Significance
Root hairs are unicellular extensions of root epidermal cells that help plants increase water and nutrient uptake and improve soil anchorage, both of which are crucial for the globally recognized goal of yield improvement with reduced fertilizer use. Previous studies have implicated numerous genes and phytohormones in the control of root hair development. This work uncovers the molecular mechanism of ethylene (ET)-promoted root hair growth and identifies a transcriptional complex consisting of EIN3/EIL1 and RHD6/RSL1 as the key regulator of root hair initiation and elongation. As ET mediates the effects of various root hair stimuli, this work also elucidates a convergent signaling network that integrates diverse environmental cues and intrinsic signals to modulate plant organ development.
Keywords: ethylene, root hair, EIN3, RHD6, RSL4
Abstract
Root hairs are an extensive structure of root epidermal cells and are critical for nutrient acquisition, soil anchorage, and environmental interactions in sessile plants. The phytohormone ethylene (ET) promotes root hair growth and also mediates the effects of different signals that stimulate hair cell development. However, the molecular basis of ET-induced root hair growth remains poorly understood. Here, we show that ET-activated transcription factor ETHYLENE-INSENSITIVE 3 (EIN3) physically interacts with ROOT HAIR DEFECTIVE 6 (RHD6), a well-documented positive regulator of hair cells, and that the two factors directly coactivate the hair length-determining gene RHD6-LIKE 4 (RSL4) to promote root hair elongation. Transcriptome analysis further revealed the parallel roles of the regulator pairs EIN3/EIL1 (EIN3-LIKE 1) and RHD6/RSL1 (RHD6-LIKE 1). EIN3/EIL1 and RHD6/RSL1 coordinately enhance root hair initiation by selectively regulating a subset of core root hair genes. Thus, our work reveals a key transcriptional complex consisting of EIN3/EIL1 and RHD6/RSL1 in the control of root hair initiation and elongation, and provides a molecular framework for the integration of environmental signals and intrinsic regulators in modulating plant organ development.
Plants, unlike animals, are sessile organisms constantly exposed to various environmental changes. Organ morphogenesis in response to different challenges is critical for plant survival. Root hairs are unicellular extensions of root epidermal cells that help increase water and nutrient uptake, and improve soil anchorage. As the outermost plant surface in the soil, root hairs rapidly and effectively respond to environmental stimuli. Their flexible nature makes them an exceptional model system for studying the organization and coordination of gene regulatory networks modulated by endogenous and exogenous signals (1, 2).
In the angiosperm model plant Arabidopsis thaliana, the two root epidermal cell fates are determined by the relative positioning of cells above the inner layer of cortical cells. Hair (H) cells overlie the junction between two cortical cells, whereas nonhair (N) cells overlie only one cortical cell (3). Although numerous genes influence root epidermal cell morphology after cell fate determination, the core regulatory network is driven by a transcriptional cascade (2, 4). GLABRA 2 (GL2), a homeodomain-leucine zipper transcription factor, plays a vital role in N cell differentiation (5, 6). GL2 maintains N cell fate mainly through direct repression of a group of basic helix–loop–helix (bHLH) family transcription factors that positively regulate root hair initiation and elongation in H cells (7). Among them, ROOT HAIR DEFECTIVE 6 (RHD6) plays a major role in promoting H cell development (8–10). RHD6 and its closest homolog, RHD6-LIKE 1 (RSL1), form class I of the bHLH group VIII subfamily. RHD6/RSL1 positively regulate four class II members of group VIII, RSL2–5, which also positively regulate root hair growth (11). However, only RSL4 has been shown to be directly regulated by RHD6 (12). RSL4 transcripts rapidly accumulate before hair cells enter the elongation stage, with the amount of accumulated RSL4 mRNA proportional to the final root hair length (13). Recently, phytohormone application, such as auxin and cytokinin treatment, and nutrient starvation were found to stimulate root hair growth in an RSL4-dependent manner (12, 14, 15), revealing that RSL4 is an essential node that integrates multiple root hair elongation signals.
The gaseous phytohormone ethylene (ET) is a well-known stress hormone that helps plants to survive under various biotic and abiotic stresses (16, 17). Upon ET binding, ET receptors deactivate the Ser/Thr kinase CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1), leading to the hypophosphorylation and proteolytic activation of ETHYLENE INSENSITIVE 2 (EIN2) (18). Downstream of EIN2, EIN3 and its functionally redundant homolog EIN3-LIKE 1 (EIL1) are two master transcription factors that activate numerous signaling cascades and ET responses (16, 17). Activated EIN2 protects EIN3/EIL1 from proteasomal degradation, at least in part, by repressing the E3 ligases EIN3-BINDING F-BOX 1 (EBF1) and EBF2 (19, 20). Thus, EIN3/EIL1 proteins accumulate in the nuclei upon ET treatment (21, 22). One notable role of ET is to stimulate root hair growth. Exogenous application of the ET biosynthesis precursor 1-aminocyclopropane-1-carboxylic acid (ACC) or a loss-of-function mutation in CTR1 leads to much longer root hairs (23, 24). In contrast, the completely abolished ET response in the ein2 mutant leads to a short-hair phenotype and reduced sensitivity to other hair growth stimuli, such as auxin, strigolactones, and low boron (23, 25, 26).
Although the roles of ET in promoting root hair growth and mediating other hair growth signals are well established, the underlying molecular mechanism remains unknown. Our findings show that EIN3/EIL1 are necessary and sufficient for ET-induced root hair elongation. EIN3 and RHD6 interact and cooperatively promote the hair elongation factor RSL4. Moreover, the functions of EIN3/EIL1 and RHD6/RSL1 are coordinated during root hair initiation. These findings elucidate the molecular mechanism of ET action during root hair initiation and elongation, and provide insight into the coordination of environmental and developmental signals during plant organ development.
Results
EIN3/EIL1 Are Critical for ET-Promoted Root Hair Elongation.
The ein3 eil1 mutants were previously found to have reduced root hair density and compromised hair growth induction by jasmonic acid, suggesting that EIN3/EIL1 positively regulate root hair development (27). Phenotypic analysis indicated that EIN3 and EIL1 appeared to function partially redundantly in regulating root hair length (Fig. S1). To investigate the role of EIN3/EIL1 in mediating ET-induced root hair growth, we examined root hair length in young Arabidopsis wild-type Col-0 and ein3 eil1 seedlings upon ACC treatment. In Col-0, a low concentration of ACC (20 nM) stimulated significant root hair elongation, with further elongation observed under a higher concentration (100 nM). The ein3 eil1 root hairs were shorter than those of Col-0 and were completely unresponsive to ACC induction, suggesting that EIN3/EIL1 are necessary for ET-induced root hair elongation (Fig. 1 A and B). We also tested the sufficiency of EIN3 protein in promoting Arabidopsis root hair elongation. In the ein3 eil1 mutant background, root hair length significantly increased with accumulated EIN3-FLAG fusion protein induced by 5 nM and 10 nM β-estrogen application (Fig. 1 C and D). These results show that EIN3/EIL1 are essential positive regulators mediating ET-promoted root hair elongation.
Fig. 1.
EIN3/EIL1 mediate ET-induced root hair elongation. (A) Representative root hairs from 6-d-old Col-0 and ein3 eil1 on ACC media. (B) Quantification of root hair length in A. (C) Representative root hairs from 6-d-old iEIN3 ein3 eil1. (D) Quantification of root hair length in C. (Scale bars: A and C, 200 μm.) Data are means ± SD (n = 10 roots). One-way ANOVA with a post hoc Tukey honest significant difference (HSD) test (**P < 0.01) was used. NS, not significantly different.
ET-Promoted Root Hair Growth Requires RSL4/RSL2.
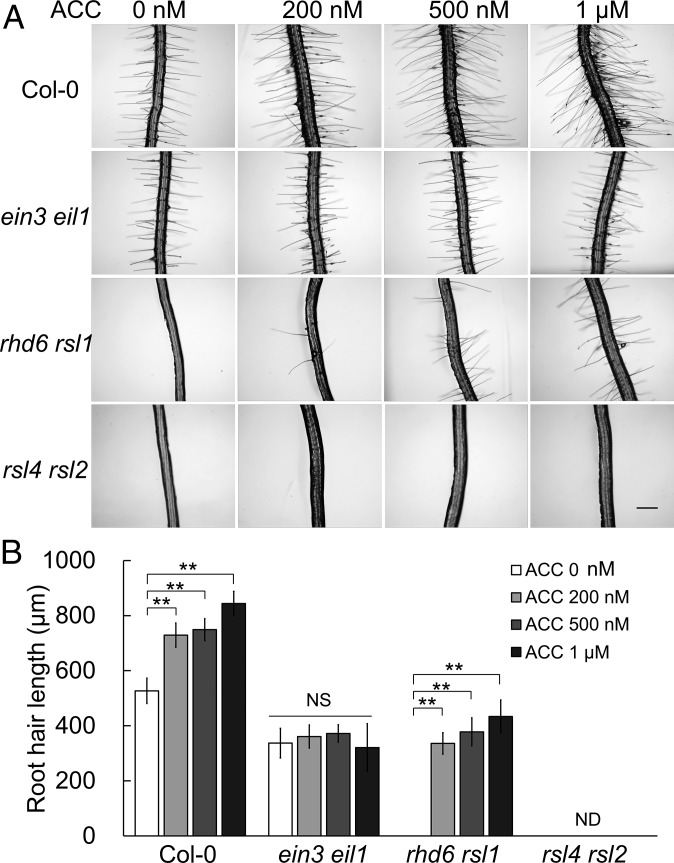
Arabidopsis root hair formation is regulated by a series of transcription factors (2, 4). We therefore assessed the interplay between EIN3/EIL1 and these transcription factors during root hair development. RHD6/RSL1 and RSL4/RSL2 positively regulate root hair differentiation downstream of GL2 (7, 12, 23). In fact, there are no visible root hairs in the rhd6 rsl1 and rsl4 rsl2 mutants (9, 12). To determine whether ET promotes hair growth through these factors, we assessed the response of these mutants to ACC treatment. In rhd6 rsl1, root hairs grew to about 400 μm in length with ACC treatment (Fig. 2), suggesting rescue of the root hair formation defect. The expression pattern of RHD6-GFP fusion protein driven by the native RHD6 promoter remained largely unchanged during ACC incubation (Fig. S2A), thereby excluding the regulation of RHD6 expression by ET. Furthermore, RHD6 overexpression under the constitutive 35S promoter in the ein3 eil1 mutant background resulted in longer hair growth, suggesting that RHD6 may function downstream of or in parallel with EIN3/EIL1 (Fig. S2 B and C).
Fig. 2.
ET-promoted root hair elongation requires RSL4/RSL2. (A) Representative root hairs from 6-d-old Col-0, ein3 eil1, rhd6 rsl1, and rsl4 rsl2 on ACC media. (Scale bar: 200 μm.) (B) Quantification of root hair length in A. Data are means ± SD (n = 10 roots). One-way ANOVA with a post hoc Tukey HSD test (**P < 0.01) was used. ND, not detectable; NS, not significantly different.
In rsl4 rsl2, the roots remained hairless even with a high ACC concentration, indicating that RSL4/RSL2 are required for ET-induced hair growth (Fig. 2). RHD6/RSL1 promotes root hair differentiation by positively regulating the RSL class II genes RSL2–5 (11). Given the requirement of RSL4/RSL2 in ET-induced root hair growth, we hypothesized that EIN3/EIL1 might also positively regulate RSL class II genes in parallel with RHD6/RSL1.
RSL4 Is a Direct Target of EIN3.
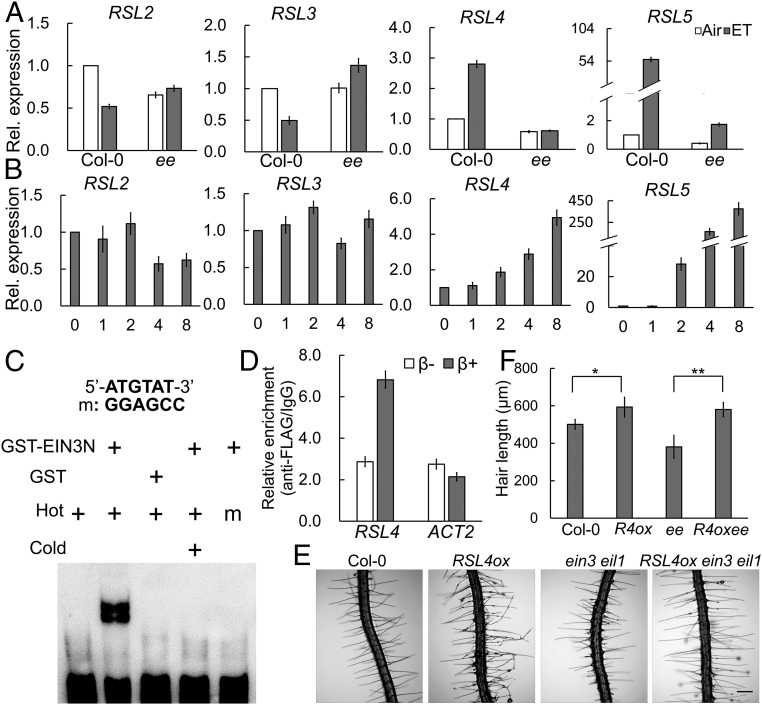
To determine whether EIN3/EIL1 regulate RSL genes, we measured RSL transcript levels in ein3 eil1 and in transgenic plants with inducible EIN3 overexpression. Exogenous ET treatment increased RSL4 and RSL5 transcript levels, but only the increase of RSL4 was completely dependent on EIN3/EIL1 (Fig. 3A). In plants with inducible EIN3 overexpression, RSL4 mRNA levels increased as the induction time increased (Fig. 3B). RSL5 levels were very low in untreated plants but dramatically increased after ET treatment and induction of EIN3 overexpression (Fig. 3 A and B). RSL4 was previously reported to positively regulate RSL5 (11); thus, the effect of ET and EIN3 induction on RSL4 transcription likely promoted RSL5 expression. In contrast, RSL2 and RSL3 transcript levels were either slightly reduced or largely unchanged upon exogenous ET application or EIN3 induction (Fig. 3 A and B).
Fig. 3.
RSL4 is a direct target of EIN3. (A) Relative (Rel.) expression levels of RSL class II genes in Col-0 and ein3 eil1 (ee) in response to ET. Six-day-old seedlings were treated with air or ET at 10 ppm for 4 h. (B) Rel. expression levels of RSL class II genes with EIN3 overexpression induction. Six-day-old iEIN3 ein3 eil1 ebf1 ebf2 (iEqm) seedlings were transferred to Murashige and Skoog medium containing 2 μM β-estrogen for the amount of time indicated on the x axis (hours). (C) In vitro EMSA showing EIN3 directly binding to the RSL4 promoter. (Upper) EIN3-binding site (ATGTAT, boldfaced) and the mutated form (GAAGCC) are shown. The GST-fused EIN3 N terminus (GST-EIN3141-352) was purified from E. coli. Unlabeled cold probe (200-fold) was added for competition. Probe containing the mutated binding site (m) was used to assess binding specificity. (D) ChIP-qPCR shows that EIN3 binds to the RSL4 promoter in vivo. The roots of 6-d-old iEqm were collected after incubation in 10 nM β-estrogen for 4 h to induce EIN3-FLAG overexpression. Fragmented chromatin was precipitated with anti-FLAG antibody. (E and F) RSL4 overexpression restored hair elongation in ein3 eil1. (E) Representative root hairs from Col-0, ein3 eil1, RSL4ox, and RSL4ox ein3 eil1. (Scale bar: 200 μm.) (F) Quantification of root hair length in E. Data are means ± SD (n = 10 roots). R4ox, RSL4ox. One-way ANOVA with a post hoc Tukey HSD test (**P < 0.01, *P < 0.05) was used.
The changes in RSL4 mRNA abundance indicated that RSL4 may be a direct target of EIN3. Analysis of the promoter region of RSL4 uncovered a putative EIN3-binding site (EBS, 5′-ATGTAT-3′, starting at -853 upstream of the RSL4 gene). In vitro electrophoretic mobility shift assays (EMSAs) confirmed the specific binding of Escherichia coli-purified EIN3 protein (DNA-binding region, 141–352 aa) to the RSL4 EBS but not to the mutated EBS (5′-GGAGCC-3′) (Fig. 3C). Accordingly, EIN3 induction of RSL4 was significantly impaired in planta when the promoter EBS motif was mutated (Fig. S3A). We also conducted an in vivo chromatin immunoprecipitation (ChIP) assay and verified the binding of EIN3-FLAG fusion protein to the RSL4 EBS-containing region in Arabidopsis roots using anti-FLAG antibody to precipitate chromatin fragments (Fig. 3D). Consistently, RSL4 was identified as an ET-responsive EIN3 target gene in a ChIP-sequencing assay using endogenous anti-EIN3 antibody (28). Furthermore, RSL4 overexpression rescued the short root hair defect of ein3 eil1 (Fig. 3 E and F). We therefore concluded that RSL4 is a direct target of EIN3 and that ET promotes root hair elongation through transcriptional activation of RSL4 by EIN3/EIL1.
EIN3 and RHD6 Exhibit Protein–Protein Interaction.
RHD6 was reported to directly activate RSL4 transcription (12), although evidence of RHD6 binding to the RSL4 promoter was lacking. We investigated whether EIN3 and RHD6 coordinately regulate RSL4 transcription. A yeast two-hybrid assay showed that EIN3 and RHD6 interacted with each other (Fig. S3B). Consistently, an in vitro pull-down assay demonstrated a direct interaction between HA-tagged EIN3 protein and His-tagged RHD6 protein (Fig. 4A). A luciferase complementation imaging (LCI) assay was also performed in tobacco leaves and Arabidopsis protoplasts, and verified the EIN3–RHD6 interaction in planta (Fig. S3 C–E). EIN3-RFP protein driven by EIN3 promoter colocalized with RHD6-GFP driven by RHD6 promoter in H cell files of Arabidopsis roots (Fig. 4B). A coimmunoprecipitation assay provided further in vivo evidence of the association between EIN3 and RHD6 (Fig. 4C).
Fig. 4.
EIN3 physically interacts with RHD6. (A) Pull-down analysis shows direct interaction between EIN3 and RHD6 in vitro. EIN3-HA and His-TF-RHD6 (TF-RHD6) were incubated for 4 h before precipitation by nickel-nitrilotriacetic acid agarose. TF, trigger factor. (B) Colocalization of EIN3 and RHD6 in H cells (white arrows). Six-day-old F1 progeny of pRHD6::GFP:RHD6 × pEIN3::EIN3:RFP were treated with ET for 2 h. (C) Coimmunoprecipitation of EIN3 and RHD6. Whole-protein extraction from root tips of pRHD6::GFP:RHD6 was immunoprecipitated by GFP-trap agarose, and endogenous anti-EIN3 antibody was used for blotting. IP, immunoprecipitation. (D) Relative (Rel.) expression level of RSL4 in ein3 eil1 rhd6 rsl1 (ee61). Seedlings were treated with air or ET for 4 h. (E) Dual-luciferase reporter assay shows the coactivation of RSL4 by EIN3 and RHD6. The indicated vectors were transformed into Arabidopsis root protoplasts. The activity ratio of firefly luciferase relative to Renilla luciferase was calculated as a metric of transcriptional activity. Error bars indicate ±SD of three biological replicates. One-way ANOVA with a post hoc Tukey HSD test (**P < 0.01, *P < 0.05) was used.
To determine whether EIN3 and RHD6 act together to regulate RSL4, the quadruple mutant ein3 eil1 rhd6 rsl1 was generated by crossing ein3 eil1 and rhd6 rsl1. Notably, due to the absence of two distinct positive regulators, RSL4 was barely expressed in ein3 eil1 rhd6 rsl1 and did not respond to ET treatment (Fig. 4D). We conducted a dual-luciferase reporter experiment (29) in Arabidopsis root cell protoplasts and found that transient expression of EIN3 together with RHD6 activated RSL4 transcription more effectively than expression of either EIN3 or RHD6 alone (Fig. 4E). Based on these results, we conclude that EIN3 and RHD6 associate with each other and coactivate RSL4 transcription.
ET Promotes Root Hair Initiation Through EIN3/EIL1 and RHD6/RSL1.
Besides promoting root hair elongation, RHD6/RSL1 are known regulators of root hair initiation (8, 9). ET also promotes root hair initiation under hairless conditions (10, 30, 31). We carefully observed the surface changes of rhd6 rsl1 and rsl4 rsl2 in response to ACC and found that ACC led to bulge formation in both mutants, a marker event of successful hair initiation (Fig. 5 A and B). When higher ACC concentrations were used to treat rhd6 rsl1, the number of root hairs and hair length both increased (Fig. 2). Nevertheless, large portions of the root epidermal regions of rhd6 rsl1 remained hairless (Fig. 5 A and B), suggesting that the effect of ET on root hair initiation partially depends on the presence of RHD6/RSL1. In rsl4 rsl2, all H positions formed bulges upon ACC treatment (Fig. 5 A and B). However, the lack of RSL4/RSL2 led to tip-growth failure of the hair bulges, and no length-measurable hair was observed (Figs. 2 and 5). We further examined hair initiation in ein3 eil1 rhd6 rsl1 and ein3 eil1 rsl4 rsl2. Neither bulge formation nor hair growth was found even in the presence of ACC, illustrating the importance of EIN3/EIL1 and RHD6/RSL1 in ET-induced root hair initiation.
Fig. 5.
ET promotes root hair initiation through EIN3/EIL1 and RHD6/RSL1. (A) Representative roots show hair bulge formation induced by 2 μM ACC in 6-d-old hairless mutants. (Scale bar: 200 μm.) (B) Root hair/bulge percentage in H cells. The percentage was calculated based on bulge and hair numbers per H cell. Two hundred cells per sample were counted. ee, ein3 eil1; ee42, ein3 eil1 rsl4 rsl2; ee61, ein3 eil1 rhd6 rsl1; 42, rsl4 rsl2; 61, rhd6 rsl1; ND, not detectable; NS, not significantly different. Error bars indicate ±SD of three biological replicates. Statistical significance was determined by a Student’s t test (***P < 0.001). (C) Summary diagram for the identification of root hair initiation genes. RNA sequencing and differential expression analysis were performed using roots of Col-0, ee, 61, and ee61 after air or ET treatment. Among the 43 H genes, 25 were identified as RSL4-regulated genes (12, 32). Nine genes, not characterized in previous publications, are closely related to the known root hair genes based on coexpression analysis (Fig. S5). These nine genes plus 18 non–RSL4-regulated H genes comprise the group of 27 hair initiation candidate genes. EPGs, ET-promoted genes.
Identification of Genes Coregulated by EIN3/EIL1 and RHD6/RSL1 in Root Hair Initiation.
For genome-wide analysis of EIN3/EIL1 and RHD6/RSL1 interaction, transcriptome profiles of Col-0, ein3 eil1, rhd6 rsl1, and ein3 eil1 rhd6 rsl1 roots were obtained by RNA sequencing. A total of 956 differentially expressed genes (DEGs) were identified in ein3 eil1 rhd6 rsl1 vs. Col-0. Biological processes involved in root hair development were statistically overrepresented among the 956 DEGs, including cell wall organization, cell tip growth, response to stimulus, and root epidermis differentiation (Fig. S4A). Moreover, the majority of the 956 genes had a greater fold change in ein3 eil1 rhd6 rsl1 vs. Col-0 than in either double mutant versus Col-0 (Fig. S4B), suggesting that gene expression coregulation by EIN3/EIL1 and RHD6/RSL1 occurs at loci throughout the genome and not only at RSL4.
Next, we identified 187 genes induced by ET in an EIN3/EIL1-dependent manner in rhd6 rsl1 (ET-promoted genes). Compared with other published genes related to root epidermis morphology, strikingly, 43 of 154 core H genes (10) were included (Fig. 5C), but none of the 54 N genes was found, strongly indicating that EIN3/EIL1 and RHD6/RSL1 positively regulate hair formation. In rhd6 rsl1, the expression level of 43 H genes induced by ET was still lower than that in Col-0 without ET. Such an expression trend was consistent with the finding that ET only partially restored the hair growth defect in rhd6 rsl1 (Figs. 2 and 5 A and B), suggesting that the coordinated activity of EIN3/EIL1 and RHD6/RSL1 is needed to fully activate root hair initiation. In light of the coactivation of the hair elongation factor RSL4 by EIN3/EIL1 and RHD6/RSL1, we found 25 of the 43 H genes to be RSL4-regulated genes (12, 32) (Fig. 5C). The remaining 18 genes not influenced by RSL4 (Fig. 5C) were considered to be potential initiation stage factors. Furthermore, coexpression analysis of the 187 genes revealed compact clustering of previously published root hair genes, including core root epidermal genes (10), RSL4-dependent genes (12, 32), and H cell-enriched genes (33) (Fig. S5). Although nine genes within the compact cluster were not found in previous studies, they may nevertheless participate in root hair development considering the similarity of their expression patterns to those of other root hair genes. These nine genes and the 18 non–RSL4-regulated H genes comprised a candidate pool of 27 putative downstream target genes of RHD6/RSL1 and EIN3/EIL1 involved in root hair initiation (Table S1). More analysis details are provided in SI Identification of Genes Coregulated by EIN3/EIL1 and RHD6/RSL1 in Root Hair Initiation.
EIN3/EIL1 and RHD6/RSL1 Mediate Diverse Root Hair Stimuli.
Transcriptome profiling revealed a more general role of EIN3/EIL1 and RHD6/RSL1 as gene expression coregulators during root hair formation. RSL4, the target gene coactivated by EIN3 and RHD6, is required for several root hair-inducing signals, including nutrient deficiency and hormone application (12, 14, 15). We therefore assessed whether EIN3/EIL1 and RHD6/RSL1 mediate the effects of these stimuli. Four root hair stimuli, application of auxin or cytokinin and depletion of phosphorus or nitrogen, were used to assess the response in rhd6 rsl1, ein3 eil1, and ein3 eil1 rhd6 rsl1. The rhd6 rsl1 was responsive to both hormone treatments but not to nutrient deficiency (Fig. 6 A–D and Fig. S7), suggesting that RHD6/RSL1 are essential for mediating the effects of nutrient depletion but not hormone application. The ein3 eil1 was responsive to all four stimuli, although root hair length in the mutant was relatively shorter than that of wild type under all conditions (Fig. 6 A–D). The ein3 eil1 rhd6 rsl1 quadruple mutant was insensitive to all four stimuli and had no visible root hairs (Fig. 6 A–D and Fig. S7), highlighting the importance of EIN3/EIL1 and RHD6/RSL1 coordination in response to these treatments.
Fig. 6.
Relative root hair length of Col-0, ein3 eil1(ee), rhd6 rsl1(61), and ein3 eil1 rhd6 rsl1(ee61) under the following stimuli: auxin [indole-3-acetic acid (IAA)] (A), cytokinin [6-benzylaminopurine (6-BA)] (B), nitrogen (N) deficiency (C), and phosphorus (P) deficiency (D). The length of Col-0 root hair was calibrated as 1.0. Data are means ± SD (n = 10 roots). The values were compared between treated and untreated samples within the same genotype. Statistical significance was determined by a Student’s t test (***P < 0.001). ND, not detectable. (E) Working model. (Top Right) In the presence of ET in Col-0, high levels of ET-induced EIN3/EIL1 protein accumulate (E3; red ovals). EIN3/EIL1 interact with RHD6/RSL1 (R6; yellow hexagons) in H cells to directly activate the transcription of RSL4, an essential positive regulator of root hair tip growth, as well as a set of genes involved in hair initiation. (Top Left) In the absence of ET, root hair initiation and elongation are mainly regulated by R6, with E3 playing a minor role. (Bottom Right) In the rhd6 rsl1 mutant, ET-induced and stabilized E3 independently promotes hair initiation and elongation to partially restore hair growth of the hairless mutant. (Bottom Left) In the absence of both ET-induced E3 and functional R6, the hairless phenotype is observed.
Discussion
Research over the past few decades has clearly established the importance of ET in regulating plant root hair development. However, the underlying molecular mechanisms were poorly understood. In this study, we found that ET promotes root hair formation at both the initiation and elongation stages. Moreover, the master regulators EIN3/EIL1 mediate the effects of ET at both stages. EIN3 directly binds the promoter region of the hair length-determining gene RSL4 and activates its transcription to promote root hair elongation. EIN3 also physically interacts with RHD6, another essential regulator of root hair development upstream of RSL4. EIN3/EIL1 and RHD6/RSL1 function in parallel and synergistically as positive regulators of root hair initiation and elongation. Based on these findings, we propose the following model for ET-induced root hair growth. In wild-type roots where EIN3/EIL1 levels are low, RHD6/RSL1 are mainly responsible for the induction of RSL4 and root hair initiation genes to maintain normal root hair growth. Upon ET treatment, EIN3/EIL1 accumulate and complex with RHD6/RSL1 to synergistically activate the expression of hair initiation genes as well as RSL4 to potently increase hair growth. In rhd6 rsl1, ET-induced EIN3/EIL1 act independently to partially rescue the hair initiation and elongation defects (Fig. 6E).
ET, a widely documented stress hormone, helps plants adapt to various environmental challenges. In the proposed model, the ET signal is integrated with the internal root hair development pathway, with EIN3/EIL1 conferring stress responsiveness to the EIN3–RHD6 transcription complex. Meanwhile, due to its strict expression pattern in H cells, RHD6 confers spatiotemporal specificity to the complex. In this way, different internal and external signals converge at key nodes through associated transcription factors that provide flexibility and adaptability to ever-changing environments. Consistent with this hypothesis, the simultaneous loss of both EIN3/EIL1 and RHD6/RSL1 led to virtual insensitivity to various root hair-inducing signals, underscoring the central role of the EIN3/EIL1–RHD6/RSL1 transcription complex in signaling integration. Notably, ET signaling is known to act upstream of auxin biosynthesis in the root tip on cell elongation and polar root hair initiation (34–38). The ein3 eil1 rhd6 rsl1 remains hairless in the presence of auxin applications (Fig. 6A and Fig. S7 A and B), revealing that ET does not promote root hair initiation and elongation simply through increasing auxin biosynthesis. Other effects on promoting hair formation by ET–auxin interplay, including transportation and signaling, still need further investigation. Similar to EIN3, ARF5 is also a direct regulator of RSL4 in mediating auxin-promoted polar growth (14). Combining findings in this study, RSL4 promoter is suggested to be a direct, central point of convergence for auxin signaling via ARF5 and for ET signaling via EIN3, and that it is regulated by the major hair cell differentiation regulator RHD6.
In addition to RSL4, EIN3/EIL1 and RHD6/RSL1 coregulate a subset of genes that likely contribute to root hair initiation and elongation. Of the 27 candidate root hair initiation genes identified in this study, some have been reported to participate in root hair formation. For example, overexpression of ROOT HAIR SPECIFIC 3 (RSH3) leads to spiral, bent, and branched hair morphologies (39). LEUCINE-RICH REPEAT/EXTENSIN 1 (LRX1) encodes a chimeric leucine-rich repeat/extensin protein, and the lrx1 mutant exhibits aberrant root hair formation, including aborted, swollen, and branched hairs (40). Lotus japonicus (Lj)RHL1-LIKE 3 (LRL3), encoding a bHLH subfamily XI protein, rescues the hairless defect of Ljrhl1 mutants (41).
As master regulators, EIN3/EIL1 associate with a group of transcriptional regulators in hormone cross-talk. All EIN3-associated proteins reported thus far repress its biological function (27, 42, 43). Intriguingly, this study uncovered a class of EIN3-associated transcription regulators that enhance its activity, although how this enhancement is achieved is unclear. Several possibilities can be considered. First, RHD6 may act as a positive regulator that enhances EIN3 transcription activity. Although RHD6 directly regulates RSL4 expression, no DNA-binding ability has been demonstrated. RHD6 may be recruited by other DNA-binding factors, such as EIN3, to the RSL4 promoter. Second, RHD6 may directly bind to a specific DNA sequence in the RSL4 promoter, while EIN3 binds to the EBS motif. In turn, the association between the two classes of transcription factors could mutually and greatly enhance their respective DNA-binding ability. Third, RHD6 may adopt a de-repression mechanism by competing with EIN3-associating repressors in H cells, and RHD6 interaction could release EIN3 from an otherwise repressed state. Further investigation of these alternative mechanisms is needed to fully understand the action and importance of the EIN3–RHD6 complex.
Materials and Methods
Plant Materials.
The A. thaliana mutant ein3 eil1 and transgenic plant iEIN3 ein3 eil1 ebf1 ebf2 and iEIN3 ein3 eil1 were described previously (43, 44). The rhd6 rsl1 and rsl4 rsl2 double mutants and transgenic pRHD6::GFP:RHD6 and RSL4ox plants were gifts from Liam Dolan, University of Oxford, Oxford, United Kingdom. Details about plant growth conditions and treatments are described in SI Materials and Methods. Plant transformation, root hair length measurement, gene expression, ChIP-qPCR, EMSA, yeast two-hybrid assay, pull-down analysis, LCI, dual-luciferase reporter assay, and sequencing analysis were carried out according to protocols described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Liam Dolan (University of Oxford) for sharing several plant materials. This work was supported by the National Natural Science Foundation of China (Grant 91017010), the Peking-Tsinghua Center for Life Sciences, and start-up funding from the Southern University of Science and Technology (to H.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE107699).
This article is a PNAS Direct Submission. J.M.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711723115/-/DCSupplemental.
References
- 1.Giehl RF, von Wirén N. Root nutrient foraging. Plant Physiol. 2014;166:509–517. doi: 10.1104/pp.114.245225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grierson C, Nielsen E, Ketelaarc T, Schiefelbein J. Root hairs. Arabidopsis Book. 2014;12:e0172. doi: 10.1199/tab.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dolan L, et al. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- 4.Schiefelbein J, Huang L, Zheng X. Regulation of epidermal cell fate in Arabidopsis roots: The importance of multiple feedback loops. Front Plant Sci. 2014;5:47. doi: 10.3389/fpls.2014.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Cristina M, et al. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10:393–402. doi: 10.1046/j.1365-313x.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- 6.Masucci JD, et al. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- 7.Lin Q, et al. GLABRA2 directly suppresses basic helix-loop-helix transcription factor genes with diverse functions in root hair development. Plant Cell. 2015;27:2894–2906. doi: 10.1105/tpc.15.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masucci JD, Schiefelbein JW. The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 1994;106:1335–1346. doi: 10.1104/pp.106.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menand B, et al. An ancient mechanism controls the development of cells with a rooting function in land plants. Science. 2007;316:1477–1480. doi: 10.1126/science.1142618. [DOI] [PubMed] [Google Scholar]
- 10.Bruex A, et al. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 2012;8:e1002446. doi: 10.1371/journal.pgen.1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pires ND, et al. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc Natl Acad Sci USA. 2013;110:9571–9576. doi: 10.1073/pnas.1305457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi K, Menand B, Bell E, Dolan L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat Genet. 2010;42:264–267. doi: 10.1038/ng.529. [DOI] [PubMed] [Google Scholar]
- 13.Datta S, Prescott H, Dolan L. Intensity of a pulse of RSL4 transcription factor synthesis determines Arabidopsis root hair cell size. Nat Plants. 2015;1:15138. doi: 10.1038/nplants.2015.138. [DOI] [PubMed] [Google Scholar]
- 14.Mangano S, et al. Molecular link between auxin and ROS-mediated polar growth. Proc Natl Acad Sci USA. 2017;114:5289–5294. doi: 10.1073/pnas.1701536114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S, et al. Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. J Exp Bot. 2016;67:6363–6372. doi: 10.1093/jxb/erw400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson PR, Ecker JR. The ethylene gas signal transduction pathway: A molecular perspective. Annu Rev Genet. 1998;32:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- 17.Ju C, Chang C. Mechanistic insights in ethylene perception and signal transduction. Plant Physiol. 2015;169:85–95. doi: 10.1104/pp.15.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ju C, et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:19486–19491. doi: 10.1073/pnas.1214848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, et al. EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell. 2015;163:670–683. doi: 10.1016/j.cell.2015.09.037. [DOI] [PubMed] [Google Scholar]
- 20.Merchante C, et al. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell. 2015;163:684–697. doi: 10.1016/j.cell.2015.09.036. [DOI] [PubMed] [Google Scholar]
- 21.Potuschak T, et al. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- 22.Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 23.Masucci JD, Schiefelbein JW. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell. 1996;8:1505–1517. doi: 10.1105/tpc.8.9.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitts RJ, Cernac A, Estelle M. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 1998;16:553–560. doi: 10.1046/j.1365-313x.1998.00321.x. [DOI] [PubMed] [Google Scholar]
- 25.Kapulnik Y, et al. Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J Exp Bot. 2011;62:2915–2924. doi: 10.1093/jxb/erq464. [DOI] [PubMed] [Google Scholar]
- 26.Martín-Rejano EM, et al. Auxin and ethylene are involved in the responses of root system architecture to low boron supply in Arabidopsis seedlings. Physiol Plant. 2011;142:170–178. doi: 10.1111/j.1399-3054.2011.01459.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Z, et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang KN, et al. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. Elife. 2013;2:e00675. doi: 10.7554/eLife.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellens RP, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao XF, Linstead P, Berger F, Kieber J, Dolan L. Differential ethylene sensitivity of epidermal cells is involved in the establishment of cell pattern in the Arabidopsis root. Physiol Plant. 1999;106:311–317. doi: 10.1034/j.1399-3054.1999.106308.x. [DOI] [PubMed] [Google Scholar]
- 31.Cho HT, Cosgrove DJ. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell. 2002;14:3237–3253. doi: 10.1105/tpc.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vijayakumar P, Datta S, Dolan L. ROOT HAIR DEFECTIVE SIX-LIKE4 (RSL4) promotes root hair elongation by transcriptionally regulating the expression of genes required for cell growth. New Phytol. 2016;212:944–953. doi: 10.1111/nph.14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 34.Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Růzicka K, et al. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell. 2007;19:2197–2212. doi: 10.1105/tpc.107.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swarup R, et al. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell. 2007;19:2186–2196. doi: 10.1105/tpc.107.052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stepanova AN, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 38.Ikeda Y, et al. Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat Cell Biol. 2009;11:731–738. doi: 10.1038/ncb1879. [DOI] [PubMed] [Google Scholar]
- 39.Won SK, et al. Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 2009;150:1459–1473. doi: 10.1104/pp.109.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baumberger N, Ringli C, Keller B. The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 2001;15:1128–1139. doi: 10.1101/gad.200201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karas B, et al. Conservation of lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol. 2009;151:1175–1185. doi: 10.1104/pp.109.143867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, et al. Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell. 2014;26:1105–1117. doi: 10.1105/tpc.113.122002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An F, et al. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012;22:915–927. doi: 10.1038/cr.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.An F, et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell. 2010;22:2384–2401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gendrel AV, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- 46.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 50.Ma C, Wang X. Application of the Gini correlation coefficient to infer regulatory relationships in transcriptome analysis. Plant Physiol. 2012;160:192–203. doi: 10.1104/pp.112.201962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.