Significance
Current cancer therapies fail to repress tumor recurrence and metastasis in triple-negative breast cancer (TNBC) because they fail to target cells that possess epithelial–mesenchymal (E-M) plasticity and acquire cancer stem cell (CSC) properties. Identifying and engaging signaling pathways that regulate E-M/CSC plasticity within TNBC therefore remains an unmet critical clinical need. Recent evidence demonstrates that presence of E-M/CSC plasticity in TNBC correlates with a repressed interferon/STAT gene signature. Our data demonstrate that exogenous IFN-β targets and represses E-M/CSC plasticity by reengaging type I IFN signaling in CSC. Our findings have clinical relevance, as IFN-β signaling correlates with improved patient survival and repressed CSC in TNBC. Thus, our work suggests a therapeutic use for IFN-β in the repression of E-M/CSC–driven tumor recurrence and metastasis in TNBC.
Keywords: triple-negative breast cancer, cancer stem cells, interferon-beta, tumor microenvironment
Abstract
Triple-negative breast cancer (TNBC), the deadliest form of this disease, lacks a targeted therapy. TNBC tumors that fail to respond to chemotherapy are characterized by a repressed IFN/signal transducer and activator of transcription (IFN/STAT) gene signature and are often enriched for cancer stem cells (CSCs). We have found that human mammary epithelial cells that undergo an epithelial-to-mesenchymal transition (EMT) following transformation acquire CSC properties. These mesenchymal/CSCs have a significantly repressed IFN/STAT gene expression signature and an enhanced ability to migrate and form tumor spheres. Treatment with IFN-beta (IFN-β) led to a less aggressive epithelial/non–CSC-like state, with repressed expression of mesenchymal proteins (VIMENTIN, SLUG), reduced migration and tumor sphere formation, and reexpression of CD24 (a surface marker for non-CSCs), concomitant with an epithelium-like morphology. The CSC-like properties were correlated with high levels of unphosphorylated IFN-stimulated gene factor 3 (U-ISGF3), which was previously linked to resistance to DNA damage. Inhibiting the expression of IRF9 (the DNA-binding component of U-ISGF3) reduced the migration of mesenchymal/CSCs. Here we report a positive translational role for IFN-β, as gene expression profiling of patient-derived TNBC tumors demonstrates that an IFN-β metagene signature correlates with improved patient survival, an immune response linked with tumor-infiltrating lymphocytes (TILs), and a repressed CSC metagene signature. Taken together, our findings indicate that repressed IFN signaling in TNBCs with CSC-like properties is due to high levels of U-ISGF3 and that treatment with IFN-β reduces CSC properties, suggesting a therapeutic strategy to treat drug-resistant, highly aggressive TNBC tumors.
Triple-negative breast cancer (TNBC), defined by its lack of estrogen receptor and progesterone receptor expression and HER2 amplification, remains the most lethal subtype of breast cancer (1–4). Metastasis and tumor recurrence are responsible for the majority of deaths in TNBC (1, 5, 6). Highly metastatic TNBC tumors are often composed of cells harboring epithelial–mesenchymal (E-M) plasticity, whereby cancer cells reversibly express epithelial or mesenchymal proteins (7–9). Importantly, our laboratory, along with others, has demonstrated that E-M plasticity can lead to the emergence of highly migratory, mesenchymal cancer stem cells (Mes/CSCs) (8–10). In TNBC patients, standard-of-care chemotherapy effectively debulks the primary tumors by eliminating the more proliferative epithelial cells, referred to here as epithelial/non-CSCs (Ep/non-CSCs), because they often lack aggressive CSC properties. In contrast, chemotherapy often fails to target the more slowly growing Mes/CSCs (10). Identifying the signaling pathways that regulate plasticity in TNBC has the potential to provide therapeutic options for this difficult disease.
Emerging evidence suggests that E-M/CSC plasticity is influenced by the tumor microenvironment (TME). Tumors consist of a heterogeneous mixture of tumor, immune, endothelial, and stromal cells (7–11). Recent metaanalyses have identified a number of TNBC subtypes, including two distinguishable by the presence or absence of tumor-infiltrating lymphocytes (TILs) and IFN/signal transducer and activator of transcription (IFN/STAT) signaling (3, 4, 12). Immune-responsive TNBCs are defined by the presence of TILs and IFN/STAT signaling, and patients with immune-responsive tumors have a decreased incidence of recurrence (3, 4, 12). In contrast, TNBCs classified as immune-repressed lack TILs and evidence of IFN/STAT signaling, are more refractory to chemotherapy, and more likely to recur.
There are three different types of IFNs: Type I (IFNs α and β) Type II (IFN-γ), and Type III (IFN-λ) (13). First identified for their ability to interfere with viral and bacterial replication, the IFNs are also potent immune-modulatory proteins (13, 14). Type I IFNs (α, β) have been most extensively studied in a variety of cancers for their antiproliferative and proapoptotic activities, including breast cancer (13). Type I IFNs bind to their receptors (IFNAR1/2) and activate the receptor-associated kinases JAK1 and TYK2, resulting in phosphorylation of cytoplasmic STAT1 and STAT2. Phosphorylated STAT1 and STAT2 bind to IRF9 to form phosphorylated IFN-stimulated gene factor 3 (P-ISGF3), the major transcription factor complex that drives the expression of hundreds of IFN-stimulated genes (ISGs). The STAT1, STAT2, and IRF9 genes are themselves targets of P-ISGF3, resulting in elevated levels of newly synthesized STAT1, STAT2, and IRF9 proteins, which, even without tyrosine phosphorylation, form the related complex, unphosphorylated-ISGF3 (U-ISGF3), which can persist for days following an initial exposure to IFN, resulting in prolonged expression of a subset of ISGs. An effective antitumorigenic response to IFN requires the ability to generate a full transcriptional response following robust P-ISGF3 activation. Unbalanced IFN signaling, involving a high level of U-ISGF3, engages an IFN-related DNA damage resistance gene signature (IRDS) that protects tumor cells from DNA-damaging chemotherapy and radiation, resulting in therapeutic resistance and poor prognosis in a variety of cancers, including TNBC (15, 16). Our laboratory previously demonstrated that even a very low level of IFN-β is sufficient to generate U-ISGF3–mediated transcription of genes that correspond to the IRDS (17).
Previous studies have shown that immune-repressed TNBC tumors lacking endogenous IFN/STAT signaling are resistant to therapy and highly recurrent and are characterized by CSC-like properties (3, 4, 14), suggesting a critical role for IFN signaling in regulating TNBC plasticity. Importantly, our work demonstrates that reengaging IFN signaling within these immune-repressed TNBC tumors is an important therapeutic strategy to suppress the aggressive, metastatic and protumorigenic properties associated with TNBC and improve patient survival. Utilizing a human mammary epithelial cell (HMEC) transformation model (8), we demonstrate that treatment with IFN-β successfully reverses CSC properties. Thus, our findings indicate that IFN-β signaling is a critical determinant of positive clinical outcomes in TNBC and provides a potential use for IFN-β in treating TNBC.
Results
IFN-Stimulated Genes Are Repressed in Mesenchymal/CSCs.
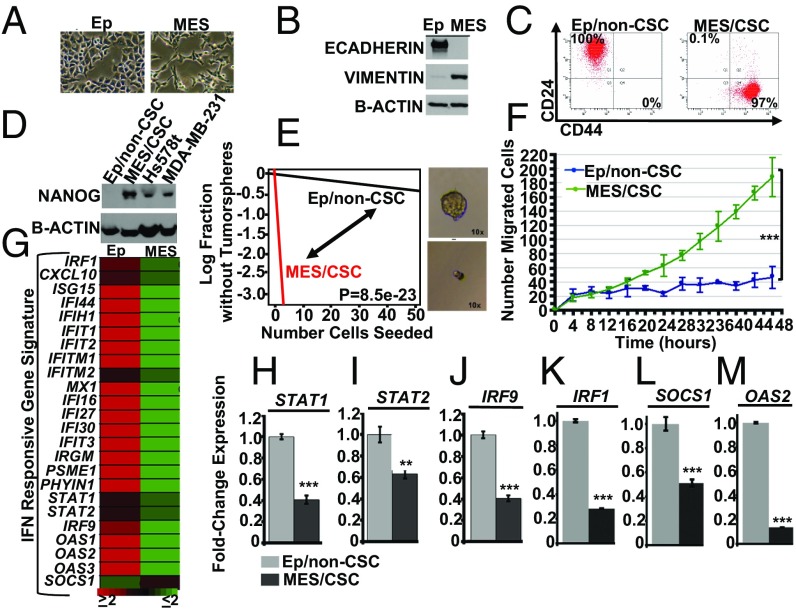
To test how IFN-β influences the mesenchymal cell state and CSC properties, we utilized our HMEC transformation model (8). Briefly, upon acute expression of transforming elements in primary HMECs, an emergent population of cells with a mesenchymal morphology and expression of mesenchymal proteins, indicative of epithelial-to-mesenchymal transition (EMT), consistently appears (8). By using differential trypsinization, we demonstrated that the transformed HMECs contain a mixed population of cells that can be separated into two distinct groups: an epithelial population, characterized by epithelial morphology (Fig. 1A) and expressing E-CADHERIN (Fig. 1B), and a mesenchymal population characterized by mesenchymal morphology (Fig. 1A) and expressing VIMENTIN (Fig. 1B). Importantly, using flow cytometry to examine CD24/CD44 cell surface marker profiles as indicators for breast CSCs, we found that the mesenchymal cells are CD24LoCD44Hi, indicative of breast CSCs (Fig. 1C), while the epithelial cells are CD24HiCD44Lo, indicative of breast non-CSCs (Fig. 1C). To further evidence that the mesenchymal cells are in fact CSCs, they express elevated levels of the pluripotent stem cell transcription factor, NANOG, have greater tumor sphere-forming capacity, and exhibit enhanced migration (Fig. 1 D–F) and elevated expression of cell motility genes (SI Appendix, Fig. S1). We refer to them as mesenchymal/CSCs (Mes/CSCs) and epithelial/non-CSCs (Ep/non-CSCs). By applying an IFN-responsive gene signature, derived from our previous publications (18, 19), to our microarray analysis, we find that numerous ISGs are basally down-regulated in Mes/CSCs relative to Ep/non-CSCs in the absence of IFN-β stimulation (Fig. 1G). The repression of several ISGs, including IRF1, SOCS1, OAS2, STAT1, STAT2, and IRF9, was confirmed by quantitative PCR (qPCR; Fig. 1 H–M). Taken together, our results suggest that ISG expression is repressed in cells that undergo EMT and acquire CSC-like properties, perhaps contributing to the more aggressive features associated with Mes/CSCs.
Fig. 1.
IFN-stimulated genes (ISGs) are repressed in mesenchymal/CSCs. (A) Transformed HMECs consist of two subpopulations, epithelial cells (epithelial morphology) and mesenchymal cells (mesenchymal morphology), as determined by bright field microscopy (10×). (B) Epithelial cells express E-CADHERIN, while mesenchymal cells express VIMENTIN, as determined by Western analysis. (C) Epithelial cells are characterized by a CD24Lo/CD44Hi cell surface profile, while mesenchymal cells are characterized by CD24LO/CD44HI profile, as determined by flow cytometry. (D) Mesenchymal cells express the pluripotent stem cell transcription factor, NANOG, relative to epithelial cells, as shown by Western analysis. Hs578t and MDA-MB-231 are TNBC cell lines used as positive controls. Mesenchymal cells (E) form robust tumor spheres at limiting dilution (stem cell frequency; P = 8.5e-23; n = 3) and (F) show enhanced migration in a cell motility assay (two-tailed t test, ***P < 0.001, n = 3). (G) An IFN-responsive gene signature derived from our previous publications (18, 19) shows ISGs (STAT1, STAT2, IRF9, IRF1, SOCS1, and OAS2) are repressed at least twofold in Mes/CSCs relative to Ep/non-CSCs, as determined by microarray analysis. (H–M) ISGs (STAT1, STAT2, IRF9, IRF1, SOCS1, and OAS2) are repressed in Mes/CSCs relative to Ep/non-CSCs, as determined by qRT-PCR. Data represent mean fold-changes ± SEM, n = 3 (**P < 0.01, ***P < 0.001).
IFN-β Reactivates Canonical Signaling and Gene Transcription in Mes/CSCs.
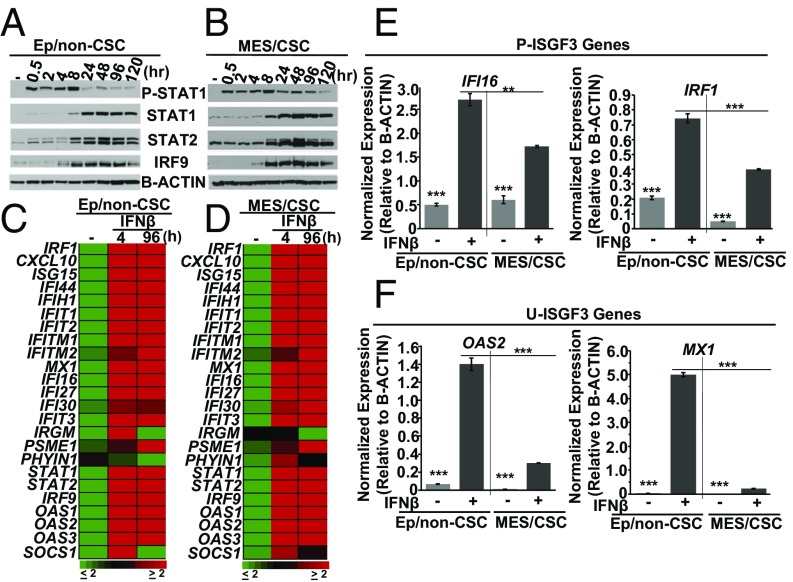
We next sought to determine whether the IFN-β–signaling pathway is fully dismantled in Mes/CSCs or whether exposure to IFN-β could reactivate the suppressed ISGs. Mes/CSCs and Ep/non-CSCs were treated with IFN-β (100 IU/mL) for 0.5–120 h, and Western and gene expression analyses were performed. Interestingly, both populations responded similarly to IFN-β treatment, with comparable kinetics, including the early induction of P-ISGF3, represented by P-STAT1, from 0.5 to 8 h (Fig. 2 A and B). Following the initial P-ISGF3 activation, increased expression of the U-ISGF3 components STAT1, STAT2, and IRF9 was observed, between 8 and 120 h (Fig. 2 A and B). In addition, microarray, using an IFN-responsive gene signature derived from our previous publications (18, 19) and qRT-PCR analyses confirmed that IFN-β induced ISGs in both Mes/CSC and Ep/non-CSC relative to their untreated controls (≥ twofold induction) (Fig. 2 C–F). Importantly, however, the total level of gene expression (ISGs driven by either P-ISGF3 or U-ISGF3) was overall reduced in Mes/CSCs compared with Ep/non-CSCs (Fig. 2 E and F) (SI Appendix, Tables S1 and S2). In our previous work (18) we have defined P-ISGF3–mediated ISGs as transiently induced following short-term IFN-β treatment (4–6 h; IRF1, IFI16). Likewise, we have defined U-ISGF3–mediated ISGs as sustained following prolonged IFN-β treatment (96 h; OAS2, MX1), when P-ISGF3 has returned to basal levels but U-ISGF3 is robust, or following ectopic expression of U-ISGF3 (Y701F STAT1, WT-STAT2, WT-IRF9; OAS2, MX1) in the absence of IFN-β. Taken together, our results demonstrate significant repression of basal ISGs (both P-ISGF3 and U-ISGF3) in Mes/CSCs, but that treatment with IFN-β does cause phosphorylation of ISGF3 and drives ISG expression.
Fig. 2.
IFN-β–mediated canonical signaling reactivates ISG expression in Mes/CSCs. (A and B) Single-dose IFN-β (100 IU/mL) induces biphasic signaling kinetics in both Ep/non-CSCs and Mes/CSCs, with rapid, transient induction of phosphorylated STAT1 (P-STAT1) and P-ISGF3 (0.5–8 h), followed by induction and sustained expression of unphosphorylated STATs 1 and 2, IRF9 (U-STAT1, U-STAT2, IRF9), and U-ISGF3 (24–120 h), as determined by Western analysis. (C and D) Single-dose IFN-β (100 IU/mL) induces ISG transcripts by at least twofold (4 and 96 h) in both Mes/CSCs and Ep/non-CSCs relative to untreated controls, as determined by microarray analysis using an IFN-responsive gene signature derived from our previous publications (18, 19). (E and F) ISG transcripts (P-ISGF3; IFI16, IRF1, U-ISGF3; MX1, OAS2) are induced in both Mes/CSCs and Ep/non-CSCs (4 h), with less total gene expression in Mes/CSCs, as determined by qRT-PCR. Data represent mean fold-changes ± SEM, n = 3 (**P < 0.01, ***P < 0.001).
Sustained IFN-β Exposure Represses Mes/CSC Properties and Inhibits Migration.
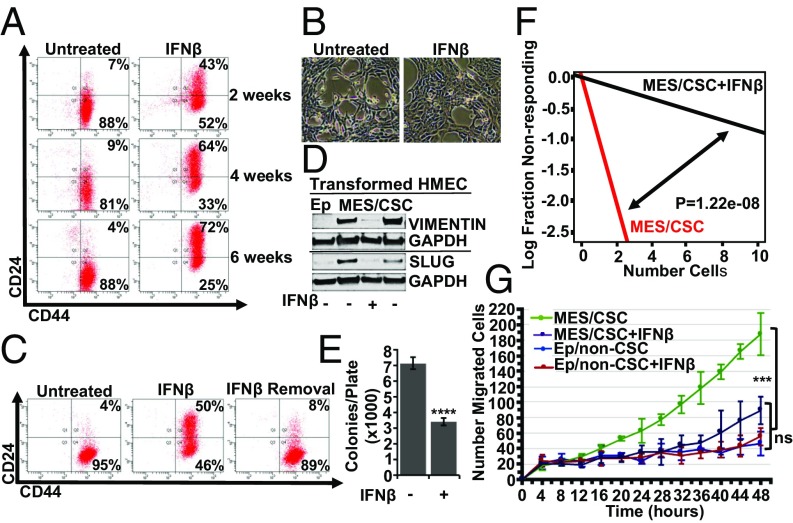
Because elevated IFN/STAT activity correlates with a better prognosis (3, 4), we hypothesized that sustained reactivation of IFN-signaling would alter at least some of the Mes/CSC properties typically associated with poor outcomes. To test this hypothesis, Mes/CSCs were exposed to IFN-β (100 IU/mL) every 48 h for up to 6 wk. Sustained IFN-β exposure reversed the Mes/CSC phenotype, as demonstrated by expression of the non-CSC surface marker CD24 and by acquisition of a pronounced epithelium-like morphology (Fig. 3 A and B). Importantly, induction of CD24 expression required sustained IFN-β exposure, as removal of IFN-β for 1 wk resulted in loss of CD24 expression (Fig. 3C). Exposure to IFN-γ (type II IFN, 1 ng/mL) did not induce CD24 expression, despite driving robust STAT1 activation (SI Appendix, Fig. S2 A and B). Furthermore, chronic IFN-β exposure repressed mesenchymal markers such as VIMENTIN and SLUG (Fig. 3D) and reduced anchorage-independent growth (Fig. 3E), tumor sphere formation (Fig. 3F), and migratory capacity (Fig. 3G). Exposure of Ep/non-CSCs to IFN-β did not alter CD24 expression or epithelial morphology [other than an increase of CD24 in a minority of cells that had spontaneously reduced CD24 expression following cell sorting along with repression of anchorage-independent growth (AIG)] (SI Appendix, Fig. S3 A–C). These results suggest that IFN-β selectively represses the more aggressive properties associated with Mes/CSCs.
Fig. 3.
Sustained exposure to IFN-β represses CSC properties and inhibits migration. Sustained exposure to IFN-β (100 IU/mL, 6 wk) induces (A) CD24 expression, as shown by flow cytometry, and (B) an epithelium-like morphology, as determined by bright field microscopy (10×). (C) IFN-β removal (1 wk) results in loss of CD24, as determined by flow cytometry. (D) Sustained IFN-β (100 IU/mL, 2–6 wk) represses mesenchymal markers (VIMENTIN, SLUG) and removal for 1 wk and reestablishes expression (VIMENTIN and SLUG), as determined by Western analysis (line indicates separate blots). (E) Sustained IFN-β (100 IU/mL, 4 wk) represses AIG (two-tailed t test, ****P < 0.0001, ±SD, n = 3). (F) Sustained IFN-β (100 IU/mL, 2–6 wk), followed by removal for 5 d, partially represses tumor sphere formation at limiting dilution (stem cell frequency; P = 1.22e-08, n = 3). (G) Sustained IFN-β (100 IU/mL, 2–6 wk), followed by removal for 2 d represses cell migration in Mes/CSCs (one-way ANOVA, ***P = 0.0004, ±SD, n = 3) without altering repressed migration in Ep/non-CSCs (one-way ANOVA, ±SD, ns; n = 3).
Mes/CSCs Have Elevated, Stable U-ISGF3 Expression Critical for Cell Migration.
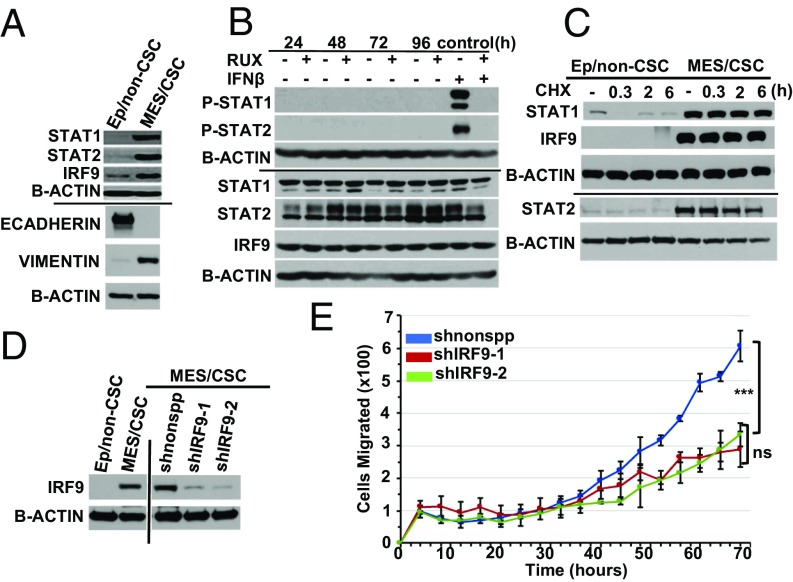
While Mes/CSCs remain responsive to IFN-β, we were interested to examine why basal ISG expression is repressed. The expression of mRNAs encoding the three ISGF3 components, STAT1, STAT2, and IRF9, is reduced in Mes/CSCs relative to Ep/non-CSCs (Fig. 1 H–M). Surprisingly, however, Western analysis demonstrated that Mes/CSCs had markedly higher levels of STAT1, STAT2, and IRF9 proteins relative to Ep/non-CSCs (Fig. 4A). We validated this finding in a second patient-derived HMEC transformation model (SI Appendix, Fig. S4A) that again harbored elevated U-ISGF3 proteins in the Mes/CSCs. Interestingly, we also found that Ep/non-CSCs induced to undergo EMT by ectopic exposure to two cytokines found in tumor microenvironments, transforming growth factor beta (TGF-β) or Oncostatin M (OSM), also acquired elevated U-ISGF3 expression (SI Appendix, Fig. S4B), together with the emergence of mesenchymal markers and a mesenchymal phenotype (8, 9).
Fig. 4.
Mes/CSCs have elevated, stable U-ISGF3 expression critical for cell migration. (A) U-ISGF3 (STAT1, STAT2, and IRF9) expression is elevated in Mes/CSCs relative to Ep/non-CSCs, along with mesenchymal markers, as determined by Western analysis. (B) Mes/CSCs have elevated U-ISGF3 expression independent of IFN-β in the presence of the JAK1/2 inhibitor, Ruxolitinib (line indicates separate blots). (C) The stability of U-ISGF3 is increased in Mes/CSCs relative to Ep/non-CSCs following cycloheximide treatment, as determined by Western analysis. (D) Efficient knockdown of IRF9 in Mes/CSCs as determined by Western analysis (1 wk after lentiviral transduction). All samples were on the same blot; vertical line indicates where the blot was cut. (E) IRF9 KD represses cell migration in Mes/CSCs over time (0–70 h). Data represent means ± SD (one-way Anova, ***P = 0.0009, n = 3), (two-tailed t test, ns; n = 3).
Our previous studies have shown that constitutive low levels of either exogenous or endogenous IFN-β can induce low levels of P-ISGF3, which in turn induce elevated expression of the STAT1, STAT2, and IRF9 proteins, and thus of U-ISGF3 (17, 18). While the repression of ISGF3 transcripts in Mes/CSCs already suggested that the increase in U-ISGF3 was not due to autocrine IFN-β signaling, we confirmed that canonical JAK/STAT signaling was dispensable for elevated U-ISGF3 protein expression by using the JAK1/2 inhibitor, Ruxolitinib. At times up to 96 h, Ruxolitinib treatment did not reduce U-ISGF3 protein expression (Fig. 4B). To confirm that Ruxolitinib was capable of suppressing JAK1/2 activity at the doses used, Mes/CSCs were challenged with IFN-β in the presence or absence of this drug. Indeed, Ruxolitinib completely inhibited IFN-β–mediated phosphorylation of STAT1 and STAT2, confirming its efficient inhibition of JAK1/2 signaling (Fig. 4B). Furthermore, despite the elevated expression of STAT1 in Mes/CSCs, no basal STAT1 phosphorylation was detected, whereas stimulation with IFN-β (100 IU/mL) for 30 min induced robust formation of P-STAT1 (SI Appendix, Fig. S4C). As an explanation for the elevated levels of STAT1, STAT2, and IRF9 in Mes/CSCs, we found that protein stability was greatly enhanced. Treatment of Mes/CSCs with cycloheximide confirmed that the half-lives of STAT1, STAT2, and IRF9 were all greater than 8 h. In contrast, the half-lives of STAT1, STAT2, and IRF9 were much shorter in epithelial/non-CSCs (less than 30 min; Fig. 4C). Taken together, our results demonstrate that elevated, stable U-ISGF3 expression, which occurs independently of IFN signaling, may be involved in promoting the aggressive properties associated with CSCs, including migratory capacity, metastatic potential, and therapeutic failure in TNBC.
We hypothesized that the elevated U-ISGF3 observed in the Mes/CSCs might be important for the phenotypes commonly associated with CSCs. To test the importance of U-ISGF3 in CSC biology, shRNAs targeting IRF9 (or a nonspecific control) were stably delivered to Mes/CSCs by lentiviral transduction, and IRF9 knockdown (KD) was confirmed by Western analysis (Fig. 4D). Because IRF9 is the critical DNA-binding component of U-ISGF3, suppression of IRF9 will inhibit overall U-ISGF3 activity. Interestingly, while IRF9 KD did not alter proliferation (SI Appendix, Fig. S4D), it did significantly decrease cell migration relative to control cells (Fig. 4E). Moreover, the addition of IFN-β to induce phosphorylation of ISGF3 components also decreased migration. The addition of IFN-β to IRF9 KD cells did not further repress their ability to migrate (SI Appendix, Fig. S4E). In contrast to the role of IRF9/U-ISGF3 in Mes/CSC migration, we noted no difference in VIMENTIN and CD44 expression and no reduction in their capacity for AIG following IRF9 KD (SI Appendix, Fig. S5 A–D). We also found that knockdown of other components of U-ISGF3, such as STAT1, did not suppress CD44 expression or inhibit AIG (SI Appendix, Fig. S6 A–C). Our findings suggest that high levels of U-ISGF3 contribute to the aggressive, migratory capacity of TNBC cells and that reducing the expression of IRF9, the DNA-binding component of U-ISGF3, or phosphorylating ISGF3 in response to IFN-β is a potential strategy for inhibiting migration and metastasis.
TNBC Cell Lines Express Elevated U-ISGF3 and Are Sensitive to IFN-β.
To test whether Mes/CSC properties are linked to U-ISGF3 in TNBCs in addition to our HMEC model, we examined two separate established TNBC cell lines (MDA-MB-231 and Hs578t), observing robust CD44 expression (SI Appendix, Fig. S7A), mesenchymal morphology (SI Appendix, Fig. S7B), and elevated U-ISGF3 (SI Appendix, Fig. S7C), along with expression of mesenchymal markers (SI Appendix, Fig. S7C) and NANOG (Fig. 1D). We next exposed MDA-MB-231 and Hs578t cells to IFN-β (100 IU/mL) every 48 h for 2 wk, finding that IFN-β partially reversed the CSC phenotype, as indicated by increased expression of CD24 in MDA-MB-231 cells (SI Appendix, Fig. S7D). Our results thus far suggest that elevated U-ISGF3 expression and the global suppression of ISGs observed in Mes/CSCs contribute to more aggressive CSC properties and that exposure to IFN-β can reverse these properties.
Clinical Relevance for IFN-β in TNBC.
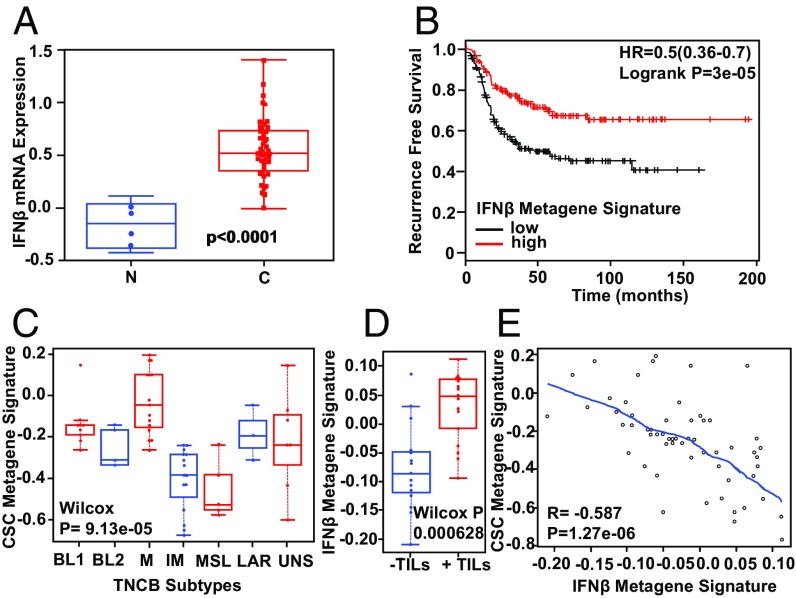
To determine whether our findings could be translated to a clinical setting, we first examined IFN-β mRNA expression levels in breast stroma by mining a publicly available gene expression dataset consisting of both normal, noncancer tissue and invasive ductal carcinoma tissue (19). Importantly, we found that IFN-β mRNA is significantly elevated in the stroma of invasive ductal carcinomas relative to normal breast (two-tailed t test, P < 0.0001) (Fig. 5A). To determine the impact of this elevated IFN-β within the breast TME, we next examined how IFN-β impacts patient survival. By applying our IFN-β metagene signature (generated from microarray analysis in which Ep/non-CSCs were treated with IFN-β for 96 h) to curated TCGA and EGA datasets publicly available through the online Kaplan Meier plotter (20), we revealed that increased IFN-β–mediated target gene expression significantly correlated with improved recurrence-free survival in basal/TNBC (logrank P = 7.1e-05) (Fig. 5B). These findings suggest that IFN-β plays a positive, critical role in TNBC outcome. To determine whether IFN-β promotes survival by impinging on CSC properties, we next evaluated our IFN-β metagene signature and a previously published CSC metagene signature (21), in a clinical trial dataset (PrECOG 0105) (22), using gene expression profiling of pretreatment TNBC tumors. Tumors were stratified according to TNBC subtype and compared for expression of a CSC metagene signature (23). We found that the mesenchymal subtype (M) had the highest degree of CSC-gene expression, while the immune-modulatory (IM) subtype had the lowest (Fig. 5C). The results obtained with our HMEC cell culture model, demonstrating that mesenchymal cells have robust CSC properties while exhibiting repressed ISGs relative to Ep/non-CSC, are consistent with these clinical findings. The IM subtype has already been linked to increased IFN/STAT activity (3, 4), consistent with the Ep/non-CSCs in our model, which express elevated ISGs and lack CSC gene expression. Moreover, an elevated IFN-β metagene signature (derived from Ep/non-CSCs treated with IFN-β) significantly correlated with the presence of total TILs by pathologic assessment in TNBC tumors, while a repressed IFN-β metagene signature significantly correlated with their absence (Wilcox P = 0.000628) (Fig. 5D), demonstrating that IFN-β plays an important immune modulatory role, specifically as it relates to TILs. In addition, elevated expression of the IFN-β metagene signature also significantly correlated with repression of the CSC metagene signature in TNBC (R = 0.587, P = 1.27e-06) (Fig. 5E), demonstrating that IFN-β within the breast TME not only has potent immune modulatory functions but also regulates CSC properties by repressing CSC-specific target genes. In line with these findings, our work here demonstrates that IFN-β represses CSCs in a tumor cell-intrinsic manner, suggesting that IFN-β can directly target CSCs in the absence of an adaptive immune response. Taken together, these data provide important clinical evidence to suggest that IFN-β–related signaling is necessary for repressing aggressive CSC-like properties in TNBC.
Fig. 5.
Clinical relevance of IFN-β in TNBC. (A) IFN-β mRNA is significantly elevated in breast tumor stroma of invasive ductal carcinomas relative to the normal noncancerous stroma (two-tailed t test, P < 0.0001). (B) An elevated, experimentally derived IFN-β metagene signature significantly correlates with improved recurrence-free survival in basal/TNBC patients (P = 3e-05). (C) TNBC subtypes are characterized by differential expression of a CSC metagene signature; the mesenchymal (M) subtype has an elevated CSC metagene signature, and the immune-modulatory (IM) subtype has a repressed CSC metagene signature (Wilcox P = 9.13e-05). (D) An elevated IFN-β metagene signature significantly correlates with the presence of TILs (Wilcox P = 0.000628). (E) An elevated IFN-β metagene signature significantly correlates with a repressed CSC metagene signature (R = 0.587, P = 1.27e-06).
Discussion
TNBC is the most lethal form of breast cancer, characterized by poorly differentiated cells, therapeutic resistance, enhanced metastatic potential, and tumor recurrence (1–13). Emerging evidence suggests that E-M plasticity is linked with CSC properties, which are important determinants of therapeutic resistance and metastasis and are largely influenced by the TME (9, 10). Several recent publications have demonstrated that the presence of immune cells within the TME strongly influences clinical outcomes in TNBC (1, 12, 15). An immune-responsive TME characterized by elevated TILs and IFN signaling has been linked to improved prognosis, while an immune-repressed TME lacking TILs and IFN signaling portends a worse prognosis. Here our work demonstrates a link between a Mes/CSC phenotype and a repressed IFN signature, suggesting that reawakening IFN signaling in these problematic Mes/CSCs may be a viable therapeutic strategy to repress the aggressive features associated with TNBC.
In our studies, Ep/non-CSCs have a basally elevated IFN signature, while Mes/CSCs have a basally repressed signature, which models immune-responsive and immune-repressed TNBC, respectively. Importantly, both spontaneous EMT and EMT induced by cytokines, such as TGF-β or OSM, correlated with comparable induction of U-ISGF3 proteins, together with basal repression of ISGs. The high levels of STAT1, STAT2, and IRF9 proteins in Mes/CSCs indicated that IFN exposure might be able to reawaken ISG transcription. Indeed, acute exposure of Mes/CSCs to IFN-β, which resulted in a prolonged P-STAT1 response relative to Ep/non-CSC (Fig. 2 A and B), reactivated ISG expression, at least partially. Importantly, sustained IFN-β exposure was required to restore an epithelial phenotype (epithelial morphology, CD24 expression) while inhibiting Mes/CSC properties (repression of VIMENTIN and SLUG, reduced migration, tumor sphere formation, and colony formation). The reemergence of CD24 and repression of SLUG suggests that IFN-β engages a differentiation program to reduce stem-like characteristics. This finding is distinct from prior studies of IFN, which attributed IFN’s antitumorigenic properties to its antiproliferative and proapoptotic functions (13). In our experiments, at the doses of IFN-β used, we observed little impact on proliferation or apoptosis.
Interestingly, recent work published by Qadir et al. (24) shows that high doses of IFN-β (1,000 IU/mL) drive cancer stemness in a luminal breast cancer cell line (MCF7), as evidenced by robust tumor sphere formation, expression of the pluripotent stem cell transcription factor SOX2, and acquisition of the breast CSC marker CD44. In this model, IFN-β also induced STAT3 phosphorylation. Both IFN-β–mediated tumor sphere formation and STAT3 phosphorylation were prevented following abrogation of STAT1 expression. As a possible explanation for the contradictory findings, we do not see activation of STAT3 in response to IFN-β in our TNBC model (SI Appendix, Fig. S9). In fact, shRNA-mediated repression of STAT1 in Ep/non-CSCs actually enhances the ability of the IL-6 family member OSM, a STAT3 activator, to drive CSC properties. Thus, in contrast to luminal breast cancers, STAT1 in TNBC appears to repress rather than promote STAT3-mediated CSC properties (SI Appendix, Fig. S10). In addition, the role of IFN-β to regulate CSC properties in patients’ tumors may also be dependent on breast cancer subtype. While the presence of an IFN-β gene signature was correlated with improved TNBC patient survival and inversely correlated with a CSC gene signature, there were no such correlations in patients with luminal breast cancers (luminal A and B) (Fig. 5 and SI Appendix, Fig. S8). Consequently, the biological impact of IFN-β signaling in regulating CSC is likely unique to TNBC. We currently envision a potential therapeutic role for IFN-β, specifically in TNBC, to be given before chemotherapy with the intent to differentiate and thereby functionally deplete therapeutically resistant Mes/CSCs. Our studies have focused on IFN-β, as we observed that type II IFNs (IFN-γ), which signal through STAT1 homodimers, did not induce a differentiation program.
The mesenchymal phenotype and high U-ISGF3 expression observed in our CSCs have both separately been shown to confer therapeutic resistance and poor clinical outcomes (3, 9, 10, 16–18). In Mes/CSCs, elevated U-ISGF3 protein expression is independent of IFN signaling, as evidenced by lack of P-ISGF3 expression, repressed mRNA transcripts (STAT1, STAT2, and IRF9), and sustained U-ISGF3 expression, despite blocking IFNAR receptor-mediated JAK activity. Rather, all three ISGF3 components appear to be highly stable in Mes/CSCs relative to Ep/non-CSCs. The mechanism for increased U-ISGF3 stability is not yet clear, and future studies will examine whether the stabilization of the U-ISGF3 proteins is caused by a posttranslational modification (for example, sumoylation, which can lead to decreased protein degradation) or whether specific protein–protein interactions disrupt normal ISGF3 turnover.
Previous work has demonstrated that elevated U-ISGF3 expression (either following IFN stimulation or ectopic expression of the three U-ISGF3 components) correlates with elevated U-ISGF3 target gene expression, including the expression of several genes that have now been linked to therapeutic resistance. We therefore anticipated that elevated U-ISGF3 in our Mes/CSCs would result in elevated U-ISGF3 target gene expression as well. In contrast, however, elevated U-ISGF3 in Mes/CSCs not only correlated with basally repressed U-ISGF3 target gene expression but also correlated with a repressed overall ISG response following IFN-β stimulation, suggesting that, in these cells, U-ISGF3 inhibits the IFN-β response and may therefore contribute to the overall suppressed immune response observed in aggressive TNBCs lacking an IFN/STAT gene signature. Thus, we have uncovered a function for U-ISGF3 in Mes/CSCs which involves repression rather than activation of ISG expression and could help explain why Mes/CSCs are less responsive to acute IFN-β exposure compared with Ep/non-CSCs, despite Mes/CSCs having a more prolonged STAT1 phosphorylation response (Fig. 2 A and B). Although the cause for this prolonged P-STAT1 response in Mes/CSCs is unclear at this time, one possible explanation is that Mes/CSCs express higher levels of stable STAT1 protein (Fig. 4 A–C), and these conditions may contribute to sustained phosphorylation (Fig. 2 A and B) and will need to be examined in future studies. We conclude therefore that either repressing U-ISGF3 or restoring IFN-β–mediated signaling in Mes/CSCs expressing elevated U-ISGF3 are important potential therapeutic strategies for repressing Mes/CSC properties in immune-repressed TNBC.
TNBC is increasingly recognized as an immunogenic tumor, where immune infiltration of T cells and an IFN signature directly correlate with improved patient survival (3, 4, 12). Significant efforts have been made to promote an immune response to TNBC, including the use of checkpoint inhibitors targeting PD1, PDL-1, and/or CTLA4 (25, 26). However, while immunotherapies have proven beneficial in treating immune-responsive TNBC, these therapies have no impact on immune-repressed tumors, which lack TILs and an IFN signature and make up the majority of TNBC tumors. Importantly, while we report here that IFN-β can reengage canonical IFN signaling and repress the migratory, chemo-resistant Mes/CSC phenotype in a tumor-cell intrinsic manner, IFN-β is also well known for its immune-modulatory functions, including the recruitment and activation of cytotoxic T lymphocytes (CD8+ T cells) (13, 15). Thus, IFN treatment may help to recruit TILs into the tumor, essentially turning a so-called immunologically “cold” tumor into a “hot” tumor. The net result of IFN treatment is likely to be a combination of cell-intrinsic and immune-modulatory functions, as cancer cells defective in critical components of IFN signaling (point mutations resulting in loss of function of IFNAR, IRF1, IRF7, TLR3, TLR9, RIGI, or downstream ISGs) are resistant to both IFN and chemotherapy (6, 13–15, 25).
Previous use of Type I IFN as a cancer therapy has been hampered due to dose-limiting toxicities, as high concentrations are required to achieve antiproliferative or proapoptotic responses (13, 15). Here, we demonstrate that a nontoxic dose of IFN-β effectively reverses Mes/CSC characteristics, indicating a role for IFN-β as a potential therapeutic strategy for targeting CSC, to undergo differentiation to a less aggressive cell state. Because immune-repressed TNBCs, which lack IFN signaling, are especially recalcitrant to standard-of-care chemotherapy, resulting in significant metastasis and tumor recurrence, our work provides evidence that reengaging IFN signaling within these tumors will not only directly repress CSC but also successfully promote an immune modulatory response, as demonstrated by our clinical data. Interestingly, several recent publications have shown that IFN-β localized to tumors in vivo can promote tumor regression. Delivery of IFN-β using (i) gene therapy (adenovirus encoding IFN-β) (13, 15); (ii) cell therapy (mesenchymal stem cells engineered to express IFN-β) (2); or (iii) targeted therapy (tumor-targeted monoclonal antibodies fused to IFN-β protein) (15) successfully reduced tumor burdens. We propose future studies to examine IFN-β delivery platforms that do not have long-term stability issues or integrate into the host genome (viral-based therapies). Recent developments in nanoparticle technology have demonstrated that nanoparticles successfully target and inhibit tumor growth and metastasis in vivo by delivering chemotherapeutic drugs or siRNAs into tumors (27). Because nanoparticle technology can be used to deliver known concentrations of drug over time followed by removal, it is a promising therapeutic strategy to explore for administration of localized, low concentrations of IFN-β to target and differentiate Mes/CSCs, before administration of chemotherapy or ionizing radiation.
Materials and Methods
Detailed materials and methods are available in SI Appendix.
HMECs (Ep/non-CSCs and Mes/CSCs) and TNBC cell lines were treated acutely (4 or 96 h) or chronically (every 48 h for several weeks) with either IFN-β (100 IU/mL) or IFN-γ (1 ng/mL) and monitored for impact on Mes/CSC properties.
Supplementary Material
Acknowledgments
This work was funded by the Cancer Biology Training [Grant CBTG T32CA198808 (to M.R.D.)], American Cancer Society [Grant RSG-CCG-122517 (to M.W.J.)], and Grant NCI R21CA198808 (to M.W.J. and G.R.S.). Research Core Facility support; Cytometry & Imaging Microscopy (Michael Sramkoski and Allison Kipling); Core Facilities Case Comprehensive Cancer Center (P30CA43703), Cleveland Clinic Lerner Research Institute Flow Cytometry Core (Eric Schultz and Joe Gerow). The prECOG 0105 clinical trial, referenced in this study, was funded by the Breast Cancer Research Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE106782).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713728114/-/DCSupplemental.
References
- 1.Stagg J, Allard B. Immunotherapeutic approaches in triple-negative breast cancer: Latest research and clinical prospects. Ther Adv Med Oncol. 2013;5:169–181. doi: 10.1177/1758834012475152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ling X, et al. Mesenchymal stem cells overexpressing IFN-β inhibit breast cancer growth and metastases through stat3 signaling in a syngeneic tumor model. Cancer Microenviron. 2010;3:83–95. doi: 10.1007/s12307-010-0041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann B-D, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burstein M-D, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen D-X, Bos P-D, Massagué J. Metastasis: From dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 6.Bidwell B-N, et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat Med. 2012;18:1224–1231. doi: 10.1038/nm.2830. [DOI] [PubMed] [Google Scholar]
- 7.Doherty M-R, Smigiel J-M, Junk D-J, Jackson M-W. Cancer stem cell plasticity drives therapeutic resistance. Cancers (Basel) 2016;8:1–13. doi: 10.3390/cancers8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Junk D-J, Cipriano R, Bryson B-L, Gilmore H-L, Jackson MW. Tumor microenvironmental signaling elicits epithelial-mesenchymal plasticity through cooperation with transforming genetic events. Neoplasia. 2013;15:1100–1109. doi: 10.1593/neo.131114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junk D-J, et al. Oncostatin M promotes cancer cell plasticity through cooperative STAT3-SMAD3 signaling. Oncogene. 2017;36:4001–4013. doi: 10.1038/onc.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mani S-A, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldman A, et al. Temporally sequenced anticancer drugs overcome adaptive resistance by targeting a vulnerable chemotherapy-induced phenotypic transition. Nat Commun. 2015;6:6139. doi: 10.1038/ncomms7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, et al. CD8+ lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 2012;14:R48. doi: 10.1186/bcr3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker B-S, Rautela J, Hertzog P-J. Antitumour actions of interferons: Implications for cancer therapy. Nat Rev Cancer. 2016;16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 14.Sistigu A, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. 2014;20:1301–1309. doi: 10.1038/nm.3708. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, et al. Targeting the tumor microenvironment with IFNβ bridges innate and adaptive immune responses. Cell. 2014;25:37–48. doi: 10.1016/j.ccr.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weichselbaum RR, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci USA. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheon H, Stark G-R. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci USA. 2009;106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheon H, et al. IFNβ-independent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013;32:2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finak G, et al. Gene expression signatures of morphologically normal breast tissue identify basal-like tumors. Breast Cancer Res. 2006;8:R58. doi: 10.1186/bcr1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szasz A-M, et al. Cross-validation of survival associated data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J-C, et al. Seventeen-gene signature from enriched Her2/Neu mammary tumor-initiating cells predicts clinical outcome for human HER2+:ERα- breast cancer. Proc Natl Acad Sci USA. 2012;109:5832–5837. doi: 10.1073/pnas.1201105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, et al. TNBCtype: A subtyping tool for triple-negative breast cancer. Cancer Inform. 2012;11:147–156. doi: 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Telli M-L, et al. Phase II study of gemcitabine, carboplatin, and iniparib as neoadjuvant therapy for triple-negative and BRCA1/2 mutation-associated breast cancer with assessment of a tumor-based measure of genomic instability: PrECOG0105. J Clin Oncol. 2015;33:1895–1901. doi: 10.1200/JCO.2014.57.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qadir A-S, et al. CD95/Fas increases stemness in cancer cells by inducing a STAT1-dependent type I interferon response. Cell Rep. 2017;18:2373–2386. doi: 10.1016/j.celrep.2017.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katlinski K-V, et al. Inactivation of interferon receptor promotes the establishment of immune privileged tumor microenvironment. Cancer Cell. 2017;31:194–207. doi: 10.1016/j.ccell.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Migali C, et al. Strategies to modulate the immune system in breast cancer: Checkpoint inhibitors and beyond. Ther Adv Med Oncol. 2016;8:360–374. doi: 10.1177/1758834016658423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parvani J-G, Gujrati MD, Mack MA, Schiemann WP, Lu ZR. Silencing β3 integrin by targeted ECO/siRNA nanoparticle inhibits EMT and metastasis of triple negative breast cancer. Cancer Res. 2015;75:2316–2325. doi: 10.1158/0008-5472.CAN-14-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.