Abstract
Cells are bombarded by extrinsic signals that dynamically change in time and space. Such dynamic variations can exert profound effects on behaviors, including cellular signaling, organismal development, stem cell differentiation, normal tissue function, and disease processes such as cancer. Although classical genetic tools are well suited to introduce binary perturbations, new approaches were necessary to investigate how dynamic signal variation may regulate cell behavior. This fundamental question is increasingly being addressed with optogenetics, a field focused on engineering and harnessing light-sensitive proteins to interface with cellular signaling pathways. Channelrhodopsins defined optogenetics; however, through recent use of light-responsive proteins with myriad spectral and functional properties, practical applications of optogenetics currently encompass cell signaling, subcellular localization, and gene regulation. Now, important questions regarding signal integration within branch points of signaling networks, asymmetric cell responses to spatially restricted signals, and effects of signal dosage versus duration can be addressed. This review summarizes emerging technologies and applications within the expanding field of optogenetics.
Keywords: light-responsive, illumination, spatiotemporal, signaling, photoswitching
INTRODUCTION
Cells reside in dynamic microenvironments where input signals or stimuli often fluctuate in both time and space. For example, highly complex spatiotemporal activation of signaling cascades during development controls the transition of stem cells into patterned specific tissues, electrophysiological communication within neuronal circuits can modulate the behavior of an organism, and misregulation of signals can lead to tumorigenesis. Signaling cascades and gene regulatory networks within such cells can be viewed as computational devices that sense and either integrate or discard extracellular input to specify downstream behaviors.
Investigation of the dynamic behavior of such signaling events would be greatly aided by the capacity to introduce dynamic perturbations. However, although classical genetic approaches (overexpression and knockdown/knockouts) to investigate signaling have led to considerable advances in cell biology, they typically involve binary gain or loss of function but offer comparatively little control over spatial or temporal variation in a target pathway (1). Similarly, precise control over signal dosage and location is difficult to achieve with small molecules or protein ligands owing to the binding kinetics of proteins, signaling pathway desensitization, and diffusion of small molecules through the cytoplasm or extracellular matrices. By comparison, light offers numerous advantages as a ligand to stimulate cell signaling (Figure 1a). Specifically, light is readily modulated in space, time, and intensity, allowing for exquisite stimulation of a system by an inexpensive, penetrating, and highly specific substrate that can be added or removed completely by the flick of a switch. Early approaches to optical control involved signaling molecule analogs that could be specifically caged or uncaged in response to light (2, 3). However, the field of optogenetics evolved toward light-activated molecules that could be genetically encoded, aided by the discovery of light-responsive proteins in the eye, plants, fungi, jellyfish, coral, bacteria, and algae (4–6). These proteins have since been engineered, as genetic fusions, into optogenetic systems (input through wavelength-specific illumination) that can control neuronal activity, direct subcellular localization of protein activity, turn protein functionality on or off, promote gene expression or repression, or induce protein degradation (output) (Figure 1b,c) (7–9). With the development of this broad toolbox, the optogenetic field is poised to address questions related to how temporal signaling dynamics impact cell function, how signals are integrated or filtered within key nodes of signaling pathways, and how highly localized activation or repression of proteins can drive cell behavior in a range of biological systems (Figure 1c). We discuss the history of optogenetics, provide an overview of the current technologies available, and comment on current and future applications.
Figure 1.
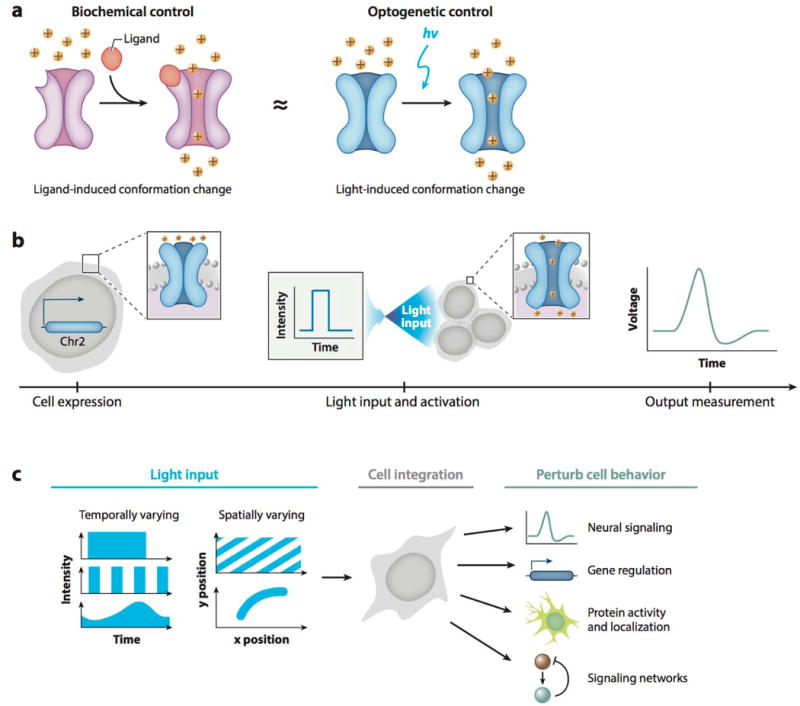
User-defined control of protein behavior using optogenetics. (a) Light-activated channelrhodopsins (blue, right) mimic neurotransmitter-activated ion channel opening (violet, left). (b) Experimental workflow of a typical optogenetic experiment. Optogenetic constructs are stably expressed in a cell (left); user-defined illumination patterns activate optogenetic proteins (middle); cell output, such as an action potential, is measured (right). (c) Biological applications of optogenetic methods. Temporally and/or spatially varying input signals (left) are integrated by a cell (middle) to control and study a variety of cell behaviors (right).
HISTORY
Neuroscience provided the initial drive toward what would eventually become the field of optogenetics. Francis Crick was one of the first to recognize the potential power of light as a highly specific method to control cellular activation in both space and time (10, 11). In particular, he observed that light could offer neuroscience the unique capability to inactivate a specific neural type within a brain at a given time, without disturbing its neighbors, to understand the function of particular subpopulations of neurons (10). The subsequent development of light-responsive caged glutamate and other neurotransmitters, where light breaks a chemical bond to free the ligand, allowed for limited control over the spatial and temporal activation of specific neuronal populations; however, this methodology is irreversible, does not permit controlled dynamic fluctuations, and is difficult to implement within living organisms (2, 12). The revolution in optogenetics truly began with the discovery and development of genetically encoded, light-sensitive proteins that could be engineered into chimera and fusion protein photoswitches that activate target signaling pathways.
Light-sensitive proteins can be found in all three kingdoms and serve diverse functions, such as directing bacteria or plants toward light, controlling the timing of growth or flowering, producing energy through photosynthesis, and organismal vision. Rhodopsins and other light-sensitive opsins, first discovered in mammalian eyes, consist of a G-protein-coupled receptor bound to a chromophore, most commonly retinal, a vitamin A aldehyde (4, 8). One of the first engineered signaling pathways controlled using the optical properties of rhodopsin was chARGed, a signaling cascade consisting of three Drosophila proteins: blue light–sensitive rhodopsin, an α G-protein subunit, and arrestin-2 (6, 11). When overexpressed in mammalian neurons, the cascade nonspecifically activated cation channels to result in neuronal firing. However, chARGed exhibited slow kinetics and high variability. To improve specificity, the intracellular loops of rhodopsin were replaced with the corresponding domains of other G-protein-coupled receptors to gain optogenetic control over specific neural signaling pathways (8). However, the resulting OptoXRs were still limited by the slow kinetics of signal activation via a multiprotein signaling cascade. A single-component, rapid-firing system was necessary, and the cation-permeable channelrhodopsins held the answer (1, 11) (Figure 1a). In a landmark work, when ChR2 was expressed in hippocampal neurons, those neurons exhibited rapid firing in response to light (Figure 1b) (4). In subsequent years, various rhodopsins with specific selective permeabilities were discovered in archea, prokaryotes, and eukaryotes and expanded the optogenetic toolkit for neuroscience research (1, 4).
In recent years, optogenetic tools have extended well past opsins. Many other proteins that contain light-responsive domains, many of which bind chromophore cofactors, have been discovered in plants, corals, bacteria, fungi, and invertebrates and have unique properties that expand the range of potential photoswitches. Specifically, photoreceptors that undergo conformational changes, reversible protein-protein interactions, and oligomerization can be engineered to drive forward specific activities, including the (de)activation of specific protein activities, transcriptional activation or repression, subcellular localization, localized reactive oxygen species production, and other functions (Table 1). The strategic use of these photoswitches to gain a deeper understanding of biological functions within individual or populations of cells, in vitro or in vivo, by merely flicking a light switch, is pushing the boundaries of experimental biology in a range of fields.
Table 1.
Summary of optogenetic system characteristics
| Opto system | Ta1/2 | Td1/2 | λon (nm) | λοff (nm) |
Size (AA) | Chromophore | Mechanism | Applications | Limitations | References |
|---|---|---|---|---|---|---|---|---|---|---|
| ChR2 | ~0.2ms | ~15 ms | 470–480 nm | – | 737 | Retinal (endogenous) | Channel opening | Neuron firing, signaling network control | Some desensitization | 4,15 |
| AsLOV2 | ~Seconds | 30–50 s | <500 nm | – | 143 | FAD (endogenous) | Caging | Protein activation/deactivation | On and off switch times depend on fusion, sensitive to fusion orientation and linker lengths, dark-state activity | 28 |
| Phy/Pif | 1.3 s | 4s | 650 nm | 750 nm | Pif: 100 PhyB: 908 |
Phycocyanobilin (exogenous) | Dimerization | Protein interactions/signaling, subcellular localization | Large PhyB, sensitive to expression level, exogenous chromophore | 30 |
| BphPl/PpsR2 | 3–30 s | 15 min (dark), 1.6× faster using λ off | 740–780 nm | 640–660 nm | PpsR2: 465 BphP1: 732 |
Biliverdin (endogenous) | Dimerization | Protein interactions/signaling, subcellular localization | Large domains | 35 |
| Cry2PIIR/CIBN | 90% complete in 10 s | 100% complete in ~12 min | 420–490 nm | – | Cry2PHR: 498 CIBN: 170 |
FAD (endogenous) | Dimerization | Protein interactions/signaling, subcellular localization | Potential background oligomerization of Cry2, sensitive to fusion orientation | 39 |
| FKFI/Gigantea | ~5–10 min | ~Hours | <500nm | – | FKF1:6I9 GI: 1,173 |
FMN (endogenous) | Dimerization | Protein interactions/signaling, subcellular localization | GI is very large, slow kinetics, sensitive to expression level | 49 |
| EL222 | <10s | <50 s | <500nm | – | 222 | FAD (endogenous) | Dimerization | DNA binding and transcription regulation | Single DNA recognition site | 52 |
| Magnets (pMagFast2(3×)/nMagHigh1) | 1.5 s | 6.8 s | <500nm | – | nMag: 152 pMag: 150 |
FMN (endogenous) | Dimerization | Protein interactions/signaling, subcellular localization | Tandem repeats of nMag/pMag needed for sufficient dynamic range | 56 |
| TULIP | ~Seconds | ~1 min | <500nm | – | AsLOV2: 125 ePDZ: 194 |
FAD (endogenous) | Dimerization | Protein interactions/signaling, subcellular localization | Potential crosstalk with endogenous pathways, sensitive to fusion orientation | 58 |
| iLID | ~30 s to 1 min | ~Minutes | <500 mn | – | iLID: 148 SspB: 113 |
FAD (endogenous) | Dimerization | Protein interactions/signaling, subcellular localization | Some background dark-state binding | 61 |
| Cry2PHR oligomerization | ~30 s | 5.5 min | 420-490 nm | – | 498 | FAD (endogenous) | Oligomerization/clustering | Protein interactions/signaling | Oligomerization mechanism unknown, large protein | 62,64 |
| UVR8/COP1 | ~Hours | Irreversible | 280-315 nm | – | UVR8:440 COP1: 675 |
Internal | Dimerization | Protein interactions/signaling | Irreversible dimerization, large proteins, slow kinetics, phototoxicity from UV illumination | 41 |
Abbreviations: AA, amino acid; FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide; GI, Gigantea; λ, wavelength; Ta1/2, half-life of association; Td1/2, half-life of dissociation.
OPTOGENETIC SYSTEMS
Ion Channels
A large number of efforts in the field of optogenetics involves the discovery, modification, and subsequent application of light-responsive ion channels to modulate various cellular activities, the majority of which belong to the microbial opsin family of proteins.
Microbial opsins/rhodopsins
Microbial (or type I) opsins are a class of light-sensitive, seven-pass transmembrane proteins that perform a variety of functions across several species (13). When an opsin incorporates the vitamin A–derived cofactor retinal, it becomes light responsive and is referred to as a rhodopsin. One of the earliest additions to the optogenetic toolbox, channel-rhodopsin, is a member of this protein family. The two widely used channelrhodopsin variants, ChR1 and ChR2, were isolated from the green freshwater alga Chlamydomonas reinhardtii (14, 15), and they mediate the flow of cations across the cell membrane upon illumination with blue light (Figure 2a). It was originally believed that ChR1 is proton selective; however, subsequent studies have shown that both ChR1 and ChR2 allow the passage of a variety of cations, including Na+, K+, and Ca2+ (16–18). A single gene encodes for both the light-sensitive and effector domains in channelrhodopsins, and they exhibit fast on and off kinetics in response to blue light, thus making them ideal for introduction in non-light-sensitive cells to achieve optical control over a range of different cellular phenomena (Table 1).
Figure 2.
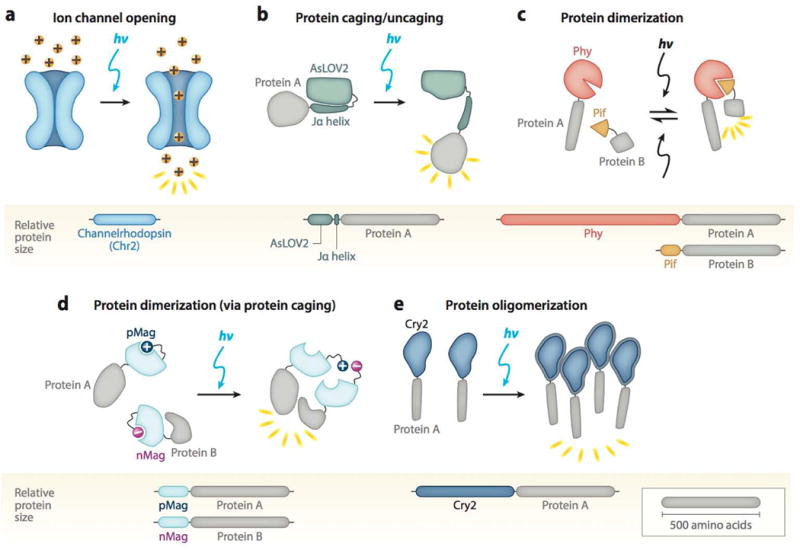
Representative schematics of the five major classes of optogenetic systems. Protein size indicated by relative size of bars, and sample proteins A/B are drawn to a scale of 500 amino acids. (a) Light-induced ion channel (blue) opens in response to blue light to allow cation passage (yellow balls). (b) Jα helix unfolds from AsLOV2 core (green) to uncage a fused protein (gray). (c) Reversible interaction between Phy (red) and Pif (orange) leads to dimerization of attached domains (gray). (d) Light-induced uncaging of affinity domains [positive domain of pMag (dark blue) and negative domain of nMag (pink)] results in dimerization of attached domains (gray). (e) Cry2 (blue) clusters an attached protein (gray) in response to light.
Other type I opsins, including halorhodopsins (chloride pumps), have been considered to inhibit neuronal firing (13). However, this process is highly inefficient, as only one Cl− ion is transported per photon absorbed by the protein. In 2014, a crystal structure–guided study of a modified channelrhodopsin protein, C1C2, guided the conversion of this cation channel into a light-responsive chloride (Cl−) channel (19). One of these variants, iC1C2, exhibited enhanced physiological inhibition in neurons without the requirement of a major membrane potential. Another variant, termed SwiChRCT, showed comparatively enhanced stability and improved light sensitivity over its predecessors in the field.
Caged Proteins
Light control over a protein’s activation state is a powerful tool for investigating and controlling cell behavior in a precise, noninvasive manner. Optogenetic systems have been developed to retain a target protein in an inactive (i.e., caged) state via steric hindrance from a specialized, light-responsive domain; however, light stimulation induces a conformational change that liberates the protein to interact with its substrate or target protein (i.e., light-induced uncaging). Such optogenetic tools, predominantly based on a plant light, oxygen, and voltage photosensory domain, allow reversible control over protein signaling.
Light, oxygen, and voltage domain caging
A widely studied and used caging domain is the light, oxygen, and voltage (LOV) domain isolated from the plant photosensor phototropin 1 (phot1). Originally studied for its involvement in light-responsive plant growth, phot1 contains two LOV domains that mediate the activity of its kinase effector domain through blue light– responsive uncaging (20, 21). The second LOV domain, LOV2, plays the dominant role in kinase activation (22) and has been engineered as an optogenetic tool for light control of mammalian protein activity (23). In the dark state, the Avena sativa (oat) LOV2 domain (AsLOV2) binds its C-terminal helix, Jα, which resides in a folded state against the LOV domain core (24) (Figure 2b). Photoactivation with blue light converts the noncovalent interaction between the LOV core and its bound flavin chromophore, FMN, into a covalent one through a conserved cysteine residue. The accompanying light-induced conformational change displaces the Jα helix away from the protein core, leading to uncaging of a fused effector domain (e.g., the kinase domain of phot1) (24, 25). The Jα helix reverts to its dark-state caged conformation within minutes owing to spontaneous decay of the protein-cofactor bond (Table 1). Optogenetic methods can mimic this light-induced uncaging of the phot1 kinase domain by fusing signaling domains of interest to AsLOV2, in place of the phot1 domain.
However, several limitations of the native AsLOV2 domain have motivated efforts to engineer improved variants. First, when fused to foreign protein domains, spontaneous undocking of the Jα helix can lead to a relatively high dark-state activity, resulting in a low dynamic range upon AsLOV2 uncaging (26). For example, the light-inducible DNA-binding system LovTAP has only a fivefold change in DNA affinity between the dark and illuminated states (27). To address this issue, Strickland et al. (26) used rational design to introduce four mutations into AsLOV2 that stabilized the docking of Jα to the LOV core. This increased the dynamic range of LovTAP from 5-fold to 70-fold, an approach that can be applied to other LOV domain optogenetic systems to reduce dark-state activity. AsLOV2 fusions are also particularly sensitive to linker lengths and the size and structure of attached domains (28, 29), and as a result, each new fusion protein switch requires optimization to achieve low dark-state and high light-state activity in mammalian cells.
Dimerizing Systems
Protein-protein interactions are a frequent driver of molecular processes. The ability to control such interactions with light is a powerful tool for controlling many aspects of cell behavior. Light-inducible dimerizers are the most diverse class of optogenetic systems, with varying properties such as homo-versus heterodimerization, binding kinetics, reversibility, and absorption spectra, which allow for a wide variety of applications to biological systems.
Phytochromes
Phytochromes are a class of proteins that regulate plant growth and development in response to red light. The mechanism of Arabidopsis thaliana phytochrome B (PhyB) is particularly well studied and has been harnessed for optogenetic control of protein-protein association (30, 31). PhyB contains two primary structural domains: a light-absorbing N-terminal photosensory domain and a C-terminal effector domain that interacts with downstream pathway components (32). The PhyB light-sensing capability arises from a tetrapyrrole chromophore, such as phycocyanobilin, covalently bound by the photosensory domain. Absorption of red (650-nm) light triggers chromophore photoisomerization, a rotation about a carbon-carbon double bond that is reversed with the absorption of far-red (750-nm) light (31). This light-induced structural change triggers a conformational change in the surrounding protein that in turn modulates the biological activity of the photoreceptor. In the dark, or after absorption of far-red light, PhyB is biologically inactive and does not bind effector proteins; however, upon illumination with red light, PhyB binds to a class of target transcription factors termed phytochrome-interacting factors (Pifs) (33) (Figure 2c). This light-induced, reversible Phy/Pif dimerization is harnessed to optogenetically stimulate protein-protein interactions in mammalian cells (30). Fusion of proteins or localization signals of interest to optimized, truncated versions of PhyB and Pif6 (the so-called Phy/Pif system) can be used to control the interaction state of the attached domains with red and far-red light (30, 34). Recently, a near-infrared-responsive alternative to the Phy/Pif system has been engineered based on the reversible interaction of bacterial phytochrome BphP1 with its partner PpsR2 (35) (Table 1).
The main advantages of the Phy/Pif and BphP1/PpsR2 systems are their triggered reversibility and speed. That is, unlike most optogenetic systems whose excited state decays spontaneously, the dissociation of these systems can be induced with light. Moreover, the fast on/off rate of Phy/Pif (with half-times on the order of seconds) allows for studies that perturb protein behavior at precise and/or short timescales. Such tools are particularly valuable for studying the dynamics of intracellular signaling networks and of signal presentation during cell development. Lastly, such phytochrome-based dimerizing systems are responsive to long-wavelength (red to near-infrared) light, whose lower-energy photons are less phototoxic to cells compared with blue or UV light and can penetrate deeper into tissues. That said, several considerations remain for Phy/Pif and BphP1/PpsR2 expression in mammalian cells. Whereas the BphP1 chromophore is endogenously present in mammalian cells, the PhyB chromophore is not and requires purification and supplementation to the cell culture medium or injection in vivo for cell uptake prior to photoactivation (30, 36, 37). Pharmacokinetic considerations could lead to variable outcomes in vivo. In addition, the cell expression levels of the phytochromes, as well as the expression ratio between them and their binding partners, have an effect on the dimerization efficiency (38). Lastly, phytochromes are large proteins that can be sensitive to fusions and linker lengths, potentially complicating the development of more complex systems (36).
Cryptochromes for dimerization
Another such dimerizing module is the cryptochrome 2 (Cry2) and CIB1 protein pair, derived from the plant A. thaliana. It consists of a basic helix-loop-helix protein, CIB1, and a blue light–sensitive protein, Cry2, that binds CIB1 in its photoexcited state. Unlike several such dimerizers, this module does not require any exogenously added cofactors, as it uses chromophores ubiquitously present in biological systems. Specifically, Cry2 becomes light responsive as it binds the flavin and pterin chromophores in its conserved N-terminal photolyase homology region (PHR). Kennedy et al. (39) showed that the PHR domain of Cry2 (Cry2PHR) alone, when used with a full-length CIB1 or CIB1 lacking the basic helix-loop-helix domain (CIBN), was sufficient to mediate robust and specific light-sensitive protein dimerization. Both Cry2/CIBN and Cry2PHR/CIBN pairs had very similar association and dissociation kinetics: The dimerization reached ∼90% completion within a matter of seconds of exposure to a 100-ms blue-light (488-nm) pulse, whereas the dissociation took place more gradually over an ∼10 min period (Table 1). The observed dimerization and dissociation kinetics were highly reproducible over successive blue light–exposure cycles, indicating stable maintenance of a high level of efficacy in the system. In addition, two-photon excitation at ∼860 nm was reported to be sufficient to activate the interaction (39). Finally, Taslimi et al. (40) recently reported two truncated versions of Cry2, Cry2(515) and Cry2(535), that exhibit significantly reduced background interaction with CIB1 in the dark. Also, two mutants of Cry2, L348F and W349R, showing slower and faster dissociation kinetics with CIB1 were reported.
Ultraviolet response locus 8
During the course of normal growth, plants are exposed to high levels of ultraviolet B (UVB) wavelengths (280–315 nm), and they have evolved UVB-specific response pathways to mitigate the risk of DNA damage. Constitutively photomorphogenic 1 (COP1), an E3 ubiquitin ligase, functions as a repressor of this UVB response under dark conditions by binding to and targeting UVB response transcription factors for degradation (41, 42); however, UV response locus 8 (UVR8) blocks COP1 in the presence of light. UVR8 exists as a homodimer in the absence of UVB (42), but adjacent tryptophan residues serve as an internal chromophore to induce UVR8 monomerization upon UVB excitation (43). The resulting monomers translocate into the nucleus and bind to COP1, specifically the C-terminal WD-40 domain, and thereby liberate COP1-bound transcription factors (44, 45). This irreversible binding of the UVR8 monomer to COP1 can be harnessed for optogenetics (Table 1).
To further expand the potential for control of multiple cellular events through different and specific excitation wavelength responses, optogenetic systems responsive to excitation wavelengths other than blue or red are necessary. For example, GFP-tagged UVR8 can translocate into the nucleus to bind to a nuclear mCherry tagged COP1 WD-40 domain (COP1C340) upon UVB excitation in U2OS cells (46). Cargo proteins fused to UVR8 or COP1C340 are then irreversibly brought into close proximity, which may promote or repress certain target protein functions. To create larger oligomeric complexes, an even-numbered scaffold of UVR8 monomers can be assembled with linkers and, upon UVB exposure, bind multiple COP1C340 monomers, potentially intensifying an optogentically induced function. In addition, gene regulation can be directly influenced by fusing a transcription factor to COP1C340 and its coactivator to UVR8 (46). Upon UVB excitation, gene expression is induced. An analogous approach could also be used for gene repression. The UVR8/COP1 system is titratable, wherein the level of UVR8 monomerization is proportional to the intensity and duration of the UVB light. UVB exposure can be targeted to specific areas of a single cell using light patterning. However, UVB exposure is known to cause extensive DNA damage. Such damage can be mitigated by short exposure time, low UVB intensity, or most practically, the use of UVB-emitting light-emitting diodes (LEDs) with narrower wavelength bands than UVB fluorescent lamps (46). However, the irreversible nature of the UVR8/COP1 binding is a significant limitation of this system.
Caged proteins for dimerization
Another optogenetic method to induce protein dimerization makes use of the light-induced uncaging of LOV domain-based proteins (see section titled Light, Oxygen, and Voltage Domain Caging). Certain photoreceptors that contain LOV domains possess the inherent ability to dimerize upon blue light exposure, whereas others have been engineered to dimerize using synthetic dimerization domains. The most prominent examples of the former are the FKF1/Gigantea, EL222, and Vivid (VVD)-based systems, while the tunable light-inducible protein (TULIP) and improved light-inducible dimerization system (iLID) systems are examples of the latter (Table 1). The unifying principal is that, upon illumination, a conformational change in the protein exposes a binding site for a specific second protein, resulting in homoor heterodimerization (Figure 2d).
FKF1 is an A. thaliana photoreceptor that regulates flowering onset by sensing blue light levels with its LOV domain (47). In particular, blue light stimulation triggers an FKF1 conformational change that stimulates heterodimerization with its binding partner Gigantea (47, 48), a capability that has been adapted for optogenetic control of protein association in mammalian cells (49). Unlike the canonical Phot1-based LOV uncaging system, however, the dark-state reversion of FKF1/Gigantea binding is on the order of tens of hours (48), allowing long-lasting dimer formation after a single light stimulation, a property that is useful for long-term activation studies but occurs at the expense of temporal and spatial resolution.
The natural homodimerization properties of the bacterial transcription factor EL222 have also been harnessed for optogenetics. Homodimerization is mediated by a LOV domain that, in the dark, binds and inhibits a helix-turn-helix DNA-binding domain (50). Upon light activation, the helix-turn-helix domain is uncaged and mediates the dimerization, yielding a complex that binds a target DNA sequence (50–52). Thus, the most direct application of the EL222 system is for transcriptional control or DNA modification (52). However, upon truncation of its target DNA-binding sequence, EL222 could also be used for the regulation of protein association with rapid on-off kinetics (Table 1).
The fungal photoreceptor VVD is the smallest LOV domain–containing protein. Its N-terminal helix undergoes a conformational change upon photoexcitation (53), leading to homodimerization (54) and thus to close association of attached domains of interest (55). However, VVD’s slow off kinetics (half-life of dissociation, Td1/2 = 2.8 h) and limitation to homodimerization prompted the development of an improved optogenetic system that incorporated either positive or negative amino acids into the VVD N-terminal helix. These so-called Magnets, one with negative (nMag) and one with positive (pMag) residues, preferentially heterodimerize because electrostatic repulsion impairs the natural homodimerization (56) (Figure 2d). Furthermore, multiple variants with faster dissociation kinetics (on the order of seconds) and optimized dimerization efficiencies were created (56, 57). Its small size and fast kinetics make Magnets one of the most robust dimerization systems and allow for precise subcellular control of protein association with high spatiotemporal resolution.
Another class of light-inducible dimerizers uses engineered synthetic peptides to mediate protein interactions. Specifically, the TULIP optogenetic system harnesses LOV domain uncaging to induce dimerization. In the dark state, a peptide epitope fused to the Jα helix of AsLOV2 (LOVpep) is sterically blocked from interaction with the coexpressed synthetic PDZ affinity clamp protein domain (ePDZ) (58, 59). After photostimulation, light-induced unwinding of the LOVpep Jα helix exposes the epitope, leading to dimerization between LOVpep and ePDZ (58). One question that remains to be explored is whether this system is orthogonal to endogenous proteins and signaling pathways, as proteins containing PDZ domains or their binding targets are ubiquitously present in yeast and mammalian cells (60). To address this issue, an iLID was generated using AsLOV2-based uncaging of SsrA, an Escherichia coli peptide that binds the SspB adapter protein (61). Through a phage display selection, this system was engineered to have low dark-state binding, a high dynamic range, and fast on/off kinetics (61).
Oligomerizing Systems
Many signal transduction receptors and effectors undergo oligomerization as part of their signal activation. Hence, optogenetic systems that form multimeric clusters in response to light are useful tools for regulating biological activities that depend on protein oligomerization.
Cryptochromes for oligomerization
In 2013, Bugaj et al. (62) demonstrated that upon blue light stimulation, Cry2PHR—a system previously explored for mediating heterodimerization with partner CIBN—can, on its own, oligomerize in mammalian cells to generate visible protein clusters within ∼10 s (Figure 2e). The number of clusters increased sigmoidally over time, with a half-maximal time of association (Ta1/2) that varied inversely with illumination intensity and had a mean of 30 s. Furthermore, upon light withdrawal, the number of Cry2 clusters decreased exponentially with a half-life of dissociation (Td1/2) of ∼5.5 min (Table 1). Like the Cry2PHR/CIBN pair, Cry2PHR exhibited very little loss in maximal response upon repeated clustering and declustering, indicating the system can be harnessed intracellularly for extended times. Researchers also reported dynamic exchange of Cry2 subunits between different clusters as well as the bulk protein, colocalization of CIB1 along with the Cry2 clusters, and a level of homooligomerization that depended on overall Cry2 expression levels (62). Recent protein purification studies along with multiangle light scattering (SEC-MALS) experiments have shown that Cry2PHR most likely forms a tetramer (63), which may be subject to higher-order oligomerization. Recently, an E490G mutation in the protein has been shown to greatly enhance the amount of clustering in the Cry2 system (64). It was shown that 40–90% of this cytosolic protein, named Cry2olig, immediately formed clusters when exposed to blue light. The clustering kinetics were again reported to be dependent on both the protein concentration in the cell and the light dose, with the half-maximal time of association (Ta1/2) varying between 15 and 75 s. In contrast, the half-life of dissociation (Td1/2 = 23.1 min) is significantly longer in Cry2olig than in wild-type Cry2PHR. Additionally, Cry2olig is capable of incorporating Cry2PHR in its cluster, and the photoresponse can be triggered by using two-photon excitation at a wavelength of 850 nm.
Recently, a Cry2-based clustering module termed CLICR (clustering indirectly using Cry2) was used to achieve endogenous transmembrane clustering and subsequent activation in mammalian and neural stem cells (65). The Cry2/CIBN and Cry2 modules have also found application in several other cellular biological phenomena, including, but not limited to, signal transduction, transcription, and protein-protein interactions (39, 62, 64, 65).
APPLICATIONS
Just as the desire to visualize smaller structures and finer interactions has spurred the progressive development of higher-resolution microscopic techniques, so the drive for a deeper understanding of cellular responses to extracellular inputs has stimulated the technological development and biological applications of optogenetics. One can view a cell as a small biological computer evaluating inputs from its surrounding environment through spatiotemporal filters to determine subsequent output behaviors. To investigate these dynamic input-output relationships, techniques are needed to perturb signaling in real time, and optogenetics offers the exquisite capacity to precisely modulate spatial, temporal, and intensity inputs into target pathways. Applications include addressing what number of a specific class of neurons must be stimulated to initiate a target organismal behavior, or at what signal dosages or durations neurons are desensitized permanently to specific neurotransmitters. Likewise, organismal development is highly dependent on spatial and temporal regulation of signaling cascades and gene expression. The location of a signaling molecule may induce asymmetric cell division or specific migration to a tissue layer. Also, specific temporal application of gene expression may specify cell fate during differentiation. Furthermore, could aberrant spatiotemporal application of signaling ligands or gene expression underlie cancers or genetic or age-related disorders? The potential applications are vast and just beginning to be fully explored. Below we describe a cross section of the current applications of optogenetics.
Synaptic Plasticity/Ion Channel Control
Achieving synaptic control is perhaps one of the most thoroughly explored applications of the optogenetic tools (4, 66). In 2005, Karl Deisseroth and colleagues (4) introduced ChR2 in mammalian neurons and were able to precisely elicit single spikes as well as spike trains over a millisecond timescale using blue light illumination, developments that have been reviewed extensively (8, 67, 68). More recent applications have included restoring vision by holographic optical stimulation of retinal ganglion cells (69), arresting spontaneous epileptic seizures (70), and the development of optogenetic tools for subcellular electrophysiological measurements in the microsecond timescale (71). A vast number of studies have also used these tools to probe different facets of behavior in animal models, e.g., anxiety response (72) or reward-seeking behavior (73).
Genome Modification and Regulation
Optogenetics has proven to be an extremely powerful tool for the spatiotemporal modulation of gene expression, including transcription, translation, and even genome editing. Unlike traditional chemical inducers, which are limited by slow rates of diffusion and removal, optogenetic gene regulatory systems can be controlled orthogonally (without any notable off-target effects) with unparalleled precision by the use of light at specific wavelengths. In seminal work on photoinducible gene expression systems, Shimizu-Sato et al. (74) fused a Gal4-DNA-binding domain to phytochrome (Phy-GBD) and a Gal4-DNA-activating domain to the partner protein PIF3 (PIF3-GAD). Upon red light illumination, Phy-GBD bound the PIF3-GAD fusion protein, thereby activating transcription of genes under Gal4 promoter control. Transcription was switched off upon exposure to far-red light (74). In another study, Hughes et al. (75) used the blue light–responsive Cry2-CIB1 dimerizer system to modulate the transcription of genes under LexA operator control. Next, Wang et al. (55) used a slightly different approach, in which they truncated the Gal4-DNA-binding domain to include only its DNA recognition element (residues 1–65) and eliminated the dimerization domain. They then fused these truncated GBD (1–65) domains to the LOV domain–based VVD system, such that blue light–induced dimerization of the GBD-VVD fusion proteins enhanced the binding of Gal4 to its DNA-binding sequence and thereby activated target transcription. The system exhibited a greatly diminished background, and transcription could be activated using blue light pulses in place of continuous irradiation, thus reducing the chances of phototoxicity (55). In another study, Motta-Mena et al. (52) fused a VP16 transcription activation domain and a nuclear localization signal sequence to the N terminus of EL222 and successfully turned on downstream gene transcription upon photostimulation. This system exhibited a high dynamic range of gene expression, a linear response to light, and rapid activation and deactivation kinetics. To modulate mRNA translation in mammalian cells, Cao et al. (76, 77) used the Cry2PHR-CIBN protein pair to localize the translational activator eIF4E to the mRNA of interest upon blue light illumination, thus activating its translation.
The main limitation associated with the above approaches was the requirement for the exogenous insertion of bulky promoter regions in the cellular genome. More recently, optogenetics has also been used to recruit effectors to endogenous genomic loci, with applications in both transcriptional modulation and genome editing. As an example of the former, Polstein et al. (78) interfaced the Gigantea and FKF1 optogenetic system with a customizable zinc finger DNA-binding protein domain (ZFP) to recruit the VP16 transcriptional activation domain to a DNA sequence of interest, and transcription was turned on upon blue light exposure. Analogously, Konermann et al. (79) used a customizable transcription activator-like effector DNA-binding domain and the transcriptional activator VP64, along with the Cry2-CIB1 optogenetic system, to induce robust transcriptional activation upon photostimulation both in primary neurons and in vivo. This optogenetic tool was also able to modify histone acetylation, thereby exhibiting its capability of potentially tuning epigenetic states and post-translational modifications.
The recent advent of the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats) genome editing system has greatly accelerated the engineering of targeted DNA-binding domains relative to other systems (80), and in 2015, Cas9-mediated blue light–responsive transcriptional modulation systems were designed to target endogenous DNA loci (81, 82). As with ZFP-mediated transcriptional regulation, CIB1 was fused to a catalytically inactive version of Cas9, e.g., dCas9. A suitable guide RNA along with the Cry2-CIB1 optogenetic system was used to localize transcriptional activators such as p65 and VP64 to the DNA sequence of interest, leading to target gene transcription upon light exposure (81, 82).
Recent studies have also developed photoresponsive CRISPR/Cas9 genome-editing methods. For example, truncated N- and C-terminal fragments of Cas9 were designed that are inactive in a monomeric form but regain activity upon dimerization, and fusing these domains to the photoswitchable proteins pMag and nMag led to Cas9 activity reconstitution upon illumination (83). In another approach, Hemphill et al. (84) expanded the genetic code to incorporate photocaged lysine (Lys) residues at specific sites of the Cas9 protein. This engineered version of Cas9 remained inactive in the dark but was activated upon light-induced uncaging of the lysine residues. In both instances, the engineered Cas9, when activated, was able to induce cleavage of specified DNA sequences as well as mediate both nonhomologous end joining and homology-directed repair (83, 84).
Protein Activity and Subcellular Localization
The subcellular localization of a protein can dictate its function, as well as restrict its activity to a desired spatial location. Traditional molecular techniques offer constitutive localization that is not tunable (e.g., fusion of localization tags to proteins of interest). However, many biological studies of cell migration, development, and neural connectivity would benefit from user-defined input of localization onset and duration, and early methodology studies indicate that optogenetics can offer a robust approach to control subcellular protein localization.
Optogenetic bait domains can be localized to specific organelles (e.g., the plasma membrane, nucleus, or peroxisomes) using well-characterized localization tags, and light stimulation can lead to the specific recruitment of effector targets (36, 85). For example, by targeting one optogenetic dimer subunit to the plasma membrane, light-induced dimerization colocalizes the second subunit to the plasma membrane in a manner that can emulate the effects of ligand binding to transmembrane receptors. Such plasma membrane recruitment is widely used for optogenetic control of Rho-family GTPases, providing a dynamic and spatiotemporally precise method to perturb and study cell morphology and movement (30, 49, 58, 61). In a similar manner, light-induced plasma membrane translocation of specific phospholipid kinase and phosphatase domains has been demonstrated to modulate endocytosis and actin dynamics (38, 56, 86–88). A particularly unique application of optogenetic plasma membrane recruitment is the control of calcium (Ca2+) influx (89–91), which has been used to study memory formation in the mouse hippocampus (91). In an optogenetic strategy similar to CLICR (65), light-induced oligomerization of Cry2-fused STIM1 activated the endogenous CRAC calcium channel family and stimulated an intracellular calcium influx. This OptoSTIM1 system was expressed and activated in hippocampal excitatory neurons of wild-type mice that subsequently received either an auditory or a foot-shock stimulus. Investigators found that this increased calcium stimulated contextual fear memory in foot-shocked mice (91).
Optogenetic approaches have also been developed to shuttle individual proteins or protein complexes between cellular organelles. For example, the gene delivery efficiency of adeno-associated virus was increased sixfold and made light tunable by a Pif-bound viral capsid whose nuclear translocation was enhanced by a nuclear localization signal–tagged PhyB (93). A range of strategies have also been developed to control intracellular membrane trafficking (64, 94–96) to study protein secretion, endocytosis, and intracellular protein trafficking. Many optogenetic methods now exist for light control of protein activity and localization, poising the field for powerful behavioral and mechanistic studies on an organismal as well as cellular level.
In a particularly innovative study, optogenetic regulation and subcellular targeting of the small GTPase Rac1 was used to study learning in the mouse motor cortex (97). A series of targeting sequences localized a photoactivatable (fused to AsLOV2) Rac1 to small regions within a neuron’s dendrite termed dendritic spines (Figure 3a,b). These small membranous protrusions receive input from neighboring axons, forming a structural connection that is a locus of synaptic plasticity and has been correlated with learning and memory. When expressed in select neurons of the mouse motor cortex (those involved in motor learning), endogenous synaptic excitation induced translocation of the engineered AS-PaRac1 (activated synapse targeting photoactivatable Rac1) to the now potentiated dendritic spines. In one application, fiber-optic illumination was found to elicit Rac1-mediated dendritic spine shrinkage specifically at the activated synapses in the motor cortex (Figure 3b), and this light-controlled disruption of active synaptic connections was used to study motor learning. Specifically, mice were trained to run on a rotating rod, and this acquired learning was erased upon AS-PaRac1 photoactivation without affecting general motor performance. Interestingly, when learning two tasks (walking on a rotating rod and crossing a balance beam), a fraction of the newly formed synaptic connections were the same between the two tasks, whereas the majority were distinct, suggesting that learning of different tasks was encoded by distinct synaptic patterns. Furthermore, retraining after optical erasure of a trained task elicited synaptic activation of a highly similar pattern of neurons, suggesting that the same task had a conserved synaptic pattern (Figure 3c). This study highlights the power of optogenetics, where gaining precise, tunable control of protein localization can advance fundamental mechanistic knowledge of complex biological processes, such as synaptic memory trace formation.
Figure 3.
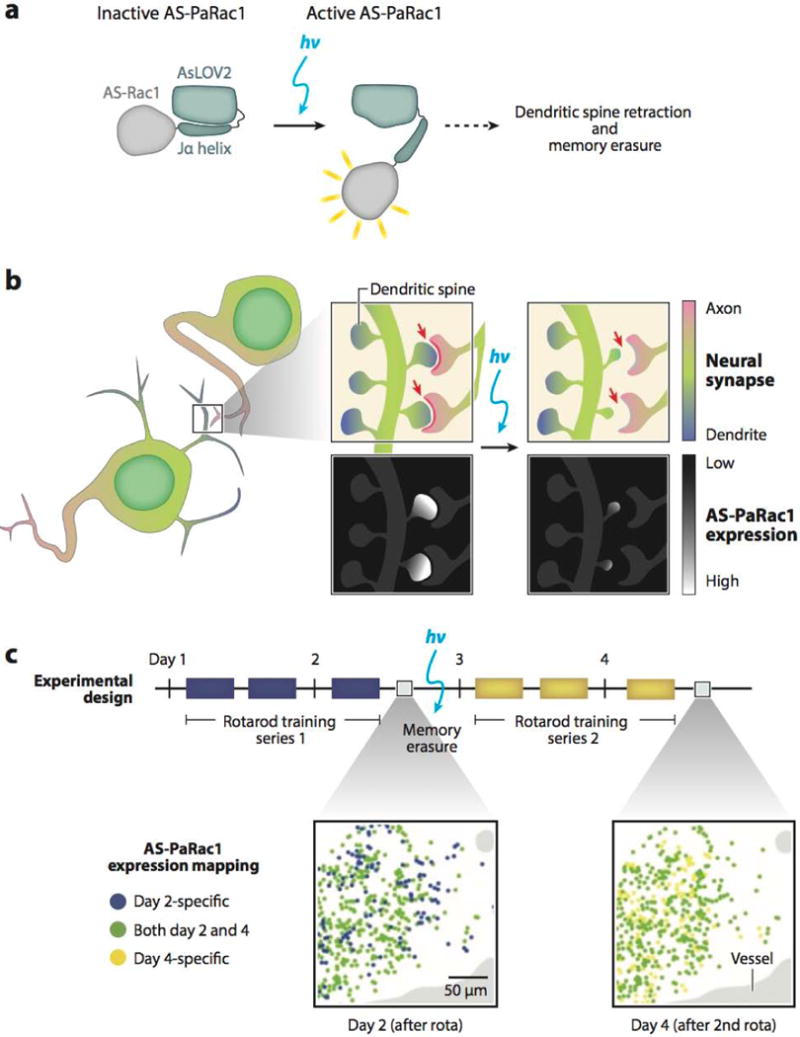
Memory erasure with photoactivatable Rac1 targeted to activated synapses (AS-PaRac1). (a) Schematic of AS-PaRac1 activation in response to a light stimulus. (b) AS-PaRac1 engineered to target active dendritic spines, where blue light stimulation triggers dendritic spine shrinkage and disruption of synaptic connection (red arrows). (c) Repeated motor tasks activate similar synaptic patterns. Expression map of AS-PaRac1 within the adult brain (left) after a series of rotarod trainings and (right) after memory erasure and retraining shows a large amount of overlap in AS-PaRac1 location. Adapted with permission from Reference 97, Macmillan Publishers Ltd., Nature ©2015.
Applications to Signaling Network Dynamics
Extracellular signals activate intracellular signaling cascades that in turn induce changes in cell behavior. Signal perturbations can vary in dosage, persist for short or long temporal periods, and/or be spatially restricted. Because such dynamic variations can impact downstream cell behaviors, several questions arise, including (a) how specific branches or nodes within signaling pathways function to integrate or filter signals, (b) how spatiotemporal modulation of key pathways within a cell determine its activity or fate, and (c) how cells integrate or arbitrate the activation of multiple signaling pathways. Small-molecule regulable signaling systems do not offer the capacity for rapid temporal or spatial modulation; however, optogenetics is extremely well suited for exploring such questions.
Numerous signaling pathways exhibit high levels of crosstalk or branching, such that differing temporal durations or intensities of signal may steer a cell down alternate behavioral responses. Therefore, it is highly desirable to develop optogenetic systems to control key junctures within signaling networks. For example, the Ras/Raf/Mek/Erk cascade is a ubiquitously important pathway that can be activated by multiple extracellular signals and induce a range of sometimes mutually exclusive outcomes (proliferation, differentiation, physiological changes, or cellular arrest/death) (7, 62, 98, 99). Although the activation of specific parallel pathways could guide the final outcome, a specific relationship between signal dynamics and its downstream response is also a strong possibility.
Zhang et al. (99) examined the effects of Raf/Erk signal strength and duration on neurite extension behavior of rat PC12 pheochromocytoma tumor cells. Cry2PHR was fused to Raf1 (Cry2-Raf1), and upon excitation with blue light this fusion translocated to plasma membrane– bound CIBN, where Raf1 is activated and stimulates downstream Mek/Erk activity. Interestingly, neurite outgrowth was proportional to the total duration of on-time light exposure, regardless of pulse interval, up to an off-time threshold of 45 min. When the off-time exceeded 45 min, neurite outgrowth decreased significantly, demonstrating a filtering effect wherein the cell is insensitive to signal oscillations up to a specific off-time threshold.
Toettcher et al. (98) developed a reversible and titratable optogenetic system (opto-SOS) to examine how information is passed from Ras to Erk. Pif was fused to the catalytic domain of SOS (a guanine nucleotide exchange factor and Ras activator dependent on membrane localization for activity), and PhyB was tagged to the plasma membrane. Upon red light stimulation, Pif translocated to the membrane to activate the Ras/Erk signaling pathway. Short (<4 min) light pulses were not transmitted by the pathway, whereas sustained activation (>10 min, i.e., a low-frequency input) elicited a cellular response. The Ras/Erk pathway thus acts as a low-pass filter, filtering away any transient signals. Furthermore, results of a proteomic screen showed that with sustained Ras/Erk activation certain downstream proteins respond quickly to any sustained stimulus (>10 min), whereas others require much longer activation (>1 h) to become active. This suggests that the nature of the elicited physiological response can be encoded by the duration of a presented signal. Investigation of these questions with conventional agonists—such as exogenous growth factors, small molecules, or chemical inductions—would have been confounded by slow agonist diffusion and binding kinetics, receptor internalization and desensitization, and slow signal reversibility.
Canonical Wnt signaling, which is initiated by Wnt3a binding to Frizzled and its coreceptors LRP5/6 and culminates in the inhibition of GSK-3β and induction of β-catenin activity as a transcriptional coactivator, is an important pathway that plays critical roles in development, homeostasis, and disease. To enable investigations of how input signals map to downstream cellular responses, our group fused the C-terminal portion of LRP6 (LRP6c) to the light-responsive domain of cryptochrome2 (Cry2PHR) (62). Upon blue light exposure, Cry2PHR-LRP6c forms homo-oligomers within 30 s that are reversible within ∼10 min of light removal (Figure 4a,b). Downstream β-catenin activity is potently induced within 16 h of blue light illumination in comparison to expression of constitutively active GSK-3β or no illumination (Figure 4c). Furthermore, illumination-induced β-catenin activity showed a dynamic range comparable to induction by Wnt3a ligand or a strong GSK-3β inhibitor (CHIR99021) (62) (Figure 4c,d). In recent work, when Cry2PHR-LRP6c was introduced into adult neural stem cells, sustained illumination led to the strong expression of β3-tubulin, a marker of neuronal differentiation (A. Rosenbloom, L. Bugaj, R. Kane & D.V. Schaffer, unpublished results). Such a potent system could be used to explore the effects of dynamic canonical Wnt signaling on neural stem cell differentiation.
Figure 4.
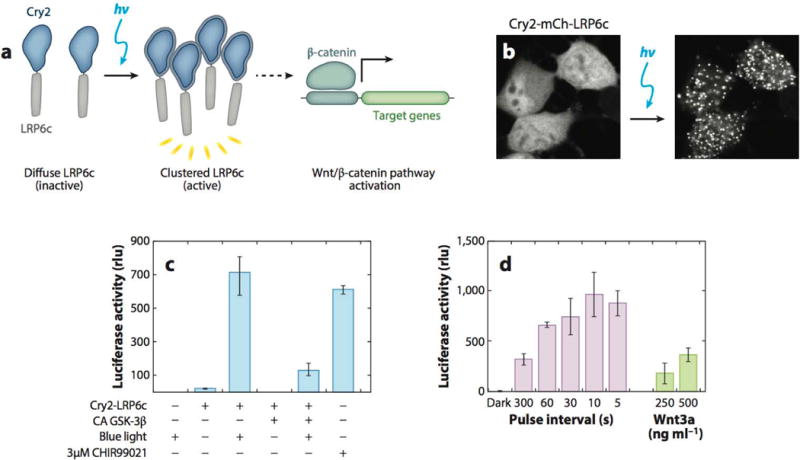
Optogenetic clustering of LRP6c induces Wnt/β-catenin pathway activation in neural stem cells. (a) Schematic of LRP6c oligomerization with Cry2 to induce the Wnt/β-catenin signaling pathway. (b) Live neural stem cell (NSC) fluorescence imaging of Cry2-mCherry-LRP6c upon blue light illumination for 2.5 min. (c) HEK 293 T cells expressing Cry2-LRP6c and a firefly luciferase reporter for β-catenin show Wnt/β-catenin pathway activation upon illumination. Activation is attenuated by constitutively active (CA) β-catenin pathway inhibitor GSK-3β and is comparable to β-catenin pathway activation with the small-molecule agonist CHIR99021. (d) Illumination of NSCs expressing Cry2-LRP6c and a luciferase β-catenin reporter shows increased reporter expression with increasing light dosages (pulse interval defined as time light-off between 500 ms light-on pulses). Wnt3a protein as positive control for Wnt/β-catenin pathway activation. Adapted with permission from Reference 62, Macmillan Publishers Ltd., Nature Methods, © 2013.
Recently, Katsura et al. (87) found the induction of the Akt-FoxO1 pathway to be dependent on the temporal dynamics rather than the total signal dosage. The kinase domain of Akt was fused to Cry2 and expressed in the cytoplasm, whereas CIBN was directed to the plasma membrane. Upon illumination, Cry2-Akt was recruited to CIBN and activated by other kinases. Akt activity was monitored through downstream suppression of atrogin-1 expression by FoxO1. Katsura exposed C2C12 cells to the same total dosage of light but varied the intensity and pulse duration, resulting in temporal patterns that compared contradictory patterns, such as high-intensity, low-frequency pulsing or low-intensity, high-frequency pulsing. Atrogin-1 was successfully repressed by low-intensity high-frequency pulses but not by high-intensity low-frequency pulses. This mechanism suggests a homeostatic feedback mechanism to prevent premature Akt activity when the cell encounters rapid and transient signal induction, preferring high-frequency or continuous signaling to induce pathway activation.
These studies demonstrate how optogenetic systems can be used to address questions previously either impossible or, at a minimum, extremely difficult to explore. The high spatial and temporal resolution as well as precise dosage calibration enable the precise characterization of how signaling circuitry translates or processes a range of dynamic inputs into key downstream behavioral outputs, which both advances basic biological knowledge and facilitates future engineering applications.
ILLUMINATION AND OPTICS TECHNOLOGY
In Vitro Applications
The illumination input for optogenetic applications needs to be shaped by optical systems to control the stimulation timing, duration, location, and intensity. The complexity of the design is wholly dependent on the application; for example, a simple LED array can illuminate a plate of cells, whereas spatial and temporal precision requires microcontrollers, spatial light modulators, scanning mirrors, or other optical devices. A commonly used method for optogenetic activation of bulk cell culture is LED illumination boards, which contain an array of LEDs that uniformly illuminates a large surface area (100) (Figure 5a). Given the variety of LEDs, they can be selected for the necessary intensity and wavelength output. A simple setup can be driven directly by a power supply but can also be connected to a microcontroller (such as an Arduino) that provides a means of regulating the intensity and timing of LEDs (Figure 5b). Intensity is controlled through programmed pulse-width modulation, where the duty cycle of the on-time of the LED is controlled at a very high frequency (much higher than the integration time of our eyes, or cellular events) to achieve a desired light intensity. The timing of illumination, such as pulse sequences or delays, can be programmed as well, providing a method for controlling light input with high temporal resolution. Furthermore, LEDs can be individually addressed by separate channels of the microcontroller, allowing independent control of wells in multiwell plates. Such LED devices are used for bulk cell culture studies, where the precise spatial localization of the light input is not important (62), as well as for high-throughput live-cell screens using optogenetic systems (101).
Figure 5.
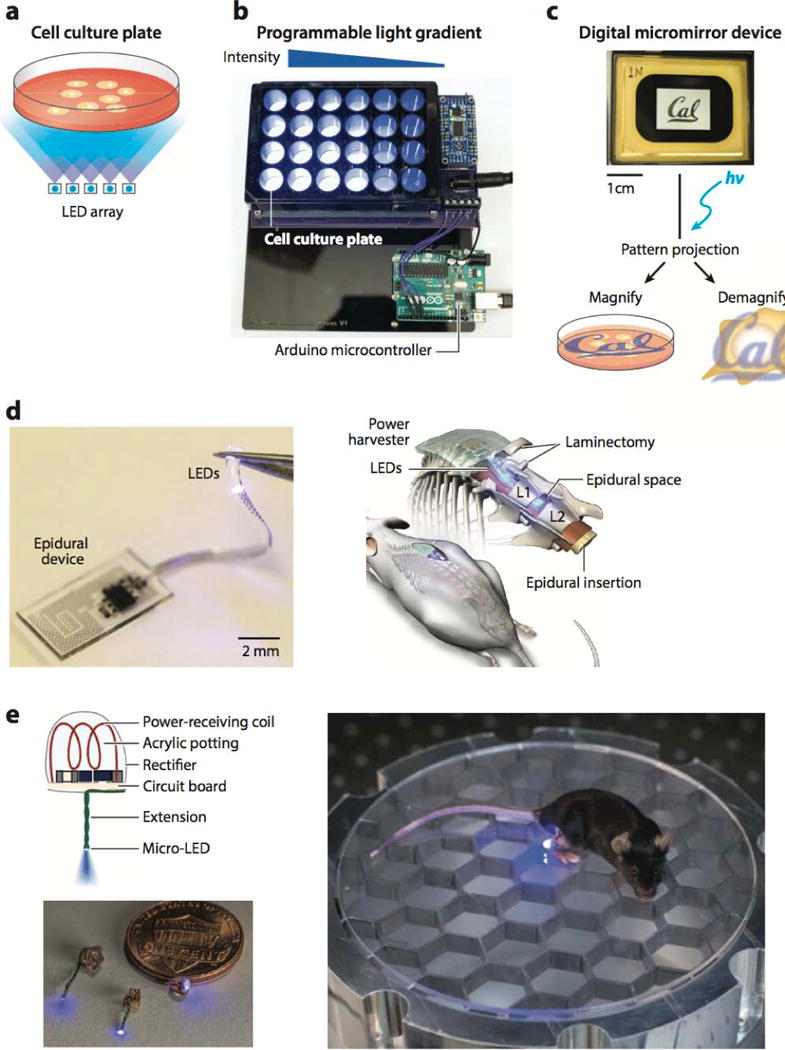
In vitro and in vivo technology for optogenetic stimulation. (a) Schematic of a cell culture plate illuminated with a light-emitting diode (LED) array. (b) Programmable LED array for multiwell cell culture stimulation. (c) Image of a digital micromirror device displaying the UC Berkeley logo. A user-defined pattern can be projected onto a sample and either magnified onto a cell culture plate for large-scale patterning or demagnified for subcellular protein control. (d) Image of assembled wireless illumination device (left) and schematic of device implantation (right) by epidural insertion into mouse spinal cord. Adapted with permission from Reference 131, Macmillan Publishers Ltd., Nature Biotechnology, © 2015. (e) Diagram and image of wireless illumination device (left) and implanted device (right) into live mouse for stimulation of peripheral nerve endings around the rear heel, powered by a resonant radio-frequency cavity below. Adapted with permission from Reference 132 Macmillan Publishers Ltd., Nature Methods, © 2015.
For control of the spatial location of illumination, a variety of existing optical methods have been used or modified. For example, confocal microscopes—the most common means used— incorporate two scanning mirrors that direct the position of a small, focused point of laser light onto the sample, which can be used for photoexcitation. As with fluorescence imaging, the laser intensity is tunable, and the spatial location of illumination can often be specified in software. Because of the small laser point size (∼250 nm in diameter), the spatial resolution of activation is high, and the resulting biological effects can be immediately imaged with confocal fluorescence imaging. For example, Yoo et al. (102) used focal illumination to control the photoactivation of caged Rac1 and thereby study the role of neutrophil migration in the zebrafish embryo. By locally perturbing Rac1, they found that Rac regulates actin polymerization and membrane protrusion at the leading edge, but not the tail edge, of neutrophils migrating toward a site of injury within the embryo.
User-defined light patterns can also be projected onto a sample using a spatial light modulator, such as a digital micromirror device (DMD). Though a scanning mirror system (e.g., a confocal microscope) can work well for focal illumination, the size and shape of the illuminated region are often difficult to control through software. There is also a time delay between the start and end of a scan, making it challenging to control the light dosage and to activate multiple regions of interest. A DMD offers a convenient, programmable alternative to combat these limitations. A DMD is an array of hundreds of thousands of small mirrors (usually ∼10 × 10 μm) that can be incorporated into a standard microscope (103) (Figure 5c, top). Each individual mirror can be programmed to reside in one of two positions: to direct light onto the sample or away from it. Thus, a user-defined pattern can be projected onto a sample with a spatial resolution up to ∼500 nm (Figure 5c, bottom). Because a DMD photoactivates all desired regions at once (as opposed to point-by-point scanning), the control of light dosage, pattern shape, and photoactivation timing is greatly improved. As a proof of concept, a DMD-based microscope has been used to illuminate the Phy/Pif system in localized areas of the plasma membrane (30, 38). One of the main limitations of any spatial control of optogenetic activation is intracellular diffusion, as most optogenetic systems have dissociation times on the order of tens of seconds to minutes that can enable significant protein diffusion away from the intended region of illumination/activation. For this reason, phytochrome-based dimerization systems are most amenable to spatial localization studies because they can be rapidly turned off with a projected negative light pattern (30).
In Vivo Applications
The advent of light-dependent control of neuronal impulses sparked widespread development of novel technologies for optical stimulation within live animals. Small, transparent model organisms (such as Caenorhabditis elegans worms and Drosophila melanogaster flies) can be stimulated with relatively simple systems that use widefield illumination from arc lamps (104, 105), lasers (106), or LEDs (52). As with in vitro methods, spatial precision can be conveyed with a spatial light modulator (107) or a scanning laser (108). Such methods can also be extended to the mammalian brain, where window craniotomies in live animal brains give light access to the superficial layers of the cortex from a microscope (widefield or point-scanning) (109, 110), mounted LED (111), or fiberoptic cable (97, 112, 113). These techniques are further enhanced by engineered red-shifted variants of rhodopsins (114, 115) and rhodopsin two-photon excitation (116, 117) to allow deeper tissue penetration. Two-photon microscopy also allows imaging neural activity deeper in the brain (up to a millimeter below the cortex surface) and, in combination with spatial light modulators, enables optogenetic excitation and imaging at single-cell resolution (118, 119).
Extension of mammalian optogenetic stimulation to deeper organs and brain regions is much more difficult, and the size, opacity, and motility of rodents and nonhuman primates demand the development of implantable optical probes. The first optical neural interface for in vivo optogenetic brain stimulation in live rodents used a laser-coupled optical fiber mounted and passed through the skull of a rat brain (120–122). The fiber was aimed at a spatially restricted neuronal population within the motor cortex to stimulate channelrhodopsin-mediated whisker deflection (120). Spatial precision (∼hundreds of micrometers) of such neural interfaces is conferred by the small fiber opening, and temporal precision (∼milliseconds) via an ultrafast shutter, pulsed laser, or LED (108, 121). Furthermore, optical fibers have been combined with recording electrodes (so-called optrodes) to allow simultaneous optical stimulation and electrophysiological readout (121, 123– 125).
In recent years, more advanced methods have been developed for wireless optical control to allow free animal movement and long-term, minimally invasive studies. Kim and colleagues (126, 127) developed a small, flexible device that incorporates an array of multicolor microLEDs and sensors (electrodes, photodetectors, and temperature sensors) that are inserted into soft tissue with a releasable injection needle. The device is wirelessly controlled and powered through radio frequency scavenging, and the small LED size (∼50 μm) improves spatial resolution and thermal management, leading to reduced tissue damage. However, a head-mountable receiver adds size and weight to the assembly, which can affect rodent behavior. In an effort to overcome this limitation, fully internal devices have been developed. Park et al. (128) also used radio frequency power transmission but engineered a device whose receiver is a stretchable, serpentine antenna that is implanted along with the LED module (Figure 5d). In addition, Montgomery et al. (129) created a fully internal device for surface stimulation of the brain, spinal cord, and peripheral nerve endings (Figure 5e). Weighing only tens of milligrams, the minimally invasive microLED device is implanted in the mouse and is wirelessly powered through induction from a resonant cavity that replaces the mouse cage floor. The device allows long-term stimulation and can be used for novel behavioral studies in which mouse stimulation is linked to defined locations within a cage or maze. Though these optical neural interface technologies are currently used mainly for controlling neuronal action potentials for electrophysiological studies, they can also be applied to future studies that use optogenetics for in vivo control of protein behavior.
SUMMARY AND FUTURE DIRECTIONS
The field of optogenetics has redefined itself since the early days of caged ligands and first-generation opsins. The influx of genetically encoded light-responsive proteins from many types of organisms with varying properties has vastly expanded the toolkit, though there is always a need for new systems, especially in wavelength ranges that are not currently well populated. With existing systems, applications have expanded into control of protein-protein interactions, investigation of signaling network dynamics, up- or downregulation of gene expression, protein activity, subcellular localization, and other applications.
However, optogenetics is not without its limitations. Standardized and improved hardware components for activating optogenetic systems that are also compatible with varying microscopic techniques are much needed, as well as illumination boards with interchangeable wavelengths and plate formats (127, 130–133). Phototoxicity is always a consideration when illuminating cells for any length of time. Lasers or LEDs with narrower-wavelength range outputs can mitigate phototoxicity by eliminating damaging wavelengths often emitted by fluorescence lamps without tight excitation filters (134). Alternately, the development of optogenetic systems with greater sensitivity to low levels of activation light would allow for longer exposures at low illumination intensities (135). Subcellular spatial resolution of optogenetic signals within single cells is limited by protein diffusion, though potentially mitigated by the combination of high-resolution illumination patterning with DMDs and directly reversible optogenetic systems, such as Phy/Pif (30). Indeed, the expansion of novel properties of light-responsive proteins is critical to broaden applications. Although optimal photoresponsive protein designs depend on the question to be answered, they share a few common characteristics: (a) They are easily genetically encoded and display (b) a large dynamic range with low or no background activity under dark conditions, (c) minimal perturbation of endogenous cargo protein function and subcellular localization, and (d) specific and rapid response to their activating wavelength.
With the advent of CRISPR technology to specifically edit fusion genes into a genome of interest at the endogenous sites, stable cell lines can be more readily established with physiological levels of fusion protein expression (80). In addition, clonal cell lines that include more than one optogenetic system integrated within the genome become possible. Cells containing multiple optogenetic systems activated by disparate wavelengths would allow for the exploration of the downstream effects of combinatorial, sequential, or orthogonal activation of two signaling pathways, two nodes within a pathway, or multiple protein (de)activation (136).
An important leap for the optogenetic field is the transition from in vitro to in vivo and, from there, toward potential biomedical translation (137–141). When moving from in vitro to in vivo, a significant concern is light penetration into tissues. One possible solution is strategic implantation of wireless, minimally invasive illumination devices into organisms to drive high-wavelength (red or two-photon) optogenetic systems (115, 116, 128, 129). Phototoxicity to surrounding tissues is a potential concern, though it is partially mitigated by illumination with narrow bandwidth and low-power LEDs. The time-consuming and expensive development of transgenic organisms for a specific optogenetic system remains a significant barrier to in vivo studies. Expanded development of cell-type-specific viral delivery systems, such as engineered adeno-associated viruses that infect specific cell tissues and/or use cell-specific promoters, can accelerate research and extend it to larger organisms (130, 142). The use of optogenetic systems to drive regeneration of tissues in vivo using the endogenous stem cell population is an attractive target for translational medicine.
In summary, the field of optogenetics has undergone a massive increase in tool development and expansion in recent years and is now primed for an explosion of novel findings in cellular responses to dynamic spatial and temporal changes. These questions lead us to fundamental questions regarding in vivo dynamics and potential translational applications. The future, one can say, looks very bright.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis-Davies GC. Caged compounds: photorelease technology for control of cellular chemistry and physiology. Nat Methods. 2007;4:619–28. doi: 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nerbonne JM. Caged compounds: tools for illuminating neuronal responses and connections. Curr Opin Neurobiol. 1996;6:379–86. doi: 10.1016/s0959-4388(96)80123-1. [DOI] [PubMed] [Google Scholar]
- 4.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–68. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 5.Hegemann P, Nagel G. From channelrhodopsins to optogenetics. EMBO Mol Med. 2013;5:173–76. doi: 10.1002/emmm.201202387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zemelman BV, Lee GA, Ng M, Miesenböck G. Selective photostimulation of genetically charged neurons. Neuron. 2002;33:15–22. doi: 10.1016/s0896-6273(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 7.Zhang K, Cui B. Optogenetic control of intracellular signaling pathways. Trends Biotechnol. 2015;33:92–100. doi: 10.1016/j.tibtech.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Tischer D, Weiner OD. Illuminating cell signalling with optogenetic tools. Nat Rev Mol Cell Biol. 2014;15:551–58. doi: 10.1038/nrm3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyden ES. A history of optogenetics: the development of tools for controlling brain circuits with light. F1000 Biol Rep. 2011;3:11. doi: 10.3410/B3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorostiza P, Isacoff E. Optical switches and triggers for the manipulation of ion channels and pores. Mol Biosyst. 2007;3:686–704. doi: 10.1039/b710287a. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–57. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagel G, Ollig D, Fuhrmann M, Kateriya S, Mustl AM, et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–98. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 15.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. PNAS. 2003;100:13940–45. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato HE, Zhang F, Yizhar O, Ramakrishnan C, Nishizawa T, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–74. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–14. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsunoda SP, Hegemann P. Glu87 of channelrhodopsin-1 causes pH-dependent color tuning and fast photocurrent inactivation. Photochem Photobiol. 2009;85:564–69. doi: 10.1111/j.1751-1097.2008.00519.x. [DOI] [PubMed] [Google Scholar]
- 19.Berndt A, Lee SY, Ramakrishnan C, Deisseroth K. Structure-guided transformation of channel-rhodopsin into a light-activated chloride channel. Science. 2014;344:420–24. doi: 10.1126/science.1252367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. PNAS. 1999;96:8779–83. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–23. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 22.Christie JM, Swartz TE, Bogomolni RA, Briggs WR. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 2002;32:205–19. doi: 10.1046/j.1365-313x.2002.01415.x. [DOI] [PubMed] [Google Scholar]
- 23.Christie JM, Gawthorne J, Young G, Fraser NJ, Roe AJ. LOV to BLUF: flavoprotein contributions to the optogenetic toolkit. Mol Plant. 2012;5:533–44. doi: 10.1093/mp/sss020. [DOI] [PubMed] [Google Scholar]
- 24.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin lightswitch. Science. 2003;301:1541–44. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 25.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jα helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–92. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 26.Strickland D, Yao X, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Rationally improving LOV domain-based photoswitches. Nat Methods. 2010;7:623–26. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed all osteric protein. PNAS. 2008;105:10709–14. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–8. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu YI, Wang X, He L, Montell D, Hahn KM. Spatiotemporal control of small GTPases with light using the LOV domain. Methods Enzymol. 2011;497:393–407. doi: 10.1016/B978-0-12-385075-1.00016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quail PH. Phytochromes. Curr Biol. 2010;20:R504–7. doi: 10.1016/j.cub.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quail PH. Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 33.Ni M, Tepperman JM, Quail PH. Binding of phytochrome Bto its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–84. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 34.Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–44. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaberniuk AA, Shemetov AA, Verkhusha VV. A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat Methods. 2016;13:591–97. doi: 10.1038/nmeth.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toettcher JE, Gong D, Lim WA, Weiner OD. Light control of plasma membrane recruitment using the Phy-PIF system. Methods Enzymol. 2011;497:409–23. doi: 10.1016/B978-0-12-385075-1.00017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buckley CE, Moore RE, Reade A, Goldberg AR, Weiner OD, Clarke J. Reversible optogenetic control of subcellular protein localization in a live vertebrate embryo. Dev Cell. 2016;36:117–26. doi: 10.1016/j.devcel.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toettcher JE, Gong D, Lim WA, Weiner OD. Light-based feedback for controlling intracellular signaling dynamics. Nat Methods. 2011;8:837–39. doi: 10.1038/nmeth.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–75. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taslimi A, Zoltowski B, Miranda JG, Pathak GP, Hughes RM, Tucker CL. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat Chem Biol. 2016;12:425–30. doi: 10.1038/nchembio.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28:591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heijde M, Ulm R. UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 2012;17:230–37. doi: 10.1016/j.tplants.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 43.Wu D, Hu Q, Yan Z, Chen W, Yan C, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–19. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 44.Cloix C, Kaiserli E, Heilmann M, Baxter KJ, Brown BA, et al. C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. PNAS. 2012;109:16366– 70. doi: 10.1073/pnas.1210898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller K, Engesser R, Schulz S, Steinberg T, Tomakidi P, et al. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 2013;41:e124. doi: 10.1093/nar/gkt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crefcoeur RP, Yin R, Ulm R, Halazonetis TD. Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nat Commun. 2013;4:1779. doi: 10.1038/ncomms2800. [DOI] [PubMed] [Google Scholar]
- 47.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–65. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zikihara K, Iwata T, Matsuoka D, Kandori H, Todo T, Tokutomi S. Photoreaction cycle of the light, oxygen, and voltage domain in FKF1 determined by low-temperature absorption spectroscopy. Biochemistry. 2006;45:10828–37. doi: 10.1021/bi0607857. [DOI] [PubMed] [Google Scholar]
- 49.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat Biotechnol. 2009;27:941–45. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 50.Nash AI, McNulty R, Shillito ME, Swartz TE, Bogomolni RA, et al. Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. PNAS. 2011;108:9449–54. doi: 10.1073/pnas.1100262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rivera-Cancel G, Motta-Mena LB, Gardner KH. Identification of natural and artificial DNA substrates for light-activated LOV-HTH transcription factor EL222. Biochemistry. 2012;51:10024–34. doi: 10.1021/bi301306t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, et al. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–57. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lamb JS, Zoltowski BD, Pabit SA, Crane BR, Pollack L. Time-resolved dimerization of a PAS-LOV protein measured with photocoupled small angle X-ray scattering. J Am Chem Soc. 2008;130:12226–27. doi: 10.1021/ja804236f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–69. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 56.Kawano F, Suzuki H, Furuya A, Sato M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun. 2015;6:6256. doi: 10.1038/ncomms7256. [DOI] [PubMed] [Google Scholar]
- 57.Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol. 2009;5:827–34. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–84. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang J, Koide A, Makabe K, Koide S. Design of protein function leaps by directed domain interface evolution. PNAS. 2008;105:6578–83. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee H-J, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal. 2010;8:1–18. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. PNAS. 2015;112:112–17. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Meth. 2013;10:249–52. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 63.Hallett RA, Zimmerman SP, Yumerefendi H, Bear JE, Kuhlman B. Correlating in vitro and in vivo activities of light-inducible dimers: a cellular optogenetics guide. ACS Synth Biol. 2016;5:53–64. doi: 10.1021/acssynbio.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, et al. An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun. 2014;5:4925. doi: 10.1038/ncomms5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bugaj LJ, Spelke DP, Mesuda CK, Varedi M, Kane RS, Schaffer DV. Regulation of endogenous transmembrane receptors through optogenetic Cry2 clustering. Nat Commun. 2015;6:6898. doi: 10.1038/ncomms7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stirman JN, Crane MM, Husson SJ, Wabnig S, Schultheis C, et al. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nat Methods. 2011;8:153–58. doi: 10.1038/nmeth.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernstein JG, Boyden ES. Optogenetic tools for analyzing the neural circuits of behavior. Trends Cogn Sci. 2011;15:592–600. doi: 10.1016/j.tics.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grosenick L, Marshel JH, Deisseroth K. Closed-loop and activity-guided optogenetic control. Neuron. 2015;86:106–39. doi: 10.1016/j.neuron.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reutsky-Gefen I, Golan L, Farah N, Schejter A, Tsur L, et al. Holographic optogenetic stimulation of patterned neuronal activity for vision restoration. Nat Commun. 2013;4:9. doi: 10.1038/ncomms2500. [DOI] [PubMed] [Google Scholar]
- 70.Krook-Magnuson E, Armstrong C, Oijala M, Soltesz I. On-demand optogenetic control of spontaneous seizures in temporal lobe epilepsy. Nat Commun. 2013;4:8. doi: 10.1038/ncomms2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hochbaum DR, Zhao Y, Farhi SL, Klapoetke N, Werley CA, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11:825–33. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adhikari A, Lerner TN, Finkelstein J, Pak S, Jennings JH, et al. Basomedial amygdala mediates top-down control of anxiety and fear. Nature. 2015;527:179–85. doi: 10.1038/nature15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferenczi EA, Zalocusky KA, Liston C, Grosenick L, Warden MR, et al. Prefrontal cortical regulation of brainwide circuit dynamics and reward-related behavior. Science. 2016;351:aac9698. doi: 10.1126/science.aac9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–44. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 75.Hughes RM, Bolger S, Tapadia H, Tucker CL. Light-mediated control of DNA transcription in yeast. Methods. 2012;58:385–91. doi: 10.1016/j.ymeth.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao JC, Arha M, Sudrik C, Bugaj LJ, Schaffer DV, Kane RS. Light-inducible activation of target mRNA translation in mammalian cells. Chem Commun. 2013;49:8338–40. doi: 10.1039/c3cc44866e. [DOI] [PubMed] [Google Scholar]
- 77.Cao JC, Arha M, Sudrik C, Schaffer DV, Kane RS. Bidirectional regulation of mRNA translation in mammalian cells by using PUF domains. Angew Chem. 2014;53:4900–4. doi: 10.1002/anie.201402095. [DOI] [PubMed] [Google Scholar]
- 78.Polstein LR, Gersbach CA. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J Am Chem Soc. 2012;134:16480–83. doi: 10.1021/ja3065667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–76. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 81.Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. CRISPR-Cas9-based photoactivatable transcription system. Chem Biol. 2015;22:169–74. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 82.Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol. 2015;33:755–60. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 84.Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A. Optical control of CRISPR/Cas9gene editing. J Am Chem Soc. 2015;137:5642–45. doi: 10.1021/ja512664v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang X, Jost AP, Weiner OD, Tang C. A light-inducible organelle-targeting system for dynamically activating and inactivating signaling in budding yeast. Mol Biol Cell. 2013;24:2419–30. doi: 10.1091/mbc.E13-03-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Idevall-Hagren O, DicksonE J, Hille B, Toomre DK, Camilli P. Optogenetic control of phosphoinositide metabolism. PNAS. 2012;109:23. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Katsura Y, Kubota H, Kunida K, Kanno A, Kuroda S, Ozawa T. An optogenetic system for interrogating the temporal dynamics of Akt. Sci Rep. 2015;5:14589. doi: 10.1038/srep14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Neill PR, Gautam N. Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration. Mol Biol Cell. 2014;25:2305–14. doi: 10.1091/mbc.E14-04-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ishii T, Sato K, Kakumoto T, Miura S, Touhara K, et al. Light generation of intracellular Ca2+ signals by a genetically encoded protein BACCS. Nat Commun. 2015;6:8021. doi: 10.1038/ncomms9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pham E, Mills E, Truong K. A synthetic photoactivated protein to generate local or global Ca2+ signals. Chem Biol. 2011;18:880–90. doi: 10.1016/j.chembiol.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 91.Kyung T, Lee S, Kim J, Cho T, Park H, et al. Optogenetic control of endogenous Ca2+ channels in vivo. Nat Biotechnol. 2015;33:1092–96. doi: 10.1038/nbt.3350. [DOI] [PubMed] [Google Scholar]
- 92.Deleted in proof
- 93.Gomez EJ, Gerhardt K, Judd J, Tabor JJ, Suh J. Light-activated nuclear translocation of adeno-associated virus nanoparticles using phytochrome B for enhanced, tunable, and spatially programmable gene delivery. ACS Nano. 2015;10:225–37. doi: 10.1021/acsnano.5b05558. [DOI] [PubMed] [Google Scholar]
- 94.Nguyen M, Kim C, Kim J, Park B, Lee S, et al. Optogenetic oligomerization of Rab GTPases regulates intracellular membrane trafficking. Nat Chem Biol. 2016;12:431–36. doi: 10.1038/nchembio.2064. [DOI] [PubMed] [Google Scholar]
- 95.Chen D, Gibson ES, Kennedy MJ. A light-triggered protein secretion system. J Cell Biol. 2013;201:631–40. doi: 10.1083/jcb.201210119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spiltoir JI, Strickland D, Glotzer M, Tucker CL. Optical control of peroxisomal trafficking. ACS Synth Biol. 2015;15:554–60. doi: 10.1021/acssynbio.5b00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hayashi-Takagi A, Yagishita S, Nakamura M, Shirai F, Wu YI, et al. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature. 2015;525:333–38. doi: 10.1038/nature15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–34. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang K, Duan L, Ong Q, Lin Z, Varman PM, et al. Light-mediated kinetic control reveals the temporal effect of the Raf/MEK/ERK pathway in PC12 cell neurite outgrowth. PLOS ONE. 2014;9:e92917. doi: 10.1371/journal.pone.0092917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tucker CL, Vrana JD, Kennedy MJ. Tools for controlling protein interactions using light0. Curr Protoc Cell Biol. 2014;64(17):16.1–20. doi: 10.1002/0471143030.cb1716s64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ingles-Prieto, Reichhart E, Muellner MK, Nowak M, Nijman SM, et al. Light-assisted small-molecule screening against protein kinases. Nat Chem Biol. 2015;11(12):952–54. doi: 10.1038/nchembio.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM. Differential regulation of protrusion and polarity by PI(3)K during neutrophil motility in live zebrafish. Dev Cell. 2010;18:226–36. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bansal V, Saggau P. 2013 Digital micromirror devices: principles and applications in imaging. Cold Spring Harb Protoc. 2013:404–11. doi: 10.1101/pdb.top074302. [DOI] [PubMed] [Google Scholar]
- 104.Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–84. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 105.Zhang F, Wang L-PP, Brauner M, Liewald JF, Kay K, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–39. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 106.Lima SQ, Miesenböck G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–52. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 107.Guo ZV, Hart AC, Ramanathan S. Optical interrogation of neural circuits inCaenorhabditis elegans. Nat Methods. 2009;6:891–96. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang F, Wang L-P, Boyden ES, Deisseroth K. Channelrhodopsin-2 and optical control of excitable cells. Nat Methods. 2006;3:785–92. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- 109.Ayling OGS, Harrison TC, Boyd JD, Goroshkov A. Automated light-based mapping of motor cortex by photoactivation of channelrhodopsin-2 transgenic mice. Nat Methods. 2009;6:219–24. doi: 10.1038/nmeth.1303. [DOI] [PubMed] [Google Scholar]
- 110.Hira R, Honkura N, Noguchi J, Maruyama Y. Transcranial optogenetic stimulation for functional mapping of the motor cortex. J Neurosci Methods. 2009;179:258–63. doi: 10.1016/j.jneumeth.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 111.Huber D, Petreanu L, Ghitani N, Ranade S, Hroma ´dka T, et al. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, et al. Invivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–18. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yazdan-Shahmorad A, Diaz-Botia C, Hanson TL, Kharazia V, Ledochowitsch P, et al. A large-scale interface for optogenetic stimulation and recording in nonhuman primates. Neuron. 2016;89:927–39. doi: 10.1016/j.neuron.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 114.Chuong AS, Miri ML, Busskamp V, Matthews GA, Acker LC, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat Neurosci. 2014;17:1123–29. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lin JY, Knutsen P, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci. 2013;16:1499–508. [Google Scholar]
- 116.Prakash R, Yizhar O, Grewe B, Ramakrishnan C, Wang N, et al. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods. 2012;9:1171–79. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Papagiakoumou E, Anselmi F, Be`gue A, de Sars V, Glu ¨ckstad J, et al. Scanless two-photon excitation of channelrhodopsin-2. Nat Methods. 2010;7:848–54. doi: 10.1038/nmeth.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Packer AM, Russell LE, Dalgleish HW, Hausser M. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat Methods. 2015;12:140–46. doi: 10.1038/nmeth.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bovetti S, Fellin T. Optical dissection of brain circuits with patterned illumination through the phase modulation of light. J Neurosci Methods. 2015;241:66–77. doi: 10.1016/j.jneumeth.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 120.Aravanis AM, Wang L-PP, Zhang F, Meltzer LA, Mogri MZ, et al. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4:56. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- 121.Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc. 2010;5:439–56. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–24. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang J, Borton DA, Zhang J, Burwell RD, Nurmikko AV. 2010 A neurophotonic device for stimulation and recording of neural microcircuits. Proc IEEE Eng Med Biol Sci. 2010:2935–38. doi: 10.1109/IEMBS.2010.5626296. [DOI] [PubMed] [Google Scholar]
- 124.Anikeeva P, Andalman AS, Witten I, Warden M, Goshen I, et al. Optetrode: a multichannel readout for optogenetic control in freely moving mice. Nat Neurosci. 2011;15:163–70. doi: 10.1038/nn.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee J, Ozden I, Song Y-K, Nurmikko AV. Transparent intracortical microprobe array for simultaneous spatiotemporal optical stimulation and multichannel electrical recording. Nat Methods. 2015;12:1157–62. doi: 10.1038/nmeth.3620. [DOI] [PubMed] [Google Scholar]
- 126.Kim T-i, McCall JG, Jung YH, Huang X, Siuda ER, et al. Injectable, cellular-scale optoelectronics with applications for wireless optogenetics. Science. 2013;340:211–16. doi: 10.1126/science.1232437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McCall JG, Kim T-i, Shin G, Huang X, Jung YH, et al. Fabrication and application of flexible, multimodal light-emitting devices for wireless optogenetics. Nat Protoc. 2013;8:2413–28. doi: 10.1038/nprot.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Park SI, Brenner DS, Shin G, Morgan CD, Copits BA, et al. Soft, stretchable, fully implantable miniaturized optoelectronic systems for wireless optogenetics. Nat Biotechnol. 2015;33:1280–86. doi: 10.1038/nbt.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Montgomery KL, Yeh AJ, Ho JS, Tsao V, Iyer S, et al. Wirelessly powered, fully internal optogenetics for brain, spinal and peripheral circuits in mice. Nat Methods. 2015;12:969–74. doi: 10.1038/nmeth.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chow BY, Boyden ES. Optogenetics and translational medicine. Sci Transl Med. 2013;5:177ps5. doi: 10.1126/scitranslmed.3003101. [DOI] [PubMed] [Google Scholar]
- 131.Wu F, Stark E, Im M, Cho IJ, Yoon ES, et al. An implantable neural probe with monolithically integrated dielectric waveguide and recording electrodes for optogenetics applications. J Neural Eng. 2013;10:056012. doi: 10.1088/1741-2560/10/5/056012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Packer AM, Roska B, Hausser M. Targeting neurons and photons for optogenetics. Nat Neurosci. 2013;16:805–15. doi: 10.1038/nn.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vaziri A, Emiliani V. Reshaping the optical dimension in optogenetics. Curr Opin Neurobiol. 2012;22:128–37. doi: 10.1016/j.conb.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 134.Wojtovich AP, Foster TH. Optogenetic control of ROS production. Redox Biol. 2014;2:368–76. doi: 10.1016/j.redox.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–65. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Muller K, Engesser R, Timmer J, Zurbriggen MD, Weber W. Orthogonal optogenetic triple-gene control in Mammalian cells. ACS Synth Biol. 2014;3:796–801. doi: 10.1021/sb500305v. [DOI] [PubMed] [Google Scholar]
- 137.Tonnesen J, Parish CL, Sorensen AT, Andersson A, Lundberg C, et al. Functional integration of grafted neural stem cell-derived dopaminergic neurons monitored by optogenetics in an in vitro Parkinson model. PLOS ONE. 2011;6:e17560. doi: 10.1371/journal.pone.0017560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zeng H, Madisen L. Mouse transgenic approaches in optogenetics. Prog Brain Res. 2012;196:193–213. doi: 10.1016/B978-0-444-59426-6.00010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Del Bene F, Wyart C. Optogenetics: a new enlightenment age for zebrafish neurobiology. Dev Neurobiol. 2012;72:404–14. doi: 10.1002/dneu.20914. [DOI] [PubMed] [Google Scholar]
- 140.Zalocusky K, Deisseroth K. Optogenetics in the behaving rat: integration of diverse new technologies in a vital animal model. Optogenetics. 2013;1:1–17. [Google Scholar]
- 141.Ruiz O, Lustig BR, Nassi JJ, Cetin A, Reynolds JH, et al. Optogenetics through windows on the brain in the nonhuman primate. J Neurophysiol. 2013;110:1455–67. doi: 10.1152/jn.00153.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet. 2014;15:445–51. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]


