Significance
We develop a network analysis approach for identifying clusters of interactions between chromosomes, which we validate experimentally. Our method integrates 1D features of the genome, such as epigenetic marks, with 3D interactions, allowing us to study spatially colocalized regions between chromosomes that are functionally relevant. We observe that clusters of interchromosomal regions fall into active and inactive categories. We find that active clusters share transcription factors and are enriched for transcriptional machinery, suggesting that chromosome intermingling regions play a key role in genome regulation. Our method provides a unique quantitative framework that can be broadly applied to study the principles of genome organization and regulation during processes such as cell differentiation and reprogramming.
Keywords: chromosome intermingling, Hi-C, network and clustering analysis, epigenetics, 3D FISH
Abstract
The 3D structure of the genome plays a key role in regulatory control of the cell. Experimental methods such as high-throughput chromosome conformation capture (Hi-C) have been developed to probe the 3D structure of the genome. However, it remains a challenge to deduce from these data chromosome regions that are colocalized and coregulated. Here, we present an integrative approach that leverages 1D functional genomic features (e.g., epigenetic marks) with 3D interactions from Hi-C data to identify functional interchromosomal interactions. We construct a weighted network with 250-kb genomic regions as nodes and Hi-C interactions as edges, where the edge weights are given by the correlation between 1D genomic features. Individual interacting clusters are determined using weighted correlation clustering on the network. We show that intermingling regions generally fall into either active or inactive clusters based on the enrichment for RNA polymerase II (RNAPII) and H3K9me3, respectively. We show that active clusters are hotspots for transcription factor binding sites. We also validate our predictions experimentally by 3D fluorescence in situ hybridization (FISH) experiments and show that active RNAPII is enriched in predicted active clusters. Our method provides a general quantitative framework that couples 1D genomic features with 3D interactions from Hi-C to probe the guiding principles that link the spatial organization of the genome with regulatory control.
The 3D structure of the genome plays a key role in regulatory control of the cell. Historically, the spatial organization of the genetic material has been probed with fluorescence in situ hybridization (FISH), and it was shown that chromosome organization is nonrandom. Each chromosome occupies its own territory with gene-dense chromosomes more likely to be in the nuclear interior (1). As an addition to FISH, chromosome conformation capture methods (3C, 4C, 5C, and Hi-C) have been designed to probe the 3D organization of the genome by measuring the genome-wide contact frequencies over a population of cells (2–5). Computational and experimental efforts have largely focused on investigating intrachromosomal contacts. Studies where these interactions have been analyzed together with epigenetic modifications as measured by chromatin immunoprecipitation sequencing (ChIP-seq) showed that epigenetic marks are tightly linked to shaping the architecture of the genome (6, 7).
Few studies have considered interchromosomal interactions. It was shown that regions on neighboring chromosome territories may loop out and intermingle with each other in a transcription-dependent manner (8, 9). In addition, a recent study has revealed that intermingling regions are enriched in both active and repressive epigenetic marks, as well as the active form of RNA polymerase II (RNAPII) and transcription factors (10). Furthermore, it was identified that genes are spatially colocalized and coregulated by sharing common transcription factors (11, 12) and epigenetic machinery like the polycomb proteins (13). For example, TNF-responsive genes (on the same and different chromosomes) have been shown to colocalize upon their stimulation. Their spatial clustering was found to be correlated with their temporal expression patterns (12). The clustering of genes, transcriptional machinery, and regulatory factors to coordinate expression, also known as transcription factories, has been proposed as a model for gene regulation (14–16). Collectively, these studies suggest that interchromosomal regions could harbor coregulated gene clusters. However, missing in this picture is a systematic analysis linking 1D epigenetic marks and 3D intermingling regions and their roles in transcription control.
Various methods have been developed to infer the spatial connectivity of the whole genome from Hi-C data. Restraint-based approaches transform Hi-C contact matrices into distances to deduce one consensus structure (17–21). However, it remains a challenge to map contact frequencies to spatial distances due to biases in Hi-C matrices (22). A different approach is to produce an ensemble of structures that could explain the experimental data (23, 24). Computational methods have largely focused on inferring the 3D genome structure based on Hi-C data alone without leveraging functional genomic data for studying its architecture. A recent study has explored this idea by superimposing ChIP-seq data of three transcription factors (TFs) on the 3D genome architecture inferred from Hi-C and determined functional hotspots in Saccharomyces cerevisiae (25). Another study used 1D epigenomic tracks to predict 3D interactions (26). But there remains a lack of a general quantitative framework that integrates 1D functional genomic features with 3D intermingling regions to determine a regulatory code for interchromosomal interactions.
In this paper, we take a unique approach by integrating Hi-C and functional genomic data to predict regions that are colocalized and coregulated in 3D. The model of gene regulation that is captured by our analysis is the spatial clustering of genomic regions for their coregulation (27). This mode of gene regulation may enable the cell to coordinate gene expression and activate or repress pathways that are important for cell function in a coordinated manner. We focus on interchromosomal interactions to study chromosome intermingling regions. Using a network analysis approach, we construct a network of chromosomal interactions weighted by correlations in their genomic features at a 250-kb resolution. We find that intermingling regions can be divided into active and inactive clusters, where active clusters are hotspots for TF binding. We validate our predictions using FISH by comparing a predicted active cluster vs. a predicted negative control and also confirm that active RNAPII is significantly enriched in the predicted active cluster.
Results
Identification of Intermingling Domains.
To identify interchromosomal regions that are both spatially colocalized and coregulated, we leveraged spatial information from Hi-C experiments and regulatory information, namely, epigenetic marks, TF ChIP-seq, DNase I hypersensitivity (DNase-seq), and RNA-seq. Our aim was to identify clusters of chromosome regions at the whole-genome scale that interact spatially due to similarities in their regulatory features and thus might be coregulated by shared regulatory factors and epigenetic marks. Our method consists of four steps outlined in Fig. 1: (i) identification of highly interacting domains by determining large average submatrices in interchromosomal Hi-C maps, (ii) superimposing regulatory marks on the interacting domains, (iii) construction of a network of interacting regions with edges weighted by the correlation of the superimposed marks as a measure of coregulation, and (iv) network clustering to obtain spatially colocalized and coregulated domains.
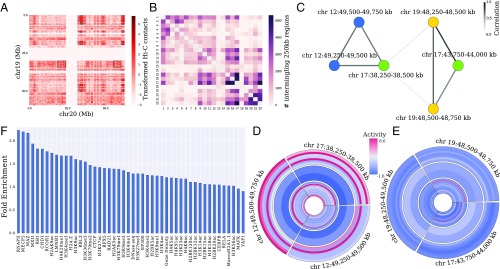
Fig. 1.
Overview of the proposed quantitative framework for detecting intermingling regions. (A) Example of an observed interchromosomal Hi-C contact matrix at 250-kb resolution after preprocessing and transformation (standardized by mean and SD after log(1 + x) transformation) for chromosomes 19 and 20 (SI Appendix). Rectangular boxes represent interacting domains for this pair of chromosomes as detected by the LAS algorithm, which finds submatrices with high average. (B) Matrix containing the number of interacting 250-kb regions identified by the LAS algorithm for each pair of chromosomes. (C) Subnetwork of the chromosome interaction network corresponding to two distinct clusters. Nodes are colored by chromosome number. Each node in the network corresponds to a 250-kb region. Edges link nodes that are found together in a submatrix (box) as determined by the LAS algorithm. The edge weights are given by the strength of correlation between the genomic features (histone modifications, TF ChIP-seq, DNase-seq, and RNA-seq as listed in SI Appendix, Table S2) of adjacent 250-kb nodes. (D and E) Activity (normalized number of peaks in a 250-kb region) of the genomic features for the two clusters obtained by weighted correlation clustering on the subnetwork in C. Each ring corresponds to one genomic feature, listed from outer ring to inner ring in SI Appendix, Table S2. Features are grouped into active (outer rings—RNA-seq, RNAPII, H3K4me1, H3K4me2, H3K4me3, H3K36me3, H3K9ac), repressive (middle rings—H3K27me3 and H3K9me3), and other (inner rings) categories. (F) Fold enrichment of each genomic feature in the intermingling regions (SI Appendix).
We analyzed Hi-C data from human lung fibroblast (IMR-90) cells at 250-kb resolution, obtained from ref. 28. After bias correction, filtering, and transforming the data (SI Appendix), we identified a stringent set of highly interacting interchromosomal regions by solving the following submatrix finding problem in Hi-C maps. We sought a contiguous submatrix that has a high average , within the real-valued data matrix , where each entry is an interchromosomal contact frequency between two 250-kb regions. We used the iterative large average submatrix (LAS) algorithm (29) that balances matrix size and average value, as outlined in SI Appendix to discover highly interacting domains. Fig. 1A shows the identified domains in the Hi-C contact map for chromosomes 19 and 20. As shown in Fig. 1A, the LAS algorithm captures the regions with high intensity in the interchromosomal matrix. Applying this procedure to all pairwise interchromosomal maps yields Fig. 1B, where each entry in the matrix corresponds to the number of 250-kb regions identified for the particular chromosome pair [false discovery rate (FDR) < , SI Appendix]. The total size of highly interacting domains across all chromosomes spanned 903.25 Mb (SI Appendix, Table S1). Consistent with previous observations (2, 30), Fig. 1B shows that gene-dense chromosomes such as 15–17 and 19–22 had a high number of intermingling 250-kb regions. In addition, as previously noted (31), we found a striking difference between chromosomes 18 and 19—although these two chromosomes are approximately equal in size, the gene-poor chromosome 18 has a low level of intermingling across most chromosomes, while the gene-rich chromosome 19 tends to intermingle more with other chromosomes.
Integration of Functional Genomic Data and Network Analysis.
We obtained functional genomic data: TF ChIP-seq, histone modifications, DNase-seq, and RNA-seq data from ENCODE (32), Roadmap Epigenomics (33), and GEO databases (SI Appendix, Table S2). We used these experimental data as a regulatory profile for all 250-kb regions that lay within the intermingling domains.
Considering each selected 250-kb region as a node, a whole-genome network of chromosomal interactions was constructed as follows. Between chromosomes, the edges in the network were placed between pairs of 250-kb regions that lay within the same submatrix as identified by the LAS algorithm. Within chromosomes, edges were placed between loci that fall within the same intrachromosomal domain, as determined in ref. 28. After establishing the skeleton of the network, the edge weights were calculated as follows. Since our goal was to determine spatially coregulated regions, we weighted the edges by Spearman’s correlation between the genomic profiles of adjacent 250-kb regions. This combined approach can mitigate some of the noise associated with using Hi-C contact frequencies alone. In addition, it allows us to identify chromosome intermingling regions with coordinated activity, which might be controlled by the same set of TFs or epigenetic marks, as opposed to domains that interact in 3D by chance. A subnetwork containing six 250-kb regions from three distinct chromosomes is shown in Fig. 1C. The edge weights in this subnetwork suggest the presence of two separate clusters.
To retrieve intermingling regions that are coregulated, the weighted network of 250-kb regions was partitioned into clusters, using weighted correlation clustering (34) (see SI Appendix). This approach can for example identify regions that are brought together for transcription, since these would have high RNAPII and low repressive epigenetic marks. This approach indeed found two clusters in the subnetwork shown in Fig. 1C. The regulatory profiles of the six regions, separated into two clusters, are illustrated in Fig. 1 D and E. As a consequence of using weighted correlation clustering, the genomic features within a cluster are more similar than across clusters. Interestingly, the particular cluster in Fig. 1D is enhanced for active genomic features (we analyzed H3K9ac, H3K36me3, H3K4me3, H3K4me2, H3K4me1, RNAPII, and RNA-seq) and depleted for repressive features (we analyzed H3K27me3 and H3K9me3), while the cluster in Fig. 1E is depleted for active features. Using this method, 446 clusters (totaling 459.5 Mb; SI Appendix, Table S1) were identified (P value under a test) that consist of at least two nodes and span multiple chromosomes (SI Appendix, Table S3 and Dataset S1). On average, 2.5 chromosomes interact within one cluster (SI Appendix, Fig. S1).
We analyzed the enrichment of regulatory marks in intermingling regions and found that these regions were most enriched for RNAPII, namely by a factor of 2.23 (Fig. 1F). We also found the active and repressive marks (e.g., H3K9ac, H3K4me3, and H3K9me3) to be enriched in intermingling clusters, which is consistent with a previous study (10).
Regulatory Features are Predictive of Intermingling.
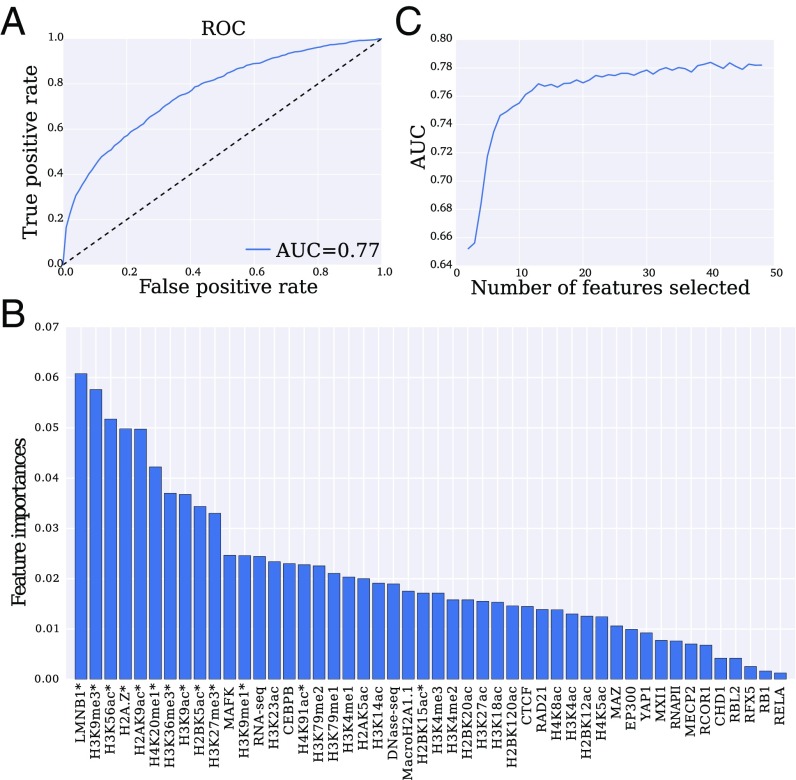
To characterize intermingling regions as a whole and evaluate whether they are distinct from nonintermingling regions on a regulatory level, we built a classifier and determined the features that contribute the most to distinguishing between these two classes. These features may represent a mechanism to spatially cluster genes for their coregulation. We annotated 250-kb regions as intermingling or nonintermingling based on the results from our network analysis and clustering. We then performed classification based on the associated regulatory profiles (SI Appendix, Table S2). We used eXtreme gradient boosting trees with 10-fold cross-validation to train our classifier. Using all features, the classifier achieves an accuracy of and the corresponding receiver operating characteristic (ROC) curve in Fig. 2A has an area under the curve (AUC) of 0.77.
Fig. 2.
Performance and feature importance for classifying intermingling regions. (A) ROC curve for eXtreme gradient boosting trees classifier that was trained on genomic features of intermingling vs. nonintermingling regions. This results in AUC of 0.77. (B) Features ranked in order of importance (relative depth of feature in the decision tree) for distinguishing intermingling domains. (C) AUC when recursively eliminating one feature at a time based on 10-fold cross-validation. Near-optimal performance is reached with 13 features, which are indicated by asterisks in B.
To quantify the importance of each feature by itself and in conjunction with all other features, we computed its univariate and multivariate rank based on its depth in the decision trees of the ensemble (Fig. 2B and SI Appendix, Fig. S2). The most important features determined by this analysis are lamin B1 (LMNB1), H3K9me3, H3K56ac, and H2A.Z. The importance of both repressive (H3K9me3, LMNB1) and active (H3K56ac, H2A.Z) marks ties with the observation that intermingling regions contain both active and repressed regions (35). Furthermore, previous mapping of LMNB1 in the genome revealed the presence of lamina-associated domains (LADs) that interact with the lamina on the nuclear envelope, spatially organize chromosomes by anchoring them to the lamina, and display coordinated gene repression (36–38). H3K9me3 is enriched in LADs and may facilitate gene silencing in LADs (37, 39). The context-dependent importance of this feature is in line with its low univariate, but high multivariate rank (SI Appendix, Fig. S2). H3K56ac is a known mark of transcriptionally active chromatin regions (40, 41). Finally, H2A.Z is enriched at transcription start sites (42), indicating its involvement in transcription initiation, and it appears to be a defining feature of intermingling on its own (SI Appendix, Fig. S2).
Performing stepwise feature elimination shows that ∼13 features are sufficient for achieving high AUC (Fig. 2C) and the corresponding features are annotated by asterisks in Fig. 2B.
Intermingling Clusters Are Divided into Active and Inactive Clusters.
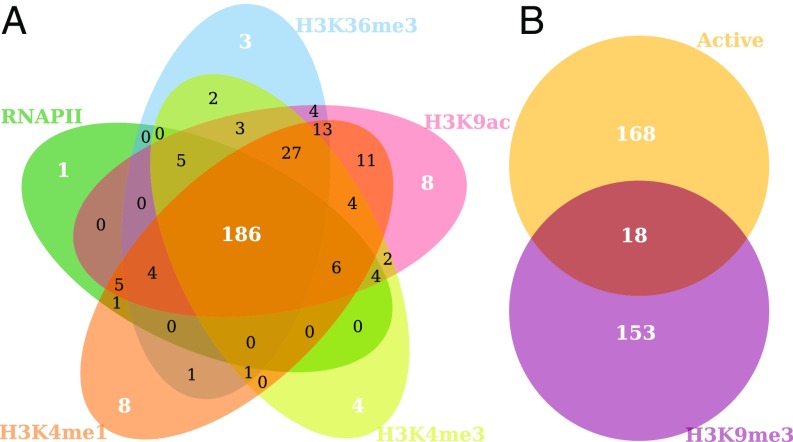
While it is interesting to evaluate intermingling regions altogether, studying these on a cluster-by-cluster level may give insights into the links between regulatory processes and spatial colocalization. Based on previous evidence (43) we hypothesized that active regions are clustered with other active regions and inactive regions with other inactive regions. To analyze the types of clusters we obtained, we computed the fold enrichment of each cluster for several regulatory features (SI Appendix, Table S3 and Dataset S1). We found that a high proportion of the clusters—41.7% (186 clusters)—was enriched for all active marks—RNAPII, H3K9ac, H3K36me3, H3K4me3, and H3K4me1 as shown in Fig. 3A (P value = under a test, SI Appendix). Notably, the majority of clusters were either enriched for all five active marks or not enriched for any active mark.
Fig. 3.
Classification of intermingling regions into active and inactive clusters. (A) Five-way Venn diagram representing the number of clusters enriched for each active epigenetic mark and RNAPII. Interestingly, many clusters (186 of 446) are enriched for all five active marks. (B) Venn diagram of the active clusters (the 186 clusters in the intersection of the five-way diagram in A) and clusters enriched for the silencing mark H3K9me3. Note that only 18 of 446 clusters are both active and silenced, showing that the clusters separate into two categories of active and inactive clusters.
The percentage of clusters enriched for the repressive/inactivating mark H3K9me3 was 38.3% (171 clusters). Interestingly, we observed a clear separation of the intermingling clusters into active and inactive, with only 4% of clusters (18 clusters) that were in both categories as shown in Fig. 3B (P value = under a test, SI Appendix). Active clusters were defined as those clusters enriched for RNAPII (fold enrichment >1) but not for H3K9me3. Inactive clusters were defined as enriched for H3K9me3 but not for RNAPII. Active clusters also had significantly higher gene expression (P value = 0.004 under a t test) in comparison with inactive clusters (SI Appendix, Fig. S3). In addition, high-occupancy target (HOT) regions, i.e., regions that are occupied by many TFs (44), were overrepresented in active clusters in comparison with low-occupancy target (LOT) regions, by a HOT:LOT ratio of 2.94 (SI Appendix, Table S4). These findings suggest that active clusters may be hotspots for TF binding.
Active Clusters Are Hotspots for TF Binding.
We probed the active clusters for shared TFs that may be involved in colocalizing and coregulating regions in a cluster by analyzing TF binding sites (TFBS). We used the JASPAR 2016 database to obtain the TFBS. These data were overlaid and then filtered using ChIP-seq peaks from all human cell lines available from ENCODE (32) (SI Appendix). This resulted in TFBS for 52 TF motifs. We performed an additional analysis to also consider a larger set of TF motifs (386) by overlaying and filtering the JASPAR 2016 database with a robust set of CAGE peaks from ref. 45, collected across 353 human tissue samples as part of the FANTOM5 project (SI Appendix). This filtering step provided us with a list of potential transcription start sites that contain motifs for the TFs under consideration.
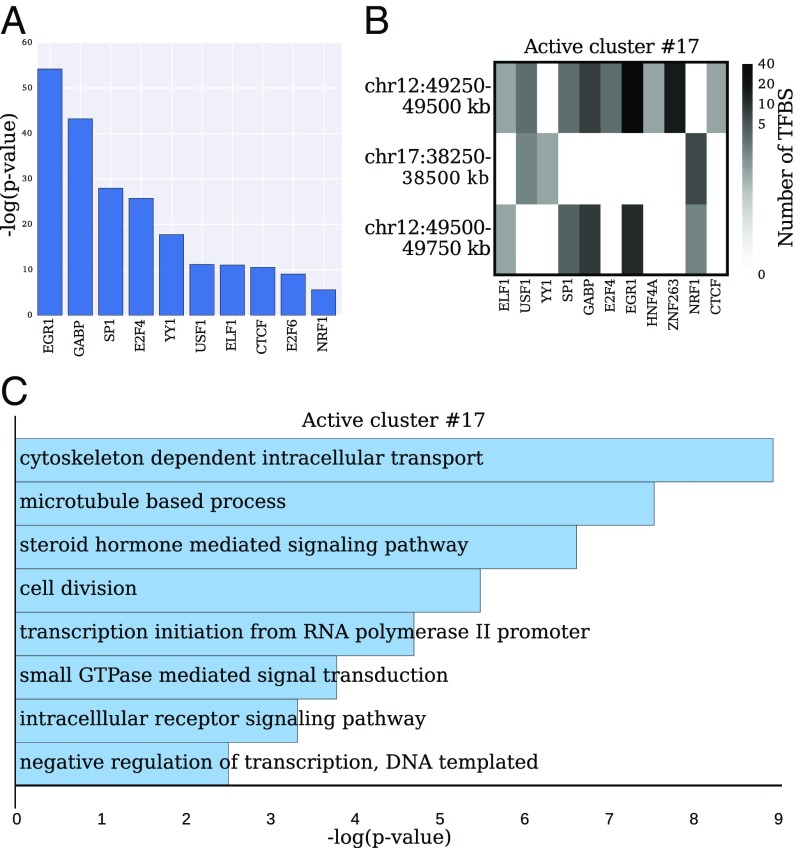
We compared the distributions of TFBS counts per 250-kb region for active clusters vs. the whole genome. Several factors, such as EGR1, YY1, CTCF, and the E2F family of proteins, showed a significant increase in TFBS counts under a Mann–Whitney U test (Fig. 4A).
Fig. 4.
TFBS and GO terms across active clusters. (A) Top 10 TFs with significantly overrepresented TFBS in active clusters compared with the whole-genome distribution (under a Mann–Whitney U test). (B) Matrix corresponding to a representative active cluster with the number of TFBS for each 250-kb region in the cluster. Only TFs containing at least one nonzero column entry are shown. A TF shared among multiple regions in the cluster may indicate its role in colocalization and coregulation of the clustered regions. (C) Significantly enriched GO terms computed from the genes that are expressed and colocalized in the intermingling cluster shown in B [ranked by P value using DAVID, SI Appendix].
The majority of active clusters contained binding sites for TFs that are shared across regions spanning multiple chromosomes (SI Appendix, Fig. S4). For example, the cluster studied in Fig. 1D involving chromosomes 12 and 17 contains binding sites for the TFs USF1 and NRF1 on regions of both chromosomes (Fig. 4B). This cluster is formed by the colocalization between two adjacent 250-kb regions on chromosome 12 and one region on chromosome 17. Gene ontology (GO) term analysis of the expressed genes (SI Appendix, Fig. S5) in this cluster revealed an enrichment for biological processes related to fibroblasts such as “cytoskeleton-dependent intracellular transport” (Fig. 4C). On the other hand, we found that inactive clusters contained a low number of TFBS (SI Appendix, Fig. S6 and Table S5), reaffirming the existence of two distinct types of cluster categories for intermingling regions.
Experimental Validation.
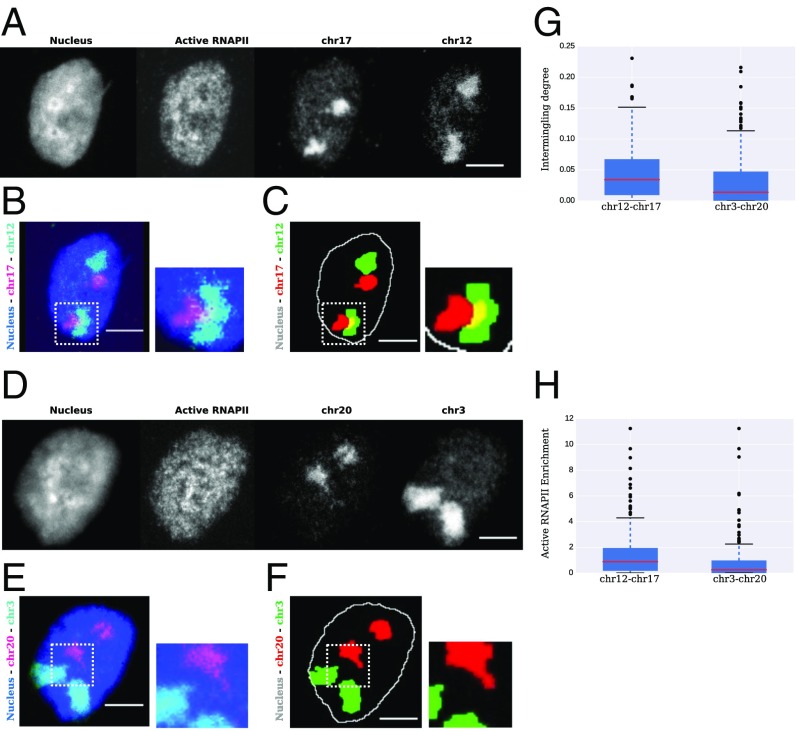
We ranked the active clusters according to the presence of binding sites for TFs that were shared across multiple chromosomes, using a permutation test (SI Appendix). The top 15 active clusters are shown in SI Appendix, Table S6. Chromosomes 12 and 17 were consistently found together among the top highly ranked clusters and were thus chosen for experimental validation (SI Appendix, Fig. S7). We compared the amount of overlap between chromosomes 12 and 17 to a negative control that we obtained by analyzing the network of least-interacting chromosomes (SI Appendix, Fig. S8). The chromosome territories were identified in human fibroblast (BJ) cells using DNA FISH and visualized using a laser scanning confocal microscope (Fig. 5 A–F). To obtain a representative sample of the population, we imaged at least 200 cells for each chromosome pair. We confirmed that chromosomes 12 and 17 consistently intermingle in a population of cells (Fig. 5C; SI Appendix, Fig. S9; and Movie S1), while the negative control chromosome pair does not (Fig. 5F; SI Appendix, Fig. S10; and Movie S2). To quantify our results, the intermingling degree, i.e., the amount of overlap between the two pairs of chromosome territories, was calculated as explained in SI Appendix. We found that the chromosome pair 12 and 17, which was predicted to interact, had a significantly higher intermingling degree than the negative control pair 3 and 20 (Fig. 5G, P value = 0.005 under a Welch two-sample t test). The percentage of nuclei that were intermingling (intermingling degree >0) was higher in the predicted pair of interacting chromosomes, 12 and 17, than in the negative control, 3 and 20 (SI Appendix, Fig. S11). In addition, we also calculated the enrichment of active RNAPII in the intermingling regions for the aforementioned pairs (SI Appendix). We found that the predicted chromosome pair, 12 and 17, which belongs to an active cluster, had significantly higher enrichment for active RNAPII in the intermingling regions compared with the negative control pair, 3 and 20 (Fig. 5H, P value = 7.125e-05 under a Welch two-sample t test), showing that the chromosome pair 12 and 17 indeed contains an active mark at the site of intermingling.
Fig. 5.
Experimental validation. (A) Representative images of the maximum-intensity Z projections of the nucleus, active RNAPII, and chromosomes 17 and 12, from Left to Right, respectively. (B) Raw image resulting from merging the nuclear (blue) and the two chromosome channels depicting the overlap between chromosomes 17 (purple) and 12 (cyan). (C) Image in B after segmentation with nucleus (white), chromosome 17 (red), and chromosome 12 (green). Yellow regions are the overlapping or intermingling regions. (C, Right) Enlargement of the region in the dotted white boxes in C, Left. (D) Representative images of the maximum-intensity Z projections of the nucleus, active RNAPII, and chromosomes 20 and 3, from Left to Right, respectively. (E) Raw image resulting from merging the nuclear (blue) and the two chromosome channels depicting the overlap between chromosomes 20 (purple) and 3 (cyan). (F) Image in E after segmentation with nucleus (white), chromosome 20 (red), and chromosome 3 (green). (F, Right) Enlargement of the region in the dotted white boxes in F, Left. (G) Boxplot depicting intermingling degree between chromosomes 12 and 17 and chromosomes 3 and 20 (P value = 0.005 under a Welch two-sample t test). (H) Boxplot depicting the enrichment of active RNAPII between chromosomes 12 and 17 and chromosomes 3 and 20 (P value = 7.125e-05 under a Welch two-sample t test). (All scale bars, 5 m.)
Discussion
Understanding the spatial organization of the chromosomes within the cell nucleus has been a major question in cell biology. A number of studies have suggested that the packing of DNA plays a critical role in regulating genomic programs (3). Earlier experiments took advantage of chromosome painting methods and revealed that chromosomes are organized nonrandomly and in a cell–type-specific manner (1, 8, 9). Analysis of gene positioning using FISH showed that coregulated genes were coclustered (11, 12). Such clusters of genes were also found to be colocalized with transcription-related machinery such as active RNAPII and TFs (11, 12). Recent developments in chromosome capture technologies further revealed that genome-wide chromosome contact maps are correlated with epigenetic marks (6, 7). The majority of studies using chromosome conformation capture focused on linking chromatin contacts with epigenetic modifications at the resolution of genes in intrachromosomal regions (6, 7). However, the coupling between the global organization of chromosomes with genome-wide epigenetic marks and the intermingling regions as an additional layer of transcriptional regulation has not been well studied.
In this paper, we developed a network analysis approach to reveal the principles of transcription-dependent chromosome intermingling by taking advantage of 3D contact maps obtained using Hi-C and 1D epigenetic marks, TF ChIP-seq, DNA accessibility, and RNA-seq. Our computational approach focuses on interchromosomal domains, since their organizational principles have been largely unknown. The proposed quantitative framework enables the prediction of chromosome intermingling regions at a genome-wide scale, thereby complementing experimental methods such as FISH that can be used to study specific clusters of interchromosomal interactions. The novelty of our method lies in leveraging 1D genomic features in combination with 3D interactions from Hi-C data. This allows us to study functionally colocalized regions: Since interactions can occur by chance in 3D, some intermingling regions may not be of biological relevance. By leveraging epigenetic marks and data from TF binding and DNA accessibility, as well as gene expression, we can determine interchromosomal regions that are colocalized and coregulated.
Our predictions reveal intriguing patterns of chromosome organization and have been validated by FISH experiments. Our findings recapitulate known principles of chromosome interactions, such as the tendency of gene-dense chromosomes to intermingle more frequently (2, 30) and the enrichment of RNAPII in intermingling regions (10), suggesting that RNAPII may play a crucial role in establishing and maintaining chromosome interactions. We observe that the clusters of interchromosomal regions fall broadly into two categories, active and inactive, where active clusters are enriched for active epigenetic marks and RNAPII and inactive clusters are enriched for H3K9me3. Interestingly, we found that active clusters are hotspots for TF binding sites, with several TFs being shared among multiple chromosomes within a cluster. These clusters contain genes with biologically relevant GO terms. We established the predictive power of our model through experimental validation. Using FISH experiments we showed that the predicted intermingling chromosomes interact consistently across a population of cells and that such intermingling regions are enriched for active RNAPII. Our quantitative analysis provides evidence that TF hotspots in active clusters are colocalized with active epigenetic modifications and with RNAPII and have a significantly higher gene expression than inactive clusters, suggesting that the relative positioning of the chromosomes in the cell nucleus is optimized to facilitate the clustering of coregulated genes, TFs, epigenetic modifications, and transcriptional machinery.
Collectively, these findings suggest that the spatial organization of the genomic material in the cell nucleus is optimized for transcription programs. The framework we present here is general and can be applied to analyze any cell type. We showed by experimentally validating the predictions from our model using single-cell imaging methods that population-level genome-wide contact and epigenetic data carry enough information to identify highly interacting regions. However, we anticipate that the power of our method will be increased as more robust single-cell genomic data become available. We believe that our quantitative approach will provide a useful framework to gain insights into the interplay between chromosome reorganization and regulation during processes such as cell differentiation, reprogramming, or the maintenance of homeostasis.
Materials and Methods
Details about the methods used for processing the raw Hi-C matrices, the LAS algorithm for identifying highly interacting regions, weighted correlation clustering, classification into intermingling and nonintermingling domains, the computation of fold enrichment of genomic features, the cell culture and chromosome FISH protocols, and the methods and settings used for confocal imaging and image analysis are provided in SI Appendix.
The code for interchromosomal network construction via LAS and for the identification and analysis of clusters is available at https://github.com/anastasiyabel/functional_chromosome_interactions. The code for performing the image analysis is available at https://github.com/SaradhaVenkatachalapathy/Chromsome-intermingling-region-indentifcation-and-characterisation-of-protein-levels.
Supplementary Material
Acknowledgments
A.B. was partially supported by NIH Predoctoral Training Grant T32GM87232 and the National Science Foundation (NSF) Graduate Research Fellowship under Grant 1122374. S.V., M.N., and G.V.S. thank the Mechanobiology Institute, National University of Singapore, and the Ministry of Education Tier-3 Grant Program for funding. C.U. was partially supported by the NSF (1651995), the Defense Advanced Research Projects Agency (W911NF-16-1-0551), and the Office of Naval Research (N00014-17-1-2147).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708028115/-/DCSupplemental.
References
- 1.Bolzer A, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieberman-aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bickmore WA, Van Steensel B. Genome architecture: Domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Schmitt AD, Hu M, Ren B. Genome-wide mapping and analysis of chromosome architecture. Nat Rev Mol Cell Biol. 2016;17:743–755. doi: 10.1038/nrm.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan X, et al. Integration of Hi-C and ChIP-seq data reveals distinct types of chromatin linkages. Nucleic Acids Res. 2012;40:7690–7704. doi: 10.1093/nar/gks501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer KV, et al. Modeling and experimental methods to probe the link between global transcription and spatial organization of chromosomes. PLoS One. 2012;7:e46628. doi: 10.1371/journal.pone.0046628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maharana S, et al. Chromosome intermingling-the physical basis of chromosome organization in differentiated cells. Nucleic Acids Res. 2016;44:5148–5160. doi: 10.1093/nar/gkw131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papantonis A, et al. TNF signals through specialized factories where responsive coding and miRNA genes are transcribed. EMBO J. 2012;31:4404–4414. doi: 10.1038/emboj.2012.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bantignies F, et al. Polycomb-dependent regulatory contacts between distant hox loci in drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 14.Papantonis A, Cook PR. Transcription factories: Genome organization and gene regulation. Chem Rev. 2013;113:8683–8705. doi: 10.1021/cr300513p. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, et al. Functional organization of the human 4D nucleome. Proc Natl Acad Sci USA. 2015;112:8002–8007. doi: 10.1073/pnas.1505822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhler C, Shivashankar GV. Chromosome intermingling: Mechanical hotspots for genome regulation. Trends Cell Biol. 2017;27:810–819. doi: 10.1016/j.tcb.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Li G, Toh KC, Sung WK. 3D chromosome modeling with semi-definite programming and Hi-C data. J Comput Biol. 2013;20:831–846. doi: 10.1089/cmb.2013.0076. [DOI] [PubMed] [Google Scholar]
- 18.Lesne A, Riposo J, Roger P, Cournac A, Mozziconacci J. 3D genome reconstruction from chromosomal contacts. Nat Methods. 2014;11:1141–1143. doi: 10.1038/nmeth.3104. [DOI] [PubMed] [Google Scholar]
- 19.Varoquaux N, Ay F, Noble WS, Vert JP. A statistical approach for inferring the 3D structure of the genome. Bioinformatics. 2014;30:i26–i33. doi: 10.1093/bioinformatics/btu268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segal MR, Bengtsson HL. Reconstruction of 3D genome architecture via a two-stage algorithm. BMC Bioinformatics. 2015;16:373. doi: 10.1186/s12859-015-0799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serra F, et al. Restraint-based three-dimensional modeling of genomes and genomic domains. FEBS Lett. 2015;589:2987–2995. doi: 10.1016/j.febslet.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Imakaev MV, Fudenberg G, Mirny LA. Modeling chromosomes: Beyond pretty pictures. FEBS Lett. 2015;589:3031–3036. doi: 10.1016/j.febslet.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Xu J, Zeng J. Inferential modeling of 3D chromatin structure. Nucleic Acids Res. 2015;43:e54. doi: 10.1093/nar/gkv100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tjong H, et al. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proc Natl Acad Sci USA. 2016;113:E1663–E1672. doi: 10.1073/pnas.1512577113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capurso D, Bengtsson H, Segal MR. Discovering hotspots in functional genomic data superposed on 3D chromatin configuration reconstructions. Nucleic Acids Res. 2016;44:2028–2035. doi: 10.1093/nar/gkw070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y, et al. Constructing 3D interaction maps from 1D epigenomes. Nat Commun. 2016;7:10812. doi: 10.1038/ncomms10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dekker J, Misteli T. Long-range chromatin interactions. Cold Spring Harb Perspect Biol. 2015;7:a019356. doi: 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shabalin AA, Weigman VJ, Perou CM, Nobel AB. Finding large average submatrices in high dimensional data. Ann Appl Stat. 2009;3:985–1012. [Google Scholar]
- 30.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Croft JA, et al. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ENCODE Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roadmap Epigenomics Consortium,et al Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elsner M, Schudy W. 2009 Bounding and comparing methods for correlation clustering beyond ILP. Proceedings of the NAACL HLT Workshop on Integer Linear Programming and Natural Language Processing (Association for Computational Linguistics, Stroudsburg, PA). Available at https://dl.acm.org/citation.cfm?id=1611638.1611641. Accessed December 4, 2017.
- 35.Pombo A, Dillon N. Three-dimensional genome architecture: Players and mechanisms. Nat Rev Mol Cell Biol. 2015;16:245–257. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- 36.Camps J, Erdos MR, Ried T. The role of lamin B1 for the maintenance of nuclear structure and function. Nucleus. 2015;6:8–14. doi: 10.1080/19491034.2014.1003510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guelen L, et al. Domain organization of human chromosomes revealed by mapping nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 38.Finlan LE, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shachar S, Voss TC, Pegoraro G, Sciascia N, Misteli T. Identification of gene positioning factors using high-throughput imaging mapping. Cell. 2015;162:911–923. doi: 10.1016/j.cell.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stejskal S, et al. Cell cycle-dependent changes in H3K56ac in human cells. Cell Cycle. 2015;14:3851–3863. doi: 10.1080/15384101.2015.1106760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Liu F, Ren C, Bo X, Shu W. Genome-wide identification and characterisation of HOT regions in the human genome. BMC Genomics. 2016;17:733. doi: 10.1186/s12864-016-3077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang DW, Lempicki Ra, Sherman BT. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.