Significance
We present evidence for the presymptomatic dysregulation of urea metabolism in Huntington’s disease (HD). We identified increased levels of a urea transporter transcript and other osmotic regulators in the striatum of our prodromal sheep model of HD and a concomitant increase in striatal and cerebellar urea. Elevated urea was also detected in brain tissue from postmortem HD cases, including cases with low-level cell loss, implying that increased brain urea in HD is not just a product of end-stage cachexia. Disruption of urea metabolism is known to cause neurologic impairment and could initiate neurodegeneration and the symptoms of HD. Our findings suggest that lowering brain levels of urea and/or ammonia would be a worthwhile therapeutic target in HD.
Keywords: Huntington’s disease, urea, prodromal, sheep, metabolism
Abstract
The neurodegenerative disorder Huntington’s disease (HD) is typically characterized by extensive loss of striatal neurons and the midlife onset of debilitating and progressive chorea, dementia, and psychological disturbance. HD is caused by a CAG repeat expansion in the Huntingtin (HTT) gene, translating to an elongated glutamine tract in the huntingtin protein. The pathogenic mechanism resulting in cell dysfunction and death beyond the causative mutation is not well defined. To further delineate the early molecular events in HD, we performed RNA-sequencing (RNA-seq) on striatal tissue from a cohort of 5-y-old OVT73-line sheep expressing a human CAG-expansion HTT cDNA transgene. Our HD OVT73 sheep are a prodromal model and exhibit minimal pathology and no detectable neuronal loss. We identified significantly increased levels of the urea transporter SLC14A1 in the OVT73 striatum, along with other important osmotic regulators. Further investigation revealed elevated levels of the metabolite urea in the OVT73 striatum and cerebellum, consistent with our recently published observation of increased urea in postmortem human brain from HD cases. Extending that finding, we demonstrate that postmortem human brain urea levels are elevated in a larger cohort of HD cases, including those with low-level neuropathology (Vonsattel grade 0/1). This elevation indicates increased protein catabolism, possibly as an alternate energy source given the generalized metabolic defect in HD. Increased urea and ammonia levels due to dysregulation of the urea cycle are known to cause neurologic impairment. Taken together, our findings indicate that aberrant urea metabolism could be the primary biochemical disruption initiating neuropathogenesis in HD.
Huntington’s disease (HD) is a dominantly inherited neurological disorder typified by chorea, psychological disturbance, and dementia. The symptoms progress and result in premature death, typically 10–15 y after onset. Currently no available treatment can delay or prevent the onset of HD. The gene responsible, Huntingtin (HTT), is ubiquitously expressed and encodes the large and multifunctional huntingtin protein. The disease-causing mutation is an expanded CAG repeat in exon 1 of HTT, coding for a glutamine tract within the protein (1). The disease-causing repeat lower length threshold is 36 units and is fully penetrant at 40 units and above (2, 3). There is an inverse correlation between expanded CAG repeat size and age at onset of symptoms (4–6). Although the mutation is well defined, the pathogenic process is not sufficiently understood to enable effective treatment. The majority of HD research focuses on the brain where there is characteristic neuropathology, primarily atrophy of the striatum (7).
Alongside the striking neurological phenotype of HD, there is a generalized metabolic disruption. HD mutation carriers weigh less on average than non-HD individuals (8). Weight loss begins presymptomatically (9, 10), and in symptomatic individuals, energy expenditure far exceeds that utilized in movement, despite high calorie intake (11, 12). Metabolic changes have been identified both systemically and within the central nervous system (CNS) that may be responsible for this metabolic phenotype. Circulating levels of branched chain amino acids are reduced in HD individuals, and this decline correlates with weight loss (11). Levels of citrulline were also reportedly elevated in blood from HD individuals (13), indicating a potential role of the urea cycle, a critical pathway for clearing ammonia generated through protein catabolism. Within the CNS, a recent study found altered levels of amino acids in multiple regions of the postmortem HD brain as well as increased concentrations of glucose, sorbitol, fructose, and urea (14). This is in line with an earlier report describing a reduced rate of glucose utilization in the HD striatum (15). These studies suggest that metabolic dysfunction contributes to the neuropathology of HD. Indeed, studies using HD patient-derived cell lines have identified CAG repeat length correlated with changes in the ratio of ATP to ADP (16), implying a direct role of huntingtin in metabolic regulation. It is unclear how the HD mutation causes these metabolic changes, particularly in the initiating stages of the disease.
To investigate pathogenesis in the prodromal phase of HD, before overt clinical signs, our laboratory has developed and characterized a sheep model of the disease. The transgenic sheep line OVT73 expresses full-length human huntingtin with a pathogenic exon 1 glutamine repeat of 73 units, under the regulatory control of a short human genomic HTT upstream sequence (17). The OVT73 sheep display some of the neuropathological changes that occur in HD, including the accumulation of intracellular huntingtin-positive inclusions (18). An alteration in circadian rhythm has been observed, a behavioral phenotype consistent with an early phase in the disease (19). We have also found that levels of specific metabolites are altered in the OVT73 sheep brain and liver. In addition, the correlation between specific metabolite pairs are different, with increased numbers of correlations in the transgenic animals, indicating that a metabolic defect exists in this prodromal model (20).
To further investigate early molecular changes in HD, we undertook a whole genome RNA-sequencing (RNA-seq) experiment comparing gene expression profiles in striatal tissue from OVT73 and control sheep. Specific gene expression differences were detected and validated. Reported functions of the altered genes support the existence of a metabolic defect in the OVT73 sheep and, specifically, a disturbance of urea handling. Concurrently, a metabolomics study revealed a very large increase in levels of the metabolite urea that was widespread in postmortem HD human brain tissue (21). Together with its precursor ammonia, urea is neurotoxic in excess and could certainly contribute to HD pathogenesis. Considering the gene expression changes in the OVT73 sheep and the urea elevation in HD brain tissue, we further investigated this phenomenon by quantifying urea in a range of tissues from the OVT73 sheep and in postmortem brain tissue from a larger sample of HD cases. A fundamental question addressed in the current study was whether the urea phenotype is a generalized phenomenon due to elevated protein catabolism as a consequence of cell death or reflects a requirement for cellular energy production due to the observed increased rate of metabolism in HD.
Results
Differential Gene Expression in the OVT73 HD Sheep Striatum.
RNA-seq was performed on ribosomal-depleted total RNA from the anterior striatum of 5-y-old sheep, and transcripts were analyzed for differential expression between OVT73 (n = 6) and controls (n = 6). A fully annotated sheep transcriptome was not available at the outset of this research, and therefore, analysis initially required the construction of a de novo sheep transcriptome (described in Materials and Methods). During the course of the analysis, Ensembl gene models derived from the most recent sheep genome (Ovis aries v3.1) were released and therefore incorporated. Differential expression of transcripts was ultimately determined through two mapping approaches. In one approach, the experimental RNA-seq reads (50 bp paired-end) were mapped directly against Ensembl gene models. Comparative analysis revealed 278 genes and 188 isoforms that were differentially expressed (q-value < 0.05; refer to Dataset S1). In a second approach, reads were mapped against a hybrid transcriptome, assembled from RNA-seq reads (100 bp) using Ensembl gene model mapping and de novo assembly tools. This enabled the quantification of an additional 42,653 de novo assembled transcripts, and comparative analysis revealed 447 genes and 261 isoforms that were differentially expressed on average between OVT73 and controls (q-value < 0.05; refer to Dataset S2).
The distribution of expression values (FPKM) for each set of nominally identified differentially expressed genes was examined to identify and remove findings due to outliers. To enable biologically meaningful interpretation, we further restricted the final datasets to annotated transcripts and removed transcripts that were clearly poorly mapped upon inspection using the Integrative Genomics Viewer (IGV) browser. This filtering resulted in a final set of 24 genes of interest with apparent differential expression in the OVT73 sheep striatum; 18 were identified through the hybrid transcriptome mapping approach and 10 through the gene models mapping, with four transcripts in common between the approaches (Table S1).
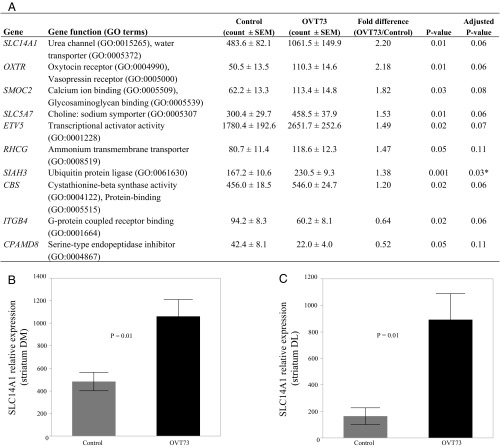
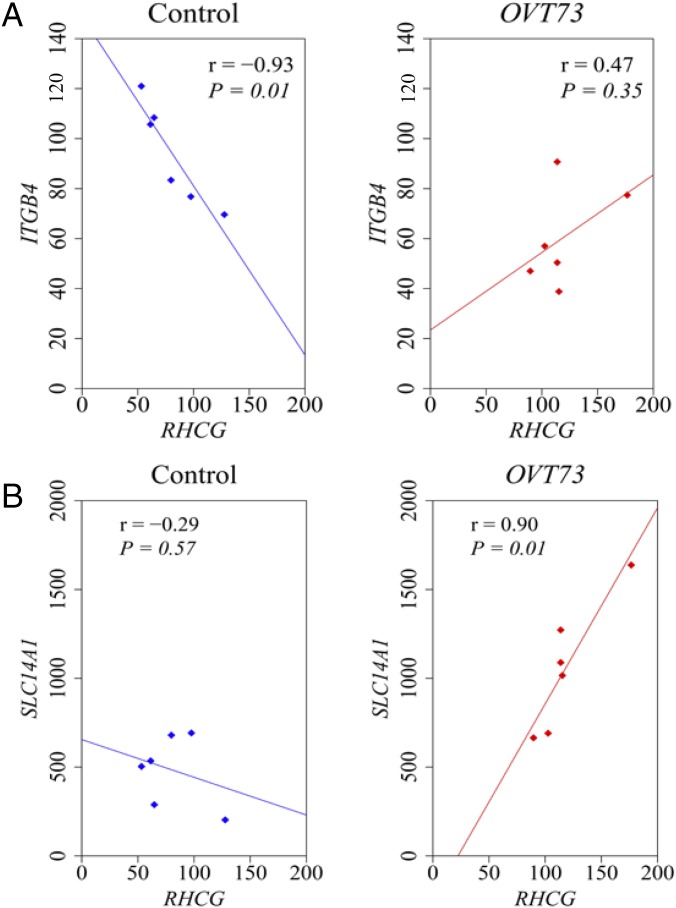
For validation, the 24 transcripts of interest identified by RNA-seq were requantified using NanoString nCounter expression analysis, in the same striatal RNA samples (dorsal-medial anterior striatum, DM) used to generate the libraries for sequencing. Ten of the 24 genes analyzed by NanoString quantification were differentially expressed at the nominal significance level (Fig. 1A; refer to Datasets S1 and S2 for RNA-seq data). Eight transcripts had higher expression levels and two had lower levels in the OVT73 samples on average compared with controls. The validated transcript with the greatest differential expression was SLC14A1, which was 2.2-fold higher in the OVT73 striatum compared with controls (P = 0.01; Fig. 1B). Three of the up-regulated genes function as solute transporters (SLC14A1, SLC5A7, and RHCG), while others are involved in protein binding (ETV5, CBS, ITGB4, and OXTR), cleavage (CPAMD8), and processing (SIAH3). To investigate possible functional relationships between the 10 validated genes, we determined the pair-wise correlation coefficient of each gene pairing and compared OVT73 with controls. Two differential correlations were detected at the nominal significance level: the loss of a negative correlation between ITGB4 and RHCG expression in OVT73 compared with controls (rOVT73 = 0.47 and rcontrol = −0.93, P = 0.01), and the gain of a positive correlation between RHCG and SLC14A1 (rOVT73 = 0.90 and rcontrol = −0.29, P = 0.03) in the OVT73 animals (Fig. 2). Correlation values for all 10 genes are provided in Table S2.
Fig. 1.
Urea transporter transcripts are elevated in OVT73 striatum. NanoString nCounter quantification of 24 target genes identified in RNA-seq analysis confirmed that 10 transcripts were altered in OVT73 striatum compared with control (P < 0.05; two-way Student’s t test). (A) Mean normalized counts ± SEM for control (n = 6) and OVT73 (n = 6), fold difference (OVT73/control), and P values (nonadjusted and false discovery rate adjusted using the Benjamini–Hochberg technique) are shown for each of the 10 validated transcripts, quantified in RNA from anterior striatum (DM portion). *P < 0.05 after false discovery rate adjustment. Key Gene Ontology (GO) terms are shown for each gene (www.geneontology.org/). Bar graphs depicting SLC14A1 expression in the DM (B) and DL (C) portions of anterior striatum are shown (±SEM). Transcript counts were normalized to the geometric mean of four reference genes: CANX, SHMT2, TRIM27, and TRIP11.
Fig. 2.
Altered correlations of gene expression in the OVT73 sheep striatum. Transcripts with validated differential expression in the OVT73 striatum (n = 6) compared with controls (n = 6) were analyzed for differences in pairwise correlation. (A) A strong negative correlation between ITGB4 and RHCG in controls is absent in transgenics (P = 0.01; Fisher r-to-z transformation). (B) A positive correlation of SLC14A1 with RHCG was significantly stronger in transgenics compared with controls (P = 0.03; Fisher r-to-z transformation). Pearson coefficients (r) and P values (P) for each correlation are provided. Axes show the nCounter analysis counts for each target gene, normalized to the geometric mean of counts for four reference genes (CANX, SHMT2, TRIM27, and TRIP11). Data points shown represent individual animals and are joined by the line of best fit.
To extend these findings, NanoString nCounter transcript quantification was also performed on new RNA samples extracted from a site in the dorsal-lateral (DL) portion of the anterior striatum from the same animals, adjacent to the RNA-seq sampling site (Table S1). Of the 24 genes assessed, only SLC14A1 was differentially expressed in the DL portion of striatum, where it was 5.4-fold higher on average in OVT73 samples compared with controls (P = 0.01; Fig. 1C). Together, the RNA-seq and NanoString nCounter data established SLC14A1, a transmembrane transporter for urea, as a high-priority gene of interest in the OVT73 sheep.
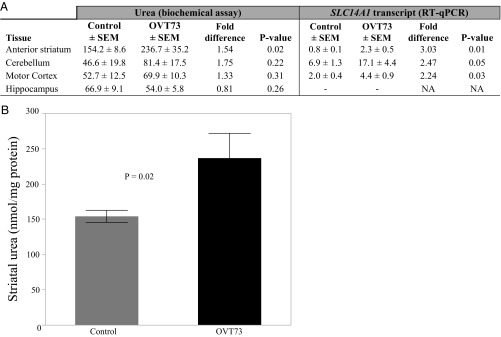
To determine whether the differential expression of SLC14A1 was potentially more widespread in the OVT73 brain, qRT-PCR was subsequently performed on RNA from striatum (DM portion), cerebellum, and motor cortex tissue from the same sheep. Comparing OVT73 and controls, SLC14A1 transcripts were more than twofold higher in all three brain regions (P ≤ 0.05, Fig. 3A).
Fig. 3.
Urea and SLC14A1 are elevated in the OVT73 sheep brain. Urea and its transporter SLC14A1 were quantified in brain tissue from 5-y-old sheep, with comparison between OVT73 (n = 6) and controls (n = 6). (A) Mean urea and SLC14A1 transcript levels in OVT73 and controls are shown for each tissue (back-transformed from log10 data where appropriate), in addition to the fold difference (OVT73/control) and P value (*P < 0.05; two-way Student’s t test). Urea (nanomole urea per milligram of protein) was quantified in anterior striatum, cerebellum, motor cortex, and hippocampus, using a biochemical urea assay. The SLC14A1 transcript was quantified by qRT-PCR in cDNA generated from anterior striatum (DM portion), cerebellum, and motor cortex tissue from the same animals. SLC14A1 transcript levels shown are relative to the geometric mean of the most stable reference gene combination for each tissue. NA, not applicable; SEM, standard error of the mean. (B) Mean urea concentration (nanomole urea per milligram of protein ± SEM) in the OVT73 anterior striatum compared with controls.
Urea Is Elevated in the OVT73 Sheep Striatum and Cerebellum.
The striking difference in brain SLC14A1 expression and its function as a urea transporter led us to consider possible effects on urea levels in the OVT73 sheep. Supporting this, in concurrent work, we identified a profound and anatomically widespread increase in urea in postmortem human brain tissue from a small number of HD cases (21). In that study, urea levels were determined by both mass spectrometry and a biochemical urea assay, with excellent concordance between methods. We applied the biochemical assay to quantify urea in brain and peripheral tissues from the same OVT73 sheep in which SLC14A1 expression was altered. Within the brain, the concentration of urea in the anterior striatum was 1.5-fold higher on average in the OVT73 sheep compared with controls (P = 0.02; Fig. 3B) but did not differ significantly overall in the other brain regions examined: cerebellum, motor cortex, and hippocampus (Fig. 3A). Interestingly, the sheep striatum RNA-seq data revealed no significant differences in the expression of transcripts encoding key enzymes of the urea cycle (Table S3), with the transcript levels of CPS1 and OTC indicating negligible expression in all of the sheep samples (Table S3).
As the sheep cohort studied was imbalanced in sexes (refer to Materials and Methods), we examined for effects of sex on urea levels. There was no effect of sex on urea concentration in the striatum or hippocampus. In the cerebellum and motor cortex, however, ewes had a significantly higher urea concentration than rams (P = < 0.0001 and 0.02, respectively). Statistically adjusting for sex in the data from these two tissues revealed a significantly higher level of urea in the OVT73 cerebellum compared with controls (P = 0.04) but not in the motor cortex (Table S4). In the peripheral tissues examined (heart, bladder, kidney, liver, muscle, urine, and serum), there were no transgene-specific differences in urea concentration (Table S4).
Evidence for Altered Serum Urea in the OVT73 Sheep.
Serum is a readily accessible bio-fluid from live subjects, and biomarkers of disease identified in serum would be invaluable for the investigation and treatment of HD. For this reason, we extended the quantification of serum urea into two larger, single-sex grazing cohorts of 4-y-old OVT73 sheep. In a cohort of 39 ewes (23 OVT73, 16 control), serum urea was modestly lower (P = 0.05) in the transgenic animals compared with controls (Fig. S1). In a cohort of 25 rams (13 OVT73, 12 controls), however, serum urea levels did not differ by transgene status (Fig. S1).
Urea Levels Are Elevated in HD Postmortem Brain Tissue.
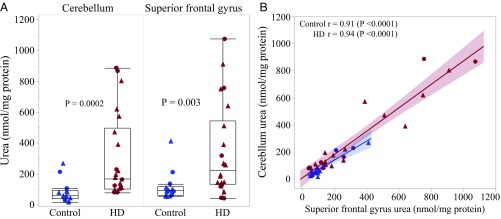
We previously reported elevated urea levels in multiple brain regions of a small number (n = 9) of postmortem HD cases with mixed (Vonsattel grades 1–4) neuropathology (21). To establish whether this urea phenotype is a generalizable phenomenon in HD, here we sought to replicate the finding in a larger cohort. Cerebellum and cortical (superior frontal gyrus, SFG) samples from 22 postmortem HD cases (Vonsattel grades 0–4) and 14 controls were kindly provided by the Neurological Foundation of New Zealand Douglas Brain Bank for this purpose (Table S5). Quantification of urea in this sample cohort using the biochemical urea assay revealed a significantly higher concentration in HD compared with controls for both the cerebellum (P = 0.0002; Fig. 4A) and SFG (P = 0.003, Fig. 4A), with no effect of sex. Pair-wise correlation analysis demonstrated that urea concentration was also highly correlated between the two tissues (Fig. 4B). For methodological validation, urea was quantified in the HD samples using gas chromatography mass spectrometry (GCMS), with excellent concordance observed between values obtained by GCMS and the biochemical assay (r > 0.96, P < 0.0001).
Fig. 4.
Brain urea is elevated in a large postmortem HD case cohort. Shown is the urea quantification in postmortem cerebellum and SFG tissue from 22 HD patients (grades 0–3) and 14 controls. (A) Box plots depict the median and quartile range of urea concentration (nanomole per milligram of protein) as measured by biochemical assay for HD case and non-HD control groups, in cerebellum and SFG, respectively (P < 0.05; Wilcoxon two-sample t test). (B) Urea concentration was highly correlated between cerebellum and SFG (r > 0.9, P < 0.0001). Data points shown represent individuals. Circles, females; triangles, males; blue, control cases; red, HD cases.
Alongside the new tissue samples assessed, we requantified urea in cerebellum and SFG tissue from four controls and eight of the nine HD cases in which we previously reported elevated urea (ref. 21, refer to Table S5 for details) to validate and extend those findings. Addition of these sample to the larger cohort (n = 30 HD and 18 control in total) improved the statistical significance of the urea elevation in both the HD cerebellum and SFG (P < 0.0001, for both biochemical assay and GCMS methods; refer to Table S6).
We find no significant relationship of HD case urea levels with disease allele CAG repeat length, Vonsattel neuropathological grade, postmortem delay, or age at death in this study (r2 < 0.02, P > 0.05).
Urea and Its Transporter Are Elevated in HD Case Brain Tissue with Low-Level Pathology.
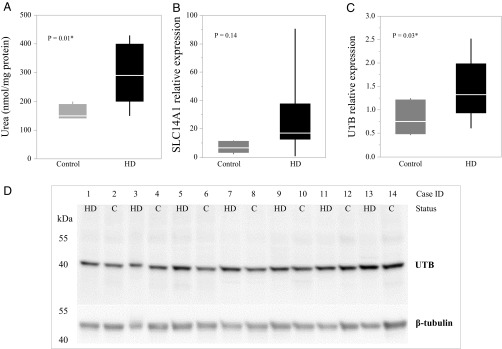
As urea is a product of protein catabolism, it was considered that the observed elevated levels in HD postmortem brain could be the consequence of cell death. To reduce the likelihood of cell death as a cause for the phenotype, urea levels were quantified specifically in brain tissue from seven postmortem HD cases with low levels of pathology and limited cell death (Vonsattel grade 0/1) and seven matched controls (Table S7). This analysis included four HD samples and three non-HD controls examined as part of the previously assessed mixed-grade cohorts (refer to Table S7 for details). Quantification by biochemical assay again demonstrated a significantly higher urea concentration in the grade 0/1 HD patient cerebellum on average compared with non-HD controls (P = 0.01; Fig. 5A). Tissue from other brain regions was not available for further assessment. We also examined the expression of the urea transporter in this sample cohort. Quantification of SLC14A1 by qRT-PCR indicated 4.2-fold higher expression on average in the grade 0/1 HD cerebellum compared with non-HD controls (Fig. 5B), although this trend was not statistically significant (P = 0.14). A polyclonal antibody raised to the urea transporter protein (UTB) encoded by SLC14A1 was used to measure human UTB in brain protein homogenates. Western blots demonstrated a highly specific single band of 42 kDa (Fig. 5D) in the cerebellar homogenates, and quantification of this band revealed 1.8-fold higher levels on average of UTB in the grade 0/1 HD cerebellum compared with controls (P = 0.03; Fig. 5C). Pairwise correlation analysis revealed no significant relationship between SLC14A1 levels and the long allele CAG repeat length of HD individuals in this cohort (r2 = 0.01 P = 0.87). These findings concur with the results from the OVT73 sheep, which show minimal neuropathology.
Fig. 5.
Urea and its transporter are elevated in low-Vonsattel grade HD human brain. Cerebellar tissue from Vonsattel pathological grade 0 or 1 HD cases (n = 7) and controls (n =7) was quantified for (A) urea (nanomole urea per milligram protein), (B) SLC14A1 transcripts (relative levels, normalized to the geometric mean of SDHA and TRIP11 reference genes), and (C) UTB protein (relative levels, normalized to beta tubulin). The Western blot underlying the data displayed in C is shown in D. The mean, quartiles, and one-way ANOVA P values are indicated. Urea and UTB protein levels were significantly higher on average in HD patient cerebellum compared with non-HD controls (*P < 0.05).
Discussion
Although the causative mutation in HD is well defined, the multifunctionality of the HTT protein and heterogeneity of symptoms present a significant challenge in defining the gain of function ultimately responsible for cell dysfunction and neurodegeneration. As cell dysfunction is initiated before onset of symptoms it is likely that an effective therapy would need to be applied early in the disease process. We must therefore gain a better understanding of the initiating pathogenic changes. Here we demonstrate that urea metabolism is disrupted in postmortem brain tissue from HD cases and the HD OVT73 sheep model, with evidence that this begins early in disease development, before overt symptoms or significant neuronal loss.
Levels of urea were elevated in a large cohort of postmortem HD brain samples compared with non-HD control samples. This finding replicates and extends a recent report by our wider laboratory of elevated urea throughout the brain in a smaller number of HD samples (21). While only two brain regions (cerebellum and SFG) were available for analysis in the present study, significantly elevated urea levels were detected in both regions, supporting the urea phenotype as a widespread phenomenon in the HD brain. Moreover, we found urea levels to be highly correlated between the brain regions in pairwise analysis. This was remarkable given that the HD tissues studied were of end-stage disease and of variable neuropathology (Vonsattel grades 0–4) and indicates that the urea phenotype may be under molecular regulation within individuals.
Urea is the final product of protein catabolism and a critical osmolyte in the mechanism for concentrating urine (reviewed in ref. 22). Excessive levels of urea and its nitrogenous precursor ammonia are neurotoxic, as evidenced by uremic encephalopathy and the urea cycle disorders (UCDs). Uremic encephalopathy typically occurs in patients with renal failure, because of urea accumulating in the blood and brain. Symptoms range from mild fatigue and generalized weakness to seizure and coma (reviewed in ref. 23). In UCDs, genetic variants in specific enzymes cause a loss of function, resulting in hyperammonemia. UCD manifestations range from lethargy and abnormal behavior such as disordered sleep and neurological posturing through to acute psychosis, weight loss, seizure, and coma (reviewed in ref. 24). Based on the symptom profile of these disorders, the accumulation of urea and/or ammonia in the brain could certainly contribute to the neuropathology and symptoms of HD. Interestingly, widespread elevation of urea has also been reported in Alzheimer’s disease brain tissue recently (25), suggesting that urea cycle disruption could also be a unifying pathogenic feature of neurodegenerative disease. Not all data are in agreement, however; Green and coworkers (26) identified 3.25-fold lower levels of urea in the postmortem HD striatum in the first 1H-NMR metabolomics study of HD tissue. The reasons for this discrepancy are not immediately clear. However, we are confident in our data because it was derived through both biochemical assay and mass spectrometry methodologies with very high concordance and employed a standard calibration curve made from an authentic urea standard.
Given that urea is the product of protein catabolism, a fundamental question is whether the urea phenotype observed in the HD brain is due to cell loss in neurodegeneration or a result of increased protein turnover as part of the underlying disease process. Regarding the latter, multiple studies report altered amino acid levels in brain tissue and blood of HD patients and animal models including the OVT73 sheep (11, 14, 20, 27). This catabolism is likely to serve as a source of energy, as HD patients appear to be in deficit based on the observation of weight loss, even in the presymptomatic phase. We also sought to address the possibility that urea may be a product of cell loss in the neurodegenerative process. We specifically analyzed postmortem tissue from individuals with low-level neuropathology or minimal cell loss (Vonsattel neuropathological grade 0/1). We found that urea levels were significantly elevated in cerebellar tissue from these cases compared with controls, implying that the urea phenotype is not likely to be a consequence of cell death. We also found no evidence of a relationship between urea levels and neuropathological grade in our large cohort of HD postmortem brain.
The current study presents evidence of a urea phenotype in HD. In agreement with the HD postmortem brain analysis, significantly elevated postmortem brain urea levels were found in the transgenic HD sheep model OVT73. The OVT73 sheep is the only large animal model that expresses full-length mutant (73 glutamines) human huntingtin (17). The OVT73 sheep are considered a prodromal model system since they display no overt symptoms of the disease or cell loss (19), despite the presence of a recognized pathologic correlate, namely huntingtin-positive aggregates (18), as well as altered metabolite levels (20), and a circadian rhythm disruption measurable by Global Positioning System tracking (19). Comparing urea changes in the postmortem brains of the prodromal OVT73 sheep model and end-stage HD individuals, our results also suggest that the urea phenotype may be progressive. While the elevation of urea in the brain from postmortem HD cases appears to be widespread (21), in the OVT73 sheep elevated urea was detected only in the striatum and the cerebellum (after adjusting for sex), not the hippocampus or motor cortex. Therefore, the urea phenotype may begin as a focal effect in the highly metabolically active striatum but progress to a generalized and widespread effect in the brain and perhaps other tissues by end-stage disease.
In addition to the striking urea phenotype, we discovered significantly increased levels of a urea transporter in postmortem brain tissue from both HD cases and OVT73 sheep. SLC14A1, which encodes the urea transporter UTB (28–30), was the most significantly differentially expressed transcript in the RNA-seq study of the OVT73 sheep striatum. SLC14A1 was subsequently found to be elevated by >twofold in two subregions of the OVT73 striatum and in the motor cortex and cerebellum. Our findings agree with an earlier report by Luthi-Carter and coworkers (31), who detected SLC14A1 up-regulation in a microarray study of the caudate nucleus from postmortem HD cases. In the present study, SLC14A1 transcript levels were unaltered in cerebellar tissue from the low-Vonsattel grade HD case cohort; cerebellum tissue lysates did, however, demonstrate significantly higher levels of UTB protein in the HD samples. Unfortunately, this antibody did not detect UTB protein in sheep brain samples, likely due to a lack of conservation in the epitope sequence. Nonetheless, taken together, this set of findings provide empirical evidence that the biology of urea transporters is altered in the HD postmortem brain, in addition to that of urea itself.
While the UTB protein is primarily expressed in erythrocytes, bladder, and kidney, it is also present in the brain, where it is expressed mainly in astrocytes (32). Urea transporter expression is known to respond directly to nitrogen and urea load (33, 34) and mediates the facilitated diffusion of urea down its concentration gradient (35, 36). The altered expression of SLC14A1 transcript/UTB protein is therefore likely to be a direct response to the elevated brain urea we observed in HD. The accumulation of urea throughout postmortem HD patient brain tissue suggests that the capacity of the up-regulated transporter may be overwhelmed in the end-stage disease. Meanwhile, the accumulation of urea only in the striatum of the prodromal HD sheep, despite SLC14A1 transcripts being higher in several regions, may indicate that elevated SLC14A1 expression is able to compensate for the urea elevation earlier in the disease.
To resolve the mechanism causing the urea phenotype, it is critical that we identify the origin of the urea that accumulates in the HD brain. One hypothesis is that excess urea is being produced in the brain, however literature is conflicting on whether a urea cycle operates in the brain. Our RNA-seq study on the OVT73 sheep striatum revealed no altered transcript expression of enzymes involved in the urea cycle to suggest altered urea cycle function in the OVT73 sheep brain. In fact, the gene expression of two enzymes (CPS1 and OTC) critical for ammonia entry to the urea cycle was absent or negligible in the sheep striatum. Levels of the key urea cycle metabolites N-acetylglutamate and ornithine are reported to be reduced in the HD brain despite urea accumulation (14), also arguing against aberrations in a urea cycle in the brain being the source of the urea.
The primary site of urea production in the body is the liver, where the urea cycle acts to dispose of toxic ammonia and amine nitrogen produced through normal metabolic processes. In the brain, a partial urea cycle appears to function primarily to dispose of two amino acids, citrulline and arginine, which are substrates of the urea cycle (37–40). Therefore, the urea could be a product of arginine metabolism. The main metabolic pathway for disposing of ammonia in the brain is the glutamate–glutamine cycle operating in astrocytes (41). Due to its instability in postmortem tissue and stored blood, ammonia was unable to be quantified in this study. We did, however, detect a significant 1.5-fold up-regulation of the transcript RHCG, which encodes an ammonia transporter (42), in the OVT73 striatum. Levels of RHCG correlated strongly with SLC14A1 in the OVT73 sheep, indicating that the expression of these genes may be coregulated or responding in concert to related stimuli, hence the possible involvement of ammonia in the urea phenotype. This RHCG–SLC14A1 correlation was not present in the control animals. As neurological disorders involving ammonia accumulation (for example, UCDs) can be well managed by a low-protein diet and therapies such as sodium phenylbutyrate (43, 44), it is imperative that we determine whether levels of ammonia are altered in HD individuals, in addition to those of urea.
As the primary site of urea production in the body is the liver, it is possible that the urea phenotype observed in the HD brain could be caused by a systemic metabolic defect. To our knowledge, urea has never been measured in peripheral tissue from HD patients. We quantified urea in several peripheral tissues from the OVT73 sheep, including the liver and blood, with no significant differences detected. However, levels of circulating urea in sheep are known to respond directly to dietary protein input, and therefore strict dietary control may be required to observe peripheral urea changes in the OVT73 sheep, remembering that they are a prodromal model. Indeed, this was highlighted recently by Morton and coworkers (45), who demonstrated a small but statistically significant increase in circulating urea in the OVT73 sheep following dietary control and hourly blood sampling over a 24-h period. The sheep utilized in that study were the same as the 4-y-old ram cohort in which circulating urea was also measured and found to be unaltered in the present study while under normal grazing conditions.
The study of OVT73 sheep blood by Morton and coworkers (45) was the first report of elevated circulating urea in HD. They also identified elevated blood citrulline and arginine, supporting systemic involvement of a urea cycle defect in HD. Increased blood citrulline has previously been described in HD cases and mouse models of HD (13, 46). Chiang et al. (13) first suggested the dysfunction of cellular processes in the liver as a pathogenic mechanism in HD, when they reported reduced activity of the urea cycle enzymes arginosuccinate-lyase, arginosuccinate synthase, and arginase in the liver of R6/2 mice, as well as hyperammonemia. As seen in UCDs, the hyperammonemia and symptom profile of these mice improved when they were treated with a low-protein diet (13). Further studies now also support HD as a systemic disease, including a breath test showing dysfunction of liver mitochondria in prodromal HD patients. Moreover, we have previously described increased levels of fatty acids in the liver of the same OVT73 sheep in which we now find elevated brain urea (20).
The corroboration of our finding of elevated urea and its transporter in the OVT73 sheep and HD human postmortem brain highlights the validity and utility of the sheep model for understanding early HD; a similar study of prodromal postmortem HD human brain tissue is clearly not feasible. To better understand urea cycle function in HD and to determine whether ammonia is the source of the urea, we now plan to measure urea cycle enzyme activity and urea and ammonia in the liver, blood, and brain of the OVT73 sheep on a controlled diet.
Further gene expression differences identified and validated in the OVT73 striatum RNA-seq data included the up-regulation of SLC5A7 encoding a sodium and chloride ion-dependent channel, which mediates choline uptake into cells for acetylcholine synthesis (47), and OXTR encoding a receptor for oxytocin and vasopressin (48). Notably, vasopressin has direct effects on the uptake of urea by cells (reviewed in ref. 49), and therefore, the altered expression of OXTR may be related to the urea phenotype. Regarding SLC5A7, striatal cholinergic interneurons are a major source of the neurotransmitter acetylcholine. In the HD brain, acetylcholine release and levels of the enzymes that synthesize it are reduced, and this cholinergic dysfunction is thought to be involved in the early symptomatology of HD (reviewed in ref. 50). The up-regulation of SLC5A7 transcript in the OVT73 sheep striatum may therefore reflect an early disruption to the cholinergic system. Together the altered expression of solute transporters (SLC14A1, RHCG, and SLC5A7) and urea is good evidence for a potential alteration in osmotic regulation in the OVT73 striatum.
The RNA-seq data also revealed increased expression of genes involved in protein binding (ETV5, CBS, ITGB4, and OXTR), cleavage (CPAMD8), and processing (SIAH3). Together these transcriptional differences suggest that protein homeostasis and interactions in the OVT73 striatum may also be altered. SIAH3 was the most significant differentially expressed gene in the RNA-seq dataset, being 1.4-fold higher in the transgenic striatum samples. SIAH3 is a ubiquitin E3 protein ligase involved in ubiquitination of proteins, tagging them for degradation by the proteasome. It is well established that ubiquitin and mutant huntingtin are colocalized in the protein aggregates that form in HD (51, 52), implying altered function of the ubiquitin–proteasome system in HD. Protein aggregates are detected in the OVT73 sheep brain (18, 53), and the up-regulation of SIAH3 in the OVT73 sheep supports this process. The up-regulation of CBS also has relevance to pathology in HD as the enzyme cystathionine-β-synthase encoded by CBS has been shown to interact directly with the huntingtin protein (54). CBS catalyzes the first step in the biosynthesis of cysteine, where cystathionine is formed from homocysteine. In a second step, the cystathionine is converted to cysteine by cystathionine-γ-lyase (CSE). Interestingly, CSE is depleted in HD patient brain tissue and mouse models of HD, and this is thought to be neurotoxic due to oxidative stress caused by the loss of cysteine (55). Indeed, N-acetylcysteine has beneficial effects on motor abnormalities and weight loss in mouse models of HD (55, 56). In contrast, we have previously reported elevated levels of cysteine in the OVT73 sheep cerebellum (20), perhaps reflecting a compensatory response of the cysteine biosynthesis pathway in this prodromal disease model.
In summary, this research provides robust evidence of widespread urea elevation in the HD postmortem brain, which occurs independently of overt neurodegeneration and disease symptoms. The cause of the urea phenotype in HD requires further investigation, including its relationship to the mutant huntingtin protein. We do not yet know what processes can produce and maintain the high concentration of urea observed in the postmortem HD brain. What we have observed are early phase processes—for example, elevated SLC14A1—potentially as a response to higher levels of urea or ammonia. One possibility is that urea is being produced via elevated protein catabolism, because of the increased rate of metabolism or energy requirement in HD. Further research should aim to discover the source of the elevated urea in HD, particularly concerning the potential involvement of ammonia and a systemic metabolic defect. Realizing this could have profound implications for our fundamental understanding of the molecular basis of HD, and its treatability, including the potential use of therapies already in use for disorders with systemic urea phenotypes.
Materials and Methods
Ovine Sample Collection.
All animals were maintained at the South Australian Research and Development Institute (SARDI) in accordance with the SARDI/PIRSA Animal Ethics Committee (approval nos. 19/02, 04/11, and 2/13). Necropsy samples were obtained from six nontransgenic controls (two ewes, four rams) and six OVT73 (three ewes, three rams) sheep aged 5 y, as previously described (20).
Ovine serum samples used in urea analysis were collected from the 5-y-old animals on May 23, 2012, a fortnight before necropsy. At the time (winter) ad libitum pasture was available to the animals. Serum samples from 4-y-old ewe (23 OVT73, 16 control) and ram (13 OVT73, 12 control) cohorts were collected on February 1, 2013 (summer) when animals were reliant on supplemental feeding as little or no pasture existed. Whole blood was collected into an EDTA tube, mixed by inversion, and centrifuged at 4 °C for 15 min at 3,000 g to remove cellular material. Serum aliquots were made into prechilled tubes and stored at −80 °C.
Human Sample Collection.
Human brain tissue samples were obtained from the Neurological Foundation of New Zealand Douglas Human Brain Bank (Centre for Brain Research, University of Auckland, Auckland, New Zealand). All procedures in this study were approved by the University of Auckland Human Participants Ethics Committee, with informed consent from all families. Fresh frozen tissue blocks were isolated postmortem and frozen as described previously (57).
For the analysis of cerebellum and SFG from a large cohort of HD cases with mixed Vonsattel neuropathology grade (1–4), tissue was dissected from frozen tissue blocks previously isolated from 30 HD cases and 18 age-matched non-HD controls. Case details are provided in Table S5. For the analysis of low-Vonsattel neuropathological grade (0–1) HD, ∼20 mg cerebellar tissue was dissected from frozen tissue blocks previously isolated from seven HD cases and seven age-matched non-HD controls. Patient details are provided in Table S7. All tissue was stored at −80 °C until analysis.
RNA-Seq and Analysis.
All RNA-seq was performed by Expression Analysis. TruSeq Stranded Total RNA Libraries were prepared after the depletion of ribosomal RNAs using a Ribo-Zero rRNA Removal Kit (Illumina), followed by sequencing using the Illumina HiSeq2000 platform (Illumina). All bioinformatic analysis was performed by AgResearch.
RNA-seq was performed on striatal RNA isolated from a cohort of 12 sheep aged 5 y (six OVT73, six controls) to facilitate differential gene expression analysis. Total RNA was isolated from ∼30-mg pieces of fresh frozen DM anterior striatum using an RNeasy mini Kit, as per the manufacturer’s instructions (Qiagen). Multiple RNA extractions were made for each animal to represent the entire DM portion of the anterior striatum, weighing 207 mg total on average. RNA quality was measured by Agilent 2100 Bioanalyzer (Agilent Technologies) and samples with an RNA Integrity (RIN) score >5.0 pooled for each animal. HiSeq2000 sequencing of the 12 prepared TruSeq Stranded Total RNA Libraries generated a total of 2.3 × 109 50 bp paired-end reads. Raw sequence data were quality-checked using FastQC (58) with standard settings. Low-quality regions and artifacts such as sequencing adaptors were removed using Flexbar (59), reducing the dataset to 2.1 × 109 paired-end reads for further analysis.
A fully annotated sheep transcriptome was not available at the outset of this research, and therefore, differential gene expression analysis of the 5-y-old sheep RNA-seq data initially required the construction of a de novo sheep transcriptome. To enable the assembly of a de novo transcriptome further, RNA-seq with longer read lengths (100 bp PE) was performed on RNA isolated from fetal/neonatal sheep liver and brain. Total RNA was extracted from fresh frozen brain and liver tissue (∼30 mg) isolated from fetal brain (63 d gestation) and neonate (1 mo old) control and OVT73 sheep using an RNeasy Lipid Tissue mini kit (Qiagen). RNA was treated with recombinant DNase (Ambion DNA-free Kit) and quality-assessed using the Agilent 2100 Bioanalyzer with an RNA 6000 Nano LabChip kit (Agilent Technologies). HiSeq2000 sequencing of the liver/brain RNA libraries generated 1.27 × 109 paired-end reads (100 bp). Raw sequence data were quality-checked using FastQC and cleaned using Flexbar as described for the 5-y-old sheep RNA-seq dataset. Cleaning reduced the liver/brain RNA-seq dataset to 3.27 × 108 read pairs, which were subsequently used for mapping and assembly. Reads were de novo assembled into transcripts using the Trinity suite (60).
During the course of this research, annotated Ensembl gene models became available (Ovis aries v3.1; sheep release 75, feb2014.archive.ensembl.org/Ovis_aries/Info/Index) and were incorporated into the analysis. Gene expression in the 5-y-old OVT73 sheep was therefore ultimately assessed by two approaches. In one approach, the cleaned read pairs (50 bp) were mapped against the Ensembl gene models using TopHat2 (61) and Cufflinks (62) resulting in 1.9 × 109 reads mapping to 101,591 genes and 161,371 isoforms. In the second approach, cleaned read pairs (50 bp) were again mapped (TopHat2/Cufflinks) to the existing Ensembl sheep gene models, with unmapped reads then mapped against the de novo sheep transcriptome. The de novo assembled transcripts that had single read counts of more than 171 (average mapping count) were then extracted. Of the extracted de novo transcripts, only those that had less than 90% identity over 90% of the length of genome-based transcripts were considered new transcripts and were appended to the genome-based transcripts. This added 42,653 de novo assembled transcripts to the 161,371 genome-based transcripts. The hybrid transcriptome approach resulted in a total of 101,547 genes and 204,024 isoforms mapped and quantified. For both mapping approaches, Cuffdiff2 (63) was subsequently used to identify differentially expressed genes and isoforms. Outputs from Cuffdiff2 are provided in Datasets S1 and S2. Those transcripts with q-values (adjusted P value) of less than 0.05 were selected for further investigation.
NanoString nCounter Transcript Quantification.
For validation purposes, 24 genes identified as differentially expressed through RNA-seq were requantified using the nCounter gene expression analysis system (NanoString Technologies). Custom CodeSet design and nCounter analysis was performed by New Zealand Genomics Ltd. Two RNA samples were analyzed for each animal: The first was from the same aliquot used for RNA-seq (DM portion of anterior striatum), and the second was obtained from an adjacent, DL portion of anterior striatum, extracted and quality-assessed using the same methodology. Each sample was assessed in duplicate, and results were averaged for statistical analysis. Transcript counts were normalized (nSolver software) against internal positive control samples and the geometric mean of four reference genes (CANX, SHMT2, TRIM27, and TRIP11). These four genes were identified as invariant in the RNA-seq dataset and so were selected to act as reference genes and quantified by nCounter analysis. CodeSet designs and a full list of gene names for the 24 target genes and 4 reference genes are provided in Table S8. The molecular function of validated genes was investigated using GO terms provided by the Gene Ontology Consortium website www.geneontology.org/.
Urea Quantification.
Biochemical quantification of urea was performed using the Urea Assay Kit (ab83362) as per the manufacturer’s recommendations (Abcam). Tissue samples were processed as described previously (21). Sample extracts were assayed in triplicate and background controls assayed in duplicate. Protein content in each sample lysate was determined by BioRad DC protein assay (BioRad), and data were presented as nanomole urea per milligram protein.
Quantification of urea by GCMS was performed as described previously (21) on ∼50 ± 5 mg of cerebellum and SFG tissue from 30 HD cases and 18 non-HD controls (Table S5). A calibration curve consisting of known concentrations of a synthetic urea analytical standard was used for quantitative estimation of urea in experimental samples, presented as nanomole urea per kilogram of fresh tissue.
qRT-PCR of SLC14A1 Transcript.
SLC14A1 transcript was quantified by RT-qPCR in anterior striatum (DM portion), cerebellum, and motor cortex from the 5-y-old sheep and in cerebellar tissue from the low neuropathological grade HD case cohort (Table S7). Sheep anterior striatum RNA was a subsample of the same aliquot used for RNA-seq analysis and the NanoString nCounter analysis. RNA from the other tissues was extracted from fresh frozen tissue (up to 100 mg per sample) using the RNeasy Lipid column kit, as per the manufacturer’s instructions (Qiagen). RNA was treated with recombinant DNase (Ambion DNA-free kit). Quantification and integrity of DNase-treated RNA were assessed by Nanodrop (NanoDrop Technologies). Synthesis of cDNA was performed using the SuperScript III First-Strand Synthesis SuperMix Kit (Life Technologies). Reactions containing 1 µL annealing buffer, 27 µM of random pentadecamer primer, and 1 µg of RNA and water to 8 µL were initially heated to 65 °C for 5 min, after which 10 µL of the reaction mix and 2 µL of the enzyme mix were added (total reaction volume 20 µL). A negative reverse transcription control reaction (RTC) was generated using 1 µg RNA pooled from the experimental samples, with the exclusion of enzyme mix. The reverse transcription reactions were then incubated at 25 °C for 10 min, 50 °C for 50 min, and heat inactivated at 85 °C for 5 min and diluted and stored at −20 °C until use.
qRT-PCR assays for SLC14A1 and appropriate reference genes were designed to publicly available gene sequences from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/gene) and using the Roche Universal ProbeLibrary Assay Design Center (https://qpcr.probefinder.com/organism.jsp). Three reference gene assays were selected for testing in sheep tissues (CANX, TRIM27, and YWHAZ) and three for human tissue (CANX, SDHA, and TRIP11). Primer and probe sequences are provided in Table S9. qRT-PCR was performed using the LightCycler Universal Probe system and the LightCycler 480 (Roche Diagnostics). Each 10 µL qRT-PCR contained 4 µL cDNA and 6 µL of master mix (0.4 µL of each 5 µM primer, 0.1 µL probe, 5 µL Roche Probes Master, and 0.1 µL water). Reactions were set up using an epMotion 5075 robot (Eppendorf). Cycling conditions on the LC480 were 95 °C for 10 min, followed by 95 °C for 10 s, 60 °C for 30 s, × 45 cycles, and 40 °C for 40 s. Samples (including RTC and no template-negative controls) were assayed on a 384-well plate in triplicate. Standard curves for each assay were generated from a cDNA dilution series (3×–2,187×) and used to normalize expression values (LightCycler 480 Software Absolute Quantification method). The expression stability of reference gene assays was assessed using GeNorm and NormFinder software (64, 65). LightCycler 480 Software Relative Quantification method was used to create an expression ratio for each sample of the target gene compared with the geometric mean of the reference gene combination considered most stable (refer to Table S9). The specificity of assays was tested by running PCR products on a 2% agarose gel. The PCR efficiency for each assay used was calculated as 1.8–2.0 (LightCycler 480 Software Absolute Quantification method).
Western Blot Analysis of UTB Protein.
For detection and quantification of UTB protein, total protein homogenates were prepared and quantified as described previously (18), from 50 to 100 mg of cerebellar tissue. Thirty micrograms of protein in 1× Laemmli buffer (Sigma-Aldrich) was loaded per lane of a 4–15% gradient TGX precast gel (Bio-Rad) and electrophoresed at 200 V for 1 h, using a Tris–Glycine SDS buffer system (Bio-Rad). Separated protein was transferred to a PVDF membrane (Millipore) at 30 mA for 1 h in a tank transfer system (Bio- Rad) in chilled Tris–Glycine buffer containing 10% methanol. Membranes were blocked in 5% nonfat milk powder in TBST (10 mM Tris, 150 mM NaCl, 0.05% vol/vol Tween-20, pH 8.0) for 1 h and subsequently incubated with the SLC14A1 polyclonal antibody ABIN310975 (Antibodies online) in 5% nonfat milk in TBST overnight at 4 °C, at a 1: 100,000 dilution. Membranes were then incubated in 1:5,000 diluted goat anti-rabbit IgG-HRP secondary antibody sc-2004 (Santa Cruz Biotechnologies) for at least 2 h. Signal was detected using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare) on an ImageQuant LAS4000 CCD imaging system (GE Healthcare). The blot was subsequently stripped by submersion in stripping buffer (10 mM 2-Mercaptoethanol, 2% SDS, 62.5 mM Tris⋅HCl, pH 6.8) for 30 min at 70 °C and reprobed with polyclonal beta tubulin antibody ab6046 (Abcam) to enable normalization of the UTB signal.
Statistical Analysis.
For all statistical analyses, replicate data points were first averaged and data normalized to appropriate reference values already described. Statistical analysis was performed using JMP Statistical Discovery software (JMP 11.2.0; SAS Institute Inc.). The normality of each set of measurements was tested using the Shapiro–Wilk W test. Measurements that failed this test (P < 0.05) were log10 transformed to approximate a normal distribution. Comparisons of means between groups were then performed using ANOVA standard least squares models. Mean ± SEM is presented for these data. Where a set of measurements did not approximate a normal distribution even after log10 transformation (Shapiro–Wilk W < 0.05), comparison between groups was performed on nontransformed data using the nonparametric Wilcoxon two-sample t test. Confidence intervals (95%) of each group are presented for nonparametric data. Differences between groups were considered statistically significant at P < 0.05. Corrections for multiple testing are reported where applied. The mean comparisons performed were between disease and control groups and between males and females. For measures where a statistically significant effect of sex was observed, this is reported in text. For these measures, the disease effect was subsequently recalculated, with sex included as a variable within the statistical model, and results reported.
To analyze the pairwise correlation between variables, Pearson’s correlation coefficient (r) for parametric data and Spearman’s correlation coefficient for nonparametric data were determined. Differences in correlation coefficients between disease and control groups were analyzed using Fisher’s r-to-z transformation. Correlations, and differences in correlations between disease and control groups, were considered statistically significant at P < 0.05. Corrections for multiple testing are reported where applied.
Supplementary Material
Acknowledgments
We thank all the families of patients with HD in New Zealand who so generously supported this research through the donation of brain tissue to the Neurological Foundation of New Zealand Douglas Human Brain Bank in the Centre for Brain Research, Faculty of Medical and Health Sciences, University of Auckland, New Zealand. We also thank the Freemasons of New Zealand for their ongoing support of this project, Marika Eszes for sourcing the required tissues, and Professor Christopher Triggs for his advice regarding statistical analysis. Special thanks to the incredible on-farm SARDI team for all animal management, in particular Ian Gollan, Kylie Chenoweth, Pene Keynes, and Nick Brown, as well as Tim Kuchel and the team at the South Australian Health and Medical Research Institute for their assistance in sample collection. This work was kindly supported by the Cure Huntington's Disease Initiative Foundation (A-8247) and Brain Research New Zealand.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711243115/-/DCSupplemental.
References
- 1.MacDonald ME, et al. The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Rubinsztein DC, et al. Phenotypic characterization of individuals with 30-40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36-39 repeats. Am J Hum Genet. 1996;59:16–22. [PMC free article] [PubMed] [Google Scholar]
- 3.Myers RH, et al. De novo expansion of a (CAG)n repeat in sporadic Huntington’s disease. Nat Genet. 1993;5:168–173. doi: 10.1038/ng1093-168. [DOI] [PubMed] [Google Scholar]
- 4.Snell RG, et al. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington’s disease. Nat Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 5.Andrew SE, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- 6.Duyao M, et al. Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nat Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- 7.Vonsattel JP, et al. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Djoussé L, et al. Weight loss in early stage of Huntington’s disease. Neurology. 2002;59:1325–1330. doi: 10.1212/01.wnl.0000031791.10922.cf. [DOI] [PubMed] [Google Scholar]
- 9.Robbins AO, Ho AK, Barker RA. Weight changes in Huntington’s disease. Eur J Neurol. 2006;13:e7. doi: 10.1111/j.1468-1331.2006.01319.x. [DOI] [PubMed] [Google Scholar]
- 10.Aziz NA, et al. EHDI Study Group Weight loss in Huntington disease increases with higher CAG repeat number. Neurology. 2008;71:1506–1513. doi: 10.1212/01.wnl.0000334276.09729.0e. [DOI] [PubMed] [Google Scholar]
- 11.Mochel F, et al. Early energy deficit in Huntington disease: Identification of a plasma biomarker traceable during disease progression. PLoS One. 2007;2:e647. doi: 10.1371/journal.pone.0000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trejo A, et al. Assessment of the nutrition status of patients with Huntington’s disease. Nutrition. 2004;20:192–196. doi: 10.1016/j.nut.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Chiang MC, et al. Dysregulation of C/EBPalpha by mutant Huntingtin causes the urea cycle deficiency in Huntington’s disease. Hum Mol Genet. 2007;16:483–498. doi: 10.1093/hmg/ddl481. [DOI] [PubMed] [Google Scholar]
- 14.Patassini S, et al. Metabolite mapping reveals severe widespread perturbation of multiple metabolic processes in Huntington’s disease human brain. Biochim Biophys Acta. 2016;1862:1650–1662. doi: 10.1016/j.bbadis.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Antonini A, et al. Striatal glucose metabolism and dopamine D2 receptor binding in asymptomatic gene carriers and patients with Huntington’s disease. Brain. 1996;119:2085–2095. doi: 10.1093/brain/119.6.2085. [DOI] [PubMed] [Google Scholar]
- 16.Seong IS, et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 17.Jacobsen JC, et al. An ovine transgenic Huntington’s disease model. Hum Mol Genet. 2010;19:1873–1882. doi: 10.1093/hmg/ddq063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reid SJ, et al. Huntington’s Disease Sheep Collaborative Research Group Further molecular characterisation of the OVT73 transgenic sheep model of Huntington’s disease identifies cortical aggregates. J Huntingtons Dis. 2013;2:279–295. doi: 10.3233/JHD-130067. [DOI] [PubMed] [Google Scholar]
- 19.Morton AJ, et al. Early and progressive circadian abnormalities in Huntington’s disease sheep are unmasked by social environment. Hum Mol Genet. 2014;23:3375–3383. doi: 10.1093/hmg/ddu047. [DOI] [PubMed] [Google Scholar]
- 20.Handley RR, et al. Metabolic disruption identified in the Huntington’s disease transgenic sheep model. Sci Rep. 2016;6:20681. doi: 10.1038/srep20681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patassini S, et al. Identification of elevated urea as a severe, ubiquitous metabolic defect in the brain of patients with Huntington’s disease. Biochem Biophys Res Commun. 2015;468:161–166. doi: 10.1016/j.bbrc.2015.10.140. [DOI] [PubMed] [Google Scholar]
- 22.Sands JM. Critical role of urea in the urine-concentrating mechanism. J Am Soc Nephrol. 2007;18:670–671. doi: 10.1681/ASN.2006121314. [DOI] [PubMed] [Google Scholar]
- 23.Mahoney CA, Arieff AI. Uremic encephalopathies: Clinical, biochemical, and experimental features. Am J Kidney Dis. 1982;2:324–336. doi: 10.1016/s0272-6386(82)80090-5. [DOI] [PubMed] [Google Scholar]
- 24.Gropman AL, Summar M, Leonard JV. Neurological implications of urea cycle disorders. J Inherit Metab Dis. 2007;30:865–879. doi: 10.1007/s10545-007-0709-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J, et al. Graded perturbations of metabolism in multiple regions of human brain in Alzheimer’s disease: Snapshot of a pervasive metabolic disorder. Biochim Biophys Acta. 2016;1862:1084–1092. doi: 10.1016/j.bbadis.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham SF, et al. Metabolic signatures of Huntington’s disease (HD): (1)H NMR analysis of the polar metabolome in post-mortem human brain. Biochim Biophys Acta. 2016;1862:1675–1684. doi: 10.1016/j.bbadis.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Underwood BR, et al. Huntington disease patients and transgenic mice have similar pro-catabolic serum metabolite profiles. Brain. 2006;129:877–886. doi: 10.1093/brain/awl027. [DOI] [PubMed] [Google Scholar]
- 28.Olives B, et al. Cloning and functional expression of a urea transporter from human bone marrow cells. J Biol Chem. 1994;269:31649–31652. [PubMed] [Google Scholar]
- 29.Timmer RT, et al. Localization of the urea transporter UT-B protein in human and rat erythrocytes and tissues. Am J Physiol Cell Physiol. 2001;281:C1318–C1325. doi: 10.1152/ajpcell.2001.281.4.C1318. [DOI] [PubMed] [Google Scholar]
- 30.Tsukaguchi H, et al. Cloning and characterization of the urea transporter UT3: Localization in rat kidney and testis. J Clin Invest. 1997;99:1506–1515. doi: 10.1172/JCI119313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodges A, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoue H, et al. Regulated expression of renal and intestinal UT-B urea transporter in response to varying urea load. Am J Physiol Renal Physiol. 2005;289:F451–F458. doi: 10.1152/ajprenal.00376.2004. [DOI] [PubMed] [Google Scholar]
- 34.Simmons NL, et al. Dietary regulation of ruminal bovine UT-B urea transporter expression and localization. J Anim Sci. 2009;87:3288–3299. doi: 10.2527/jas.2008-1710. [DOI] [PubMed] [Google Scholar]
- 35.Sands JM, Gargus JJ, Fröhlich O, Gunn RB, Kokko JP. Urinary concentrating ability in patients with Jk(a-b-) blood type who lack carrier-mediated urea transport. J Am Soc Nephrol. 1992;2:1689–1696. doi: 10.1681/ASN.V2121689. [DOI] [PubMed] [Google Scholar]
- 36.Fröhlich O, Macey RI, Edwards-Moulds J, Gargus JJ, Gunn RB. Urea transport deficiency in Jk(a-b-) erythrocytes. Am J Physiol. 1991;260:C778–C783. doi: 10.1152/ajpcell.1991.260.4.C778. [DOI] [PubMed] [Google Scholar]
- 37.Sporn MB, Dingman W, Defalco A, Davies RK. The synthesis of urea in the living rat brain. J Neurochem. 1959;5:62–67. doi: 10.1111/j.1471-4159.1959.tb13333.x. [DOI] [PubMed] [Google Scholar]
- 38.Sporn MB, Dingman W, Defalco A, Davies RK. Formation of urea from arginine in the brain of the living rat. Nature. 1959;183:1520–1521. doi: 10.1038/1831520a0. [DOI] [PubMed] [Google Scholar]
- 39.Kemp JW, Woodbury DM. Synthesis of urea-cycle intermediates from citrulline in brain. Biochim Biophys Acta. 1965;111:23–31. doi: 10.1016/0304-4165(65)90469-1. [DOI] [PubMed] [Google Scholar]
- 40.Sadasivudu B, Rao TI. Studies on functional and metabolic role of urea cycle intermediates in brain. J Neurochem. 1976;27:785–794. doi: 10.1111/j.1471-4159.1976.tb10409.x. [DOI] [PubMed] [Google Scholar]
- 41.Martinez-Hernandez A, Bell KP, Norenberg MD. Glutamine synthetase: Glial localization in brain. Science. 1977;195:1356–1358. doi: 10.1126/science.14400. [DOI] [PubMed] [Google Scholar]
- 42.Weiner ID, Verlander JW. Ammonia transport in the kidney by Rhesus glycoproteins. Am J Physiol Renal Physiol. 2014;306:F1107–F1120. doi: 10.1152/ajprenal.00013.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iannitti T, Palmieri B. Clinical and experimental applications of sodium phenylbutyrate. Drugs R D. 2011;11:227–249. doi: 10.2165/11591280-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leonard JV, Morris AAM. Urea cycle disorders. Semin Neonatol. 2002;7:27–35. doi: 10.1053/siny.2001.0085. [DOI] [PubMed] [Google Scholar]
- 45.Skene DJ, et al. Metabolic profiling of presymptomatic Huntington’s disease sheep reveals novel biomarkers. Sci Rep. 2017;7:43030. doi: 10.1038/srep43030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C-M, et al. High protein diet and Huntington’s disease. PLoS One. 2015;10:e0127654. doi: 10.1371/journal.pone.0127654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okuda T, et al. Identification and characterization of the high-affinity choline transporter. Nat Neurosci. 2000;3:120–125. doi: 10.1038/72059. [DOI] [PubMed] [Google Scholar]
- 48.Kimura T, et al. Molecular characterization of a cloned human oxytocin receptor. Eur J Endocrinol. 1994;131:385–390. doi: 10.1530/eje.0.1310385. [DOI] [PubMed] [Google Scholar]
- 49.Sands JM, Blount MA, Klein JD. Regulation of renal urea transport by vasopressin. Trans Am Clin Climatol Assoc. 2011;122:82–92. [PMC free article] [PubMed] [Google Scholar]
- 50.D’Souza GX, Waldvogel HJ. Targeting the cholinergic system to develop a novel therapy for Huntington’s disease. J Huntingtons Dis. 2016;5:333–342. doi: 10.3233/JHD-160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 52.Sieradzan KA, et al. Huntington’s disease intranuclear inclusions contain truncated, ubiquitinated huntingtin protein. Exp Neurol. 1999;156:92–99. doi: 10.1006/exnr.1998.7005. [DOI] [PubMed] [Google Scholar]
- 53.Patassini S. 2014. Discovery and validation of relevant markers of Huntington’s disease progression using a transgenic sheep model. PhD thesis (University of Auckland, Auckland)
- 54.Boutell JM, Wood JD, Harper PS, Jones AL. Huntingtin interacts with cystathionine beta-synthase. Hum Mol Genet. 1998;7:371–378. doi: 10.1093/hmg/7.3.371. [DOI] [PubMed] [Google Scholar]
- 55.Paul BD, Snyder SH. Neurodegeneration in Huntington’s disease involves loss of cystathionine γ-lyase. Cell Cycle. 2014;13:2491–2493. doi: 10.4161/15384101.2014.950538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wright DJ, et al. N-Acetylcysteine improves mitochondrial function and ameliorates behavioral deficits in the R6/1 mouse model of Huntington’s disease. Transl Psychiatry. 2015;5:e492. doi: 10.1038/tp.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waldvogel HJ, Curtis MA, Baer K, Rees MI, Faull RL. Immunohistochemical staining of post-mortem adult human brain sections. Nat Protoc. 2006;1:2719–2732. doi: 10.1038/nprot.2006.354. [DOI] [PubMed] [Google Scholar]
- 58.Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data (Babraham Bioinformatics, Cambridge, UK)
- 59.Dodt M, Roehr JT, Ahmed R, Dieterich C. FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grabherr MG, et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.