Significance
The ability to identify protein targets for different ligands would have enormous impact on drug discovery, both in the repurposing of known drugs and in the identification of off-targets. Moreover, identifying small-molecule binding sites and associating potential ligands with those sites is important in protein function annotation. The methods described in this paper are designed to accomplish these goals. They rely heavily on the use of protein structural alignment to detect relationships not available from sequence alone and use a measure of protein–ligand interaction similarity to determine whether proteins with similar shapes are likely to bind similar ligands. We present results that compare favorably with existing methods using an approach that can be applied on a genome-wide scale.
Keywords: protein–ligand interactions, drug off-targets, machine learning, structure-based prediction
Abstract
We report a template-based method, LT-scanner, which scans the human proteome using protein structural alignment to identify proteins that are likely to bind ligands that are present in experimentally determined complexes. A scoring function that rapidly accounts for binding site similarities between the template and the proteins being scanned is a crucial feature of the method. The overall approach is first tested based on its ability to predict the residues on the surface of a protein that are likely to bind small-molecule ligands. The algorithm that we present, LBias, is shown to compare very favorably to existing algorithms for binding site residue prediction. LT-scanner’s performance is evaluated based on its ability to identify known targets of Food and Drug Administration (FDA)-approved drugs and it too proves to be highly effective. The specificity of the scoring function that we use is demonstrated by the ability of LT-scanner to identify the known targets of FDA-approved kinase inhibitors based on templates involving other kinases. Combining sequence with structural information further improves LT-scanner performance. The approach we describe is extendable to the more general problem of identifying binding partners of known ligands even if they do not appear in a structurally determined complex, although this will require the integration of methods that combine protein structure and chemical compound databases.
Computational methods that match small ligands to specific proteins they bind have many practical applications including protein function annotation and drug discovery/repurposing. Underlying these goals are related but distinct algorithmic challenges including the following: (i) given a protein, where on its surface does it bind small molecules; (ii) given a protein, what small molecules will it bind; (iii) given a small molecule, what proteins will it bind. There is a large literature on some of these subjects, and this paper is intended to add to this literature. However, as we discuss below, the methods we introduce have distinct features that enable us to account for protein–ligand interactions in the binding site while still allowing large-scale, genome-wide predictions to be made in a relatively limited amount of time on a modern computer cluster.
Problem i, the prediction of residues on a protein surface that bind ligands, has been widely studied. Predicted ligand-binding residues can be used to guide in silico screening of chemical libraries using docking or other approaches. Existing structure-based methods for binding site prediction fall into distinct categories. One involves the identification of binding pockets on the protein surface based for example on surface curvature (1, 2). However, since there can be concave regions on a protein surface that do not bind small molecules, or conversely, convex/flat regions that do, programs such as ConCavity (3) and LIGSITECSC (4) combine pocket finding algorithms with sequence conservation information. FTsite (5) uses docking to probe a protein surface with various types of chemical groups and uses an empirical scoring function to identify surface patches that might favorably interact with those groups. MetaPocket 2.0 (6) and COACH (7) are “metaservers” that combine results from a range of structure-based approaches using machine learning.
Although pocket finding and sequence-based methods are often highly successful, they may miss binding sites that do not display the expected curvature or sequence characteristics. Template-based methods rely on the observation (8) that two proteins that share structural similarity will likely bind ligands at similar geometric locations on their surfaces. This is true even for remotely related proteins (i.e., different SCOP fold) (9), thus enabling the exploitation of both close and distant structural homologs in binding site prediction. Similar observations for protein–protein binding sites led to the development of the PredUs server, which has been shown to be extremely effective in predicting regions on a protein surface that bind other proteins (10–12).
A number of template-based programs that predict ligand-binding site residues have been reported in the past few years. A common feature is the use of geometric alignments to superimpose the structure of a template with a bound ligand (“holo” structure) on a query structure without ligands (“apo” structure). Algorithms such as 3DLigandsite (13) and FINDSITE (14, 15) score residues based in part on the number of superimposed ligands within a fixed distance from that residue. Hybrid methods have also been developed; in particular, the COACH metaserver (7) combines a number of template-based methods, sequence conservation information, and ConCavity.
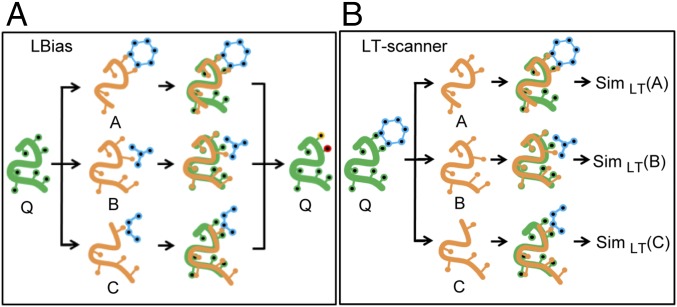
Here, we report a template-based method, “ligand binding site analysis” (LBias). As in other template-based methods, LBias first identifies proteins structurally similar to a query protein that contains a ligand and then places these structures and their ligands in the coordinate system of the query (Fig. 1A). LBias has a unique scoring function that reflects whether the specific types of interactions between the template and its ligand could also form with the query. As will be discussed, LBias performance is found to compare very favorably to existing state-of-the-art methods, in part due to its use of binding site similarity in weighting the contribution of a given template.
Fig. 1.
Overview of LBias and LT-scanner methods. (A) For a given query protein (shown in green) LBias collects and superposes structure neighbors A, B, and C (shown in yellow) that are cocrystalized with their bound ligands (shown in blue). Then LBias predicts the most likely ligand-binding residues (shown in red and yellow) on the query protein based on collective contact information that the superposed ligands make. (B) For a given template cocrystal structure of a drug (shown in blue) and a template protein (shown in green), LT-scanner scans through protein A, B, and C (shown in orange) for by superposing the template structure onto each protein so as to create interaction models. Then LT-scanner calculates the SimLT-scanner interaction similarity score (shown as SimLT) between the interaction models of the query–drug complex and the interactions in the binding site of the template.
The success of LBias suggests that its representation of specific types of protein–ligand interactions might be effective in the prediction of the proteins that bind to a particular ligand (the ligand’s “targets”). With this goal in mind, we developed ligand–target scanner (LT-scanner) a method to predict, on a genome-wide scale, target proteins for a given ligand based on the LBias scoring function. LT-scanner takes a ligand–protein complex structure as input and scans through a protein structure database to identify proteins that might bind to that ligand (Fig. 1B). Several computational approaches have been developed previously for target protein prediction. A number of methods use binding site similarities to predict targets (16, 17). Others involve ligand-based quantitative structure–activity relationships (18–22), although a recently developed approach, FINDSITEcomb (23), combines both template-based and chemical similarity-based approaches. LT-scanner was used to predict known target human proteins of 200 Food and Drug Administration (FDA)-approved drugs that were extracted from drug–target databases (24–27). Its encouraging performance and its ability to account for binding specificity among closely related proteins suggests that the method can be used effectively for both drug repurposing and “off-target” prediction (i.e., unintended targets of a given drug). Notably, using a naive Bayesian network to combine LT-scanner with a sequence-based approach yielded further improvement in performance.
Results
Ligand-Binding Residue Prediction.
A “structural BLAST” approach (8) is used to predict potential ligand-binding residues. A schematic of the LBias workflow is shown in Fig. 1A and is described in detail in Materials and Methods. Briefly, for a given apo query protein, Q, a database of protein structures is searched to identify a set of template proteins, {Ni}, where each Ni is structurally similar to Q and is bound a ligand, Li. (Note that, here and for the purposes of ligand target prediction described below, template proteins with >96% sequence identity are excluded from the database.) Each Ni is superposed on Q, and each Li is also placed in the coordinate system of Q using the same transformation. Interactions that Li makes with Ni [van der Waals (vdW) contacts, hydrogen bonds, aromatic interactions, and ion pairs] are identified, and a score, SIMi, is calculated that reflects whether residues in Q could make the same types of interactions (see Materials and Methods and Eqs. 1–3 for details). For each Ni, SIMi is added to a counter associated with each residue of Q, if any atom of that residue is within 5 Å of Li.
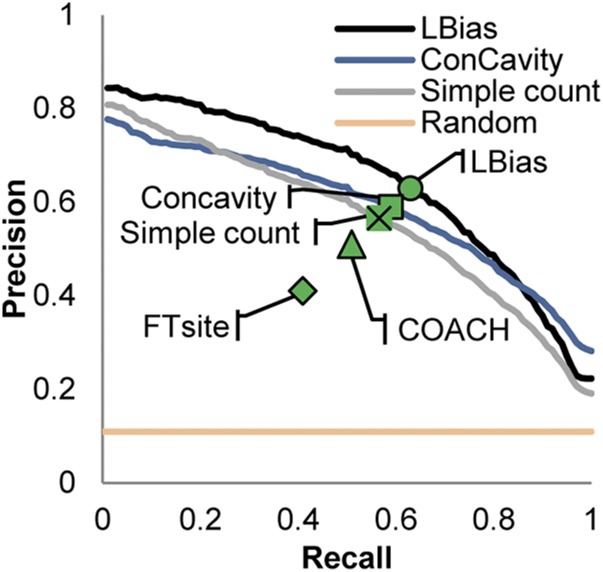
Fig. 2 displays performance measured in terms of precision and recall for ligand-binding residue prediction using a range of benchmark datasets and methods. We first compare LBias performance to a simplified version of the algorithm (“simple count”) using the LigASite (28) benchmark of experimentally determined protein/ligand structures (Materials and Methods). The simple count approach does not account for specific types of protein–ligand interactions and instead reflects only the frequency with which the set of Ni bind their associated Li’s at geometrically similar locations on their surfaces. (In the procedure described above, this is done by setting SIMi = 1 for all i.) As can be seen in the figure (compare the black and gray lines), LBias considerably outperforms “simple count,” demonstrating that there is much added value in the more detailed description of interatomic interactions contained in the SIM score. The blue line in Fig. 2 shows the performance of ConCavity on the LigASite benchmark. As can be seen, LBias outperforms ConCavity over most of the recall range (<0.8).
Fig. 2.
Precision–recall curves for ligand-binding residue prediction. Precision–recall curves are shown for LBias (black line), “simple count” (gray line), ConCavity (blue line), and random prediction (yellow line) in precision–recall curve space. Precision–recall points (PR-point; Results) are shown for LBias, ConCavity, simple count, COACH, and FTsite.
We also compared LBias performance on the LigASite benchmark to other widely used methods: COACH (7), which is template-based, and FTsite (5), which uses docking. These two approaches do not report a score for all residues in a given query protein so it was not possible to plot full precision–recall curves for them (i.e., if a score is not reported for a true ligand-binding residue, a recall of 1 may never be achieved for some queries). Instead, we show precision–recall points (“PR-points”). A PR-point is another way to compare performance and is defined as the precision and recall if the number of predicted ligand-binding residues is equal to the number of true ligand-binding residues (and therefore precision and recall values become the same). As shown in the figure, LBias outperforms the other methods based on this criterion using the LigASite benchmark.
In Table 1, we show the numerical values of the PR-points and Matthews correlation coefficients for all methods applied to the LigASite benchmark and several other benchmarks. These include (i) a benchmark of experimentally determined protein/ligand structures created by the COACH developers and, to test the sensitivity of LBias to possible inaccuracies in a given structure, two benchmarks composed of homology models (Materials and Methods) of the proteins in (ii) COACH and (iii) LigASite. As can be seen from the table, LBias slightly outperforms COACH on its experimental benchmark. As expected, LBias performance is worse on the COACH and LigASite benchmarks consisting of homology models, but its performance on both is nearly identical. Surprisingly, COACH performance is better on its homology model benchmark relative to the LigASite crystal structures and it slightly outperforms LBias on the COACH models. On the other hand, COACH performance degrades significantly on the LigASite models. It is not clear why this is the case.
Table 1.
PR-point (PRP) and Matthews correlation coefficient (MCC) for ligand-binding residue prediction
| Method | Benchmark datasets | |||||||
| LigASite | COACH | |||||||
| Experimental | Modeled | Experimental | Modeled | |||||
| PRP | MCC | PRP | MCC | PRP | MCC | PRP | MCC | |
| LBias | 0.61 | 0.55 | 0.51 | 0.52 | 0.67 | 0.65 | 0.52 | 0.47 |
| COACH | 0.51 | 0.54 | 0.38 | 0.40 | 0.63 | 0.60 | 0.55 | 0.51 |
| ConCavity | 0.57 | 0.52 | ||||||
| FTsite | 0.41 | 0.46 | ||||||
The digits in bold are for the best-performing method in each column.
Genome-Wide Ligand Target Protein Prediction.
A “structural BLAST” approach is used to identify potential targets of a given small molecule using LT-scanner (Materials and Methods). Fig. 1B describes a schematic of the LT-scanner workflow. In contrast to LBias, which starts with a protein structure without ligands, LT-scanner starts with a protein cocrystalized with a ligand, and searches a structure database for structurally similar human apo structures to find other potential targets of that ligand. For each structurally similar protein found in the database, the SIM score is calculated which, as discussed above, reflects whether or not the interactions made by the ligand in the starting holo structure might also be formed in the structurally similar protein.
To evaluate LT-scanner performance, we collected a set of 853 protein structures containing drugs from the Protein Data Bank (PDB) (29). These represented 622 unique proteins (195 human and 427 nonhuman) and 200 unique drugs. For each of the 853 protein/drug structures, we tested the ability of LT-scanner to identify other targets of those drugs in a database of apo human protein structures. This database, termed the human protein structure set (HPSS), contains ∼300,000 crystal structures and homology models for ∼15,000 human proteins (more than one model was used for each protein as described in Materials and Methods).
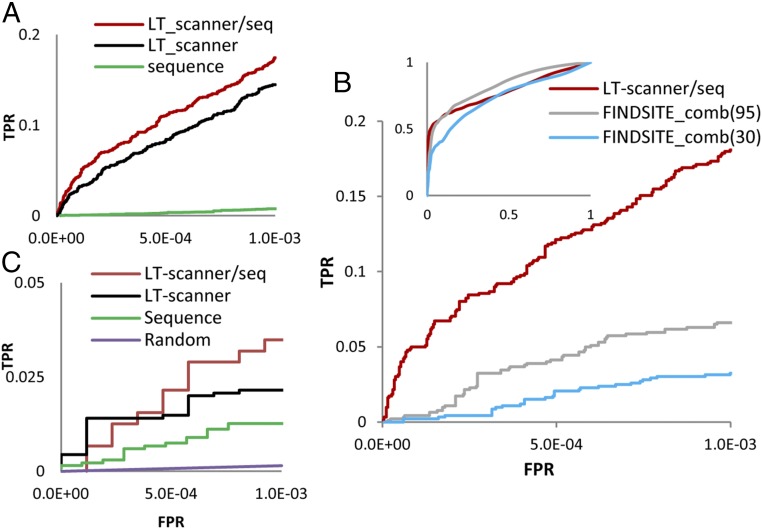
Fig. 3A plots receiver operating characteristic (ROC) curves based on a true positive set consisting of 1,887 known drug/target pairs available in the DrugBank4.0 (24), BindingDB (25), DGIdb (26), and ZINC15 (27). The true negative set consists of any protein/ligand pair where the protein is not a known target of the ligand. In the figure, we compare LT-scanner performance with an approach that uses sequence rather than structural relationships to identify targets for a given drug. Here, BLAST e-values between the template and human proteins in HPSS were used to rank the potential target of a given drug (Materials and Methods). Fig. 3A also shows results obtained from a naive Bayesian network which combines sequence with LT-scanner (the LT-scanner/seq algorithm). At a false-positive rate (FPR) of 10−3, sequence, LT-scanner and LT-scanner/seq recover 14, 272, and 328 known drug–target interactions, respectively.
Fig. 3.
ROC curves for drug target protein predictions. (A) ROC curves for LT-scanner (black line), LT-scanner/seq(red line), and Sequence (green line) were shown to evaluate performance for prediction of drug targets in the full set of ∼15,000 human proteins in HSSP. (B) ROC curves for LT-scanner/seq (red line), FINDSITE_comb(30) (blue line), and FINDSITE_comb(95) (gray line) for drugs and proteins used in both the FINDSITE study and this study. (C) ROC curves for LT-scanner/seq (red line), LT-scanner (black line), Sequence (green line), and random (purple line). Curves calculated for 21 FDA-approved kinase inhibitors and 600 human kinases.
In Fig. 3B, we compare performance of LT-scanner/seq to FINDSITEcomb, a recently developed template-based target prediction algorithm that has been shown to compare favorably to other widely used approaches (23). To carry out a meaningful comparison, we apply LT-scanner/seq to the subset of 923 drug/target pairs common to both our study and the study describing the FINDSITEcomb method (169 unique drugs and 421 unique proteins). We also limit the potential targets that LT-scanner/seq searches to the set of 1,588 proteins used in the FINDSITEcomb study and also available in HPSS. Hence, the negative set in this context is any of the 169 × 1,588 protein ligand pairs where the protein is not known to be a target of the ligand. The red ROC curve in Fig. 3B describes LT-scanner/seq performance, and the two gray curves describe FINDSITEcomb performance using two criteria: excluding drug/protein structures that have >95% [FINDSITEcomb(95)] or >30% [FINDSITEcomb(30)] sequence identity to a given potential target. As is evident from the figure, LT-scanner/seq outperforms FINDSITEcomb(30) and but not FINDSITEcomb(95) over the full FPR range. However, in the FPR region of 10−3, LT-scanner/seq yields the best results and recovered 166 known drug–target interactions followed by FINDSITEcomb(95) and FINDSITEcomb(30), which recovered 61 and 29 known drug–target interactions, respectively.
Drug Target Specificity: Kinases.
Template-based methods might be expected to encounter difficulties in dealing with specificity differences between closely related proteins since global alignments of such proteins would not necessarily identify subtle differences in their binding sites. To determine whether the SIMLT-scanner scoring function can account for such differences, we carried out a separate study on protein kinases that are quite similar in their sequences and structures, especially, at active sites.
We extracted a set of 59 kinases from the PDB cocrystalized with one of 21 kinase inhibitors and used LT-scanner and LT-scanner/seq to identify other kinases that those inhibitors might bind from a set of 600 proteins in the known human kinome for which structures are available in HPSS. The ROC curves in Fig. 3C show that both versions of LT-scanner recover interaction specificities much better than sequence, indicating that our scoring function is capturing subtle differences among the active sites of different kinases.
The human kinome has been classified into eight families based on sequence similarities within and outside of their catalytic domains, and their known biological functions (30). Four of the 21 kinase inhibitors are known to target serine/threonine kinases (which are distributed among different families) as well as 17 members of the tyrosine kinase family. In Fig. S1, we plot a radar diagram that shows how kinase targets predicted by LT-scanner are distributed throughout the eight kinase families. Most of the tyrosine kinase inhibitors are predicted to target the tyrosine kinase and tyrosine kinase-like groups, as expected. However, ∼50% and 44% of the Ca2+/calmodulin-dependent kinases and some of the serine/threonine kinases, respectively, are also predicted as targets for tyrosine kinase inhibitors, suggesting that they may be off-targets.
We further analyzed the predicted targets of the inhibitors of members of the BCR-ABL kinase family. As shown in Fig. S1 C–G, these inhibitors are predicted to primarily target the tyrosine kinase group, again as expected. However, LT-scanner also predicts that two tyrosine kinase inhibitors (Bosutinib and Dasatinib) bind to the protein salt-inducible kinases (SIKs), a serine/threonine kinase. This computational result is consistent with recent experimental evidence (31, 32). Although we are not aware of additional experiments that pertain to the other potential off-targets, this result, as well as the identification of targets in the correct kinase families, suggests that at least some of the off-targets identified may also be correct. The list of predicted kinase inhibitor targets (FPR < 10−3) is provided in Table S1 as a set of testable hypotheses.
Discussion
We have presented a template-based method, LBias, to predict ligand binding site residues for a given protein. Like related methods, it is based on the superposition of a set of template proteins that bind ligands onto the query protein. Scoring is based in part on the number times a superimposed ligand from that set contact a given residue. However, LBias incorporates a score that measures whether the protein/ligand interactions in the template can be reproduced for the same ligand when bound to the query using an adaptation of the algorithm first introduced in refs. 33–35. This of course favors contributions from templates that have binding sites most compatible with that of the query and appears responsible for much of the effective performance exhibited by LBias.
Another contributing factor to the success of LBias and LT-scanner appears to be the use of structural alignments and the resulting incorporation of information from remote structural homologs. We have shown previously that information from remote homologs is crucial to the success of the PredUs program for the prediction of protein–protein interfacial residues (10). As is the case of protein–protein interaction sites, the existence of functionally meaningful structural similarities among protein substructures greatly expands the coverage of template-based target prediction. Fig. S2 shows the distribution sequence identities between known human targets and template proteins used in our predictions. The average sequence identity in Fig. S2 is 26% and the corresponding average for kinases is 37%. This indicates that LT-scanner is able to exploit distantly homologous proteins as templates in its prediction.
The results reported here show that LBias compares favorably to the COACH metaserver. Of course, it is often the case that programs appear to be most successful in the hands of their developers, but we have carried out a series of unbiased tests on datasets that include (i) crystal structures in both the LigASite database and the one used by COACH, (ii) homology models used by COACH, and (iii) homology models we constructed for the LigASite set. COACH, which was independently trained on its own set of models, slightly outperforms LBias on these models. Interestingly, its performance is much weaker on the models we constructed for the LigASite proteins. It is not unreasonable to expect that every set of models has unique features and that scoring functions trained on such models will perform the best when those models are used. The success of COACH on blind predictions indicates that, when used together with its own set of models, it is highly effective. In contrast, LBias performance appears to be the same on both sets (ii and iii above) of models and, not surprisingly, below that obtained from crystal structures.
LT-scanner was developed to predict target proteins for a given ligand based on structures of known protein–ligand complexes. We used it in this work to scan human protein structures, both experimentally determined and homology modeled, for target protein predictions of FDA-approved drugs. LT-scanner performed similarly to the state-of-the-art FINDSITEcomb program, although somewhat better in the low FPR range of 10−3. The ROC curves shown in Fig. 3 underrepresent actual performance since every prediction of a complex not involving a known protein–drug interaction is counted as a false positive. Nevertheless, even based on the performance reported here, LT-scanner appears to be an effective means of predicting targets in repurposing drugs and in generating hypotheses as to potential off-targets.
One striking example is the KIT-kinase inhibitor, ponatinib, binds to LRRK2, a protein kinase that has mutations associated with Parkinson’s disease (36). The prediction was not trivial because (i) the template was cocrystal structure of ponatinib and KIT while the structure of the LRRK2 was obtained from a homology model based on VEGFR2. (ii) The sequence identity between KIT and LRRK2 is 8% with whole protein sequence and 23% with kinase domain alone (BLAST e-value of 1 × e−8). LT-scanner predicted that ponatinib would bind to LRRK2 based on a Sim score of 0.31, which is associated with an FPR < 10−3. This score reflects the fact that four of the five hydrogen bonds made in KIT are also found in LRRK2 and many of the nonpolar contacts are also present, but in some cases with different side chains (Fig. S3 and Table S2).
The fact that the templates used in our predictions often bear only a weak sequence relationship to the query protein indicates that many of our predictions are nontrivial in that the template and query belong to different protein families. The ability of LT-scanner to detect cross-family relationships is due in part to the use of the SKA structural alignment algorithm, which is tuned to detect local structural similarities even in the absence of a strong sequence relationship or a good global structural alignment. Moreover, LT-scanner uses multiple templates that cover different protein families. For example, 79 different template structures were used for target protein prediction for the FDA-approved drug Adenosine. Those 79 templates have a wide range of pairwise protein structure distances (PSDs) as shown in the Fig. S4. PSD > 1 for a given pair of proteins generally implies that they belong to different SCOP folds. Indeed, the 79 proteins used as templates for Adenosine cover 27 different SCOP folds (37) (listed in Table S3) and 25 different ECOD X-groups (38) (listed in Table S4).
While LT-scanner uses protein structural similarity to identify drug targets, it does not exploit chemical similarity to derive relationships between proteins based on the ligands they bind as is done for example in FINDSITEcomb and SEA from the Shoichet group (21, 23). We thus expect that combining LT-scanner with ligand-based approaches will yield both improved performance including greatly expanded coverage of potential targets. The strategy used in LT-scanner can also be applied to the problems mentioned in the introduction—the prediction of ligands that bind to a given query protein. This would involve searching a database of protein–ligand structures for proteins that align with the query and then using the LBias SIM score to identify potential small-molecule binders. Finally, we note the close relationship between LBias to the PredUs (11) program that predicts interfacial residues in protein–protein complexes, and between LT-scanner and PrePPI (39) that predicts protein–protein interactions. Both sets of programs are based on structural alignment and both utilize scoring functions that allow predictions to be made on a genome-wide scale. Their integration, combined with chemical similarity measures applied to compound databases, will offer a structure-informed genome-wide view of protein–protein and protein–ligand interactions, which in turn will enable numerous biomedical applications.
Materials and Methods
Ligand-Binding Residue Prediction.
Benchmark datasets.
The LigASite (28) database (release 9.7) was downloaded from ligasite.org/. LigASite contains structural information for 391 proteins and their ligands. For each protein, the database has two experimentally determined structures corresponding to the apo and holo forms. Only the apo form was used to predict ligand-binding residues. We downloaded the ConCavity ligand-binding residue predictions and scores for 317 of these structures from compbio.cs.princeton.edu/concavity/. The results described above are for this subset of LigASite. For this and other benchmarks sets, we define ligand-binding residues as those that make contacts (distance cutoff ≤ 5 Å) with ligands within the same chain in the holo form of the protein.
The COACH experimental benchmark was downloaded from https://zhanglab.ccmb.med.umich.edu/COACH/benchmark/receptor.tar.bz2 and contains information on 814 proteins and their ligands. As discussed below, LBias requires that ligands have molecular weight between 200 and 1,000 Da, so that results reported in Fig. 1 are for the COACH subset of 522 protein/ligand structures (425 unique proteins) where the ligand meets that criterion.
The benchmark composed of homology models of the LigASite proteins was constructed using the same protocol described in ref. 39, except that the proteins used to construct the models were required to have less than 30% sequence identity to a given LigASite protein to be consistent with the 30% sequence identity cutoff (7) The modeled COACH benchmark was downloaded from zhanglab.ccmb.med.umich.edu/COACH/benchmark/I-TASSER.tar.bz2, and, to the best of our knowledge, the same sequence identity criterion was used in its construction. The COACH software was also downloaded from the website (https://zhanglab.ccmb.med.umich.edu/I-TASSER/download/) and run locally. Results for FTsite were obtained from the web server (ftsite.bu.edu/).
LBias method.
A total of 105,646 holo forms of protein chains with bound ligands of molecular weight between 200 and 1,000 Da was collected from the PDB (May 2016). The Open Babel Package (40) was used to add hydrogens to all titratable groups on these proteins and their ligands assuming a pH of 7 and standard pKa values. The Cd-hit (41) program was used to cluster these proteins based on their sequence at a cutoff of 60% identity, resulting in 10,292 clusters. For a given query protein, Q, a query-specific protein structure database was constructed from the members of each cluster that has the highest sequence identity (but less than 96%) with the query protein. To maximize diversity in the set of ligands, if other proteins in each cluster with >60% identify to proteins already selected from that cluster were also available in the PDB but that were cocrystalized with structurally different ligands, these structures were also included in the database. Structurally different ligands were defined arbitrarily as having a Tanimoto coefficient (42) (see Supporting Information for details) less than 0.3. Proteins structurally similar to the query are then identified in this database using the program SKA (43, 44) using a protein structural distance cutoff of 0.8. These structural “neighbors” {Ni} are then used in the prediction of ligand-binding residues of the query protein as follows.
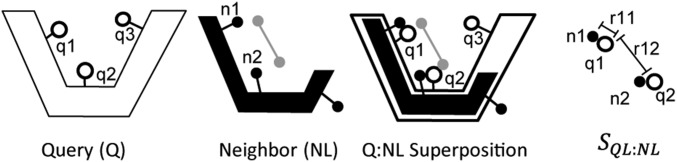
Each Ni is superimposed on Q and the same transformation is applied to the Ni’s associated ligand, Li, so as to place Ni, Q, and Li in the same coordinate system (Fig. 4). We identify four types of interactions between atoms in Ni and Li: (i) hydrogen bonds (distance Å and angle > 120°) (45), (ii) aromatic–aromatic interaction (distance Å) (46), (iii) ion pairs (distance Å) (46), and (iv) vdW contacts () (46), where is the sum of the vdW radii for given pair of atoms, taken from the Open Babel parameter set (40). To measure the degree to which interactions between Ni and ligand Li could also form between Q and Li, a protein–ligand interaction similarity score, , is calculated as follows (adapted from refs. 33–35):
| [1] |
| [2] |
Fig. 4.
Calculating LBias SIM score. A query protein Q is shown at Left with three atoms, q1, q2, and q3, identified specifically. The second panel shows a ligand-containing protein NL, structurally similar to Q with the ligand shown as a gray line connecting two gray atoms. Ligand-binding residues, n1 and n2, are identified. The NL complex is superposed onto Q. Residues from Q that interact with the superposed ligand are identified (q1 and q2 in the above). A protein–ligand similarity score between QL and NL (SQL:NL) is then calculated, where . SQL:NL is a function of all of the pairwise distances (e.g., r11, r12) between the atoms from Q interacting with the superposed ligand and the atoms from N interacting with the native ligand if the two atoms in question make chemically similar contacts with the ligands. For example, if n1 makes a hydrogen bond with the ligand, but q1 is hydrophobic, m11 would be zero and there would be no contribution to SQL:NL from this pair of atoms (Materials and Methods).
where is the distance between an atom of Q and an atom of Ni forming an interaction of a given type with Li. is a matching index (equal to 1 if these atoms are involved in at least one identical type of interaction among the four types or 0 otherwise), and is a scaling factor that attenuates distant interatomic interactions (35) and is set to 0.7 in this work [the attenuation effect of different scaling factors, on distance ranges between (0 ∼ 3 Å) is shown in Fig. S5]. is zero when there is no relationship between the two protein–ligand interactions and is equal to 1 for identical interacting binding sites where all ligand-binding atoms in both structures superimpose perfectly. For each residue of the query protein, the final LBias score, R, reflecting the likelihood of interacting with a ligand is calculated as follows:
| [3] |
where the n is the number of ligands in {Li} that have an atom within 5 Å of an atom of that residue.
Precision–recall curves.
For a given query protein, all its residues were sorted based on the value of R described above. The list then was used to calculate precision and recall to obtain a precision–recall curve (11, 47) (see Supporting Information for details).
Ligand Target Prediction.
Protein structures.
Three-dimensional models for full-length human proteins and their subdomains in HPSS were constructed using the same protocol described in ref. 39, with the exception that multiple models for each protein were constructed from up to 10 different templates if the e-values for those templates were <10−12. Approximately 340,000 models for 14,964 proteins were constructed. The use of multiple models is a way to account both for conformational variability and to reduce the general uncertainty associated with homology models.
The LT-scanner algorithm and performance.
To identify potential targets of a ligand/drug, an experimentally determined cocrystal of a protein containing that ligand is needed. This structure is superimposed on all structurally similar protein structures in HPSS (as defined by a SKA protein structural distance cutoff of 0.8) and SIM(QL,NL) is calculated for each as described above. For a given protein, we define SimLT-scanner to be the highest value of SIM(QL,NL) obtained for all available models of that protein. ROC curves were obtained using a procedure similar to that described above precision–recall curves. All human proteins were put into a list sorted based on SimLT-scanner. This list is scanned in order and TPR [TP/(TP+FN)] and FPR [FP/(TP+FP)] were calculated for each true-positive protein/target pair encountered.
Sequence similarity was independently used to identify targets related to a given template–ligand complex. Template protein sequences were used as query sequences and e-values were calculated for human proteins in HPSS. The ROC curve for sequence was obtained by ordering the sequence hits based on BLAST e-value.
We used a naive Bayes approach to combine LT-scanner and Sequence (see Supporting Information for details). The naive Bayes, LT-scanner/seq, was trained based on 1,342 positives taken from drug–target interactions in DrugBank (24), BindingDB (25), and DGIdb (26) and 638,855 negatives that do not appear in those databases [545 known drug–target interactions from ZINC15 (27) were not included for training]. The LT-scanner/seq likelihood ratio (LR) is the product of the LT-scanner LR and Sequence LR.
Supplementary Material
Acknowledgments
We thank Jose Ignacio Garzon and Diana Murray for valuable discussions and suggestions. This work was funded by NIH Grants GM030518 (to B.H.), S10OD012351 high-performance compute cluster for biomedical computing (August 2012), and S10OD021764 storage system for high-performance computing (March 2016).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705381114/-/DCSupplemental.
References
- 1.Hendlich M, Rippmann F, Barnickel G. LIGSITE: Automatic and efficient detection of potential small molecule-binding sites in proteins. J Mol Graph Model. 1997;15:359–363, 389. doi: 10.1016/s1093-3263(98)00002-3. [DOI] [PubMed] [Google Scholar]
- 2.Laskowski RA. SURFNET: A program for visualizing molecular surfaces, cavities, and intermolecular interactions. J Mol Graph. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 3.Capra JA, Laskowski RA, Thornton JM, Singh M, Funkhouser TA. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput Biol. 2009;5:e1000585. doi: 10.1371/journal.pcbi.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang B, Schroeder M. LIGSITEcsc: Predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ngan CH, et al. FTSite: High accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics. 2012;28:286–287. doi: 10.1093/bioinformatics/btr651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Li Y, Lin B, Schroeder M, Huang B. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics. 2011;27:2083–2088. doi: 10.1093/bioinformatics/btr331. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Roy A, Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dey F, Cliff Zhang Q, Petrey D, Honig B. Toward a “structural BLAST”: Using structural relationships to infer function. Protein Sci. 2013;22:359–366. doi: 10.1002/pro.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrey D, Fischer M, Honig B. Structural relationships among proteins with different global topologies and their implications for function annotation strategies. Proc Natl Acad Sci USA. 2009;106:17377–17382. doi: 10.1073/pnas.0907971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang QC, Petrey D, Norel R, Honig BH. Protein interface conservation across structure space. Proc Natl Acad Sci USA. 2010;107:10896–10901. doi: 10.1073/pnas.1005894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang H, Petrey D, Honig B. A hybrid method for protein-protein interface prediction. Protein Sci. 2016;25:159–165. doi: 10.1002/pro.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maheshwari S, Brylinski M. Predicting protein interface residues using easily accessible on-line resources. Brief Bioinform. 2015;16:1025–1034. doi: 10.1093/bib/bbv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wass MN, Kelley LA, Sternberg MJ. 3DLigandSite: Predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010;38:W469–W473. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci USA. 2008;105:129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinstein WP, Brylinski M. eFindSite: Enhanced fingerprint-based virtual screening against predicted ligand binding sites in protein models. Mol Inform. 2014;33:135–150. doi: 10.1002/minf.201300143. [DOI] [PubMed] [Google Scholar]
- 16.Liu T, Altman RB. Using multiple microenvironments to find similar ligand-binding sites: Application to kinase inhibitor binding. PLoS Comput Biol. 2011;7:e1002326. doi: 10.1371/journal.pcbi.1002326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie L, Li J, Xie L, Bourne PE. Drug discovery using chemical systems biology: Identification of the protein-ligand binding network to explain the side effects of CETP inhibitors. PLoS Comput Biol. 2009;5:e1000387. doi: 10.1371/journal.pcbi.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansch C, Maloney PP, Fujita T, Robert MM. Correlation of biological activity of phenoxyacetic acids with Hammett substituent constants and partition coefficients. Nature. 1962;194:178–180. [Google Scholar]
- 19.Klopman G. Artificial-intelligence approach to structure activity studies–Computer automated structure evaluation of biological-activity of organic-molecules. J Am Chem Soc. 1984;106:7315–7321. [Google Scholar]
- 20.Cramer RD, Patterson DE, Bunce JD. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc. 1988;110:5959–5967. doi: 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- 21.Keiser MJ, et al. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 22.Cappel D, Dixon SL, Sherman W, Duan J. Exploring conformational search protocols for ligand-based virtual screening and 3-D QSAR modeling. J Comput Aided Mol Des. 2015;29:165–182. doi: 10.1007/s10822-014-9813-4. [DOI] [PubMed] [Google Scholar]
- 23.Zhou H, Gao M, Skolnick J. Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Sci Rep. 2015;5:11090. doi: 10.1038/srep11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Law V, et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilson MK, et al. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffith M, et al. DGIdb: Mining the druggable genome. Nat Methods. 2013;10:1209–1210. doi: 10.1038/nmeth.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sterling T, Irwin JJ. ZINC 15–Ligand discovery for everyone. J Chem Inf Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dessailly BH, Lensink MF, Orengo CA, Wodak SJ. LigASite—a database of biologically relevant binding sites in proteins with known apo-structures. Nucleic Acids Res. 2008;36:D667–D673. doi: 10.1093/nar/gkm839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 31.Sundberg TB, et al. Small-molecule screening identifies inhibition of salt-inducible kinases as a therapeutic strategy to enhance immunoregulatory functions of dendritic cells. Proc Natl Acad Sci USA. 2014;111:12468–12473. doi: 10.1073/pnas.1412308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozanne J, Prescott AR, Clark K. The clinically approved drugs dasatinib and bosutinib induce anti-inflammatory macrophages by inhibiting the salt-inducible kinases. Biochem J. 2015;465:271–279. doi: 10.1042/BJ20141165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majeux N, Scarsi M, Apostolakis J, Ehrhardt C, Caflisch A. Exhaustive docking of molecular fragments with electrostatic solvation. Proteins. 1999;37:88–105. [PubMed] [Google Scholar]
- 34.Dey F, Caflisch A. Fragment-based de novo ligand design by multiobjective evolutionary optimization. J Chem Inf Model. 2008;48:679–690. doi: 10.1021/ci700424b. [DOI] [PubMed] [Google Scholar]
- 35.Kearsley SK, Smith GM. An alternative method for the alignment of molecular structures: Maximizing electrostatic and steric overlap. Tetrahedron Comput Methodol. 1990;3:615–633. [Google Scholar]
- 36.Li JQ, Tan L, Yu JT. The role of the LRRK2 gene in Parkinsonism. Mol Neurodegener. 2014;9:47. doi: 10.1186/1750-1326-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandonia JM, Fox NK, Brenner SE. SCOPe: Manual curation and artifact removal in the structural classification of proteins–Extended database. J Mol Biol. 2017;429:348–355. doi: 10.1016/j.jmb.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaeffer RD, Liao Y, Cheng H, Grishin NV. ECOD: New developments in the evolutionary classification of domains. Nucleic Acids Res. 2017;45:D296–D302. doi: 10.1093/nar/gkw1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garzón JI, et al. A computational interactome and functional annotation for the human proteome. Elife. 2016;5:e18715. doi: 10.7554/eLife.18715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Boyle NM, et al. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 42.Tanimoto TT. 1957. Internal Report, 17th Nov. (IBM Corp., Armonk, NY), IBM Technical Report.
- 43.Petrey D, Honig B. GRASP2: Visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- 44.Yang AS, Honig B. An integrated approach to the analysis and modeling of protein sequences and structures. I. Protein structural alignment and a quantitative measure for protein structural distance. J Mol Biol. 2000;301:665–678. doi: 10.1006/jmbi.2000.3973. [DOI] [PubMed] [Google Scholar]
- 45.Xu D, Tsai CJ, Nussinov R. Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Eng. 1997;10:999–1012. doi: 10.1093/protein/10.9.999. [DOI] [PubMed] [Google Scholar]
- 46.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 47.Davis J, Goadrich M. Proceedings of the 23rd International Conference on Machine Learning. ACM; New York: 2006. The relationship between precision–recall and ROC curves; pp. 233–240. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.