Significance
This work provides biophysical insights into how BRAF mutant melanoma cells adapt to the stress of MAPK inhibition via a series of reversible phenotypic transitions toward drug-tolerant or drug-resistant cell states enriched for neural-crest factors and mesenchymal signatures. This adaptation is influenced by cell phenotype-specific drug selection and cell state interconversion, but not selection of genetically resistant clones. A panel of functional proteins, analyzed at the single-cell level, pointed to signaling network hubs that drive the initiation of the melanoma cell adaptive transition. Targeting those hubs halted the transition and arrested resistance development.
Keywords: single-cell analysis, cell state transition, adaptive resistance, Markov chain model, melanoma
Abstract
Continuous BRAF inhibition of BRAF mutant melanomas triggers a series of cell state changes that lead to therapy resistance and escape from immune control before establishing acquired resistance genetically. We used genome-wide transcriptomics and single-cell phenotyping to explore the response kinetics to BRAF inhibition for a panel of patient-derived BRAFV600-mutant melanoma cell lines. A subset of plastic cell lines, which followed a trajectory covering multiple known cell state transitions, provided models for more detailed biophysical investigations. Markov modeling revealed that the cell state transitions were reversible and mediated by both Lamarckian induction and nongenetic Darwinian selection of drug-tolerant states. Single-cell functional proteomics revealed activation of certain signaling networks shortly after BRAF inhibition, and before the appearance of drug-resistant phenotypes. Drug targeting those networks, in combination with BRAF inhibition, halted the adaptive transition and led to prolonged growth inhibition in multiple patient-derived cell lines.
The high rate of both response and resistance to BRAF inhibitors (BRAFi) has made BRAF mutant melanomas and derived cell lines into paradigmatic models for exploring the challenges of targeted inhibitors. Epigenetic alterations and associated cell state transitions along the melanocytic lineage toward drug-tolerant states appear to precede the emergence of clones that are genetically resistant to BRAF inhibition (1–5). Specifically, brief BRAFi exposure (1–3 d) can trigger melanocytic differentiation in certain cell lines. This is accompanied by increased MITF (a melanocytic transcription factor) and up-regulation of downstream melanosomal antigens MART-1 and gp100 (1, 2). Slightly longer exposure (2–9 d) can induce de-differentiation toward a slow-cycling neural crest-like phenotype, with a characteristic increase of Nerve Growth Factor Receptor (NGFR) and loss of MART-1 (5). Extended exposure (>2 wk) can yield an invasive mesenchymal-like state with both MART-1 and NGFR loss (3). The kinetics and molecular details of these cell state changes depend on the drug treatment duration and dose, as well as the plasticity of the cancer cells (3–5). For some patient-derived cell lines, these drug-induced cell state transitions are reversed on drug release (5, 6). Observations on patient-derived cells have been shown to correlate with what is seen in patient tumors (3–5).
Several studies have explored the biology of nongenetic BRAFi resistance in BRAF mutant melanomas (1–5), but the biophysical picture of this process is less resolved. Biophysical studies can yield predictive insights, but may lack the mechanistic detail of a biological investigation. Of particular interest here is the nature of the cell-state changes observed over the course of drug resistance development. We consider two scenarios. The first involves the enrichment of drug-resistant cancer cell genotypes, or epigenotypes, with growth advantage on drug exposure, akin to the Darwinian-type selection (7). The second, Lamarckian induction (8), is when the drug treatment itself induces cell state changes toward a more drug-tolerant state that can persist across cell generations through transcriptional reprogramming and signaling network rewiring. The second scenario is often loosely (and imprecisely) termed an adaptive response to drugging.
In principle, experimental measures of the trajectories of many single melanoma cells would discriminate between the different scenarios for drug resistance development. However, such trajectories are not feasible for the full, few-month, reversible melanocyte-to-mesenchymal transition, although subregions of this cell state space may be so mapped (5). Here, we use whole-transcriptome analysis and single-cell phenotype profiling to investigate the responses of a series of patient-derived BRAF mutant melanoma cell lines to BRAFi. Certain cell lines exhibit the full range of adaptive responses, and data from those cells are computationally modeled to investigate the transition kinetics and the nature of the BRAFi-triggered cell state changes. We further explore the signaling pathways associated with the induction of various subphenotypes via a kinetic single-cell functional proteomic study (9). These single-cell assays uncover the emergence of drug-activated signaling before the appearance of drug-tolerant phenotypes, and identify strategies for arresting the cell state transitions and prolonging cell growth inhibition. We demonstrate that our findings extend to less-plastic cell lines.
Results
The Cellular Transition Trajectories of Phenotypically Plastic Melanoma Cells in Adaptation to BRAFi.
We interrogated 18 patient-derived BRAFV600E mutant melanoma cell lines by exposing them to BRAFi (vemurafenib) for periods of 3 d and 3 wk at a concentration of 2 × IC50 for each cell line (SI Appendix, Table S1). Phenotypic changes were characterized by flow cytometry, using well-established markers for melanocytic (MART-1) and neural crest-like cells (NGFR), respectively (5, 6, 10) (SI Appendix, Figs. S1 and S2 and Table S2). Clustering of flow cytometry data classified the 18 cell lines into four clusters according to their phenotypic plasticity to BRAFi. Cluster C cells exhibited the highest plasticity on drug treatment (SI Appendix, Fig. S1A).
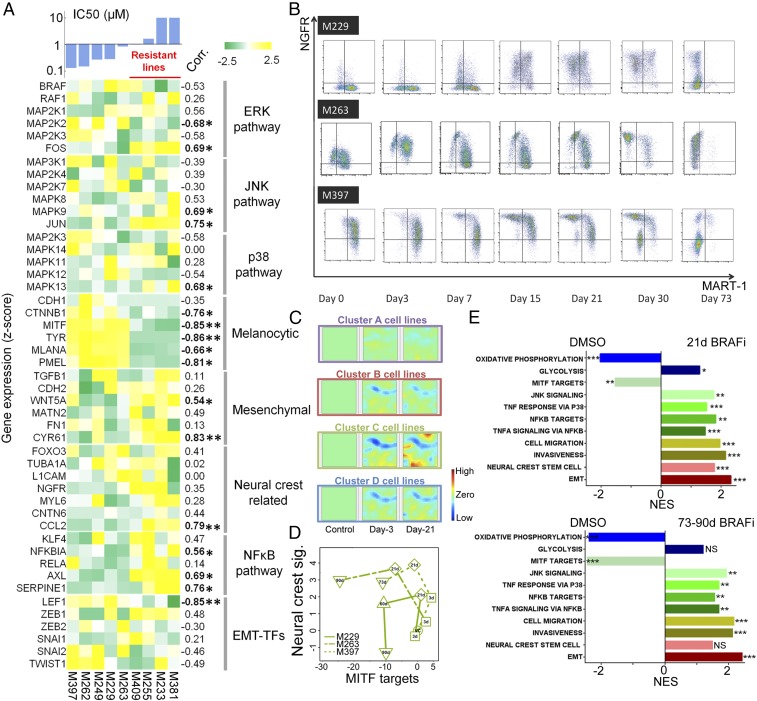
We further characterized these phenotypic changes at the whole-transcriptome level. We randomly selected nine cell lines from the four clusters for RNA-seq analysis at baseline, plus after 3 d and 3 wk of drug exposure (Fig. 1 and SI Appendix, Fig. S1B). Consistent with previous findings, the most drug-resistant cell lines (defined by IC50 > 1 μM) showed a relatively low melanocytic signature, elevated expression of neural crest and mesenchymal-related genes, and activated signaling in JNK and NFκB pathways (3, 5). AXL and c-JUN strongly correlate with the IC50 values, whereas LEF1 and MITF are anticorrelated (Fig. 1A) (3, 11, 12). A clustering analysis of genome-wide expression of the nine cell lines yielded partitions identical to those generated from two-marker flow cytometry data (SI Appendix, Fig. S1B). This confirmed NGFR and MART-1 as robust markers for characterizing the BRAFi-induced phenotype transitions. The differentially expressed genes (DEGs) on BRAFi treatment, as analyzed using Gene Expression Dynamic Inspector (13), pointed to cluster C again, which exhibited the largest transcriptional alterations on drugging (Fig. 1C). A common group of proliferation-related genes showed varying degrees of down-regulation in all clusters (SI Appendix, Fig. S3).
Fig. 1.
Phenotypic and transcriptomic characterization of a panel of patient-derived melanoma cell lines while adapting to BRAF inhibition. (A) Heat map of baseline expression levels for critical genes involved in the adaptive BRAFi resistance across a panel of melanoma cell lines, ordered by averaged IC50 to BRAFi. An IC50 of 1 µM was set to be the threshold for resistant lines. Correlation coefficients between each gene’s expression and IC50 across cell lines were evaluated with statistically significant correlations listed in bold font (*P < 0.05 and **P < 0.005). (B) Phenotypic kinetics screened by flow cytometry shows how the three plastic cell lines from cluster C experienced a phenotypic transition with a counterclockwise trajectory over a 73-d BRAF inhibition. (C) Visualization of DEGs for all of the cell line clusters relative to control by Gene Expression Dynamic Inspector. Each mosaic map represents averaged genome-wide expression profile for a specific cluster of cell lines at a time as labeled. Each pixel in the same location within the mosaic maps represents the same minicluster of genes (red, up-regulation; blue, down-regulation; green, no change). (D) Plot of signature scores of MITF and neural crest for the three plastic cell lines across different times upon BRAFi treatment. Counterclockwise trajectories appear for all three lines (VC, DMSO control). (E) Gene Set Enrichment Analysis normalized enrichment scores (NES) show significant enrichment of curated gene sets in the relevant categories associated with melanoma-adaptive cell state transition for 21 d and 73–90 d (nominal P values: *P < 0.05, **P < 0.005, ***P < 0.0005, NS: not significant).
We selected the highly plastic cluster C cell lines (M397, M229, and M263) for a time-course analysis for either a brief (3 d) or a prolonged (71–90 d) BRAF inhibition. The three lines followed similar trajectories on the flow cytometry plots (Fig. 1B), starting with transient melanocytic differentiation (increased MITF/MART-1 expression) for 3 d treatment (Fig. 1B and SI Appendix, Table S2), followed by the appearance of the slow-cycling neural crest-like (NGFRhigh/MART-1neg) state after 1–3 wk of drug treatment. Prolonged BRAF inhibition yielded a double-negative (NGFRlow/MART-1neg) state characterized by a sharp increase in cell proliferation (5). The interpretation of these trajectories (Fig. 1B) was validated at the transcriptional level by scoring MITF activity (14) and neural crest signatures (15) of each cell line, using curated gene sets (Fig. 1D and SI Appendix, Fig. S4 and Table S3). The trajectories also correlated with the development of adaptive resistance (SI Appendix, Fig. S5). Similarly, Gene Set Enrichment Analysis of the averaged gene expression of the three cell lines revealed significant enrichment in neural crest stem cell and epithelial–mesenchymal transition (EMT)-related genes with elevated invasiveness and migration signatures on continuous BRAFi exposure (Fig. 1E). Meanwhile, TNF/NF-κB-, JNK-, and p38 MAPK signaling-related genes were significantly enriched, with down-regulation of MITF signature (Fig. 1E and SI Appendix, Fig. S6). These findings were confirmed by inspecting the expression levels (SI Appendix, Fig. S7A) and mining the gene ontology terms of these DEGs (SI Appendix, Fig. S7B). The enrichment of neural crest-related genes was insignificant after prolonged drug exposure, whereas the enrichment of EMT-associated genes held steady (Fig. 1E). Thus, the double-negative state carried mesenchymal signatures and exhibited the loss of neural crest state signatures. Analysis of other clusters showed similar enrichment in neural crest and mesenchymal signatures except for cluster A, which contained innately resistant cells with MART-1neg/NGFRneg baseline expression (SI Appendix, Fig. S8). Similar enrichment patterns of the transcriptional programs were also found in published transcriptomic datasets involving 39 patient samples from pretreatment, on-treatment, and/or emergence of acquired resistance to RAF/MEK inhibitors (16). This suggests similar adaptive resistance in these BRAFi-treated patient tumors (SI Appendix, Fig. S9). Moreover, when cells were treated with BRAFi for 3 wk or longer, we identified significant enrichment in most innate anti-PD-1 resistance signatures (17) (SI Appendix, Table S4), pointing to a potential rise of resistance to PD-1 blockade for cells adapted to the run-in BRAFi therapy. The BRAFi-induced cell state changes in cluster C cells reflected a summation of the various types of adaptive transitions reported for melanomas in response to BRAFi (1–6). Thus, cluster C cell lines were carried forward for a detailed biophysical investigation of BRAFi-induced cell state transitions.
Adaptive Transition Proceeds Through Drug-Mediated Cell State Interconversion and Phenotypic Selection.
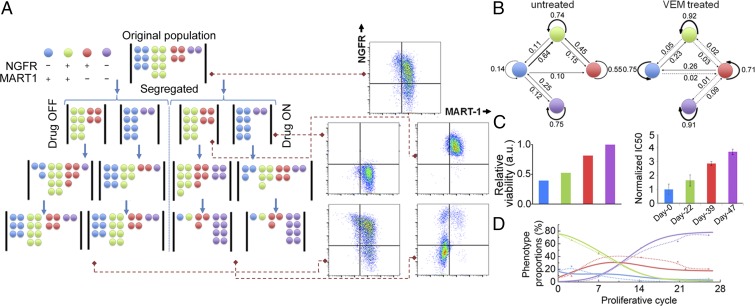
Three factors can account for the drug-induced cell state transition kinetics. The first is a deterministic factor that arises from cell state changes determined by the underlying causative gene regulatory networks. The second factor arises from the stochastic nature of gene expression. Stochastic cell state changes may be considered as a random walk diffusion process across the epigenetic landscape (18). The third factor accounts for phenotype-specific cell proliferation and survival on drug exposure. To account for all three factors, we modified a published Markov kinetic framework (19) to model the BRAFi-induced cell state transitions (Fig. 2). Our model assumed that the cell population is composed of four discrete cell states (Fig. 2A) and contains a nonsymmetric cell state interconversion matrix to account for both deterministic (Lamarckian induction) and stochastic cell state transitions. For drug-treated cells, a drug susceptibility matrix is introduced to quantify the third factor (SI Appendix, SI Materials and Methods). Experimental inputs into the model are short-time measures of the cell state composition dynamics. Computational outputs are state-to-state transition probabilities (Fig. 2B) and, for drug-treated samples, state-dependent drug susceptibility (Fig. 2C).
Fig. 2.
Markov model of cell state transition predicts phenotypic evolution of melanoma cells on BRAF inhibition. (A) Schematic illustration of phenotypic segregation and treatment conditions in the Markov model. The cells were sorted to NGFR+ and NGFR− subpopulations and treated with or without vemurafenib. (B) Cell state transition probabilities of M397 at untreated and vemurafenib-treated conditions. (C) Relative viability of different phenotypes for M397 inferred by the model (Left), and measured IC50 values (normalized to control) at different times (Right) across the transition at which one phenotype is enriched, as indicated by its respective color code (error bars: ±SD). (D) Model prediction of the phenotypic kinetics (solid lines) versus experimental data (dots connected with dashed lines) for M397 with continuous exposure to vemurafenib.
We sorted NGFRpos and NGFRneg subpopulations of two cluster C cell lines (M397 and M229) at baseline, and seeded them separately in Petri dishes for in vitro expansion (Fig. 2A and SI Appendix, Figs. S10–S12), with and without BRAFi, thus providing four initial conditions for each cell line. For untreated samples, flow cytometry analysis of the phenotype compositions during the first three cycles of expansion provided input into the Markov model. The fitted Markov model parameters (Fig. 2B) were then used to predict longer-term phenotypic evolution. Without BRAFi, the NGFRpos and NGFRneg subpopulations of both cell lines were predicted and experimentally shown to return to their original unsorted phenotypic composition after several cell cycles (5, 18–20) (SI Appendix, Fig. S10B).
BRAFi clearly altered the cell state interconversion probabilities (Fig. 2B and SI Appendix, Fig. S13). The Markov model fits from phenotype dynamics of sorted populations (Fig. 2B) under BRAFi were used to predict the drug susceptibility of each cell state. The neural crest-like and double-negative states were predicted, and experimentally shown, to be more resistant to drug treatment compared with melanocytic and double-positive states (Fig. 2C). We further used the interconversion and drug susceptibility matrices inferred from the cell state kinetics of segregated populations to predict the phenotypic evolution of unsorted cells on prolonged BRAFi exposure. The model predictions were in good agreement with the experiments (Fig. 2D and SI Appendix, Fig. S13C and Tables S5 and S6). Furthermore, the Markov model suggests that, on drug removal, the cells should return to the original phenotypic compositions characteristic of drug-naive cells. In fact, such a reversible transition was observed in both the cluster C cells and the less plastic cluster B cells (SI Appendix, Fig. S14A), and the reverted cells also exhibited a similar IC50 to BRAFi as the untreated cells (SI Appendix, Fig. S14B).
The Markov model predictions provide evidence for Lamarckian induction, as they show that BRAFi influences the specific rates of cell state interconversion. The predictions also provide evidence of phenotype-dependent Darwinian selection, as cell states with the lowest drug susceptibility eventually win out. If only the drug susceptibility matrix is included in the Markov model, the resultant prediction is in poor agreement with the experiments (SI Appendix, Fig. S13D). Therefore, the combined actions of both cell state interconversion and drug selection contribute to the observed phenotypic kinetics.
Single-Cell Functional Proteomics Reveals the Emergence of Drug-Activated Signaling at the Initiation of the Adaptive Transition.
We used the microfluidics-based single-cell barcode chip (SCBC) technology to carry out single-cell analysis of the M397 cells so as to interrogate those signaling pathways directly targeted by BRAFi (9). Guided by the transcriptional analysis, we designed a protein panel that covers critical phenotypic markers, transcription factors, and signaling effectors involved in the adaptive transition (SI Appendix, Tables S7 and S8). Each SCBC has 320 microchambers (1.5-nanoliter volume) engineered for cell lysis, and each is equipped with an antibody array for protein capture. After cell lysis, specific proteins are captured, and each array element is developed as a sandwich immunofluorescent assay (SI Appendix, Fig. S15).
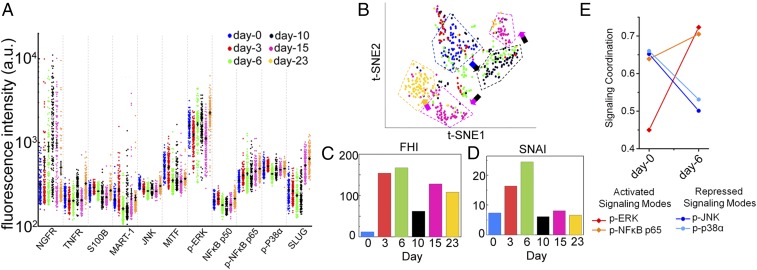
Scatter plots of single-cell data are shown in Fig. 3A, with averaged protein abundance indicated by the black horizontal lines. The time-series data are projected onto a 2D plane, using the t-distributed stochastic neighbor embedding (t-SNE) algorithm (21) (Fig. 3B). SCBC assays from different times cluster into different groups. The dispersion within a group and the distance between groups provides functional measures of how different the single cells are. Such dispersion can be quantified by the functional heterogeneity index defined in our previous study (9). Notably, BRAFi treatment increases the cellular heterogeneity in days 3 and 6, indicating an elevated plasticity (Fig. 3C). This is reflected in the sharply increased dispersion of day 6 cells in the t-SNE plane. This increase in heterogeneity between days 3 and 6 is reminiscent of the attractor destabilization and bifurcation before a cell state transition in other systems (22, 23).
Fig. 3.
Single-cell proteomic profiling of the M397 cell line during the course of adaptive cell state transition. (A) Background subtracted SCBC data represented as 1D scatter plot (mean ± SEM was overlaid for each protein by the black horizontal bar). Statistical uniqueness is evaluated by Kruskal-Wallis test comparison among multiple points. All 13 markers are statistically significant with P < 0.0005. (B) Application of t-SNE algorithm to the single-cell data separates the cells into spatially distinct clusters based on their proteomic profiles. Each point in the t-SNE plane represents a single-cell measurement, and its color is coded by time. (C) Quantification of the functional heterogeneity (FHI) of M397 cells across different times, along with the transition. (D) The SNAI across different times, along with the transition, extracted from single-cell proteomic measurements of M397 cells. (E) Change in signaling coordination quantified as correlations between key functional proteins and the PC1 for control and day 6, in which a bifurcation of signaling proteins is identified.
We used the SCBC data to assess protein–protein correlations at each point (SI Appendix, Fig. S16). The emergence of strong negative correlations between NGFR and MITF/MART-1 at day 3 points to the initiation of the cell state transition toward the neural crest-like phenotype with elevated NGFR and loss of MITF/MART-1. The overall activity of the network may be quantified by the signaling network activity index (SNAI) that accounts for both the numbers and the strengths of statistically significant correlations (SI Appendix, SI Materials and Methods). The SNAI is highest at day 6 (Fig. 3D), with particularly strong signaling participation of p-ERK and p-NFκB p65 (SI Appendix, Fig. S16), suggesting a potential gain of function through MEK/ERK and NFκB p65 signaling that might lead to BRAFi drug tolerance by promoting the adaptive transition. The SNAI drops sharply between day 6 and day 10, but then recovers to an activity similar to that seen for untreated cells (Fig. 3D), albeit with altered active network components (SI Appendix, Fig. S16). This general behavior is reminiscent of cell state transitions seen in other model systems (23) and indicates a possible cell state switch between days 6 and 10.
To quantify the influence of the functional proteins on the overall signaling coordination at the initiation of the transition, we performed Principal Component Analysis, using an analytical approach previously reported (9). We calculated the correlations between the assayed proteins and first principal component (PC1) for untreated and 6 d BRAFi treated samples. PC1 captures the most essential feature of the signaling network, and thus identifies those proteins that participate most strongly in the signaling coordination. For the untreated sample, PC1 is populated by p-ERK, p-NFκB p65, p-JNK, and p-P38α (Fig. 3E). At day 6, these four proteins bifurcate into two groups, with the influence of p-JNK and p-P38α (blue group) signaling repressed, and MEK/ERK and NFκB p65 signaling (red group) elevated (Fig. 3E). This behavior suggests that combining BRAFi with MEK and NFκB p65 inhibition might arrest the adaptive cell state transition toward drug-resistant phenotypes.
Combined MEK/ERK and NFκB p65 Inhibition with BRAFi Arrests the Adaptive Transition and Induced a Sustained Growth Inhibition.
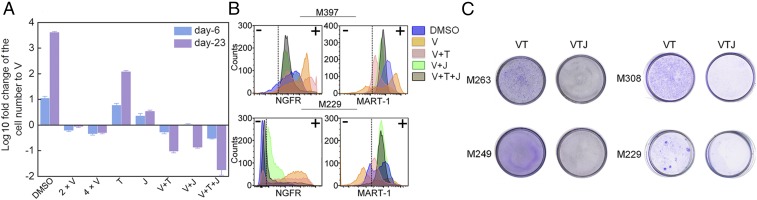
The single-cell analysis suggests that inhibiting the NFκB p65 and MEK/ERK signaling axes might keep the cells in the BRAFi sensitive state. To test this hypothesis, we used trametinib (T, MEK inhibitor) and JSH-23 (J, NFκB p65 translocation inhibitor) (24) in combination with vemurafenib (V) to treat the M397 cells in vitro and compared the results against monotherapies (V, T, and J) or dual combinations (V+T and V+J) over the course of 23 d. Consistent with our prediction, all combination therapies induced considerable cell growth inhibition (Fig. 4A). The monotherapies T and J were much less potent compared with V, but both dual combinations and the triple combination significantly outperformed the monotherapies. Consistent with previous finding (5), although V+T yielded enhanced therapeutic effect relative to V, it did not halt the phenotypic transition toward the neural crest-like drug-tolerant state (Fig. 4B and SI Appendix, Fig. S17), and therapy resistance emerged after prolonged treatment (Fig. 4C and SI Appendix, Fig. S19A). However, both V+J and V+T+J successfully arrested the transition and kept the cells in the drug-sensitive state (MART-1pos; Fig. 4B), indicating NFκB p65 nuclear translocation is necessary for the adaptive transition toward the drug-tolerant phenotypes. Drug dose–response assay under the Bliss independence assumption further confirmed that the sustained growth inhibition of the triple combination was not simply a result of drug synergy, but a result of the arrest of cell state transition (SI Appendix, Fig. S18). We further performed clonogenic assays to assess whether the combination can induce sustained growth inhibition on four other BRAFV600E mutated melanoma cell lines. The results indicated that the V+T+J combination outperforms V+T over prolonged periods for all cell lines tested, whereas treatment with J alone showed no significant cytotoxicity (Fig. 4C and SI Appendix, Fig. S19).
Fig. 4.
In vitro validation of the monotherapy and combination therapies predicted by SCBC analysis. (A) In vitro cell proliferation assay of M397 for the monotherapy and combination therapies based on the predictions from the SCBC data analysis. At each time, cell number of each test condition is normalized to the number of vemurafenib monotherapy and plotted as log 10 fold change (error bars: ±SD). (B) Flow cytometry analysis of MART-1 and NGFR levels at single-cell resolution for monotherapy and combination therapies on both M397 (23 d treatment) and M229 (28 d treatment). (C) Clonogenic assays of long-term drug treated samples confirm V+T+J induced a sustained growth inhibition (see also Fig. S19A).
Discussion
Adaptive resistance resulting from transcriptomic reprogramming permits BRAF mutant melanoma cells to survive BRAF inhibition. BRAFi exposure triggers a transient melanocytic differentiation program, followed by multiple de-differentiation programs that terminate in a drug-resistant mesenchymal-like state. Cells with distinct innate drug sensitivities may follow all or part of this transition trajectory on BRAFi exposure (SI Appendix, Fig. S2). The full transition trajectory was observed within a highly plastic set of patient-derived cells (SI Appendix, Fig. S1A, cluster C), accompanied by an elevated innate anti-PD-1 resistance gene signature associated with anti-PD-1 therapy-resistant melanomas (17). These results challenge the notion of protocols with a run-in period of targeted BRAFi therapy followed by immune checkpoint therapy, as the adaptive response to BRAFi may also create a less friendly environment to immunotherapies in certain BRAFi-sensitive tumors (25).
The kinetics of the cell state transitions that occur from the drug-naive, melanocytic state to the drug-resistant mesenchymal state was well-captured by a Markov model. This analysis revealed the critical role of drug induction, relative to the selection of drug-tolerant phenotypes, in the adaptive response to BRAFi. Drug induction can enable the de novo generation of new cell states, whereas selection accelerates the enrichment of the drug-tolerant states. The cell state interconversion probabilities over long periods of drug treatment were inferred from snapshots of flow cytometry data. However, the Markov model does have limitations. For example, the phenotype-dependent drug susceptibility determined from the Markov model is incomplete, as it groups the net effect of cell proliferation and cell death of each phenotype into a single diagonal term in the matrix. The relative contribution of each factor is not distinguished (20, 26). Similarly, on drug removal, the relative contribution of the cell state reversion versus regrowth of specific cellular phenotypes is also not resolved. In addition, the Markov model oversimplifies the continuous cell state transitions by categorizing cells into discrete phenotypes on the basis of their marker expression levels. Continuous kinetic models (18), coupled with experimental measures of single-cell trajectories (26), may prove attractive for investigating these transitions at finer resolution.
Transcriptional analysis revealed several critical signaling pathways associated with cellular adaptation to BRAFi. This, in turn, informed the design of a second kinetic investigation of the transition via single-cell functional proteomics. That study unveiled the BRAFi-induced activation of MEK/ERK and NFκB p65 signaling before the emergence of the drug-resistant phenotype. NFκB can synergize with c-Jun in the transcriptional response to TNF-α (12). Further, the c-Jun/JNK signaling axis has been reported to play a critical role in melanoma-adaptive resistance to BRAFi (3, 4), and was also found here to be associated with the emergence of adaptive resistance when the cells were transitioning to the neural crest or mesenchymal-like states (SI Appendix, Fig. S20). However, TNF/NFκB signaling was activated at the beginning of the adaptive transition (before the c-Jun/JNK activation), implying that TNF/NFκB signaling might enable the neural crest transition program. This is further supported by the report that neural crest de-differentiation can be triggered by proinflammatory cytokine TNF-α secreted from tumor-infiltrating lymphocytes (10). It echoes the observation that the MITFlow/NFκBhigh transcriptional state of melanomas is resistant to inhibition of BRAF and MEK, singly or in combination (11). NFκB inhibitors are normally used as adjuvant to chemotherapy or targeted therapies in clinical trials (27). However, because of the host toxicity, identifying more clinically actionable targets downstream of the NFκB pathway may be an appealing option (28) for arresting BRAFi adaptive resistance. As similar enrichment patterns were recurrent in more than half the on-treatment patient biopsies (SI Appendix, Fig. S9), we anticipate that combination therapy with inhibitors of this pathway may improve the durability of BRAFi therapy.
Materials and Methods
Human melanoma cell lines (M series) were established from patient’s biopsies under University of California, Los Angeles, institutional review board approval #11–003254. Please refer to SI Appendix, SI Materials and Methods for cell lines and reagents used, experimental protocols, and statistical analysis.
Supplementary Material
Acknowledgments
We acknowledge the following agencies and foundations for support: NIH Grants U54 CA199090 (to J.R.H., W.W., and A.R.), P01 CA168585 (to T.G.G. and A.R.), and R35 CA197633 (to A.R.); the Dr. Robert Vigen Memorial Fund, the Garcia-Corsini Family Fund, the Ressler Family Fund, and the Grimaldi Family Fund (A.R.); the Jean Perkins Foundation (J.R.H.); University of California, Los Angeles, Broad Stem Cell Research Center Seed Fund for Small Cell Cancer Pilot Studies, and the Phelps Family Foundation (W.W.); and ACS Research Scholar Award (RSG-12-257-01-TBE), MRA Established Investigator Award (20120279), and University of California, Los Angeles, Clinical and Translational Science Institute Grant UL1TR000124 (to T.G.G.). We acknowledge University of California, Los Angeles, Jonsson Comprehensive Cancer Center (JCCC) membership (NIH/NCI P30CA016042) for using the JCCC Flow Cytometry Core and Genomics Shared Resource. L.R. was supported by the V Foundation-Gil Nickel Family Endowed Fellowship and a scholarship from SEOM. J.T. was supported by NIH T32-CA009120.
Footnotes
Conflict of interest statement: J.R.H. and A.R. are affiliated with Isoplexis, which is seeking to commercialize the single-cell barcode chip technology.
This article is a PNAS Direct Submission.
Data deposition: The RNA-seq data reported in this paper have been deposited in the ArrayExpress database (accession no. E-MTAB-5493).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712064115/-/DCSupplemental.
References
- 1.Boni A, et al. Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res. 2010;70:5213–5219. doi: 10.1158/0008-5472.CAN-10-0118. [DOI] [PubMed] [Google Scholar]
- 2.Frederick DT, et al. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin Cancer Res. 2013;19:1225–1231. doi: 10.1158/1078-0432.CCR-12-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsdale R, et al. The transcription cofactor c-JUN mediates phenotype switching and BRAF inhibitor resistance in melanoma. Sci Signal. 2015;8:ra82. doi: 10.1126/scisignal.aab1111. [DOI] [PubMed] [Google Scholar]
- 4.Titz B, et al. JUN dependency in distinct early and late BRAF inhibition adaptation states of melanoma. Cell Discov. 2016;2:16028. doi: 10.1038/celldisc.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fallahi-Sichani M, et al. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol Syst Biol. 2017;13:905. doi: 10.15252/msb.20166796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 7.Huang S. Tumor progression: Chance and necessity in Darwinian and Lamarckian somatic (mutationless) evolution. Prog Biophys Mol Biol. 2012;110:69–86. doi: 10.1016/j.pbiomolbio.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Pisco AO, Huang S. Non-genetic cancer cell plasticity and therapy-induced stemness in tumour relapse: ‘What does not kill me strengthens me’. Br J Cancer. 2015;112:1725–1732. doi: 10.1038/bjc.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei W, et al. Single-cell phosphoproteomics resolves adaptive signaling dynamics and informs targeted combination therapy in glioblastoma. Cancer Cell. 2016;29:563–573. doi: 10.1016/j.ccell.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landsberg J, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490:412–416. doi: 10.1038/nature11538. [DOI] [PubMed] [Google Scholar]
- 11.Konieczkowski DJ, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4:816–827. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riesenberg S, et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat Commun. 2015;6:8755. doi: 10.1038/ncomms9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichler GS, Huang S, Ingber DE. Gene expression dynamics inspector (GEDI): For integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- 14.Hoek KS, et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 15.Bell GW, Yatskievych TA, Antin PB. GEISHA, a whole-mount in situ hybridization gene expression screen in chicken embryos. Dev Dyn. 2004;229:677–687. doi: 10.1002/dvdy.10503. [DOI] [PubMed] [Google Scholar]
- 16.Kwong LN, et al. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. J Clin Invest. 2015;125:1459–1470. doi: 10.1172/JCI78954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugo W, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sisan DR, Halter M, Hubbard JB, Plant AL. Predicting rates of cell state change caused by stochastic fluctuations using a data-driven landscape model. Proc Natl Acad Sci USA. 2012;109:19262–19267. doi: 10.1073/pnas.1207544109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 20.Zhou JX, Pisco AO, Qian H, Huang S. Nonequilibrium population dynamics of phenotype conversion of cancer cells. PLoS One. 2014;9:e110714. doi: 10.1371/journal.pone.0110714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amir el-AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mojtahedi M, et al. Cell fate decision as high-dimensional critical state transition. PLoS Biol. 2016;14:e2000640. doi: 10.1371/journal.pbio.2000640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bargaje R, et al. Cell population structure prior to bifurcation predicts efficiency of directed differentiation in human induced pluripotent cells. Proc Natl Acad Sci USA. 2017;114:2271–2276. doi: 10.1073/pnas.1621412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin HM, et al. Inhibitory action of novel aromatic diamine compound on lipopolysaccharide-induced nuclear translocation of NF-kappaB without affecting IkappaB degradation. FEBS Lett. 2004;571:50–54. doi: 10.1016/j.febslet.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 25.Cooper ZA, Frederick DT, Ahmed Z, Wargo JA. Combining checkpoint inhibitors and BRAF-targeted agents against metastatic melanoma. Oncoimmunology. 2013;2:e24320. doi: 10.4161/onci.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pisco AO, et al. Non-Darwinian dynamics in therapy-induced cancer drug resistance. Nat Commun. 2013;4:2467. doi: 10.1038/ncomms3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Y, Bai L, Chen W, Xu S. The NF-kappaB activation pathways, emerging molecular targets for cancer prevention and therapy. Expert Opin Ther Targets. 2010;14:45–55. doi: 10.1517/14728220903431069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tornatore L, et al. Cancer-selective targeting of the NF-κB survival pathway with GADD45β/MKK7 inhibitors. Cancer Cell. 2014;26:495–508. doi: 10.1016/j.ccr.2014.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.