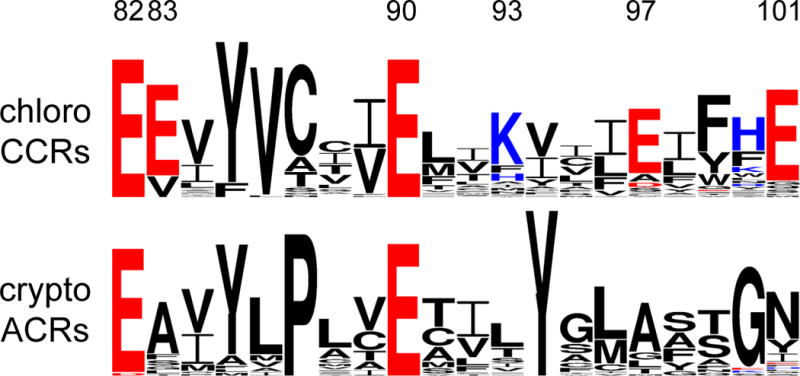
Helix 2 sequence logos of chlorophyte CCRs and cryptophyte ACRs created by WebLogo 3 as in (153). The overall height of each letter stack is proportional to the sequence conservation at that position, and the height of each letter is proportional to the frequency of the corresponding amino acid at that position. Acidic residues are red, and basic residues, blue. The residue numbers correspond to CrChR2 sequence.
In contrast to the central gate residues, only one of the five residues that form the inner gate in CCRs (Glu82) is found in all ACRs, but none of the other four (Tyr70, Glu83, His134 and His265) is conserved (Figure 4, right). Whereas replacement of Glu82 with Ala caused a strong reduction of photocurrents in CrChR2, the influence of the corresponding mutation in GtACR1 was much milder (139), which suggests that this conserved residue also plays different roles in ACRs and CCRs, as does the homolog of Glu90.
Sequence comparison with engineered Cl−-conducting mutants of CCRs
A need for more efficient inhibitory optogenetic tools than rhodopsin proton and chloride pumps instigated molecular engineering efforts to confer anion conductance to CCRs. One variant named ChloC was created by introducing an Arg at the position of the central gate Glu (the E90R mutation) in CrChR2 (138). Although permeant for Cl−, ChloC also conducted protons, but its H+ permeability could be eliminated by introducing two additional mutations (140). The second variant (iC1C2) was created by introducing nine mutations along the putative cation permeation path of C1C2 to minimize its negative charge (141). This version also showed residual H+ permeability, but further mutations resulted in iC++ that could track Cl− gradients more faithfully (142).
Although engineering of anion conductance in CCRs was a notable achievement that confirmed fundamental predictions of a structure-informed electrostatic model for CCR pore selectivity, comparison of the mutations introduced in CCRs to convert them into Cl−-conducting channels with the corresponding positions in natural ACRs reveals dramatic differences. The most revealing difference is universal conservation of Glu90 (CrChR2 numbering) in natural ACRs, whereas in all engineered Cl−-conducting variants this Glu needed to be replaced with a neutral or even positively charged residue. Furthermore, out of two positions at which positive charges were introduced in iC++, one (Gln117 in CrChR2) is occupied with a neutral residue, and another (Val242 in CrChR2), with a negatively charged residue in all natural ACRs. These mismatches show that, unlike artificial Cl−-conducting mutants, natural ACRs are not CCRs with just a few mutations conferring anion selectivity (143), but a truly distinct family of channelrhodopsins (Figure 6).
Figure 6.
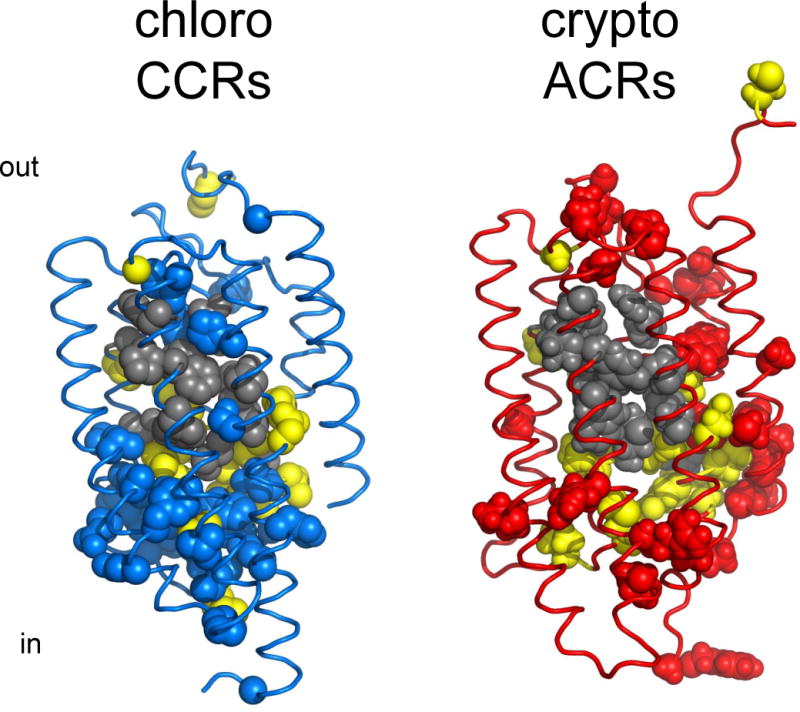
Structural comparison of chlorophyte CCRs and ACRs. Gray, residues of the retinal binding pocket of BR conserved in each of the two types of channelrhodopsins; yellow, residues conserved in both CCRs and ACRs; blue, residues conserved only in CCRs; red, residues conserved only in ACRs. The residue conservation pattern is shown using the C1C2 crystal structure (3ug9; left) and a GtACR1 homology model built on the 3ug9 template (right).
Gating mechanisms
Kinetic analysis of photocurrents generated by GtACR1 under single turnover conditions revealed that its conductance comprises two different mechanisms, one characterized by a fast rise and slow decay of photocurrents, and another, with a slow rise and fast decay (139). The two mechanisms of GtACR1 gating exhibited opposite dependencies on the membrane voltage and the bath pH. Mutant screening identified Glu68, the homolog of Glu90 in CrChR2, as a residue deprotonation of which to the extracellular side of the membrane is involved in fast closing of the channel.
Remarkably, when a positive charge was introduced at this site by the E68R mutation, channel gating was reversed, i.e., the channel was open in the dark and closed in the light (139). No such form of a channelrhodopsin had been reported previously, but a similar functional inversion (from attractant to repellent signaling) by a single point mutation either of the photoreceptor itself or of its cognate transducer has been observed in haloarchaeal sensory rhodopsin I (SRI) (144–148). In this case, a switch from the C (retinylidene Schiff base accessible from the cytoplasm) to E (Schiff base accessible from the extracellular space) conformer is responsible for the functional inversion. Similarly, the inverted function of GtACR1_E68R is likely to result from a mutation-induced inversion of its slow opening/fast closing gate.
Replacement of Cys102 with Ala in GtACR1 has very little effect on the fast phase of the current decay, but dramatically slows the slow phase, converting GtACR1 into a “step-function” channel (139). Cys102 of GtACR1 corresponds to Cys128 of CrChR2, mutation of which leads to a similarly large decrease of the current decay rate (44). In CrChR2 the C128X mutations presumably cause a disruption of the hydrogen bond (“DC-gate”) that Cys128 is proposed to form with Asp156 (101) supported by the observation that mutation of Asp156 yielded comparable or even greater extension of the channel open time, as did that of Cys128 (102). However, in contrast to CrChR2, mutation of Ser130, which in GtACR1 corresponds to Asp156, has little effect on the current decay rate, which suggests that the effect of the C102A mutation in GtACR1 is not caused by disruption of the putative hydrogen bond (139).
Photochemical conversions
Photoactive GtACR1, GtACR2 and PsuACR1 could be produced in Pichia, extracted in non-denaturing detergent, and studied in vitro. A resonance Raman study of GtACR1 showed that the retinal chromophore exists in an all-trans configuration with a protonated Schiff base very similar to that of BR (149). The most striking difference between the photocycle of all three so far tested ACRs and other type 1 rhodopsins is an extremely slow appearance and decay of a blue-shifted M-like intermediate with a deprotonated retinylidene Schiff base (133, 136). In CCRs M formation occurs within microseconds to tens of microseconds and precedes channel opening (91, 95–96). In contrast, M formation in GtACR1 is >50 times slower than channel opening, showing that the latter does not require Schiff base deprotonation.
In ACRs the open state is represented by the earlier L-like intermediate that appears on a submillisecond time scale, which decays to form M, although a rapid equilibrium between the L and red-shifted N/O-like intermediates cannot be excluded. The fast phase of channel closing temporally corresponds to the depletion of the L state and consequently generation of M because of the reversible reaction between the L and M intermediates, whereas slow channel closing corresponds to the irreversible decay of M (and hence, of L). When Cys102 was mutated to Ala in GtACR1, both M decay and recovery of the unphotolyzed state became ~100-fold slower than in the WT (136), which matched the influence of this mutation on the slow phase of the photocurrent decay.
In HRs, which have a non-carboxylate residue in the position of Asp85 of BR as do ACRs, Cl− acts as the protonated Schiff base counterion (150). However, deionization of purified pigment or substitution of SO42− for Cl− in the buffer changed neither the position of the absorption maximum nor the photocycle of GtACR1, which argues against Cl− being a Schiff base counterion in this rhodopsin (136). Patch clamp and flash photolysis analysis of the GtACR1_E68Q mutant suggests that Glu68 likely serves as a counterion and an acceptor of the proton from the Schiff base at neutral and high pH, or at least facilitates the proton transfer to the acceptor (136). Resonance Raman spectroscopy data are not consistent with this residue acting as a Schiff base counterion at neutral pH, but it cannot be excluded that Glu68 deprotonates early in the photocycle and accepts a proton from the Schiff base during formation of the M intermediate (149). A similar “two-step” process has been shown by resonance Raman and FTIR-difference spectroscopy for the Asp85 homolog in the cation channelrhodopsin CaChR1 (98). The role of Glu68 as a proton acceptor in GtACR1 is supported by the Glu68-dependence of an outward proton transfer current evident in a mutant in which the second photoactive site carboxylate, Asp234, is neurtralized (136).
Utility for optogenetics
As of this writing, the most frequently used inhibitory optogenetic tools are rhodopsin proton and chloride pumps such as Arch (14) or NpHR (28). However, they transport only one charge across the membrane per captured photon, and therefore are of limited capacity. Their use as optogenetic silencing tools requires high expression levels and light intensities which can bring about undesired side effects on the health of target cells. In contrast, ACRs (as well as Cl−-conducting CCR mutants) facilitate ion passage along a water-filled cavity that is formed within the protein upon photoexcitation, and thus are intrinsically more efficient than rhodopsin ion pumps. Furthermore, they bring the membrane potential to the Nernst equilibrium potential for Cl−, as do endogenous neuronal ionotropic GABA receptors, and in this sense are more physiological silencing tools than rhodopsin pumps.
Hyperpolarizing photocurrents generated by GtACR2 at less than a thousandth lower light intensity were equal to the maximal currents generated by Arch (81). Full suppression of spiking in cultured hippocampal neurons expressing GtACR2 was achieved at 20-times lower light intensities than that required by the slow ChloC variant, despite the latter being made more light-sensitive at the expense of a dramatically slower kinetics that required extended illumination for full activation (81). Similarly, robust inhibition of action potential firing has been demonstrated in GtACR1-expressing neurons (151). However, photoactivation of GtACR1 triggered neurotransmitter release and failed to attenuate the evoked response at the presynaptic terminals (151), consistent with the finding that the Cl− concentration maintained in the axon terminals is four to five times higher than that in the parent cell soma. Therefore, for inhibition of synaptic release ACRs will need to be targeted exclusively to somatodendritic membrane domains. Alternatively, outwardly rectifying ACR variants need to be engineered to prevent Cl− efflux at membrane potentials below the Nernst equilibrium potential for Cl−.
Another research area in which ACRs may find application as optogenetic tools is cardiology. Whereas cardiac pacing by light requires membrane-depolarizing, excitatory optogenetics tools, there is also a need for optogenetic inhibition to study pathologies of the heart conduction system or tachyarrhythmias. GtACRs have been found more efficient than Arch for silencing of electrical activity in cultured cardiomyocytes (152). Moreover, GtACRs enabled precise termination of cardiomyocyte action potentials at any time during their repolarization phase by threshold-based closed-loop optogenetics, which can potentially be used for the development of new treatments of the life-threatening long QT syndrome (152).
Cryptophyte CCRs – Independently Evolved Cation Channels from Haloarchaeal Ancestors
A distinct branch on the phylogenetic tree of G. theta rhodopsins consists of nine protein models the closest homologs of which in the global non-redundant protein database are haloarchaeal rhodopsin proton pumps (153). In particular, Asp residues in the positions of the Schiff base proton acceptor and donor (respectively, Asp85 and Asp96 in BR) are conserved (Figure 7). The presence of these carboxylates in microbial rhodopsins in general are considered a strong indicator of proton pumping ability, although counterexamples have been described, such as a rhodopsin from the fungus Neurospora crassa (154). Despite their similarity of sequence to light-driven proton pumps, when three transcripts from this G. theta cluster and a close homolog from P. sulcata were expressed in cultured animal cells, they behaved as light-gated cation channels (153).
Figure 7.
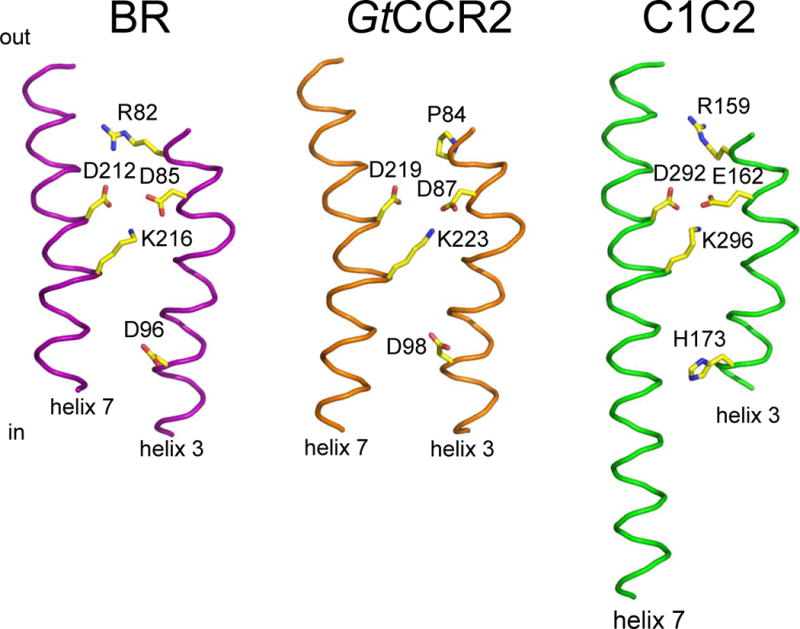
The active site residues of cryptophyte CCRs. A GtCCR2 homology model built on the 2ksy template (middle) in comparison with those of the proton pump BR (1c3w; left) and chlorophyte CCR C1C2 (3ug9; right). For clarity only helices 3 and 7 are shown.
As discussed in previous sections, helix 2 is critically important for channel gating in chlorophyte CCRs. Helix 2 contains up to 5 highly conserved Glu residues, one of which, Glu90 in CrChR2, plays a crucial role in both channel gating and cation selectivity (99, 138). However, none of these Glu residues is conserved in cryptophyte CCRs, and their overall helix 2 sequence is highly divergent from that of CCRs from green algae.
Two unusual representatives of this group are two G. theta CCRs in which homologs of Arg82 (BR numbering), nearly universally conserved in microbial rhodopsins, are substituted by Pro. Functional characteristics of GtCCR1 and GtCCR2 are very different from other characterized CCRs. Two processes contribute to the photocurrents generated by these pigments: (i) sodium channel conductance with strong inward rectification of the current-voltage dependence, and (ii) active outward proton transfer with large negative reversal potentials that is strongly suppressed by an increase in the external proton concentration (Sineshchekov O.A., Govorunova E.G. and Spudich J.L., manuscript in preparation).
The Schiff base donor position in cryptophyte CCRs is occupied by Asp instead of His in chlorophyte CCRs. Neutralization of this residue caused acceleration of proton photocurrent decay, which may indicate that reprotonation of the Schiff base occurs from this residue as in BR. Most importantly, it caused full suppression of passive channel activity, demonstrating another crucial difference between the two families of cation channelrhodopsins (Sineshchekov O.A., Govorunova E.G. and Spudich J.L., manuscript in preparation).
The cryptophyte CCRs reveal that cation channel function can be conferred on the rhodopsin scaffold in structurally different ways. These proteins have not yet been characterized in detail, but their identification has already shown that our current view of channelrhodopsins needs to be updated. At least one of the four so far examined cryptophyte CCRs generated photocurrents comparable to those of chlorophyte CrChR2, the most often used optogenetic tool (153). The ongoing transcriptome sequencing projects (134–135) have already uncovered >60 of their homologs in various cryptophyte species (Morelle, M., Melkonian, M., and Wong, G.-K., unpublished observations). Some of them may have even higher conductance, such as their ACR cousins, and offer advantages for optogenetic neural activation.
CONCLUSIONS AND FUTURE PROSPECTS
The surprising discovery in the past two years that there are three structurally and functionally distinct families of channelrhodopsins, when only one family, chlorophyte CCRs, had been known for the prior 15 years, have expanded research opportunities and enable overcoming some prior limitations to structure/function analysis of channel mechanism. It is evident from the early investigations of the two new classes of channelrhodopsins, ACRs and cryptophyte CCRs, that their selectivity, conductance, and gating mechanisms differ greatly from those of chlorophyte CCRs. Hence their elucidation along with further advances on chlorophyte CCRs is likely to give us a deep understanding of light-gated channel function and evolution. Natural ACRs offer two clear advantages for channelrhodopsin research. First, one of the main limitations to the study of chlorophyte CCRs has been their very low conductance, and ACRs are the most conductive light-gated channels known, providing a practical advantage for structure/function analysis. Second, the availability of an inverted ACR mutant, GtACR1_E68R, open in the dark and closed by illumination, provides a valuable complement to the wild-type ACR for structure/function analysis. X-ray crystal structures of both would be fascinating to compare, and almost certainly necessary for an atomic understanding of the gating mechanism. The cryptophyte CCRs have converged on cation channel function via a different evolutionary route than their distant chlorophyte cousins. Therefore, the mechanistic features shared by these two very different cation channels will help us understand the core requirements for light-gated cation conductance.
The physiological function of the cryptophyte channelrhodopsins, and of the large variety of other type 1 rhodopsins found in individual cryptophyte genomes, remain mysterious. The spectral sensitivity of photomotility responses in cryptophyte algae is consistent with the spectral range of rhodopsin absorption (131, 155), but, given the large number of rhodopsin genes in individual cryptophyte genomes, probing the cellular roles of rhodopsins including ACRs in cryptophyte algae will require the development of methods for their molecular genetic manipulation, similar to those used in C. reinhardtii (77).
In addition to the mystery of light-gated anion channel conductance as a previously unknown phenomenon in nature, ACRs have generated much interest as optogenetic tools because of their unprecedented photoefficiency to silence neurons by light-gated chloride conduction. As discussed above, due to their potency and the variation in the Cl− electrochemical potential in neurons, work on targeting ACRs to neuronal compartments and engineering of outwardly rectified variants would be useful to expand their utility as optogenetic tools. Cryptophyte CCRs, in their infancy as subjects of investigation, also may offer new properties for optogenetic use based on their different origins. Enzymerhodopsins, also little studied, are expected to provide new ways to use light for control of cell signaling and metabolism, expanding optogenetics with microbial rhodopsins beyond control of membrane electrical potential.
Supplementary Material
Acknowledgments
The University of Texas Health Science Center at Houston listing J.L.S., E.G.G., and O.A.S. as inventors has filed patent applications regarding ACRs, PsChR2, and related channelrhodopsins. J.L.S. acknowledges financial support from the NIH National Institute of General Medical Sciences, the NIH BRAIN Initiative, and endowed chair AU-0009 from the Robert A. Welch Foundation.
Footnotes
SUPPLEMENTARY MATERIAL
Follow the Supplemental Material link in the online version of this article or at http://www.annualreviews.org/
LITERATURE CITED
- 1.Spudich JL, Yang C-S, Jung K-H, Spudich EN. Retinylidene proteins: structures and functions from archaea to humans. Annu Rev Cell Dev Biol. 2000;16:365–92. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 2.Devine EL, Oprian DD, Theobald DL. Relocating the active-site lysine in rhodopsin and implications for evolution of retinylidene proteins. Proc Natl Acad Sci USA. 2013;110:13351–5. doi: 10.1073/pnas.1306826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyden ES. Optogenetics and the future of neuroscience. Nat Neurosci. 2015;18:1200–01. doi: 10.1038/nn.4094. [DOI] [PubMed] [Google Scholar]
- 4.Deisseroth K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci. 2015;18:1213–25. doi: 10.1038/nn.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem Rev. 2014;114:126–63. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Essen LO. Halorhodopsin: light-driven ion pumping made simple? Curr Opin Struct Biol. 2002;12:516–22. doi: 10.1016/s0959-440x(02)00356-1. [DOI] [PubMed] [Google Scholar]
- 7.Lanyi JK. Proton transfers in the bacteriorhodopsin photocycle. Biochim Biophys Acta. 2006;1757:1012–18. doi: 10.1016/j.bbabio.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Balashov SP, Lanyi JK. Xanthorhodopsin: Proton pump with a carotenoid antenna. Cell Mol Life Sci. 2007;64:2323–28. doi: 10.1007/s00018-007-7167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spudich JL, Spudich EN. In: Animal Models in Eye Research. Tsonis PA, editor. Amsterdam: Elsevier; 2008. pp. 6–14. [Google Scholar]
- 10.Lorenz-Fonfria VA, Heberle J. Channelrhodopsin unchained: structure and mechanism of a light-gated cation channel. Biochim Biophys Acta. 2014;1837:626–42. doi: 10.1016/j.bbabio.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Inoue K, Kato Y, Kandori H. Light-driven ion-translocating rhodopsins in marine bacteria. Trends Microbiol. 2015;23:91–98. doi: 10.1016/j.tim.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature. 1971;233:149–52. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- 13.Henderson R, Unwin P. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 14.Chow BY, Han X, Dobry AS, Qian X, Chuong AS, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, et al. Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–06. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 16.Beja O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–89. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 17.Venter JC, K R, Heidelberg JF, H AL, R D, et al. Environmental genome shotgun sequencing of the Sargasso sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 18.Sabehi G, Loy A, Jung KH, Partha R, Spudich JL, et al. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 2005;3:e273. doi: 10.1371/journal.pbio.0030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ran T, Ozorowski G, Gao Y, Sineshchekov OA, Wang W, et al. Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes. Acta Crystallogr D Biol Crystallogr. 2013;69:1965–80. doi: 10.1107/S0907444913017575. [DOI] [PubMed] [Google Scholar]
- 20.Hussain S, Kinnebrew M, Schonenbach NS, Aye E, Han S. Functional consequences of the oligomeric assembly of proteorhodopsin. J Mol Biol. 2015;427:1278–90. doi: 10.1016/j.jmb.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma AK, Zhaxybayeva O, Papke RT, Doolittle WF. Actinorhodopsins: proteorhodopsin-like gene sequences found predominantly in non-marine environments. Environ Microbiol. 2008;10:1039–56. doi: 10.1111/j.1462-2920.2007.01525.x. [DOI] [PubMed] [Google Scholar]
- 22.Balashov SP, Imasheva ES, Boichenko VA, Anton J, Wang JM, Lanyi JK. Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science. 2005;309:2061–4. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waschuk SA, Bezerra AGJ, Shi L, Brown LS. Leptosphaeria rhodopsin: Bacteriorhodopsin-like proton pump from a eukaryote. Proc Natl Acad Sci USA. 2005;102:6879–83. doi: 10.1073/pnas.0409659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuno-Yagi A, Mukohata Y. Two possible roles of bacteriorhodopsin; a comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem Biophys Res Commun. 1977;78:237–43. doi: 10.1016/0006-291x(77)91245-1. [DOI] [PubMed] [Google Scholar]
- 25.Mukohata Y, Kaji Y. Light-induced membrane-potential increase, ATP synthesis, and proton uptake in Halobacterium halobium, R1mR catalyzed by halorhodopsin: Effects of N,N′-dicyclohexylcarbodiimide, triphenyltin chloride, and 3,5-di-tert-butyl-4-hydroxybenzylidenemalononitrile (SF6847) Arch Biochem Biophys. 1981;206:72–76. doi: 10.1016/0003-9861(81)90067-9. [DOI] [PubMed] [Google Scholar]
- 26.Schobert B, Lanyi JK. Halorhodopsin is a light-driven chloride pump. J Biol Chem. 1982;257:10306–13. [PubMed] [Google Scholar]
- 27.Sasaki J, Brown LS, Chon YS, Kandori H, Maeda A, et al. Conversion of bacteriorhodopsin into a chloride ion pump. Science. 1995;269:73–5. doi: 10.1126/science.7604281. [DOI] [PubMed] [Google Scholar]
- 28.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–39. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshizawa S, Kumagai Y, Kim H, Ogura Y, Hayashi T, et al. Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria. Proc Natl Acad Sci USA. 2014;111:6732–7. doi: 10.1073/pnas.1403051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kandori H. Ion-pumping microbial rhodopsins. Front Mol Biosci. 2015;2:52. doi: 10.3389/fmolb.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue K, Koua FH, Kato Y, Abe-Yoshizumi R, Kandori H. Spectroscopic study of a light-driven chloride ion pump from marine bacteria. J Phys Chem B. 2014;118:11190–99. doi: 10.1021/jp507219q. [DOI] [PubMed] [Google Scholar]
- 32.Hosaka T, Yoshizawa S, Nakajima Y, Ohsawa N, Hato M, et al. Structural mechanism for light-driven transport by a new type of chloride ion pump, Nonlabens marinus rhodopsin-3. J Biol Chem. 2016;291:17488–95. doi: 10.1074/jbc.M116.728220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasemi T, Kikukawa T, Kamo N, Demura M. Characterization of a cyanobacterial chloride-pumping rhodopsin and its conversion into a proton pump. J Biol Chem. 2016;291:355–62. doi: 10.1074/jbc.M115.688614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue K, Ono H, Abe-Yoshizumi R, Yoshizawa S, Ito H, et al. A light-driven sodium ion pump in marine bacteria. Nat Commun. 2013;4:1678. doi: 10.1038/ncomms2689. [DOI] [PubMed] [Google Scholar]
- 35.Kwon SK, Kim BK, Song JY, Kwak MJ, Lee CH, et al. Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome Biol Evol. 2013;5:187–99. doi: 10.1093/gbe/evs134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balashov SP, Imasheva ES, Dioumaev AK, Wang JM, Jung KH, Lanyi JK. Light-driven Na+ pump from Gillisia limnaea: a high-affinity Na+ binding site is formed transiently in the photocycle. Biochemistry. 2014;53:7549–61. doi: 10.1021/bi501064n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Sineshchekov OA, da Silva GF, Spudich JL. In vitro demonstration of dual light-driven Na+/H+ pumping by a microbial rhodopsin. Biophys J. 2015;109:1446–53. doi: 10.1016/j.bpj.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.da Silva GF, Goblirsch BR, Tsai AL, Spudich JL. Cation-specific conformations in a dual-function ion-pumping microbial rhodopsin. Biochemistry. 2015;54:3950–59. doi: 10.1021/bi501386d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato HE, Inoue K, Abe-Yoshizumi R, Kato Y, Ono H, et al. Structural basis for Na+ transport mechanism by a light-driven Na+ pump. Nature. 2015;521:48–53. doi: 10.1038/nature14322. [DOI] [PubMed] [Google Scholar]
- 40.Gushchin I, Shevchenko V, Polovinkin V, Kovalev K, Alekseev A, et al. Crystal structure of a light-driven sodium pump. Nat Struct Mol Biol. 2015;22:390–5. doi: 10.1038/nsmb.3002. [DOI] [PubMed] [Google Scholar]
- 41.Spudich EN, Spudich JL. Control of transmembrane ion fluxes to select halorhodopsin-deficient and other energy-trunsduction mutants of Halobacterium halobium. Proc Natl Acad Sci USA. 1982;79:4308–12. doi: 10.1073/pnas.79.14.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogomolni R, Spudich JL. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc Natl Acad Sci USA. 1982;79:6250–54. doi: 10.1073/pnas.79.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spudich JL, Bogomolni RA. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984;312:509–13. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–34. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 45.Yao VJ, Spudich JL. Primary structure of an arhaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc Natl Acad Sci USA. 1992;89:11915–19. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudolph J, Oesterhelt D. Deletion analysis of the che operon in the archaeon Halobacterium salinarium. J Mol Biol. 1996;258:548–54. doi: 10.1006/jmbi.1996.0267. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi T, Tomioka H, Kamo N, Kobatake Y. A photosystem other than PS370 also mediates the negative phototaxis of Halobacterium halobium. FEMS Microbiol Lett. 1985;28:161–64. [Google Scholar]
- 48.Tomioka H, Takahashi T, Kamo N, Kobatake Y. Flash spectrophotometric identification of a fourth rhodopsin-like pigment in Halobacterium halobium. Biochem Biophys Res Commun. 1986;139:389–95. doi: 10.1016/s0006-291x(86)80003-1. [DOI] [PubMed] [Google Scholar]
- 49.Spudich EN, Sundberg SA, Manor D, Spudich JL. Properties of a second sensory receptor protein in Halobacterium halobium phototaxis. Proteins: Structure, Function, & Bioinformatics. 1986;1:239–46. doi: 10.1002/prot.340010306. [DOI] [PubMed] [Google Scholar]
- 50.Luecke H, Schobert B, Lanyi JK, Spudich EN, Spudich JL. Crystal structure of sensory rhodopsin II at 2.4 angstroms: insights into color tuning and transducer interaction. Science. 2001;293:1499–503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Royant A, Nollert P, Edman K, Neutze R, Landau EM, et al. X-ray structure of sensory rhodopsin II at 2.1-A resolution. Proc Natl Acad Sci USA. 2001;98:10131–36. doi: 10.1073/pnas.181203898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gordeliy VI, Labahn J, Moukhametzianov R, Efremov R, Granzin J, et al. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature. 2002;419:484–7. doi: 10.1038/nature01109. [DOI] [PubMed] [Google Scholar]
- 53.Vogeley L, Sineshchekov O, Trivedi V, Sasaki J, Spudich J, Luecke H. Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 A. Science. 2004;306:1390–93. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spudich JL. Protein-protein interaction converts a proton pump into a sensory receptor. Cell. 1994;79:747–50. doi: 10.1016/0092-8674(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 55.Schmies G, Luttenberg B, Chizhov I, Engelhard M, Becker A, Bamberg E. Sensory rhodopsin II from the haloalkaliphilic Natronobacterium pharaonis: Light-activated proton transfer reactions. Biophys J. 2000;78:967–76. doi: 10.1016/S0006-3495(00)76654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sudo Y, Spudich JL. Three strategically placed hydrogen-bonding residues convert a proton pump into a sensory receptor. Proc Natl Acad Sci USA. 2006;103:16129–34. doi: 10.1073/pnas.0607467103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inoue K, Nomura Y, Kandori H. Asymmetric functional conversion of eubacterial light-driven ion pumps. J Biol Chem. 2016;291:9883–93. doi: 10.1074/jbc.M116.716498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spudich JL, Sineshchekov OA, Govorunova EG. Mechanism divergence in microbial rhodopsins. Biochim Biophys Acta. 2014;1837:546–52. doi: 10.1016/j.bbabio.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jung K-H, Trivedi VD, Spudich JL. Demonstration of a sensory rhodopsin in eubacteria. Mol Microbiol. 2003;47:1513–22. doi: 10.1046/j.1365-2958.2003.03395.x. [DOI] [PubMed] [Google Scholar]
- 60.Sineshchekov OA, Trivedi VD, Sasaki J, Spudich JL. Photochromicity of Anabaena sensory rhodopsin, an atypical microbial receptor with a cis-retinal light-adapted form. J Biol Chem. 2005;280:14663–8. doi: 10.1074/jbc.M501416200. [DOI] [PubMed] [Google Scholar]
- 61.Irieda H, Morita T, Maki K, Homma M, Aiba H, Sudo Y. Photo-induced regulation of the chromatic adaptive gene expression by Anabaena sensory rhodopsin. J Biol Chem. 2012;287:32485–93. doi: 10.1074/jbc.M112.390864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim SY, Yoon SR, Han S, Yun Y, Jung KH. A role of Anabaena sensory rhodopsin transducer (ASRT) in photosensory transduction. Mol Microbiol. 2014;93:403–14. doi: 10.1111/mmi.12635. [DOI] [PubMed] [Google Scholar]
- 63.Wand A, Gdor I, Zhu J, Sheves M, Ruhman S. Shedding new light on retinal protein photochemistry. Annu Rev Phys Chem. 2013;64:437–58. doi: 10.1146/annurev-physchem-040412-110148. [DOI] [PubMed] [Google Scholar]
- 64.Brown LS. Eubacterial rhodopsins - unique photosensors and diverse ion pumps. Biochim Biophys Acta. 2014;1837:553–61. doi: 10.1016/j.bbabio.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 65.Kateriya S, Nagel G, Bamberg E, Hegemann P. “Vision” in single-celled algae. News Physiol Sci. 2004;19:133–37. doi: 10.1152/nips.01517.2004. [DOI] [PubMed] [Google Scholar]
- 66.Kianianmomeni A, Hallmann A. Algal photoreceptors: in vivo functions and potential applications. Planta. 2014;239:1–26. doi: 10.1007/s00425-013-1962-5. [DOI] [PubMed] [Google Scholar]
- 67.Luck M, Mathes T, Bruun S, Fudim R, Hagedorn R, et al. A photochromic histidine kinase rhodopsin (HKR1) that is bimodally switched by ultraviolet and blue light. J Biol Chem. 2012;287:40083–90. doi: 10.1074/jbc.M112.401604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Penzkofer A, Luck M, Mathes T, Hegemann P. Bistable retinal Schiff base photo-dynamics of histidine kinase rhodopsin HKR1 from Chlamydomonas reinhardtii. Photochem Photobiol. 2014;90:773–85. doi: 10.1111/php.12246. [DOI] [PubMed] [Google Scholar]
- 69.Schmidt M, Gessner G, Luff M, Heiland I, Wagner V, et al. Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell. 2006;18:1908–30. doi: 10.1105/tpc.106.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saranak J, Foster K-W. Rhodopsin guides fungal phototaxis. Nature. 1997;387:465–66. doi: 10.1038/387465a0. [DOI] [PubMed] [Google Scholar]
- 71.Avelar GM, Schumacher RI, Zaini PA, Leonard G, Richards TA, Gomes SL. A rhodopsin-guanylyl cyclase gene fusion functions in visual perception in a fungus. Curr Biol. 2014;24:1234–40. doi: 10.1016/j.cub.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao S, Nagpal J, Schneider MW, Kozjak-Pavlovic V, Nagel G, Gottschalk A. Optogenetic manipulation of cGMP in cells and animals by the tightly light-regulated guanylyl-cyclase opsin CyclOp. Nat Commun. 2015;6:8046. doi: 10.1038/ncomms9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Avelar GM, Glaser T, Leonard G, Richards TA, Ulrich H, Gomes SL. A cGMP-dependent K+ channel in the blastocladiomycete fungus Blastocladiella emersonii. Eukaryot Cell. 2015;14:958–63. doi: 10.1128/EC.00087-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scheib U, Stehfest K, Gee CE, Korschen HG, Fudim R, et al. The rhodopsin-guanylyl cyclase of the aquatic fungus Blastocladiella emersonii enables fast optical control of cGMP signaling. Sci Signal. 2015;8:rs8. doi: 10.1126/scisignal.aab0611. [DOI] [PubMed] [Google Scholar]
- 75.Litvin FF, Sineshchekov OA, Sineshchekov VA. Photoreceptor electric potential in the phototaxis of the alga Haematococcus pluvialis. Nature. 1978;271:476–78. doi: 10.1038/271476a0. [DOI] [PubMed] [Google Scholar]
- 76.Foster K-W, Saranak J, Patel N, Zarrilli G, Okabe M, et al. A rhodopsin is the functional photoreceptor for phototaxis in the unicelullar eukaryote Chlamydomonas. Nature. 1984;311:756–59. doi: 10.1038/311756a0. [DOI] [PubMed] [Google Scholar]
- 77.Sineshchekov OA, Jung K-H, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2002;99:8689–94. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–8. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 79.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–5. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–8. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 81.Govorunova EG, Sineshchekov OA, Liu X, Janz R, Spudich JL. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science. 2015;349:647–50. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sineshchekov OA, Spudich JL. Handbook of Photosensory Receptors. Weinheim: Wiley-VCH; 2005. pp. 25–42. [Google Scholar]
- 83.Sineshchekov OA, Litvin FF, Keszthelyi L. Two components of photoreceptor potential of the flagellated green alga Haematococcus pluvialis. Biophys J. 1990;57:33–39. doi: 10.1016/S0006-3495(90)82504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sineshchekov OA, Govorunova EG, Spudich JL. Photosensory functions of channelrhodopsins in native algal cells. Photochem Photobiol. 2009;85:556–63. doi: 10.1111/j.1751-1097.2008.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Govorunova EG, Jung K-W, Sineshchekov OA, Spudich JL. Chlamydomonas sensory rhodopsins A and B: Cellular content and role in photophobic responses. Biophys J. 2004;86:2342–49. doi: 10.1016/S0006-3495(04)74291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–46. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schneider F, Grimm C, Hegemann P. Biophysics of channelrhodopsin. Annu Rev Biophys. 2015;44:167–86. doi: 10.1146/annurev-biophys-060414-034014. [DOI] [PubMed] [Google Scholar]
- 88.Kato HE, Zhang F, Yizhar O, Ramakrishnan C, Nishizawa T, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–74. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bruun S, Stoeppler D, Keidel A, Kuhlmann U, Luck M, et al. Light-dark adaptation of channelrhodopsin involves photoconversion between the all-trans and 13-cis retinal isomers. Biochemistry. 2015;54:5389–400. doi: 10.1021/acs.biochem.5b00597. [DOI] [PubMed] [Google Scholar]
- 90.Lorenz-Fonfria VA, Schultz BJ, Resler T, Schlesinger R, Bamann C, et al. Pre-gating conformational changes in the ChETA variant of channelrhodopsin-2 monitored by nanosecond IR spectroscopy. J Am Chem Soc. 2015;137:1850–61. doi: 10.1021/ja5108595. [DOI] [PubMed] [Google Scholar]
- 91.Ernst OP, Sanchez Murcia PA, Daldrop P, Tsunoda SP, Kateriya S, Hegemann P. Photoactivation of channelrhodopsin. J Biol Chem. 2008;283:1637–43. doi: 10.1074/jbc.M708039200. [DOI] [PubMed] [Google Scholar]
- 92.Verhoefen MK, Bamann C, Blocher R, Forster U, Bamberg E, Wachtveitl J. The photocycle of channelrhodopsin-2: Ultrafast reaction dynamics and subsequent reaction steps. Chemphyschem. 2010;11:3113–22. doi: 10.1002/cphc.201000181. [DOI] [PubMed] [Google Scholar]
- 93.Szundi I, Li H, Chen E, Bogomolni R, Spudich JL, Kliger DS. Platymonas subcordiformis Channelrhodopsin-2 Function: I. The photochemical reaction cycle. J Biol Chem. 2015;290:16573–84. doi: 10.1074/jbc.M114.631614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bamann C, Kirsch T, Nagel G, Bamberg E. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J Mol Biol. 2008;375:686–94. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 95.Ritter E, Stehfest K, Berndt A, Hegemann P, Bartl FJ. Monitoring light-induced structural changes of Channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J Biol Chem. 2008;283:35033–41. doi: 10.1074/jbc.M806353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sineshchekov OA, Govorunova EG, Wang J, Li H, Spudich JL. Intramolecular proton transfer in channelrhodopsins. Biophys J. 2013;104:807–17. doi: 10.1016/j.bpj.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lorenz-Fonfria VA, Resler T, Krause N, Nack M, Gossing M, et al. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc Natl Acad Sci USA. 2013;110:E1273–81. doi: 10.1073/pnas.1219502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ogren JI, Yi A, Mamaev S, Li H, Spudich JL, Rothschild KJ. Proton transfers in a Channelrhodopsin-1 studied by FTIR-difference spectroscopy and site-directed mutagenesis. J Biol Chem. 2015;290:12719–30. doi: 10.1074/jbc.M114.634840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kuhne J, Eisenhauer K, Ritter E, Hegemann P, Gerwert K, Bartl F. Early formation of the ion-conducting pore in channelrhodopsin-2. Angew Chem Int Ed Engl. 2014;54:4953–57. doi: 10.1002/anie.201410180. [DOI] [PubMed] [Google Scholar]
- 100.Resler T, Schultz BJ, Lorenz-Fonfria VA, Schlesinger R, Heberle J. Kinetic and vibrational isotope effects of proton transfer reactions in channelrhodopsin-2. Biophys J. 2015;109:287–97. doi: 10.1016/j.bpj.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nack M, Radu I, Gossing M, Bamann C, Bamberg E, et al. The DC gate in channelrhodopsin-2: crucial hydrogen bonding interaction between C128 and D156. Photochem Photobiol Sci. 2010;9:194–98. doi: 10.1039/b9pp00157c. [DOI] [PubMed] [Google Scholar]
- 102.Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–78. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 103.Ritter E, Piwowarski P, Hegemann P, Bartl FJ. Light-dark adaptation of channelrhodopsin C128T mutant. J Biol Chem. 2013;288:10451–58. doi: 10.1074/jbc.M112.446427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stehfest K, Hegemann P. Evolution of the channelrhodopsin photocycle model. Chemphyschem. 2010;11:1120–6. doi: 10.1002/cphc.200900980. [DOI] [PubMed] [Google Scholar]
- 105.Radu I, Bamann C, Nack M, Nagel G, Bamberg E, Heberle J. Conformational changes of channelrhodopsin-2. J Am Chem Soc. 2009;131:7313–19. doi: 10.1021/ja8084274. [DOI] [PubMed] [Google Scholar]
- 106.Neumann-Verhoefen MK, Neumann K, Bamann C, Radu I, Heberle J, et al. Ultrafast infrared spectroscopy on channelrhodopsin-2 reveals efficient energy transfer from the retinal chromophore to the protein. J Am Chem Soc. 2013;135:6968–76. doi: 10.1021/ja400554y. [DOI] [PubMed] [Google Scholar]
- 107.Krause N, Engelhard C, Heberle J, Schlesinger R, Bittl R. Structural differences between the closed and open states of channelrhodopsin-2 as observed by EPR spectroscopy. FEBS Lett. 2013;587:3309–13. doi: 10.1016/j.febslet.2013.08.043. [DOI] [PubMed] [Google Scholar]
- 108.Sattig T, Rickert C, Bamberg E, Steinhoff HJ, Bamann C. Light-induced movement of the transmembrane helix B in channelrhodopsin-2. Angew Chem Int Ed Engl. 2013;52:9705–8. doi: 10.1002/anie.201301698. [DOI] [PubMed] [Google Scholar]
- 109.Volz P, Krause N, Balke J, Schneider C, Walter M, et al. Light and pH-induced changes in structure and accessibility of transmembrane helix B and its immediate environment in Channelrhodopsin-2. J Biol Chem. 2016;291:17382–93. doi: 10.1074/jbc.M115.711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Müller M, Bamann C, Bamberg E, Kuhlbrandt W. Light-induced helix movements in channelrhodopsin-2. J Mol Biol. 2015;427:341–9. doi: 10.1016/j.jmb.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 111.Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sasaki J, Tsai AL, Spudich JL. Opposite displacement of helix F in attractant and repellent signaling by sensory rhodopsin-Htr complexes. J Biol Chem. 2011;286:18868–77. doi: 10.1074/jbc.M110.200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wegener AA, Chizhov I, Engelhard M, Steinhoff HJ. Time-resolved detection of transient movement of helix F in spin-labelled pharaonis sensory rhodopsin II. J Mol Biol. 2000;301:881–91. doi: 10.1006/jmbi.2000.4008. [DOI] [PubMed] [Google Scholar]
- 114.Lorenz-Fonfria VA, Bamann C, Resler T, Schlesinger R, Bamberg E, Heberle J. Temporal evolution of helix hydration in a light-gated ion channel correlates with ion conductance. Proc Natl Acad Sci USA. 2015;112:E5796–804. doi: 10.1073/pnas.1511462112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Eisenhauer K, Kuhne J, Ritter E, Berndt AE, Wolf S, et al. In channelrhodopsin-2 E90 is crucial for ion selectivity and is deprotonated during the photocycle. J Biol Chem. 2012;287:6904–11. doi: 10.1074/jbc.M111.327700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Feldbauer K, Zimmermann D, Pintschovius V, Spitz J, Bamann C, Bamberg E. Channelrhodopsin-2 is a leaky proton pump. Proc Natl Acad Sci USA. 2009;106:12317–22. doi: 10.1073/pnas.0905852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Govorunova EG, Sineshchekov OA, Li H, Janz R, Spudich JL. Characterization of a highly efficient blue-shifted channelrhodopsin from the marine alga Platymonas subcordiformis. J Biol Chem. 2013;288:29911–22. doi: 10.1074/jbc.M113.505495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schneider F, Gradmann D, Hegemann P. Ion selectivity and competition in channelrhodopsins. Biophys J. 2013;105:91–100. doi: 10.1016/j.bpj.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao M, Alleva R, Ma H, Daniel AG, Schwartz TH. Optogenetic tools for modulating and probing the epileptic network. Epilepsy Res. 2015;116:15–26. doi: 10.1016/j.eplepsyres.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cohen AE. Optogenetics: Turning the microscope on its head. Biophys J. 2016;110:997–1003. doi: 10.1016/j.bpj.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wietek J, Prigge M. Enhancing channelrhodopsins: An overview. Methods Mol Biol. 2016;1408:141–65. doi: 10.1007/978-1-4939-3512-3_10. [DOI] [PubMed] [Google Scholar]
- 122.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci. 2013;16:1499–508. [Google Scholar]
- 123.Kato HE, Kamiya M, Sugo S, Ito J, Taniguchi R, et al. Atomistic design of microbial opsin-based blue-shifted optogenetics tools. Nat Commun. 2015;6:7177. doi: 10.1038/ncomms8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kleinlogel S, Feldbauer K, Dempski RE, Fotis H, Wood PG, et al. Ultra light-sensitive and fast neuronal activation with the Ca2+-permeable channelrhodopsin CatCh. Nat Neurosci. 2011;14:513–18. doi: 10.1038/nn.2776. [DOI] [PubMed] [Google Scholar]
- 125.Lin JY. A user’s guide to channelrhodopsin variants: features, limitations and future developments. Exp Physiol. 2010;96:19–25. doi: 10.1113/expphysiol.2009.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 127.Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O’Shea DJ, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2011;9:159–72. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mohanty SK, Reinscheid RK, Liu X, Okamura N, Krasieva TB, Berns MW. In-depth activation of channelrhodopsin 2-sensitized excitable cells with high spatial resolution using two-photon excitation with a near-infrared laser microbeam. Biophys J. 2008;95:3916–26. doi: 10.1529/biophysj.108.130187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Grubb MS, Burrone J. Channelrhodopsin-2 localised to the axon initial segment. PLoS One. 2010;5:e13761. doi: 10.1371/journal.pone.0013761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hochbaum DR, Zhao Y, Farhi SL, Klapoetke N, Werley CA, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;8:825–33. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sineshchekov OA, Govorunova EG, Jung K-H, Zauner S, Maier U-G, Spudich JL. Rhodopsin-mediated photoreception in cryptophyte flagellates. Biophys J. 2005;89:4310–19. doi: 10.1529/biophysj.105.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Govorunova EG, Sineshchekov OA, Spudich JL. Proteomonas sulcata ACR1: A fast anion channelrhodopsin. Photochem Photobiol. 2016;92:257–63. doi: 10.1111/php.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wietek J, Broser M, Krause BS, Hegemann P. Identification of a natural green light absorbing chloride conducting channelrhodopsin from Proteomonas sulcata. J Biol Chem. 2016;291:4121–27. doi: 10.1074/jbc.M115.699637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Matasci N, Hung LH, Yan Z, Carpenter EJ, Wickett NJ, et al. Data access for the 1,000 Plants (1KP) project. Gigascience. 2014;3:17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sineshchekov OA, Li H, Govorunova EG, Spudich JL. Photochemical reaction cycle transitions during anion channelrhodopsin gating. Proc Natl Acad Sci USA. 2016;113:E1993–2000. doi: 10.1073/pnas.1525269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–57. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wietek J, Wiegert JS, Adeishvili N, Schneider F, Watanabe H, et al. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344:409–12. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- 139.Sineshchekov OA, Govorunova EG, Li H, Spudich JL. Gating mechanisms of a natural anion channelrhodopsin. Proc Natl Acad Sci USA. 2015;112:14236–41. doi: 10.1073/pnas.1513602112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wietek J, Beltramo R, Scanziani M, Hegemann P, Oertner TG, Simon Wiegert J. An improved chloride-conducting channelrhodopsin for light-induced inhibition of neuronal activity in vivo. Sci Rep. 2015;5:14807. doi: 10.1038/srep14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Berndt A, Lee SY, Ramakrishnan C, Deisseroth K. Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science. 2014;344:420–24. doi: 10.1126/science.1252367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Berndt A, Lee SY, Wietek J, Ramakrishnan C, Steinberg EE, et al. Structural foundations of optogenetics: Determinants of channelrhodopsin ion selectivity. Proc Natl Acad Sci USA. 2015;113:822–29. doi: 10.1073/pnas.1523341113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Berndt A, Deisseroth K. Expanding the optogenetics toolkit. Science. 2015;349:590–1. doi: 10.1126/science.aac7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Olson KD, Zhang XN, Spudich JL. Residue replacements of buried aspartyl and related residues in sensory rhodopsin I: D201N produces inverted phototaxis signals. Proc Natl Acad Sci USA. 1995;92:3185–89. doi: 10.1073/pnas.92.8.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jung KH, Spudich JL. Suppressor mutation analysis of the sensory rhodopsin I-transducer complex: insights into the color-sensing mechanism. J Bacteriol. 1998;180:2033–42. doi: 10.1128/jb.180.8.2033-2042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sineshchekov OA, Sasaki J, Phillips BJ, Spudich JL. A Schiff base connectivity switch in sensory rhodopsin signaling. Proc Natl Acad Sci USA. 2008;105:16159–64. doi: 10.1073/pnas.0807486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sineshchekov OA, Sasaki J, Wang J, Spudich JL. Attractant and repellent signaling conformers of sensory rhodopsin-transducer complexes. Biochemistry. 2010;49:6696–704. doi: 10.1021/bi100798w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sasaki J, Takahashi H, Furutani Y, Kandori H, Spudich JL. Sensory rhodopsin-I as a bidirectional switch: opposite conformational changes from the same photoisomerization. Biophys J. 2011;100:2178–83. doi: 10.1016/j.bpj.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yi A, Mamaeva NV, Li H, Spudich JL, Rothschild KJ. Resonance Raman study of an anion channelrhodopsin: Effects of mutations near the retinylidene Schiff base. Biochemistry. 2016;55:2371–80. doi: 10.1021/acs.biochem.6b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kolbe M, Besir H, Essen LO, Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 angstrom resolution. Science. 2000;288:1390–96. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- 151.Mahn M, Prigge M, Ron S, Levy R, Yizhar O. Biophysical constraints of optogenetic inhibition at presynaptic terminals. Nat Neurosci. 2016;19:554–56. doi: 10.1038/nn.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Govorunova EG, Cunha CR, Sineshchekov OA, Spudich JL. Anion channelrhodopsins for inhibitory cardiac optogenetics. Sci Rep. 2016;6:33530. doi: 10.1038/srep33530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Govorunova EG, Sineshchekov OA, Spudich JL. Structurally distinct cation channelrhodopsins from cryptophyte algae. Biophys J. 2016;110:2302–04. doi: 10.1016/j.bpj.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Brown LS, Dioumaev AK, Lanyi JK, Spudich EN, Spudich JL. Photochemical reaction cycle and proton transfers in Neurospora rhodopsin. J Biol Chem. 2001;276:32495–505. doi: 10.1074/jbc.M102652200. [DOI] [PubMed] [Google Scholar]
- 155.Watanabe M, Furuya M. Action spectrum of phototaxis in a cryptomonad alga, Cryptomonas sp. Plant Cell Physiol. 1974;15:413–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


