Significance
Metal homeostasis is critical to numerous biological processes, and metalloregulators play key roles in its regulation. In transcriptional regulation, which is allosterically controlled by metalloregulators, reorganization of their metal-binding residues and/or related hydrogen bonding networks is usually utilized, while the coordination atoms on the same metal-binding residues remain seldom changed. Our study shows an example whereby the zinc-induced transcriptional regulator ZitR switches one of its histidine nitrogen atoms for zinc coordination in response to zinc fluctuation. This histidine-switch process facilitates conformational change of ZitR protein, allowing allosteric and fine-tuned control of DNA binding and transcriptional regulation.
Keywords: histidine switch, zinc homeostasis, allostery, transcription factors, X-ray crystallography
Abstract
Metalloregulators allosterically control transcriptional activity through metal binding-induced reorganization of ligand residues and/or hydrogen bonding networks, while the coordination atoms on the same ligand residues remain seldom changed. Here we show that the MarR-type zinc transcriptional regulator ZitR switches one of its histidine nitrogen atoms for zinc coordination during the allosteric control of DNA binding. The Zn(II)-coordination nitrogen on histidine 42 within ZitR’s high-affinity zinc site (site 1) switches from Nε2 to Nδ1 upon Zn(II) binding to its low-affinity zinc site (site 2), which facilitates ZitR’s conversion from the nonoptimal to the optimal DNA-binding conformation. This histidine switch-mediated cooperation between site 1 and site 2 enables ZitR to adjust its DNA-binding affinity in response to a broad range of zinc fluctuation, which may allow the fine tuning of transcriptional regulation.
The zinc transcriptional regulator ZitR in Lactococcus lactis, along with its close homolog AdcR in Streptococcus pneumoniae, are among the first metal-responsive members of the widely distributed multiple antibiotic resistance regulator (MarR) family of transcriptional factors (1–5). Most MarR family members serve as transcriptional repressors that dissociate from their promoter DNA upon the stimulation from cognate effector molecules, thus causing the derepression of their downstream genes (6–10). In contrast, ZitR and AdcR have been shown to undergo a unique corepression mechanism in which excess Zn(II) ions significantly increase their DNA-binding affinity, thus leading to the repression of the downstream zinc-responsive genes such as zitSQP for ZitR and adcCBA, phtABDE, and adcAII for AdcR (5, 11–13). Both ZitR and AdcR contain two closely located Zn(II)-coordination sites per monomer, with site 1 possessing a Zn(II)-binding affinity (Kd ∼ 10−13 M) that is four orders of magnitude higher than site 2 (Kd ∼ 10−9 M) (14, 15). Recent studies on AdcR indicate that site 1 is the primary zinc regulatory site, while site 2’s functional roles remain elusive (14, 15). Herein we show that Zn(II) binding in site 2 triggers the histidine 42 residue (H42) in site 1 to switch its Zn(II)-coordination atom from Nε2 to Nδ1, which acts as a key allosteric regulation mechanism to convert ZitR from the non–DNA-binding conformation to the DNA-binding conformation that is optimal for transcriptional corepression. This histidine switch-mediated site-1 and site-2 cooperation represents an example for coupling the metal ion coordination with allosteric control of metalloregulator conformation and DNA-binding affinity.
Results
Di-Zn(II)-ZitRWT Adopts an Optimal DNA-Binding Conformation.
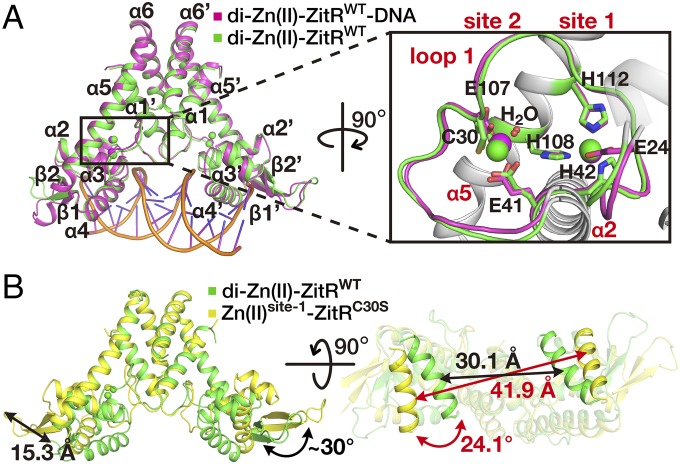
MarR family transcriptional regulators usually undertake conformational changes upon interaction with substrates (e.g., small molecules) that will affect their DNA binding (6–10). We speculate that a similar mechanism may be employed by ZitR and we adopted a structural biology approach to elucidate the mechanism underlying ZitR-mediated transcriptional regulation. We started by obtaining the crystal structures of di-Zn(II)-ZitRWT (wild-type ZitR with both zinc sites occupied) in the presence and absence of DNA. The structure of di-Zn(II)-ZitRWT was solved by molecular replacement and refined to 2.4-Å resolution (Fig. 1A and SI Appendix, Fig. S1A and Tables S1 and S2), which revealed a symmetrical dimer with each monomer, consisting of six α-helices, two antiparallel β-strands, one long loop (loop 1) connecting α1 and α2 (residues 20–37), as well as two coordinated Zn(II) ions in site 1 and site 2, respectively (Fig. 1A and SI Appendix, Fig. S1 A and B). This structure is highly similar to the previously reported triangle-shaped di-Zn(II)-AdcRWT structure (15). For example, ZitR’s dimerization domain (α1 and α6) is connected by the winged helix-turn-helix (wHTH) DNA-binding domain (α2, α3, α4, β1, and β2) through loop 1 and helix α5 (SI Appendix, Fig. S1 C and D). The Zn(II) ion in site 1 is coordinated by four residues (E24 Oε1, H42 Nδ1, H108 Nε2, and H112 Nε2) while the Zn(II) ion in site 2 is coordinated by residues C30 Sγ, E41 Oε1, E107 Oε1, and a water molecule (14) (SI Appendix, Fig. S1E). The DNA-binding helices α4 and α4′ (Cα atoms of A71 and A71′) are separated by 30.1 Å in di-Zn(II)-ZitRWT (Fig. 1B and SI Appendix, Fig. S1D and Table S2), representing a suitable conformation ready for DNA binding. To confirm that what we observed is indeed the optimal DNA-binding conformation, the structure of di-Zn(II)-ZitRWT in complex with its operator DNA was solved by molecular replacement and refined to 2.6-Å resolution (SI Appendix, SI Text, Fig. S2, and Tables S1 and S2). The overall structure of di-Zn(II)-ZitRWT including the two zinc-binding sites are highly similar with and without DNA (rmsd = 0.85 Å for 275 Cα atoms) (Fig. 1A and SI Appendix, Fig. S2B), indicating that di-Zn(II)-ZitRWT prelocks an optimal DNA-binding conformation even in the absence of DNA.
Fig. 1.
Zn(II) coordination in site 2 alters ZitR’s conformation. (A) Superposition of crystal structures of di-Zn(II)-ZitRWT protein in the presence (purple) and absence (green) of DNA. Closeup view of the dual Zn(II)-binding pocket is on the Right. Zn(II) ion at site 1 is coordinated by residues E24, H42, H108, and H112, while site 2 contains residues C30, E41, E107, and a water molecule for Zn(II) binding. Residues E24 from site 1 and C30 from site 2 reside on loop 1, while the rest of the residues reside on helix α2 (E41 and H42) and helix α5 (E107, H108, and H112), respectively. (B) Structural superposition of dimeric di-Zn(II)-ZitRWT (green) and Zn(II)site-1-ZitRC30S (yellow) proteins. The change of distance and rotation angles on helices α4s, the tip of the wings as well as the whole wHTH domains between Zn(II)site-1-ZitRC30S and di-Zn(II)-ZitRWT are indicated by arrows.
Lack of Zn(II) Coordination in Site 2 Results in a Nonoptimal Conformation for DNA Binding.
Whereas site 1 has been shown as the primary zinc regulatory site, the role of site 2 remains elusive. We next set out to elucidate the structural and functional roles of site 2 in ZitR. To obtain a site 2-deprived ZitR protein, we mutated one of the Zn(II)-coordination residues in site 2 (C30S). Inductively coupled plasma MS analysis confirmed that this mono-Zn(II)-ZitR variant binds to only one equivalent of Zn(II) per monomer [termed as Zn(II)site-1-ZitRC30S] (SI Appendix, Table S2). The crystal structure was solved by molecular replacement and refined to 1.65-Å resolution, which confirmed that Zn(II) was coordinated at site 1 but not site 2 (SI Appendix, Fig. S3A and Tables S1 and S2). Despite the high structural similarity between di-Zn(II)-ZitRWT and Zn(II)site-1-ZitRC30S when comparing their dimerization domains or DNA-binding domains separately, significant movements were observed regarding the relative positions of these domains (Fig. 1B and SI Appendix, Fig. S3B). The winged-HTH DNA-binding domain swings ∼30° with the tip of the wing (Cα atom of N88) translocated 15.3 Å in the Zn(II)site-1-ZitRC30S structure compared with di-Zn(II)-ZitRWT. In addition, helix α4 rotates about 24.1° and the distance between α4 and α4′ expands to 41.9 Å, generating a “widened” conformation not suitable for DNA binding (Fig. 1B and SI Appendix, Table S2). We further mutated one of the Zn(II)-coordination residues in site 1 (H42A) and the resulting apo form of the double mutant protein apo-ZitRC30AH42A shared a similar structure as that of Zn(II)site-1-ZitRC30S (SI Appendix, SI Text, Fig. S3 C–G, and Tables S1 and S2). Therefore, in contrast to the optimal DNA-binding conformation observed in di-Zn(II)-ZitRWT, both Zn(II)site-1-ZitRC30S and apo-ZitRC30AH42A adopt a nonoptimal DNA-binding conformation.
Functional Role of Site 2 in ZitR’s Zn(II) Binding and DNA Binding.
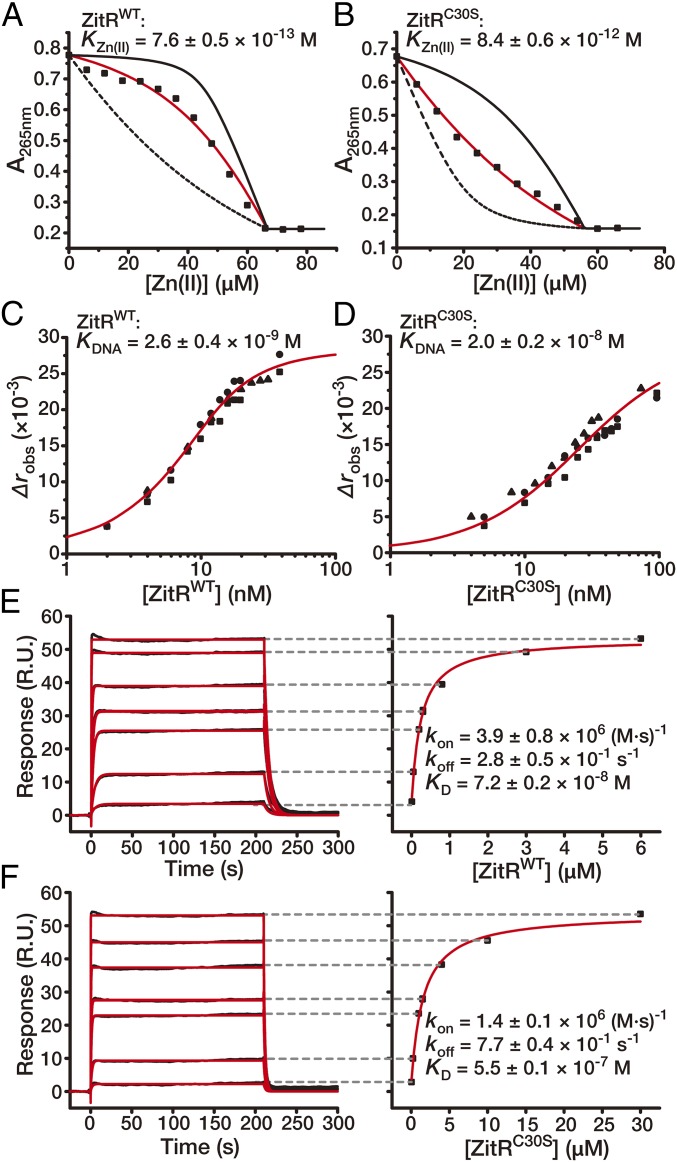
The apparent differences in ZitR structures with and without the second coordinated Zn(II) prompted us to examine the potential contribution of site 2 to site 1’s Zn(II) affinity as well as to ZitR’s DNA-binding affinity. Site 1’s Zn(II) affinities were determined with competition against the Zn(II) chelator quin-2 [KZn(II) = 3.7 × 10−12 M] and indo-1 [KZn(II) = 1.6 × 10−10 M] (14, 16). Analysis of the titration data confirmed that ZitRWT has a subpicomolar Zn(II) affinity at pH 8.0 and subnanomolar Zn(II) affinity at pH 6.0 in site 1 [KZn(II) = 7.6 ± 0.5 × 10−13 M at pH 8.0 and KZn(II) = 3.1 ± 0.5 × 10−10 M at pH 6.0] (Fig. 2A and SI Appendix, SI Text and Fig. S4A), which is similar to that of AdcR [KZn(II) = 7.1 × 10−13 M at pH 8.0 and KZn(II) < 1 × 10−9 M at pH 6.0] (14). Mutation in site 2 caused a ∼10-fold reduction of Zn(II) affinity in site 1 [KZn(II) = 8.4 ± 0.6 × 10−12 M] (Fig. 2B). The DNA-binding affinity was measured by fluorescence anisotropy (FA) with the fluorescently labeled zit promoter DNA. In comparison with ZitRWT, which possesses an apparent DNA affinity of 2.6 ± 0.4 × 10−9 M, the site-2 mutant ZitRC30S showed a near 10-fold decrease of apparent DNA affinity (apparent KDNA = 2.0 ± 0.2 × 10−8 M for ZitRC30S) (Fig. 2 C and D and SI Appendix, Fig. S4B). As expected, additional mutations in site 1 caused a more dramatic reduction of ZitR’s zinc-binding and DNA-binding affinities [KZn(II) = 2.3 ± 0.2 × 10−9 M and apparent KDNA = 1.2 ± 0.2 × 10−4 M for ZitRC30AH42A, which is ∼300-fold decreased for site 1’s Zn(II) affinity and ∼104-fold decreased for DNA affinity in comparison with ZitRC30S] (SI Appendix, Fig. S3 H and I). Therefore, with site 1 being the primary site for regulating ZitR’s DNA binding, we found that Zn(II) coordination in site 2 further enhanced its DNA-binding affinity, which may fine tune the transcriptional regulation under Zn(II) stress conditions [e.g., a transient increase of free Zn(II) level to the nanomolar range as a Zn(II) pulse] (17).
Fig. 2.
DNA-binding affinity and kinetics of ZitR with and without the second coordinated Zn(II). (A and B) Representative binding isotherms in measuring site-1 affinity by titrating Zn(II) into a mixture of ZitR protein and quin-2. The titration utilized (A) 45.0 μM ZitRWT monomer and 21.7 μM quin-2 and (B) 36.2 μM ZitRC30S monomer and 19.9 μM quin-2. The red solid lines are fitting to a model for 1:1 stoichiometric binding, while the black solid and dashed lines are simulated curves with Zn(II) affinities of ZitR proteins 10-fold tighter and weaker than the fitted value, respectively. (C and D) Binding isotherms of titrating ZitRWT (C) and ZitRC30S (D) into the fluorescently labeled zit promoter DNA. The x axes are plotted on logarithmic scales. The red solid lines are simulated curves of the mean apparent KDNA values in a 1:1 ZitR dimer:DNA binding model. Indicated affinity values are mean ± SEM of three independent experiments (different symbol shapes). (E and F) SPR sensorgrams of di-Zn(II)-ZitRWT (E, 10–6,000 nM) and Zn(II)site-1-ZitRC30S (F, 50–30,000 nM) proteins binding to their cognate DNA. Fitted curves are shown as red lines. Indicated rate constants and affinity values are mean ± SEM of n = 3 independent experiments.
Since FA only offers affinity constants under the steady state, we next conducted surface plasmon resonance (SPR) experiment to determine ZitR’s DNA-binding kinetics that are affected by Zn(II) coordination. Under the nonequilibrium condition during SPR analysis, Zn(II)site-1-ZitRC30S exhibited a near threefold increase of apparent DNA-dissociation rate constant (koff) over di-Zn(II)-ZitRWT, while its apparent DNA-association rate constant (kon) was nearly threefold lower than that of di-Zn(II)-ZitRWT (Fig. 2 E and F and SI Appendix, Fig. S4 C and D). These results suggest that Zn(II)site-1-ZitRC30S needs to readjust its conformation to fit α4 and α4′ helices into the DNA major grooves, while α4 and α4′ helices of di-Zn(II)-ZitRWT can simultaneously bind both DNA major grooves in the prelocked optimal DNA-binding conformation. Taken together, these results indicate that the lack of Zn(II) in site 2 can cause ZitR to undergo the induced-fit rearrangement for DNA binding with a lower binding affinity. This is similar to the induced-fit mechanism observed in many nonmetal-responsive MarR family proteins (7, 9, 18–20). The subsequent Zn(II) coordination in site 2 then favors ZitR to adopt the more optimal conformation with enhanced DNA-binding affinity.
Histidine Switch as an Allosteric Mechanism for Controlling ZitR Conformation.
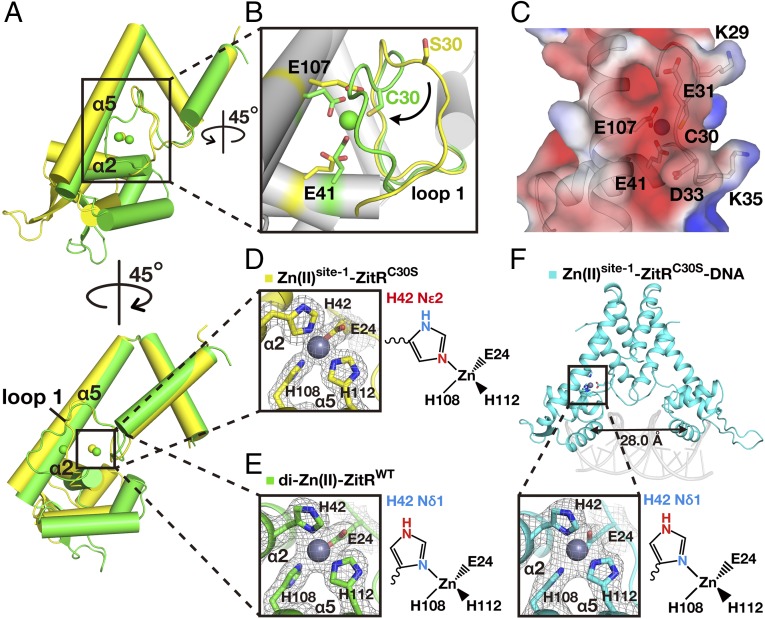
To understand molecular details underlying the conformational change between the dual-Zn(II)-occupied and the mono-Zn(II)-occupied forms of ZitR, we closely inspected and compared the structures between di-Zn(II)-ZitRWT and Zn(II)site-1-ZitRC30S. A dramatic conformational change was observed on loop 1 with and without the second zinc coordination: loop 1 is positioned toward the central region of ZitR dimer without Zn(II) ion in site 2, whereas the presence of Zn(II) ion in site 2 brings loop 1 into close proximity with helix α5 (Fig. 3 A and B). As residues surrounding site 2 are heavily negatively charged (e.g., E31, D33, E41, and E107), we speculate that electrostatic repulsion may contribute to the conformational change with and without Zn(II) binding in site 2. Indeed, four of the total seven charged residues in loop 1 showed large movement upon Zn(II) binding at site 2: the side chains of E31 and D33 are moved away from the helix α5 in Zn(II)site-1-ZitRC30S, while the side chains of K29 and K35 are positioned within the space between helix α5 and loop 1. In contrast, upon Zn(II) binding in site 2, the side chains of E31 and D33 were moved toward helix α5, while the side chains of K29 and K35 were flipped away, which is consistent with our assumptions (SI Appendix, Fig. S5 A and B). Further electrostatic analysis also verified electrostatic repulsion between loop 1 and helix α5 when both zinc sites are occupied (Fig. 3C and SI Appendix, Fig. S5C). Notably, when contoured at 1.0σ, the 2Fo-Fc density map of loop 1 is clear in di-Zn(II)-ZitRWT on the amino acid side chains, but is missing at several amino acid side chains and even a portion of the main chain in the Zn(II)site-1-ZitRC30S structure. This indicates that the flexibility of loop 1 is lower in di-Zn(II)-ZitRWT than that of Zn(II)site-1-ZitRC30S (SI Appendix, Fig. S5 D and E). Therefore, Zn(II) coordination in site 2 forces loop 1 to adopt a more rigid, higher energy conformation in close proximity with helix α5, which further causes a rearrangement of helix α2 that ultimately propagated to the DNA-binding sites (SI Appendix, Fig. S5 F and G).
Fig. 3.
A histidine-switch mediated site-1 and site-2 cooperation for optimal DNA binding. (A) Superposition of crystal structures of monomers from di-Zn(II)-ZitRWT (green) and Zn(II)site-1-ZitRC30S (yellow) proteins. Coordinated Zn(II) ions are shown as spheres. (B) Closeup view of A shows that coordination of Zn(II) at site 2 brings loop 1 to close proximity with helix α5. (C) Electrostatic potential surface presentation of site 2 in the di-Zn(II)-ZitRWT structure displayed at the +3 kT/e (blue) and −3 kT/e (red) levels. Zn(II)-occupied site 2 is surrounded by negatively charged residues. Side chains of residues from site 2 (C30, E41, and E107) and the four charged residues (K29, E31, D33, and K35) from loop 1 are labeled and shown as sticks. (D and E) Closeup view of the site-1 Zn(II)-binding pockets of Zn(II)site-1-ZitRC30S (D) and di-Zn(II)-ZitRWT (E). Different nitrogen atoms on the imidazole ring are used in coordination by H42 at site 1 in the presence and absence of Zn(II) at site 2. (F) Crystal structure of the Zn(II)site-1-ZitRC30S-DNA complex, with the distances between α4 and α4′ labeled. Closeup view of the site-1 Zn(II)-binding pocket is shown below. The Zn(II)-coordination atom on H42 is now switched from the Nε2 atom back to the Nδ1 atom in the presence of DNA. All electron density maps (2Fo-Fc) are contoured at 1.0σ.
We found that Zn(II)-binding residue H42 in site 1 on helix α2 uses different nitrogen atoms from its imidazole ring for zinc coordination: the Zn(II)-binding atom switches from Nδ1 in di-Zn(II)-ZitRWT to Nε2 in Zn(II)site-1-ZitRC30S (Fig. 3 D and E and SI Appendix, Fig. S6 A and B). This switch of Zn(II)-coordinating nitrogen on H42 (termed “histidine switch”) is triggered by the movement of loop 1 and helix α2 upon Zn(II) binding in site 2. The distance between Zn(II) and the Cβ atom of H42 decreases from 5.5 Å to 3.6 Å and causes a 16.4° rotation of helix α2, which ultimately drives the swing of wHTH DNA-binding motif (SI Appendix, Figs. S5G and S6C). To further verify the structural and biochemical differences between Zn(II)site-1-ZitRC30S and di-Zn(II)-ZitRWT that are dependent on Zn(II) coordination at site 2, we mutated E41, another Zn(II)-binding residue in site 2, and solved the crystal structure of this ZitR variant (ZitRE41A) by molecular replacement. The structure was refined to 1.9-Å resolution and also exhibited the non–DNA-binding conformation with the distance between α4 and α4′ at 36.0 Å (SI Appendix, Fig. S7 A and B and Tables S1 and S2). Notably, similar to Zn(II)site-1-ZitRC30S, the Zn(II)-coordination atom on H42 was also flipped to Nε2 in this ZitR variant (SI Appendix, SI Text and Fig. S7C). Meanwhile, similar to Zn(II)site-1-ZitRC30S, ZitRE41A also exhibited decreased apparent DNA affinity (apparent KDNA = 5.4 ± 0.1 × 10−8 M, 20-fold lower than ZitRWT), while its Zn(II) affinity was still in the picomolar range [KZn(II) = 3.1 ± 0.8 × 10−12 M, 4-fold lower than ZitRWT] (SI Appendix, Fig. S7 D and E). We thus speculate that the switch of Zn(II)-coordination nitrogen on H42 may serve as an allosteric mechanism that regulates ZitR between the nonoptimal and optimal DNA-binding conformations.
To further verify the switchable feature of H42, we solved the crystal structure of the Zn(II)site-1-ZitRC30S protein in complex with DNA by molecular replacement and refined to 2.9-Å resolution (SI Appendix, Fig. S8A and Tables S1 and S2). Zn(II)site-1-ZitRC30S protein within this complex exhibits an optimal DNA-binding conformation that is highly similar to ZitRWT from the di-Zn(II)-ZitRWT-DNA complex (SI Appendix, Fig. S8 B–D). In particular, the coordination environment of its site 1 resembles that of di-Zn(II)-ZitRWT, with the Zn(II)-coordination atom on H42 now switched back to the Nδ1 atom (Fig. 3 D and F and SI Appendix, Fig. S8 E and F). Therefore, whereas the mono-Zn(II)-occupied ZitR adopts a non–DNA-binding conformation with Zn(II) coordinated to H42’s Nε2 atom in site 1, this ZitR variant can be reversed to adopt the DNA-binding conformation via the induced-fit mechanism, which switched H42’s Zn(II)-coordination atom back to Nδ1 upon DNA binding. Interestingly, we found no hydrogen bonding networks within these crystal structures that may contribute to ZitR’s allosteric control of DNA recognition, which is different from the previously reported AdcR protein (15) (SI Appendix, SI Text and Fig. S1C). Therefore, it remains to be verified whether AdcR uses a similar mechanism for allosteric control of its DNA binding to ZitR presented here.
Genetic Incorporation of a Nonswitchable Histidine Analog to Verify the Histidine-Switch Mechanism.
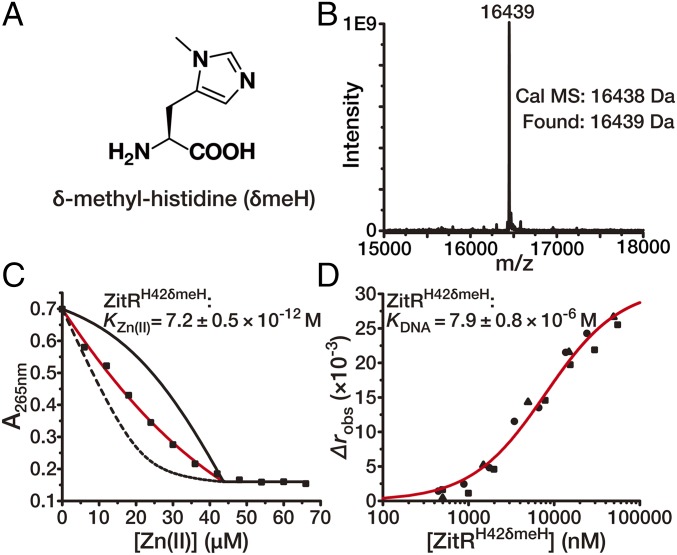
To further probe the distinct switchable feature of H42 in allosteric control of ZitR, we used a nonswitchable unnatural histidine analog to site-specifically replace H42 on ZitR. We envisioned that the blockage of Nδ1 on H42 may have neglected effects on Zn(II)-binding affinity in site 1 since ZitR can utilize H42’s Nε2 for Zn(II) binding and adopt a non–DNA-binding conformation as shown by Zn(II)site-1-ZitRC30S. However, because the “switching back” of H42’s Zn(II)-binding atom to Nδ1 is essential for ZitR to adopt the optimal DNA-binding conformation, blockage of this Nδ1 atom will significantly disrupt ZitR’s DNA binding. Previous work has incorporated Nε2-methylated histidine into metalloregulatory proteins via native chemical ligation to investigate their allostery (21, 22). Here we introduced the Nδ1-blocked histidine, termed δ-methyl-histidine (δmeH) (Fig. 4A), as an unnatural histidine analog to replace H42 on ZitR via the genetic code expansion strategy (23, 24). The pyrrolysyl-based aminoacyl-tRNA synthetase (PylRS)/tRNA pair from the archaea species Methanosarcina barkeri and Methanosarcina mazei has recently been employed to genetically incorporate δmeH into proteins at an in-frame amber codon (25, 26). By using a mutant PylRS and its cognate tRNA pair, we site-specifically incorporated δmeH at residue 42 on ZitRWT, which was verified by mass spectrometry (Fig. 4B). As expected, the generated ZitR variant ZitRH42δmeH has a Zn(II)-binding affinity of 7.2 ± 0.5 × 10−12 M in site 1, which is only approximately eightfold lower than that of ZitRWT and is at a similar level as that of ZitRC30S (Figs. 2 A and B and 4C). This result suggests that replacing H42 with δmeH has limited effects on ZitR’s metal coordination property in site 1 because Nε2 on H42δmeH can be used as the Zn(II)-coordination atom under this condition. In contrast, when H42 becomes nonswitchable, dramatically decreased DNA-binding affinity (apparent KDNA = 7.9 ± 0.8 × 10−6 M) was observed on ZitRH42δmeH, which is three magnitudes lower than that of ZitRWT (Figs. 2C and 4D). This result verified our speculation that Zn(II) coordination with H42’s Nδ1 atom is essential for ZitR to adopt the optimal DNA-binding conformation. Therefore, the switchable feature of H42 is critical for controlling the transition between ZitR’s DNA-binding and non–DNA-binding conformations.
Fig. 4.
Allosteric control of the broad-range transcriptional regulator ZitR via a histidine-switch mechanism. (A–D) Verifying the histidine-switch mechanism by a genetically encoded histidine analog δ-methyl-histidine (δmeH, A). δmeH was incorporated at residue H42 in ZitRWT by the MbPylRS- pair. The efficiency and fidelity of this incorporation on the resulting protein ZitRH42δmeH was confirmed by mass spectrometry (B). (C) Representative binding isotherms of titrating Zn(II) into a mixture of 23.0 μM ZitRH42δmeH monomer and 20.7 μM quin-2. Data are presented in the same way as in Fig. 2 A and B. (D) Binding isotherms of titrating ZitRH42δmeH into the fluorescently labeled zit promoter fragment. The x axis is plotted on a logarithmic scale. Data are presented in the same way as in Fig. 2 C and D. Indicated affinity values are mean ± SEM of three independent experiments.
Investigating the Site 2-Mediated ZitR Response to Zn(II) Stress Inside Cells.
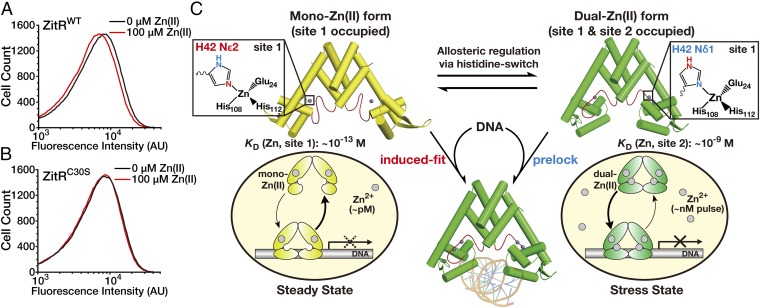
Finally, we directly tested the site 2-mediated ZitR response to Zn(II) stress inside cells. Since Zn(II) coordination at this nanomolar-affinity site would enhance ZitR’s DNA-binding affinity, we reasoned that Zn(II) binding at site 2 may facilitate ZitR’s transcription regulation under the nanomolar Zn(II) stress conditions. Previous work has shown that the addition of a large excess of Zn(II) ions (micromolar level) to bacterial culture can rapidly increase the intracellular Zn(II) to the nanomolar range (within 10 min) (17). We investigated whether site 2’s coordination would enhance ZitR’s transcriptional repression upon such a Zn(II) pulse treatment. To develop a cell-based assay to monitor the transcriptional regulation of ZitR, we engineered a ZitR-inducible GFP reporter that encodes the gfp gene under the control of zit promoter (zitR-gfp reporter, SI Appendix, Fig. S9). The reporter plasmid was cotransformed with a ZitR-expressing plasmid (pBAD-ZitR) into the Escherichia coli BW25113 strain. This allowed us to quantify the ZitR-induced transcriptional repression via flow cytometric analysis of GFP expression. Upon the addition of 100 μM Zn(II), the ZitRWT-expressing bacterial strain harboring the zitR-gfp reporter showed a decreased fluorescence compared to without Zn(II) treatment, indicating that the transcription of GFP was further repressed under this Zn(II) stress condition (Fig. 5A). The ZitRC30S-expressing strain harboring the same zitR-gfp reporter was used as a control. Because ZitRC30S loses Zn(II) binding at site 2, this bacterial strain showed negligible change with and without 100 μM Zn(II) (Fig. 5B). Together, this cell-based analysis agrees with our aforementioned in vitro results that Zn(II) coordination at site 2 may enhance and fine tune ZitR’s DNA-binding affinity and thus transcriptional repression when cells are under Zn(II) stress conditions [e.g., nanomolar Zn(II) pulse].
Fig. 5.
ZitR fine tunes cell’s transcriptional response to Zn(II) fluctuation. (A and B) Site 2-mediated ZitR response to Zn(II) stress inside cells. Flow cytometric analysis of the zitR-gfp reporter-harbored E. coli cells expressing ZitRWT (A) or ZitRC30S (B) proteins with and without the Zn(II) pulse treatment. Since previous work has shown that the addition of the micromolar level of Zn(II) ions can increase the intracellular free Zn(II) to the nanomolar range within 10 min, the bacteria were treated with (red lines) and without (black lines) 100 μM Zn(II) ions for 1 h before the fluorescence intensity of expressed GFP was recorded. (C) The proposed working model for histidine switch-mediated allosteric control of ZitR for the broad-range and fine-tuned transcriptional response to Zn(II) fluctuation. Zn(II) binding at site 1 under steady state [subpicomolar range of Zn(II)] allows ZitR to undergo an induced-fit mechanism for DNA binding, which can be used to control the steady-state zinc homeostasis. Elevated Zn(II) levels would produce a dual-Zn(II) form of ZitR with both sites occupied, which prelocks an optimal DNA-binding conformation for strong transcriptional response under zinc stress conditions [e.g., nanomolar Zn(II) pulse]. A histidine-switch mechanism on site 1’s Zn(II)-coordination residue H42 mediates the transition between these two ZitR forms to fine tune the cell’s transcriptional response to intracellular Zn(II) fluctuation.
Discussion
We report here a unique histidine switch-mediated allosteric regulation mechanism for the zinc transcriptional factor ZitR (Fig. 5C). Metalloproteins are specialized allosteric proteins in which the metal binding-induced conformational changes are frequently employed for allosteric control of binding with substrates, cofactors, partner proteins, as well as additional metal ions (27–32). In particular, additional unique features of metal ions and/or ligands have been harnessed by metalloproteins for allosteric regulation. The former example includes hemoglobin, which uses the spin-state alteration between the heme-bound Fe(II) and Fe(III) ions to control oxygen binding (33, 34). For the latter case, the two nitrogen atoms on the same histidine side chain of Mycobacterium tuberculosis CsoR are engaged in Cu(I) binding and hydrogen-binding network formation, respectively, which permits the coupling of metal coordination with the allosteric control of DNA binding (21, 35). The unique histidine-switch mechanism we observed here represents an unprecedented example in which the heteroaromatic feature of histidine’s imidazole ring is exploited to coordinate two neighboring metal-binding sites for allosteric control and fine tuning of metalloregulator’s DNA-binding affinity (Fig. 5C). The multifunctional properties of histidine may be employed as a general allosteric strategy to couple metal ion coordination with protein conformational change.
Instead of being gratuitous, our study in vitro and inside bacterial cells suggests that Zn(II) binding at site 2 is involved in the allosteric regulation of ZitR’s transcriptional corepression. Interestingly, the zinc affinities in site 1 (subpicomolar) and site 2 (nanomolar) fall in the range of the intracellular Zn(II) concentrations under steady-state and stress-state conditions, respectively (17, 30, 36–38). The large reduction (∼104-fold) of the DNA affinity induced by site-1 mutation on ZitR protein shows that the primary role of ZitR is regulating steady-state zinc homeostasis via Zn(II) coordination in site 1. Meanwhile, the higher DNA-binding affinity (∼10-fold higher) observed on dual-Zn(II)-occupied ZitR seems to indicate that ZitR may mount an enhanced transcriptional response (e.g., repression of zinc uptake genes) through site 2 upon a nanomolar Zn(II) pulse (Fig. 5C). When lacking Zn(II) coordination in site 2, ZitR adopts a non–DNA-binding conformation that may undergo the induced-fit mechanism for DNA binding as well as the control of steady-state zinc homeostasis with a weaker repression. Together, a single metalloregulator ZitR may maintain broad as well as fine-tuned responses to steady-state and stress-state Zn(II) fluctuations via an allosteric histidine switch-mediated dual Zn(II)-site cooperation. Previous work on S. pneumoniae bacteria harboring different AdcR variants showed that although site 1 plays a major role in AdcR function in vivo, bacterial strains harboring a site-2 mutant AdcR (e.g., adcR-C30A) indeed showed a slightly higher but statistically significant amount of cellular zinc than the parent adcR+ strain (14). Similarly, this small difference may indeed account for the physiological role of site 2 in ZitR, which is expected to be a fine-tuned response. In addition, another Zn(II) transcription regulator Zur contains multiple Zn(II)-binding sites and is previously reported to mount a stepwise repression of different promoters with altered Zn(II) levels (39–41). Considering the fact that both ZitR and Zur proteins have enhanced DNA affinity upon additional Zn(II) coordination besides their primary sites, ZitR might also adopt such a “stepwise” regulation. For example, a subset of promoters might be more dependent on site 2 and undergo enhanced repression at highly elevated Zn(II) levels [e.g., nanomolar Zn(II) pulse] to cope with transient Zn(II) stress. This speculation requires further investigation, particularly under in vivo settings (14).
Methods
Detailed materials and methods are available in SI Appendix. See SI Appendix, Table S1 for data collection and refinement statistics of ZitR variants and SI Appendix, Table S2 for the structural information of ZitR variants. SI Appendix, Table S3 shows conditions for crystallization of ZitR variants. SI Appendix, Table S4 reports the oligonucleotides used in this study.
Supplementary Material
Acknowledgments
We thank S. F. Reichard for editing the manuscript and staff members of the Shanghai Synchrotron Radiation Facility, the National Center for Protein Science Shanghai, and the Beijing Synchrotron Radiation Facility. This work was supported by research grants from the National Key Research and Development Program of China (2016YFA0501500 to P.R.C.), the National Natural Science Foundation of China (21432002 and 21521003 to P.R.C.), and the E-Institutes of Shanghai Municipal Education Commission (project number E09013 to P.R.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5YHX for ZitRWT, 5YI2 for ZitRWT-DNA, 5YHY for ZitRC30S, 5YI3 for ZitRC30S-DNA, 5YHZ for ZitRE41A, 5YI0 for ZitRC30AH42A, and 5YI1 for apo-ZitRC30AH42A, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708563115/-/DCSupplemental.
References
- 1.Claverys JP. A new family of high-affinity ABC manganese and zinc permeases. Res Microbiol. 2001;152:231–243. doi: 10.1016/s0923-2508(01)01195-0. [DOI] [PubMed] [Google Scholar]
- 2.Bolotin A, et al. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001;11:731–753. doi: 10.1101/gr.169701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hantke K. Bacterial zinc uptake and regulators. Curr Opin Microbiol. 2005;8:196–202. doi: 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llull D, et al. Lactococcus lactis ZitR is a zinc-responsive repressor active in the presence of low, nontoxic zinc concentrations in vivo. J Bacteriol. 2011;193:1919–1929. doi: 10.1128/JB.01109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alekshun MN, Levy SB. The mar regulon: Multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 1999;7:410–413. doi: 10.1016/s0966-842x(99)01589-9. [DOI] [PubMed] [Google Scholar]
- 7.Wilkinson SP, Grove A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol. 2006;8:51–62. [PubMed] [Google Scholar]
- 8.Ellison DW, Miller VL. Regulation of virulence by members of the MarR/SlyA family. Curr Opin Microbiol. 2006;9:153–159. doi: 10.1016/j.mib.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Perera IC, Grove A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol. 2010;2:243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 10.Hao Z, et al. The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli. Nat Chem Biol. 2014;10:21–28. doi: 10.1038/nchembio.1380. [DOI] [PubMed] [Google Scholar]
- 11.Panina EM, Mironov AA, Gelfand MS. Comparative genomics of bacterial zinc regulons: Enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc Natl Acad Sci USA. 2003;100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loisel E, et al. AdcAII, a new pneumococcal Zn-binding protein homologous with ABC transporters: Biochemical and structural analysis. J Mol Biol. 2008;381:594–606. doi: 10.1016/j.jmb.2008.05.068. [DOI] [PubMed] [Google Scholar]
- 13.Ogunniyi AD, et al. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. 2009;23:731–738. doi: 10.1096/fj.08-119537. [DOI] [PubMed] [Google Scholar]
- 14.Reyes-Caballero H, et al. The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J Mol Biol. 2010;403:197–216. doi: 10.1016/j.jmb.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerra AJ, Dann CE, 3rd, Giedroc DP. Crystal structure of the zinc-dependent MarR family transcriptional regulator AdcR in the Zn(II)-bound state. J Am Chem Soc. 2011;133:19614–19617. doi: 10.1021/ja2080532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jefferson JR, Hunt JB, Ginsburg A. Characterization of indo-1 and quin-2 as spectroscopic probes for Zn2(+)-protein interactions. Anal Biochem. 1990;187:328–336. doi: 10.1016/0003-2697(90)90465-l. [DOI] [PubMed] [Google Scholar]
- 17.Wang D, Hosteen O, Fierke CA. ZntR-mediated transcription of zntA responds to nanomolar intracellular free zinc. J Inorg Biochem. 2012;111:173–181. doi: 10.1016/j.jinorgbio.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Kumaraswami M, Schuman JT, Seo SM, Kaatz GW, Brennan RG. Structural and biochemical characterization of MepR, a multidrug binding transcription regulator of the Staphylococcus aureus multidrug efflux pump MepA. Nucleic Acids Res. 2009;37:1211–1224. doi: 10.1093/nar/gkn1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolan KT, Duguid EM, He C. Crystal structures of SlyA protein, a master virulence regulator of Salmonella, in free and DNA-bound states. J Biol Chem. 2011;286:22178–22185. doi: 10.1074/jbc.M111.245258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Z, et al. Unnatural amino acid substitution as a probe of the allosteric coupling pathway in a mycobacterial Cu(I) sensor. J Am Chem Soc. 2009;131:18044–18045. doi: 10.1021/ja908372b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campanello GC, et al. Allosteric inhibition of a zinc-sensing transcriptional repressor: Insights into the arsenic repressor (ArsR) family. J Mol Biol. 2013;425:1143–1157. doi: 10.1016/j.jmb.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Schultz PG. Expanding the genetic code. Angew Chem Int Ed Engl. 2004;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 24.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 25.Xiao H, et al. Genetic incorporation of histidine derivatives using an engineered pyrrolysyl-tRNA synthetase. ACS Chem Biol. 2014;9:1092–1096. doi: 10.1021/cb500032c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma V, Wang Y-S, Liu WR. Probing the catalytic charge-relay system in alanine racemase with genetically encoded histidine mimetics. ACS Chem Biol. 2016;11:3305–3309. doi: 10.1021/acschembio.6b00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giedroc DP, Arunkumar AI. Metal sensor proteins: Nature’s metalloregulated allosteric switches. Dalton Trans. 2007:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- 28.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 29.Reyes-Caballero H, Campanello GC, Giedroc DP. Metalloregulatory proteins: Metal selectivity and allosteric switching. Biophys Chem. 2011;156:103–114. doi: 10.1016/j.bpc.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guerra AJ, Giedroc DP. Metal site occupancy and allosteric switching in bacterial metal sensor proteins. Arch Biochem Biophys. 2012;519:210–222. doi: 10.1016/j.abb.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattle D, et al. On allosteric modulation of P-type Cu(+)-ATPases. J Mol Biol. 2013;425:2299–2308. doi: 10.1016/j.jmb.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Peralta FA, Huidobro-Toro JP. Zinc as allosteric ion channel modulator: Ionotropic receptors as metalloproteins. Int J Mol Sci. 2016;17:1059. doi: 10.3390/ijms17071059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perutz MF. Stereochemistry of cooperative effects in haemoglobin: Haem–haem interaction and the problem of allostery. Nature. 1970;228:726–734. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 34.Perutz MF, Wilkinson AJ, Paoli M, Dodson GG. The stereochemical mechanism of the cooperative effects in hemoglobin revisited. Annu Rev Biophys Biomol Struct. 1998;27:1–34. doi: 10.1146/annurev.biophys.27.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Liu T, et al. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 36.Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 37.Wang D, Hurst TK, Thompson RB, Fierke CA. Genetically encoded ratiometric biosensors to measure intracellular exchangeable zinc in Escherichia coli. J Biomed Opt. 2011;16:087011. doi: 10.1117/1.3613926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foster AW, Osman D, Robinson NJ. Metal preferences and metallation. J Biol Chem. 2014;289:28095–28103. doi: 10.1074/jbc.R114.588145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin J-H, et al. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc Natl Acad Sci USA. 2011;108:5045–5050. doi: 10.1073/pnas.1017744108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma Z, Gabriel SE, Helmann JD. Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res. 2011;39:9130–9138. doi: 10.1093/nar/gkr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin J-H, Helmann JD. Molecular logic of the Zur-regulated zinc deprivation response in Bacillus subtilis. Nat Commun. 2016;7:12612. doi: 10.1038/ncomms12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.